the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Clouds influence the functioning of airborne microorganisms
Raphaëlle Péguilhan
Florent Rossi
Muriel Joly
Engy Nasr
Bérénice Batut
François Enault
Barbara Ervens
Airborne microorganisms can remain at altitude for several days, exposed to multiple environmental constraints that prevent or limit microbial activity, the most important of which is probably the lack of available liquid water. Clouds, i.e., air masses containing liquid water, could offer more favorable conditions. In order to investigate the influence of clouds on the functioning of airborne microorganisms, we captured aerosols in a nucleic acid preservation buffer from a high-altitude mountain meteorological station under cloudy and clear-atmosphere conditions and examined the metatranscriptomes. The specificities of aeromicrobiome's functioning in clouds and a clear atmosphere were then decrypted using differential expression analysis (DEA). The data reveal a higher RNA : DNA content in clouds than in the clear atmosphere, suggesting higher metabolic activity, and the overrepresentation of microbial transcripts related to energy metabolism, the processing of carbon and nitrogen compounds, intracellular signaling, metabolic regulations, and transmembrane transports. Stress response in clouds tends towards responses to osmotic shocks and starvation rather than oxidants in a clear atmosphere. Autophagy processes in eukaryotes (macropexophagy, i.e., the recycling of peroxