the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Maximum summer temperatures predict the temperature adaptation of Arctic soil bacterial communities
Mark Dekker
Rien Aerts
James T. Weedon
Rapid warming of the Arctic terrestrial region has the potential to increase soil decomposition rates and form a carbon-driven feedback to future climate change. For an accurate prediction of the role of soil microbes in these processes, it will be important to understand the temperature responses of soil bacterial communities and implement them into biogeochemical models. The temperature adaptation of soil bacterial communities for a large part of the Arctic region is unknown. We evaluated the current temperature adaption of soil bacterial communities from 12 sampling sites in the sub- to High Arctic region. Temperature adaptation differed substantially between the soil bacterial communities of these sites, with estimates of optimal growth temperature (Topt) ranging between 23.4 ± 0.5 and 34.1 ± 3.7 °C. We evaluated possible statistical models for the prediction of the temperature adaption of soil bacterial communities based on different climate indices derived from soil temperature records or on bacterial community composition data. We found that highest daily average soil temperature was the best predictor for the Topt of the soil bacterial communities, increasing by 0.63 °C °C−1. We found no support for the prediction of temperature adaptation by regression tree analysis based on the relative abundance data of the most common bacterial species. Increasing summer temperatures will likely increase Topt of soil bacterial communities in the Arctic. Incorporating this mechanism into soil biogeochemical models and combining it with projections of soil temperature will help to reduce uncertainty in assessments of the vulnerability of soil carbon stocks in the Arctic.
- Article
(3823 KB) - Full-text XML
-
Supplement
(1197 KB) - BibTeX
- EndNote
The Arctic terrestrial biome has the potential to undergo particularly large losses of soil organic carbon, and this controls the potential loss or gain of global carbon stocks (Crowther et al., 2016; Wieder et al., 2019). This is because of the large soil organic carbon stocks in Arctic soils (Tarnocai et al., 2009) and the strong response of soil respiration rates to warming in these cold ecosystems (Carey et al., 2016). Bacterial soil communities in the Arctic terrestrial region are adapted to perform well at low temperatures (Bååth, 2018). However, these bacterial communities are likely to be exposed to increasing soil temperatures in this century (Post et al., 2019), and it remains uncertain whether these soil bacterial communities will adapt their response to temperature when exposed to warmed conditions (Rinnan et al., 2011; Weedon et al., 20232; Rousk et al., 2012). Knowledge of the climate conditions under which such an adaption takes place will help in the estimations of the potential vulnerability of Arctic soil carbon stocks to warmer climate conditions (Bååth, 2018; Bradford et al., 2019; García-Palacios et al., 2021).
The temperature adaptation of soil bacterial communities is most often characterized in relation to respiration, growth, or enzymatic activity. A commonly applied method is to estimate the relationship between whole community growth and temperature with an assay that measures 3H-leucine uptake (Bååth et al., 2001). This relationship between temperature and bacterial growth can be described by the Ratkowsky model, which has the following three cardinal points: the (theoretical) minimum growth temperature (Tmin), optimal growth temperature (Topt), and maximum growth temperature (Tmax; Ratkowsky et al., 1983). Previous research has shown that the temperature–growth relationships of soil bacterial communities adapt to their local environment, such that there is a positive correlation between mean annual air temperature (MAAT) and the parameters describing the temperature–growth relationships of soil bacterial communities (cardinal points; Bååth, 2018). For example, it has recently been found that, across an elevation gradient in the Peruvian Andes, Tmin increased, with a 0.2 °C °C−1 increase in MAAT (Nottingham et al., 2019), and a similar correlation was found between MAAT and Topt across a natural climate gradient in Europe (Cruz Paredes et al., 2021). This correlation has also been shown in the Antarctic, where the temperature–growth relationships of soil bacterial communities show higher values of Tmin with higher mean annual soil temperature (Rinnan et al., 2009). However, no comparable large-scale study on the temperature–growth relationships of soil bacterial communities in the Arctic has been performed yet. Such a large-scale study is needed for Arctic soil bacterial communities, as the Arctic differs from lower-latitudinal regions in terms of its current climate (Convey, 2013), predicted climate changes (Post et al., 2019), and importance for the global soil carbon stock (Wieder et al., 2019).
Despite strong correlations over large spatial scales, an increase in the mean annual soil temperature does not necessarily lead to a shift in temperature–growth relationships of bacterial communities when soils are experimentally warmed in lab incubation and field studies (Pietikäinen et al., 2005; Birgander et al., 2013, 2018; Rinnan et al., 2011; Weedon et al., 2023). Instead, a common observation is a rapid change in the temperature–growth relationships driven by a community turnover when soils are incubated above the optimal growth temperature of the in situ soil bacterial community (Birgander et al., 2013; Donhauser et al., 2020). This suggests that the maximum soil temperature is an important predictor of the temperature–growth relationships of bacterial communities. Supporting evidence for this comes from a study in the Antarctic, where coastal water bacterial communities are adapted to lower temperatures (lower Tmin) than soil bacterial communities in the same region, despite the mean annual temperature of Antarctic water being higher than that of Antarctic soils (van Gestel et al., 2020). The Antarctic soils are exposed to higher summer temperatures than the Antarctic marine environment, leading to the hypothesis that the maximum temperature, rather than the annual average, is a more important driver for the temperature adaptation of bacterial communities across different habitats (Birgander et al., 2013; van Gestel et al., 2020).
Analogous to the maximum temperature, the coldest soil temperature could also influence temperature–growth relationships. In desert soils, the upper layer (0–5 cm) is characterized by relatively large-amplitude fluctuations in temperature over both diurnal and annual timescales. Consequently, the bacterial communities of these upper layers tend to have lower Tmin values and higher Topt values than deeper soil layers that are exposed to more moderate and stable soil temperatures (van Gestel et al., 2013). These studies show that, while the mean annual temperature might correlate strongly with the cardinal points of the temperature–growth relationships of soil bacterial communities, the temperature adaption might be more directly related to other selective pressures of the thermal regime such as the highest or lowest soil temperature.
To predict the future temperature–growth relationships of soil bacterial communities in the Arctic, more knowledge is needed on (1) the current temperature adaptation of soil bacterial communities in the Arctic and (2) the specific mechanisms driving temperature adaptation. Bacterial communities from polar ecosystems are hypothesized to be adapted to low temperatures, as shown by a low Tmin (Baath, 2018). For example, sub-Arctic bacterial communities exhibit lower cardinal points for their temperature–growth relationships compared to bacterial communities of temperate ecosystems, with a Tmin of −9.6 to −7.0 °C and Topt 25 to 30 °C (Cruz-Paredes et al., 2021a; Rinnan et al., 2011). It is likely that soil warming will shift the temperature–growth relationships of sub-Arctic soil bacterial communities (Weedon et al., 2023; Rijkers et al., 2022). However, the in situ temperature–growth relationships of soil bacterial communities in the mid- to High Arctic are so far unknown and will need to be evaluated to understand the current temperature adaptation of soil bacterial communities and drivers of temperature adaptation under future climate conditions.
It is important to evaluate which soil thermal parameters are the most accurate predictor for soil bacterial communities in the sub- to High Arctic, as this might not be accurately predicted from the mean soil annual temperature alone. In these (sub-) Arctic regions, the maximum and minimum daily soil temperatures are only weakly correlated with the mean annual soil temperature, due to the influence of local environmental parameters on the soil climate extremes. For example, winter soil temperatures also vary greatly on the meter scale in the Arctic, due to the influence of snow cover on winter microclimate (Karjalainen et al., 2019). On the other hand, summer soil temperature is more closely related to the air temperature, which varies less between (sub-) Arctic soils (Fig. 1). Implementing knowledge about these possible drivers of the temperature adaptation of soil bacterial communities at these high northern latitudes will support accurate predictions of soil decomposition of the large carbon stock present in the Arctic under future climates.
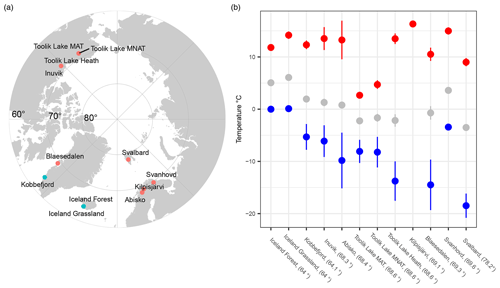
Figure 1(a) Map, with the polar projection, showing the 12 sampling sites across the Arctic and (b) the average values of soil thermal regimes for each site, including the maximum (red), mean (gray), and minimum (blue) soil temperature. Error bars indicate the standard deviation across the years.
Due to the possible influence of multiple soil thermal parameters, the accurate prediction of temperature adaptation by soil bacterial communities will likely require high-resolution soil temperature data. However, soil-temperature-logger data are particularly scarce in the Arctic region (Lembrechts et al., 2021), leading to a need for potential alternative predictors of soil microbial temperature adaptation. DNA-based bacterial community composition measures have recently been shown to correlate with shifts in the temperature–growth relationships of a soil bacterial communities (Donhauser et al., 2020; Rijkers et al., 2022; Weedon et al., 2023). More generally, temperature traits differ between members of bacterial communities from Arctic soils (Wang et al., 2021), and specific bacterial taxa have been associated with warming in forest soils across North America (Oliverio et al., 2017). The aggregated community response, such as the temperature–growth relationship, might therefore be predictable when using the abundance of specific species that are associated with a warm- or cold-adapted community (Hicks et al., 2021). In a pan-Arctic survey, the soil bacterial community showed a large diversity of species, with 15 common taxa shared between all soils (Malard et al., 2019). Therefore, there are potentially bacterial species that could indicate the current temperature adaptation of Arctic soil bacterial communities. If so, this provides opportunities to determine the temperature adaptation of soil bacterial communities in the Arctic in areas where long-term soil temperature logging is absent.
In this study, we tested which soil thermal parameters best predict the cardinal points of the temperature–growth relationships of bacterial communities from 12 soils collected in the sub- to High Arctic region. We hypothesized that the highest and lowest daily soil temperatures would be the best predictor of the corresponding cardinal points of the temperature–growth relationships. We also compared the DNA-based compositional profiles across soil types and explored whether such compositional data can be used as an alternative predictor for the temperature–growth relationships of the soil bacterial communities in Arctic soils.
2.1 Sample collection
In the summers of 2018 and 2020, soil samples were collected from 12 soil types at nine sites ranging from the sub- to High Arctic (Fig. 1). The 2018 sampling at Toolik Field Station, Svalbard, Abisko, and Iceland has previously been described in Rijkers et al. (2022). In brief, soil cores of 10 cm depth were collected from the following locations: Toolik Field Station, USA (68°38′ N, 149°36′ W), at the Long-Term Ecological Research (LTER) heath site, LTER Moist Acidic Tussock (MAT), and LTER Non-Acidic Tussock (MNAT), Svalbard, from the Bjørndalen site (78°13′ N, 15°19′ E), which is dominated by Carex sp. vegetation, and the ForHot site in Iceland (64°00′ N, 21°11′ W), which is a grassland (Agrostis capillaris) and forest site (Picea sitchensis). Last, soil samples were collected from the blanket bog (Sphagnum sp.) where the International Tundra Experiment (ITEX) warming experiment is located, close to the Abisko Scientific Research Station in Sweden (68°21′ N, 18°49′ E).
In 2020, a second sampling campaign collected triplicate soil cores to a depth of 10 cm at sites in Greenland (two sites), Canada, Norway, and Finland. On Disko Island, Greenland soil cores were collected near the AWS-2 logger at Østerlien site of the Greenland Ecosystem Monitoring (GEM; 69°15′′′ N, 53°30′ W), which was covered by Vaccinium sp. At Kobbefjord, Greenland soil samples with Empetrum sp. cover were collected near the SoilEmp logger of GEM (64°08′ N 51°22′ W). At Inuvik, Canada, soil cores were sampled at the Inuvik Airport bog (68°18.9342′ N, 133°26.0214′ W), which is characterized by low shrubs (Nixon et al., 2003). In Finland, samples were collected directly next to the ITEX site in Kilpisjärvi (69.4° N, 20.490° E), for which the vegetation cover is dominated by Vaccinium and Empetrum sp. (Ylänne et al., 2015). Last, soil samples were collected at Petersfjellet in Norway (69°35.5277′ N, 29°55.1939′ E), which was covered by Empetrum nigrum.
2.2 Soil temperature data
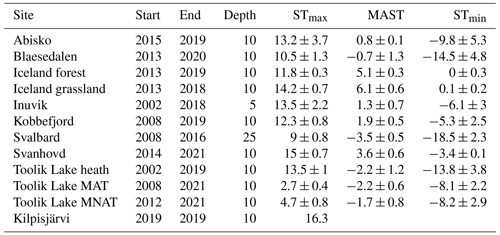
Soil temperature records were collected from the involved research stations. These stations include Abisko (Dorrepaal et al., 2004), Svanhovd (BioForsk Svanhovd, http://lmt.bioforsk.no/agrometbase/getweatherdata_new.php?weatherStationId=36, last access: 1 January 2022), Inuvik (National Resources Canada), Svalbard (Global Terrestrial Network for Permafrost database, http://gtnpdatabase.org/boreholes/view/166, last access: 1 January 2022), Toolik Lake (Hobbie and Laundre, 2021), the ForHot research site in Iceland (Sigurdsson et al., 2016), the Kilpisjärvi ITEX site (Sari Stark, personal communication, 2022), and various Greenland sites (Green Ecosystem monitoring database, https://data.g-e-m.dk/, last access: 1 January 2022). To overcome differences in the time intervals for the data collection between sites, we calculated the mean daily temperature for each day that soil temperature records were available (all records >3 years, except for Kilpisjärvi; Table 1). Based on the daily soil temperature records of each soil, we determined the mean annual daily temperature (MAST), mean warmest day (STmax), and mean coldest day (STmin), based on the annual records for the warmest day, coldest day, and mean daily temperature per year (Table 2).
Table 1Thermal regimes of the 12 sampling sites in °C. STmax depicts the warmest day of the year, MAST is the mean annual temperature, and STmin is the coldest day of the year. ± indicates standard deviation of the mean value recorded of the temperature record from the first year (Start) till the last year (End). The fourth column (Depth) indicates the soil temperature logger depth in centimeters.
2.3 Soil analysis
After collection, soils were shipped on ice and cooled upon arrival at 4 °C. The upper 10 cm was sampled, and the density of the soil samples was determined by rapid submersion in a water-filled cylinder. The water content was calculated based on the weight before and after drying the soil at 70 °C for 48 h. The dried samples were ground with a Retsch MM200 ball mill (Retsch GmbH, Haan, Germany) for 1 min at 30 rounds per second. A subsample was then ashed at 600 °C for 6 h. The carbon and nitrogen contents of ashed and non-ashed subsamples were measured on a Flash elemental analyzer (EA) 1112 (Thermo Fisher Scientific, Waltham, USA). For the calculation of organic carbon in the soil, the carbon content of the ashed samples was subtracted from the total amount of carbon content. Soil pH was measured by adding 5 g of soil to 25 mL deionized water, after which the slurries were shaken for 1 h at 100 rpm (revolutions per minute). The soil pH was then measured on a WTW inoLab® level 2 pH meter (Xylem Analytics, Rye Brook, New York, USA). The slurry was then centrifuged at 200 rpm for 1 h and then filtered on a 0.45 µm nylon filter. The filtered solution was used for the measurement of the extractable dissolved organic carbon content on a total organic carbon analyzer (TOC-L CPH/CPN; Shimadzu, Columbia, USA), with the NPOC method (to determine the dissolved organic carbon) as per the manufacturer's protocol. For the soil samples of Svalbard, only pH measurements were performed due to limited amounts of soil.
2.4 Temperature–growth relationships of soil bacterial communities
For the assessment of the temperature sensitivity of bacterial growth, 1 g of soil was subsampled for a leucine incorporation assay using methods adjusted from Bååth et al. (2001). Briefly, 20 mL of sterilized deionized water was added to the soil samples, and these slurries were vortexed for 2 min at full speed. After 10 min centrifugation at 1000 g, the 1 mL aliquots of the supernatant were suspended in 2 mL screw-top Eppendorf tubes. A 20 µL mixture of 3H-labeled and unlabeled leucine was added, resulting in a final concentration of 401 nM and 72.5 kBq mL−1 in the assay tube. The sample aliquots were incubated either for 24 h at 0 °C, 8 h at 4 °C, 4 h at 10 and 15 °C, or 2 h at 24.5, 28.5, 33, or 40 °C. Trichloroacetic acid was added to the assay tubes to terminate the leucine incorporation. Washing steps for removal of non-incorporated leucine were followed, as described in Bååth et al. (2001). For scintillation 1 mL OptiPhase HiSafe 3 (PerkinElmer, Waltham, Massachusetts, USA) was finally added to the biomass pellet after the washing steps. The 3H activity was measured on a Tri-Carb 2800T (PerkinElmer), Waltham, MA, USA) with 5 min measurement for 3H. Finally, the leucine incorporation rate, nanomolarity (nM) leucine per hour per gram of dry weight soil, was calculated based on the 3H activity measured.
2.5 Bacterial community composition
For the characterization of the soil microbial community, 0.2 g of soil were subsampled for DNA extraction and amplicon sequencing of the 16S rRNA gene. DNA was extracted by the use of PowerSoil kit (QIAGEN, Hilden, Germany), following the manufacturer's protocol, with the elution of the purified DNA into 60 µL sterile Millipore water. Amplicons were generated by a two-step polymerase chain reaction (PCR) of the 16S V4 rRNA gene, with primers designed by Caporaso et al. (2012). An initial PCR consisted of 24 cycles, with an initial denaturation step of 1 min at 98 °C, followed by 25 cycles of denaturation for 10 s at 98 °C, annealing for 30 s at 55 °C, elongation for 30 s at 72 °C, and followed by a final extension of 5 min at 72 °C. Amplicons were then 50× diluted in σ-purified water and then indexed by a PCR with unique barcoded primers for eight cycles, with the same steps as for the initial PCR amplification. Purification of the PCR product was done with AMpure XP beads (Beckman Coulter, Inc., Brea, California, USA), following the manufacturer's protocol. The indexed PCR products were then sequenced using paired-end Illumina MiSeq runs with V3 2x300 cycle chemistry. In total, 1 243 600 sequences were generated for 39 samples (median sequencing depth; 32 089 sequences per sample). Sequences were truncated at 250 nucleotides on the forward reads and 220 nucleotides on the reverse reads due to the deteriorating quality scores for later cycles (average Phred score <30). Raw sequences are available in the National Center for Biotechnology Information (NCBI) Sequence Archive (under BioProject accession no. PRJNA857550). Amplicon sequence variants (ASVs) were generated by dereplication and chimera removal of the truncated sequences, using the DADA2 algorithm and allowing a maximum expected error of 2 and consensus chimera removal mode. Phylogenetic distances between the ASVs were estimated using a MAFFT alignment (Katoh and Standley, 2013) and FastTree (Price et al., 2009). Taxonomic classification of the ASVs was performed based on the SILVA v138 database (Yilmaz et al., 2014), using a scikit-learn Naive Bayes machine-learning classifier (Bokulich et al., 2018) with a confidence threshold for limiting taxonomic depth at 70 %. ASVs identified as mitochondria, or chloroplasts or singletons were discarded prior to further statistical analyses.
2.6 Statistical analyses
All statistical analyses were performed in R (v4.0.2; R Core Team, 2020). Soil daily temperature records were filtered for data points between 2002 and 2021. Leucine incorporation rates were fitted to a Ratkowsky model for bacterial growth (Ratkowsky et al., 1983) by the use of the R package nls.multistart (Padfield and Matheson, 2018). The Ratkowsky model is based on the following equation:
where Leu is the rate of leucine incorporation, a is the coefficient below optimal growth temperature, T is the assay temperature, Tmin is the theoretical minimum growth temperature, and b is the coefficient above the optimal growth temperature and Tmax is maximum growth temperature. The optimal temperature was determined by numerical interpolation. All figures were made with the ggplot2 R package. To test for the effects of soil thermal parameters on the temperature adaptation of soil bacterial communities, we performed a linear regression between the cardinal points of the temperature–growth relationships and minimum (STmin), mean (MAST), and maximal annual soil temperature (STmax). These linear regression models tested the relationship between Tmin and minimum soil temperature, Tmax and the maximum soil temperature, and Topt with minimum, mean, and maximum soil temperature as an independent variable. We fitted a linear regression for the relationship between the temperature range (Tmin–Tmax) of the temperature–growth relationships of the soil bacterial communities and amplitude of thermal soil regime (minimum STmin to maximum soil temperature STmax) with a linear regression model.
Processing the microbial community data was done using the R package phyloseq (McMurdie and Holmes, 2013). Samples were rarified to depth of 23 687 reads. Permutational multivariate analysis of variance (PERMANOVA; Anderson, 2001) was performed on the weighted unique fraction metric (UniFrac) distances (Lozupone and Knight, 2005) of the sample of the 11 sites, which exclude Svalbard due to lack of data (Table S1 in the Supplement), using the mean annual soil temperature, pH, organic carbon content, organic nitrogen content, community Topt, and one of the three soil temperature variables (mean, maximum, or minimum daily soil temperature) as independent variables in the vegan R package. We determined the common ASVs by filtering for mean relative abundance above 0.001 % in at least two, three, or four soil types. The relative abundance of the common ASVs in the three datasets was used to predict the Topt of the soil bacterial communities. The relative abundance of these common ASVs was then used to perform three types of a regression tree analysis on the Topt of soil bacterial communities, using the R package caret (Kuhn, 2008). Data were randomly split into training (nine soils) and validation (three soils) datasets, after which a regression tree analysis was performed with the rpart1SE function, using the control settings (maxdepth = 4, minsplit = 4, and minbucket = 2). We also built a regression tree with cross validation (10 folds; 10 repeats), using the rpart function with the same control settings. Additionally, we used the Rborist function with the default setting to calculate a random forest regression tree to predict Tmin based on the relative abundance of common ASVs in the training soils. For direct comparison with regression models, we performed an additional linear regression using Topt as an independent variable and STmax as a dependent variable, using the nine soils of the training dataset and three soils in the validation dataset. Due to the small datasets that these models were based on, the random division into training and validation datasets had a strong influence on the computed RMSE (root mean square error) value. Therefore, we trained each of the four models on all 220 possible combinations of soils in the training and validation dataset of the dataset with common ASVs found in four soil types (with a 9 : 3 split between soil for training and testing, respectively). We then compared the performance of the four different models based on median RMSE over the 220 simulations.
3.1 Temperature adaptation of soil bacterial communities
From the sub- to High Arctic, mean annual soil temperatures at 10 cm depth varied between −3.5 and 6.1 °C (Table 1, Fig. 1). The sampled bacterial communities varied in Tmin between −11.1 ± 4 (SD) in Østerlien and −5.5 ± 2.1 in the Icelandic grassland. Topt varied between 23.4 ± 0.5 in Toolik Lake MAT and 34.1 ± 3.7 in Kilpisjärvi (Fig. 2). Tmax varied between 42.2 ± 1.0 in Svalbard and 57.8 + 9.3 at Toolik Lake heath. The temperature range of growth (Tmin–Tmax) varied between 48.7 and 65.2 °C.
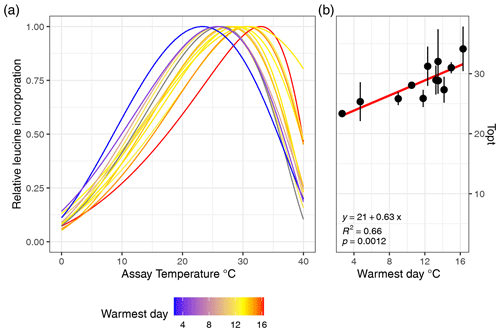
Figure 2(a) Estimated growth curves for each soil type depicted by the normalized leucine incorporation over incubation temperature. Colors indicate the maximum soil temperature of each sampling site. (b) Linear relationships between the optimal growth temperature and maximum soil temperature are shown, and the error bars indicate the standard error.
The MAST of soils was not significantly related to Topt (P=0.5) nor was Tmin (P=0.78; adjusted ). However, Topt did relate significantly to STmax, which increased by 0.63 °C °C−1 (Fig. 2; P<0.01; adjusted R2=0.63) and also showed a significant but weaker relationship to the mean summer temperature (P<0.05; adjusted R2=0.34). In contrast, Topt was not significantly correlated to the number of days above 0 °C (P=0.9). Tmin showed no significant correlation to mean winter soil temperature and coldest daily soil temperature (P>0.05). The temperature range of growth was significantly related to the amplitude of the temperature soil temperature (linear regression; adjusted R2=0.3; ). We computed the optimal growth temperature of soil bacterial communities across the Arctic based on combining the SoilTemp database (Lembrechts et al., 2021) with our estimates of the relationship between soil temperature (STmax) and Topt (Fig. 4).
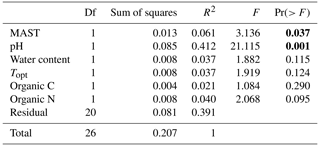
3.2 Bacterial community composition
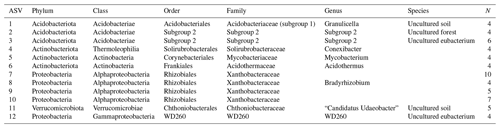
After filtering for singletons, we retrieved a total of 967 146 reads across the samples, which belonged to 12 692 ASVs. PERMANOVA analyses showed that the bacterial community composition was significantly influenced by pH and MAST of the sampling sites (Table 3; Fig. 5) but showed no significant correlation with maximum nor minimum daily soil temperature (P>0.05; Tables S2 and S3). The bacterial community composition was not significantly related to the Topt of the bacterial communities (P=0.124). Proteobacteria (25.9 %), Acidobacteriota (21.9 %), Actinobacteriota (18.4 %), Verrucomicrobiota (7 %), Bacteroidota (6.7 %), Chloroflexi (5.2 %), Planctomycetota (5.1 %), and Myxococcota (2.1 %) were the most abundant phyla across all samples (Fig. 3).
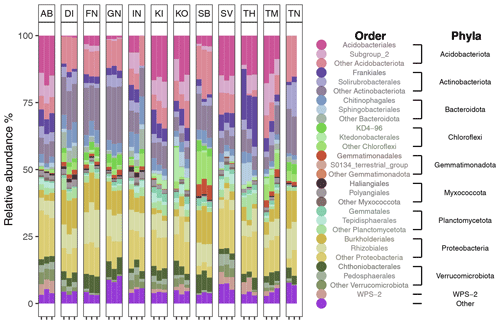
Figure 3Bar plot showing the relative abundance (%) of the top 10 most abundance phyla across all soil samples. Color shades indicate the two most abundant orders for each of these phyla.
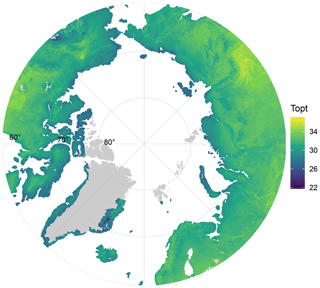
Figure 4Map of the predicted Topt of soil bacterial communities across the Arctic, based on the linear relationship between maximum soil temperature (from SoilTemp database) and Topt.
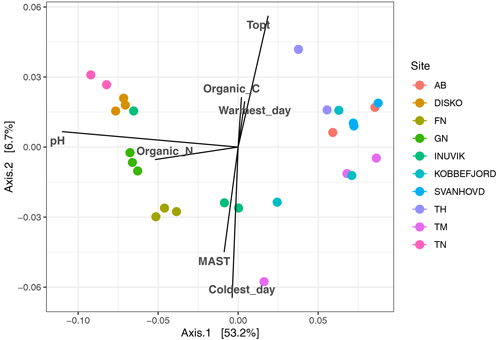
Figure 5Principal coordinate analysis of bacterial community composition (16s rRNA amplicon sequencing), based on weighted UniFrac distances with the correlation of measured environmental factors.
Table 2Description of the abbreviations used to described temperature–growth relationships and soil thermal regimes.
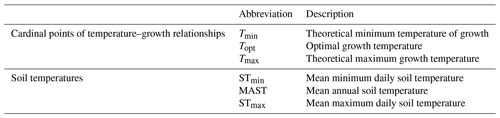
Table 3Permutational multivariate analysis of variance (PERMANOVA), showing the influence of mean annual soil temperature and other environmental factors on the weighted unique fraction metric (UniFrac) distance between 16 rRNA gene sequence from soil bacterial communities across the 11 sampled sites. Bold values indicate the significance (P<0.05).
We found 118 ASVs and 32 ASVs that were observed in two or three soils with a relative abundance greater than 0.001 %. Additionally, we observed only 12 ASVs that occurred at relative abundance greater than 0.001 % in four or more sites (Table 4). The common ASV datasets differing in cutoffs showed similar RMSE values for predicting the Topt of the bacterial community (Table S4). We therefore focused on the dataset with ASVs that occurred in at least four soils for further analysis, as these ASVs are most likely useful in soil types outside of this study. Both regression tree and random forest analyses based on the relative abundance of these common ASVs in at least four soils showed that the relative abundance of ASV11 was the best predictor of the corresponding community of Topt (Fig. S1 in the Supplement), in which it differentiated between the ASV11 absence from the community and relative abundance of >0.055 %. The pruned regression tree showed a RMSE lower than the full tree on the validation dataset (Fig. S2; Table S4). The linear regression model based on the STmax, as a dependent variable, showed the larger predictive power of Topt than the pruned regression tree and random forest. Since it had been summarized across the 220 possible training sets, the median RMSE of the linear model was lower than that median RMSE of the pruned tree and random forest for common ASVs found in at least four soil types (2.17, 4.14, and 3.51, respectively; Fig. S3).
4.1 Temperature adaptation across the Arctic
In this study, we explored the role of soil thermal parameters on the temperature adaptation of soil bacterial communities in the Arctic. The cardinal points estimated from bacterial communities sampled at 12 Arctic locations were comparable to other bacterial communities from polar soils and showed a large variety between sites and soil types. We found Tmin to vary between −11.1 and −5.5 °C, which is comparable to soils sampled from sub-Arctic soils (Cruz-Paredes et al., 2021b; Rinnan et al., 2011; Weedon et al., 20223). Tmin was lowest at the low Arctic site Østerlien, which is lower than any the Tmin of those previously described for Arctic soil bacterial community but fits within the range of Tmin of bacterial communities previously described in Antarctic soils (Rinnan et al., 2009). In contrast to Tmin, Topt is hypothesized to vary less between thermal environments (Rinnan et al., 2009). At the Toolik Lake moist acidic tundra site, estimated Topt was 23.5 °C, which is so far the lowest Topt described for a soil bacterial community in the Arctic (Rinnan et al., 2011; Weedon et al., 2023; Cruz-Paredes et al., 2021b) and is also comparable to soil bacterial communities from Antarctica (Donhauser et al., 2020; Rinnan et al., 2009, 2011; van Gestel et al., 2020). This site was characterized by relatively low summer temperatures and moderate annual mean temperatures compared to the other sites (Table 1; Fig. 1).
4.2 Temperature adaptation is influenced by mean daily maximum soil temperature
Of the soil thermal parameters we tested, only STmax had a significant correlation with temperature–growth relationships of Arctic bacterial communities (Fig. 2). Temperatures above the optimum growth temperature can induce heat-related death of bacterial cells, which results in a strong selective pressure by the maximum soil temperature on the bacterial community (Bárcenas-Moreno et al., 2009; Birgander et al., 2013; Donhauser et al., 2020). Consequently, the optimal growth temperature of soil bacterial communities is always observed to greatly exceed the maximum soil temperatures at a given location (Bárcenas-Moreno et al., 2009; Birgander et al., 2018; van Gestel et al., 2013; Rinnan et al., 2009). Our results show that, even in cold biome environments, the maximum soil temperature is an important determinant of the temperature physiology of soil bacterial communities. For two of our sites (Inuvik and Svalbard), soil temperatures were recorded at a different depths where the soil samples were taken (Table 1). Excluding these sites from our analysis, we still found the same relationship between Topt and the maximum soil temperature, with a slope coefficient and intercept differing by less than 2 % (File S1, Methods, in the Supplement). While samples in this study were collected in summer, the temperature–growth relationships are not affected by seasonal dynamics (van Gestel et al., 2013; Birgander et al., 2018), making it likely that the STmax is the most important predictor of thermal adaptation amongst those we measured. All in all, the evidence collected in this study provides further support for the idea that the temperature adaptation of soil microbial communities is best explained by the optimum-driven hypothesis (Alster et al., 2020). According to this hypothesis, temperature–growth relationships are driven by the maximum soil temperatures, and this was previously proposed, as temperature adaptation could only be induced after the exposure of communities to conditions above a certain threshold temperature (Bárcenas-Moreno et al., 2009; Birgander et al., 2013, 2018).
4.3 No evidence for influence of soil thermal parameters on Tmin or Tmax
In contrast to the clear relationship between STmax and Topt, we found no evidence for a relationship between soil thermal parameters, neither for the minimum and maximum cardinal points nor with the thermal breadth of the bacterial temperature–growth relationships. This non-significance could, in principle, be a result of statistical artifacts since, for the estimation of Tmin and Tmax, both cardinal points are extrapolated beyond the assay temperatures, which could cause a large standard deviation of the mean and increase the chance of type II errors. Indeed, the mean of site-level standard deviations across sites was relatively high for both Tmin and Tmax (respectively, with a mean SD of 1.94 and 2.8). However, this variation was of the same order as that observed for Topt estimates amongst the sampled soil bacterial communities (mean SD of 2.06), implying that the lack of significance is most likely not due to limited power of the statistical analysis.
Given the importance of Tmin for determining activity at low temperatures, we expected that the Tmin of communities would be related to site STmin. However, we did not detect a significant influence of STmin on the Tmin of soil bacterial communities. There is a general consensus that constantly frozen subsoils (permafrost) are an unlikely environment for the proliferation of soil microbial life (Abramov et al., 2021). Due to this limited growth, cold-adapted (low Tmin) species might not necessarily thrive at subzero temperatures but are likely to be better equipped to survive the winter conditions. Consequently, winter temperatures might not pose an environmental filter for the community assembly. Soil temperatures above freezing might have a larger influence on the temperature adaption of soil bacterial communities, when soil bacteria are most metabolically active (van Gestel et al., 2020). Therefore, the high soil temperatures in summer might induce a large environmental influence on the assembly of the bacterial communities. Additionally, strategies to survive subzero temperatures might not necessarily be indicative of the optimal growth temperature, as many microbial species that can cope with subzero temperature still grow best at relatively high temperature and are best described as psychrotolerant rather than as true psychrophiles (Cavicchioli, 2015). These factors might therefore be the reason why we are unable to make predictions of Tmin based on the temperature parameters measured in this study.
Since STmax influenced the Topt of the soil bacterial communities, we expected that this parameter would also correlate with the Tmax value of the soil bacterial community. Tmax has been hypothesized to increase with higher soil temperatures (Rinnan et al., 2009; Birgander et al., 2013), but to date, this has not been directly tested. In our results, Tmax was not influenced by any of the measured soil thermal parameters. As noted above, Topt was far above the maximum soil temperatures, which suggests that the measured growth rates of bacterial communities above Topt are rarely relevant in the soil environment. Therefore, it is likely that Tmax is less relevant for the performance of soil bacterial species and, consequently, not subject to the selection in the sense of Vellend (2010).
4.4 What will happen in response to warming?
Since STmax was found to be the most important predictor, it follows that changes to summer temperatures are likely to be the most important factor determining the temperature–growth relationships of bacterial communities in Arctic soils under a changing climate. Arctic summer air temperatures are predicted to increase by less than the mean annual and winter temperature (Karjalainen et al., 2019). While it has been estimated that the mean annual soil temperature will rise by ∼ 2–4 °C around the Arctic by 2100 under RCP4.5 (Representative Concentration Pathway 4.5; Aalto et al., 2018), accurate predictions of summer soil temperature in the Arctic are complicated by a variety of environmental factors that influence soil temperature. At the local scale, soil temperatures are largely influenced by air temperature, solar radiation, and precipitation (Karjalainen et al., 2019), leading to >5 °C variation on the microscale (Aalto et al., 2013; Karjalainen et al., 2019). Increasing air temperatures in the Arctic can also lead to changes in vegetation height and shrub expansion (Mekonnen et al., 2021), which moderately increase the soil temperature by shading during the summer season (Paradis et al., 2016; Blok et al., 2011). Furthermore, it is likely that the Arctic terrestrial region will experience more frequent and extreme heat waves, which could induce rapid change in the temperature–growth relationships if soil temperatures exceed historical maximum soil temperatures and/or the Topt of the soil bacterial communities (Bárcenas-Moreno et al., 2009; Birgander et al., 2013; Donhauser et al., 2020). These complicated local-scale effects imply that more microclimatic data will be needed for more accurate assessments of temperature adaptation of soil bacterial communities in the Arctic.
Our study covered a large portion of the range of maximum soil temperature within the Arctic region, as these temperatures currently vary between −0.4 and 20.6 °C (Lembrechts et al., 2021). Figure 4 shows that the Topt of Arctic soil bacterial communities likely varies between 22 and 35 °C. A combination of this pan-Arctic projection, predicted future summer (soil) temperatures, and other spatial databases such as soil C maps, could be useful to identify locations in where soil bacterial communities will be sensitive to future warming, where potential shifts in the temperature–growth relations can occur, and where these may have disproportionate impacts on regional biogeochemistry, e.g., by identifying regions in which local soil temperatures are expected to rise rapidly and soil organic stocks are large.
4.5 Can we use microbial community data for predicting temperature adaption?
Predicting the temperature adaptation of soil bacterial communities across the Arctic might be limited by the lack of long-term soil temperature data across the Arctic, as most Arctic research has focused on only few research sites (Metcalfe et al., 2018). To explore the potential use of microbial bio-indicators for predicting the temperature–growth relationships of in situ soil bacterial communities (Hicks et al., 2021), we evaluated whether microbial community data can reveal the temperature adaptation of microbial communities. We found that regression tree analysis using bacterial ASVs as potential predictors (Fig. S4) produced larger estimation errors in the prediction of the Topt of soil bacterial communities when compared with the linear regression against STmax (Table S4). This can be partially attributed to the low effective sample sizes resulting from the use of cross-validation methods to prune the regression trees but likely also reflects a lack of consistent signal in the bacterial composition data. Although these results do not refute the potential for using compositional data to predict community-wide temperature–growth relationships (Hicks et al., 2021), it implies that such methods would need a larger training dataset with more sample sites for proper validation and more accurate predictions. The full regression tree used a low number of ASVs (Fig. S2; Table S5), which were not observed in all soil types and which might indicate limited use for other datasets. This suggests that indicator species, if they exist, might be indicative of the temperature adaptation of bacterial communities for only certain, particular soil types or climatic regions. Despite these caveats, it is notable that the pruned regression tree and random forest model both identified the abundance of ASV11 as effective for the prediction of the Topt (Fig. S1). ASV11 matches 100 % to ASV that is the most commonly observed bacterial in Arctic soils (Malard et al., 2019). The genus of “Candidatus Udaeobacter”, to which ASV11 matches, is commonly found in soil environments globally (Brewer et al., 2016). It has been proposed to be a small oligotrophic and resilient soil bacteria characterized by aerobic heterotrophic metabolism, with a small genome size (2.8–3.2 Mbp), large diversity of antibiotic resistance genes, and a preference for acidic soils (Brewer et al., 2016; Willms et al., 2020, 2021). However, so far no study has successfully cultivated the any lineage of the genus “Candidatus Udaeobacter”, and traits related to temperature preferences have not been recorded. In the pruned tree (Fig. S4), the presence or absence of ASV11 was indicative of a Topt of 26.3 or 31.4 °C, respectively. As this taxon was absent in 7 out of 12 soils, the utility as an indicator of temperature adaption is quite limited. In summary, although there is some potential utility in using community data to estimate and predict aspects of soil bacterial temperature physiology, our results suggest that more accurate predictions can be made from soil temperature records.
In this study, we showed a large variety in the temperature adaptation of soil bacterial communities from the sub- to High Arctic region. Due to the large influence of maximum soil temperatures, we predict that summer warming, to the extent that it leads to higher maximum soil temperatures, will lead to a community-level increase in the Topt of these bacterial communities under future climate conditions in the Arctic. The influence of shifting optimal growth temperature for soil bacterial communities on soil carbon cycling will need further investigation to evaluate the contribution to the vulnerability of soil carbon stocks in the Arctic under future climate conditions.
The data used for statistical analysis and visualization are available from Figshare (https://doi.org/10.6084/m9.figshare.20977588, Rijkers, 2023a; https://doi.org/10.6084/m9.figshare.20977582, Rijkers, 2023b; and https://doi.org/10.6084/m9.figshare.20977585, Rijkers, 2023c). The raw sequences of the 16S rRNA gene amplicon sequences have been deposited on NCBI's Sequence Read Archive (under BioProject PRJNA857550).
The supplement related to this article is available online at: https://doi.org/10.5194/bg-20-767-2023-supplement.
RR and JTW designed the study. MD and RR performed soil analysis and bacterial community analysis. RR performed leucine assays and statistical analysis. All authors contributed to writing of the paper.
The contact author has declared that none of the authors has any competing interests.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the special issue “Global change effects on terrestrial biogeochemistry at the plant–soil interface”. It is not associated with a conference.
We thank all involved partners in the INTERACT network, for the collaboration for the (remote) field campaign 415 during the COVID-19 pandemic, especially Katrine Raundrup, Bo Elberling, Birgitte Kortegaard Danielsen, Cornelia Klutsch, Juho Vuolteenaho, Erika Hille, Caroline Duchesne, Joel McAlister, Sari Stark, Rauni Partanen, and Oula M. Kalttopää. We also thank Lucie Malard, for providing information on the bacterial ASVs commonly found across the Arctic.
The research leading to these results has been funded by the European Union's Horizon 2020 project INTERACT (grant no. 730938). Ruud Rijkers and James T. Weedon have been supported by the NWO Netherlands Polar Program (project ID 866.16.042).
This paper was edited by Emily Solly and reviewed by Natasja van Gestel and Aline Frossard.
Aalto, J., Le Roux, P. C., and Luoto, M.: Vegetation Mediates Soil Temperature and Moisture in Arctic-Alpine environtments, Arct. Antarct. Alp. Res., 45, 429–439, 2013.
Aalto, J., Karjalainen, O., Hjort, J., and Luoto, M.: Statistical Forecasting of Current and Future Circum-Arctic Ground Temperatures and Active Layer Thickness, Geophys. Res. Lett., 45, 4889–4898, https://doi.org/10.1029/2018GL078007, 2018.
Abramov, A., Vishnivetskaya, T., and Rivkina, E.: Are permafrost microorganisms as old as permafrost?, FEMS Microbiol. Ecol., 97, 1–12, https://doi.org/10.1093/femsec/fiaa260, 2021.
Alster, C. J., von Fischer, J. C., Allison, S. D., and Treseder, K. K.: Embracing a new paradigm for temperature sensitivity of soil microbes, Glob. Change Biol., 26, 3221–3229, https://doi.org/10.1111/gcb.15053, 2020.
Anderson, M. J.: A new method for non-parametric multivariate analysis of variance, Austral. Ecol., 26, 32–46, https://doi.org/10.1046/j.1442-9993.2001.01070.x, 2001.
Bååth, E.: Temperature sensitivity of soil microbial activity modeled by the square root equation as a unifying model to differentiate between direct temperature effects and microbial community adaptation, Glob. Change Biol., 24, 2850–2861, https://doi.org/10.1111/gcb.14285, 2018.
Bååth, E., Pettersson, M., and Söderberg, K. H.: Adaptation of a rapid and economical microcentrifugation method to measure thymidine and leucine incorporation by soil bacteria, Soil Biol. Biochem., 33, 1571–1574, https://doi.org/10.1016/S0038-0717(01)00073-6, 2001.
Bárcenas-Moreno, G., Brandón, M. G., Rousk, J., and Bååth, E.: Adaptation of soil microbial communities to temperature: Comparison of fungi and bacteria in a laboratory experiment, Glob. Change Biol., 15, 2950–2957, https://doi.org/10.1111/j.1365-2486.2009.01882.x, 2009.
Birgander, J., Reischke, S., Jones, D. L., and Rousk, J.: Temperature adaptation of bacterial growth and 14C-glucose mineralisation in a laboratory study, Soil Biol. Biochem., 65, 294–303, https://doi.org/10.1016/j.soilbio.2013.06.006, 2013.
Birgander, J., Olsson, P. A., and Rousk, J.: The responses of microbial temperature relationships to seasonal change and winter warming in a temperate grassland, Glob. Change Biol., 24, 3357–3367, https://doi.org/10.1111/gcb.14060, 2018.
Blok, D., Schaepman-Strub, G., Bartholomeus, H., Heijmans, M. M. P. D., Maximov, T. C., and Berendse, F.: The Response of Arctic Vegetation to the Summer Climate: relation between shrub cover, NVDI, surface albedo and temperature, Environ. Res. Lett., 6, 035502, https://doi.org/10.1088/1748-9326/6/3/035502, 2011.
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., Huttley, G. A., and Gregory Caporaso, J.: Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin, Microbiome, 6, 1–17, https://doi.org/10.1186/s40168-018-0470-z, 2018.
Bradford, M. A., McCulley, R. L., Crowther, T. W., Oldfield, E. E., Wood, S. A., and Fierer, N.: Cross-biome patterns in soil microbial respiration predictable from evolutionary theory on thermal adaptation, Nat. Ecol. Evol., 3, 223–231, https://doi.org/10.1038/s41559-018-0771-4, 2019.
Brewer, T. E., Handley, K. M., Carini, P., Gilbert, J. A., and Fierer, N.: Genome reduction in an abundant and ubiquitous soil bacterium “Candidatus Udaeobacter copiosus,” Nat. Microbiol., 2, 16198, https://doi.org/10.1038/nmicrobiol.2016.198, 2016.
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., Owens, S. M., Betley, J., Fraser, L., Bauer, M., Gormley, N., Gilbert, J. A., Smith, G., and Knight, R.: Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms, ISME J., 6, 1621–1624, https://doi.org/10.1038/ismej.2012.8, 2012.
Carey, J. C., Tang, J., Templer, P. H., Kroeger, K. D., Crowther, T. W., Burton, A. J., Dukes, J. S., Emmett, B., Frey, S. D., Heskel, M. A., Jiang, L., Machmuller, M. B., Mohan, J., Panetta, A. M., Reich, P. B., Reinsch, S., Wang, X., Allison, S. D., Bamminger, C., Bridgham, S., Collins, S. L., de Dato, G., Eddy, W. C., Enquist, B. J., Estiarte, M., Harte, J., Henderson, A., Johnson, B. R., Larsen, K. S., Luo, Y., Marhan, S., Melillo, J. M., Peñuelas, J., Pfeifer-Meister, L., Poll, C., Rastetter, E., Reinmann, A. B., Reynolds, L. L., Schmidt, I. K., Shaver, G. R., Strong, A. L., Suseela, V., and Tietema, A.: Temperature response of soil respiration largely unaltered with experimental warming, P. Natl. Acad. Sci. USA, 113, 13797–13802, https://doi.org/10.1073/pnas.1605365113, 2016.
Cavicchioli, R.: On the concept of a psychrophile, ISME J., 10, 793–795, https://doi.org/10.1038/ismej.2015.160, 2015.
Convey, P.: Antarctic Ecosystems, Encyclopedia of Biodiversity, 2nd Edn., Elsevier, 179–188, https://doi.org/10.1016/B978-0-12-384719-5.00264-1, 2013.
Crowther, T., Todd-Brown, K. E. O., Rowe, C., Wieder, W., Carey, J., Machmuller, M., Snoek, L., Fang, S., Zhou, G., Allison, S., Blair, J., Bridgham, S., Burton, A., Carrillo, Y., Reich, P., Clark, J., Classen, A., Dijkstra, F., Elberling, B., Emmett, B., Estiarte, M., Frey, S., Guo, J., Harte, J., Jiang, L., Johnson, B., Kröel-Dulay, G., Larsen, K., Laudon, H., Lavallee, J., Luo, Y., Lupascu, M., Ma, L., Marhan, S., Michelsen, A., Mohan, J., Niu, S., Pendall, E., Penuelas, J., Pfeifer-Meister, L., Poll, C., Reinsch, S., Reynolds, L., Schmidth, I., Sistla, S., Sokol, N., Templer, P., Treseder, K., Welker, J., and Bradford, M.: Quantifying global soil C losses in response to warming, Nature, 540, 104–108, https://doi.org/10.1038/nature20150, 2016.
Cruz-Paredes, C., Tájmel, D., and Rousk, J.: Can moisture affect temperature dependences of microbial growth and respiration?, Soil Biol. Biochem., 156, 108223, https://doi.org/10.1016/j.soilbio.2021.108223, 2021a.
Cruz Paredes, C., Tajmel, D., and Rousk, J.: Microbial temperature adaptation across a European gradient, EGU General Assembly 2021, online, 19–30 Apr 2021, EGU21-4867, https://doi.org/10.5194/egusphere-egu21-4867, 2021b.
Donhauser, J., Niklaus, P. A., Rousk, J., Larose, C., and Frey, B.: Temperatures beyond the community optimum promote the dominance of heat-adapted, fast growing and stress resistant bacteria in alpine soils, Soil Biol. Biochem., 148, 107873, https://doi.org/10.1016/j.soilbio.2020.107873, 2020.
Dorrepaal, E., Aerts, R., Cornelissen, J. H. C., Callaghan, T. V., and Van Logtestijn, R. S. P.: Summer warming and increased winter snow cover affect Sphagnum fuscum growth, structure and production in a sub-arctic bog, Glob. Change Biol., 10, 93–104, https://doi.org/10.1111/j.1365-2486.2003.00718.x, 2004.
García-Palacios, P., Crowther, T. W., Dacal, M., Hartley, I. P., Ye, S., and Bradford, M. A.: Evidence for large microbial-mediated losses of soil carbon under anthropogenic warming, Nat. Rev. Earth Environ., 2, 507–517, https://doi.org/10.1038/s43017-021-00178-4, 2021.
Hicks, L. C., Frey, B., Kj, R., Lukac, M., Moora, M., Weedon, J. T., and Rousk, J.: Towards a function-first framework to make soil microbial ecology predictive, Ecology, 103, e03594, https://doi.org/10.1002/ecy.3594, 2021.
Hobbie, S. and Laundre, J.: Hourly temperature and humidity data from the LTER Moist Non-acidic Tussock Experimental plots (MNT), Environmental Data Initiative [data set], https://doi.org/10.6073/pasta/a48892da5bc9eab27b18d2364dea6998, 2021.
Karjalainen, O., Luoto, M., Aalto, J., and Hjort, J.: New insights into the environmental factors controlling the ground thermal regime across the Northern Hemisphere: a comparison between permafrost and non-permafrost areas, The Cryosphere, 13, 693–707, https://doi.org/10.5194/tc-13-693-2019, 2019.
Katoh, K. and Standley, D. M.: MAFFT multiple sequence alignment software version 7: Improvements in performance and usability, Mol. Biol. Evol., 30, 772–780, https://doi.org/10.1093/molbev/mst010, 2013.
Kuhn, M.: Building predictive models in R using the caret package, J. Stat. Softw., 28, 1–26, https://doi.org/10.18637/jss.v028.i05, 2008.
Lembrechts, J. J., van den Hoogen, J., Aalto, J., et al.: Global maps of soil temperature, Glob. Change Biol., 28, 3110–3144, https://doi.org/10.1111/gcb.16060, 2021.
Lozupone, C. and Knight, R.: UniFrac: A new phylogenetic method for comparing microbial communities, Appl. Environ. Microbiol., 71, 8228–8235, https://doi.org/10.1128/AEM.71.12.8228-8235.2005, 2005.
Malard, L. A., Anwar, M. Z., Jacobsen, C. S., and Pearce, D. A.: Biogeographical patterns in soil bacterial communities across the Arctic region, FEMS Microbiol. Ecol., 95, 1–13, https://doi.org/10.1093/femsec/fiz128, 2019.
McMurdie, P. J. and Holmes, S.: Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data, PLoS One, 8, https://doi.org/10.1371/journal.pone.0061217, 2013.
Mekonnen, Z. A., Riley, W. J., Berner, L. T., Bouskill, N. J., Torn, M. S., Iwahana, G., Breen, A. L., Myers-Smith, I. H., Criado, M. G., Liu, Y., Euskirchen, E. S., Goetz, S. J., Mack, M. C., and Grant, R. F.: Arctic tundra shrubification: a review of mechanisms and impacts on ecosystem carbon balance, Environ. Res. Lett., 16, 053001, https://doi.org/10.1088/1748-9326/abf28b, 2021.
Metcalfe, D. B., Hermans, T. D. G., Ahlstrand, J., Becker, M., Berggren, M., Björk, R. G., Björkman, M. P., Blok, D., Chaudhary, N., Chisholm, C., Classen, A. T., Hasselquist, N. J., Jonsson, M., Kristensen, J. A., Kumordzi, B. B., Lee, H., Mayor, J. R., Prevéy, J., Pantazatou, K., Rousk, J., Sponseller, R. A., Sundqvist, M. K., Tang, J., Uddling, J., Wallin, G., Zhang, W., Ahlström, A., Tenenbaum, D. E., and Abdi, A. M.: Patchy field sampling biases understanding of climate change impacts across the Arctic, Nat. Ecol. Evol., 2, 1443–1448, https://doi.org/10.1038/s41559-018-0612-5, 2018.
Nottingham, A. T., Bååth, E., Reischke, S., Salinas, N., and Meir, P.: Adaptation of soil microbial growth to temperature: Using a tropical elevation gradient to predict future changes, Glob. Change Biol., 25, 827–838, https://doi.org/10.1111/gcb.14502, 2019.
Oliverio, A. M., Bradford, M. A., and Fierer, N.: Identifying the microbial taxa that consistently respond to soil warming across time and space, Glob. Change Biol., 23, 2117–2129, https://doi.org/10.1111/gcb.13557, 2017.
Padfield, D. and Matheson, G.: nls. multstart: robust non-linear regression using AIC scores, R Package version, 1, 1–5, https://cran.r-project.org/package=nls.multstart (last access: 12 February 2023), 2018.
Paradis, M., Lévesque, E., and Boudreau, S.: Greater effect of increasing shrub height on winter versus summer soil temperature, Environ. Res. Lett., 11, 085005, https://doi.org/10.1088/1748-9326/11/8/085005, 2016.
Pietikäinen, J., Pettersson, M., and Bååth, E.: Comparison of temperature effects on soil respiration and bacterial and fungal growth rates, FEMS Microbiol. Ecol., 52, 49–58, https://doi.org/10.1016/j.femsec.2004.10.002, 2005.
Post, E., Alley, R. B., Christensen, T. R., Macias-Fauria, M., Forbes, B. C., Gooseff, M. N., Iler, A., Kerby, J. T., Laidre, K. L., Mann, M. E., Olofsson, J., Stroeve, J. C., Ulmer, F., Virginia, R. A., and Wang, M.: The polar regions in a 2 °C warmer world, Sci. Adv., 5, eaaw9883, https://doi.org/10.1126/sciadv.aaw9883, 2019.
Price, M. N., Dehal, P. S., and Arkin, A. P.: Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix, Mol. Biol. Evol., 26, 1641–1650, https://doi.org/10.1093/molbev/msp077, 2009.
Ratkowsky, D. A., Lowry, R. K., McMeekin, T. A., Stokes, A. N., and Chandler, R. E.: Model for bacterial culture growth rate throughout the entire biokinetic temperature range, J. Bacteriol., 154, 1222–1226, https://doi.org/10.1002/14651858.CD002209.pub2, 1983.
R Core Team: R: A Language and Environment for Statistical Computing, 2020.
Rijkers, R.: Soil Temperature Arctic Sample Sites.csv, figshare [data set], https://doi.org/10.6084/m9.figshare.20977588, 2023a.
Rijkers, R.: Temperature-growth relationships Arctic soil bacterial communities – Cardinal points, figshare [data set], https://doi.org/10.6084/m9.figshare.20977582, 2023b.
Rijkers, R.: Soil Characteristics, figshare [data set], https://doi.org/10.6084/m9.figshare.20977585, 2023c.
Rijkers, R., Rousk, J., Aerts, R., and Weedon, J. T.: Optimal growth temperature of Arctic soil bacterial communities increases under experimental warming, Glob. Change Biol., 28, 6050–6064, https://doi.org/10.1111/gcb.16342, 2022.
Rinnan, R., Rousk, J., Yergeau, E., Kowalchuk, G. A., and Bååth, E.: Temperature adaptation of soil bacterial communities along an Antarctic climate gradient: Predicting responses to climate warming, Glob. Change Biol., 15, 2615–2625, https://doi.org/10.1111/j.1365-2486.2009.01959.x, 2009.
Rinnan, R., Michelsen, A., and Bååth, E.: Long-term warming of a subarctic heath decreases soil bacterial community growth but has no effects on its temperature adaptation, Appl. Soil Ecol., 47, 217–220, https://doi.org/10.1016/j.apsoil.2010.12.011, 2011.
Rousk, J., Frey, S. D., and Bååth, E.: Temperature adaptation of bacterial communities in experimentally warmed forest soils, Glob. Change Biol., 18, 3252–3258, https://doi.org/10.1111/j.1365-2486.2012.02764.x, 2012.
Sigurdsson, B. D., Leblans, N. I. W., Dauwe, S., Gudmundsdóttir, E., Gundersen, P., Gunnarsdóttir, G. E., Holmstrup, M., Ilieva-Makulec, K., Kätterer, T., Marteinsdóttir, B., Maljanen, M., Oddsdóttir, E. S., Ostonen, I., Peñuelas, J., Poeplau, C., Richter, A., Sigurdsson, P., Van Bodegom, P., Wallander, H., Weedon, J., and Janssens, I.: Geothermal ecosystems as natural climate change experiments: The ForHot research site in Iceland as a case study, Iceland. Agr. Sci., 29, 53–71, https://doi.org/10.16886/IAS.2016.05, 2016.
Tarnocai, C., Canadell, J. G., Schuur, E. A. G., Kuhry, P., Mazhitova, G., and Zimov, S.: Soil organic carbon pools in the northern circumpolar permafrost region, Global Biogeochem. Cy., 23, 1–11, https://doi.org/10.1029/2008GB003327, 2009.
van Gestel, N. C., Reischke, S., and Bååth, E.: Temperature sensitivity of bacterial growth in a hot desert soil with large temperature fluctuations, Soil Biol. Biochem., 65, 180–185, https://doi.org/10.1016/j.soilbio.2013.05.016, 2013.
van Gestel, N. C., Ducklow, H. W., and Bååth, E.: Comparing temperature sensitivity of bacterial growth in Antarctic marine water and soil, Glob. Change Biol., 26, 2280–2291, https://doi.org/10.1111/gcb.15020, 2020.
Vellend, M.: Conceptual synthesis in community ecology, Q. Rev. Biol., 85, 183–206, https://doi.org/10.1086/652373, 2010.
Wang, C., Morrissey, E. M., Mau, R. L., Hayer, M., Piñeiro, J., Mack, M. C., Marks, J. C., Bell, S. L., Miller, S. N., Schwartz, E., Dijkstra, P., Koch, B. J., Stone, B. W., Purcell, A. M., Blazewicz, S. J., Hofmockel, K. S., Pett-Ridge, J., and Hungate, B. A.: The temperature sensitivity of soil: microbial biodiversity, growth, and carbon mineralization, ISME J., 15, 2738–2747, https://doi.org/10.1038/s41396-021-00959-1, 2021.
Weedon, J. T., Bååth, E., Rijkers, R., Reischke, S., Sigurdsson, B. D., Oddsdóttir, E. S., Van Hal, J., Aerts, R., Janssens, I. A., and Van Bodegom, P. M.: Community adaptation to temperature explains abrupt soil bacterial community shift along a geothermal gradient on Iceland, Soil Biol. Biochem., 177, 108914, https://doi.org/10.1016/j.soilbio.2022.108914, 2023.
Wieder, W. R., Sulman, B. N., Hartman, M. D., Koven, C. D., and Bradford, M. A.: Arctic Soil Governs Whether Climate Change Drives Global Losses or Gains in Soil Carbon, Geophys. Res. Lett., 46, 14486–14495, https://doi.org/10.1029/2019GL085543, 2019.
Willms, I. M., Rudolph, A. Y., Göschel, I., Bolz, S. H., Schneider, D., Penone, C., Poehlein, A., Schöning, I., and Nacke, H.: Globally Abundant “Candidatus Udaeobacter” Benefits from Release of Antibiotics in Soil and Potentially Performs Trace Gas Scavenging, mSphere, 5, e00186-20, https://doi.org/10.1128/msphere.00186-20, 2020.
Willms, I. M., Bolz, S. H., Yuan, J., Krafft, L., Schneider, D., Schöning, I., Schrumpf, M., and Nacke, H.: The ubiquitous soil verrucomicrobial clade “Candidatus Udaeobacter” shows preferences for acidic pH, Env. Microbiol. Rep., 13, 878–883, https://doi.org/10.1111/1758-2229.13006, 2021.
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., Schweer, T., Peplies, J., Ludwig, W., and Glöckner, F. O.: The SILVA and “all-species Living Tree Project (LTP)” taxonomic frameworks, Nucleic Acids Res., 42, 643–648, https://doi.org/10.1093/nar/gkt1209, 2014.
Ylänne, H., Stark, S., and Tolvanen, A.: Vegetation shift from deciduous to evergreen dwarf shrubs in response to selective herbivory offsets carbon losses: Evidence from 19 years of warming and simulated herbivory in the subarctic tundra, Glob. Change Biol., 21, 3696–3711, https://doi.org/10.1111/gcb.12964, 2015.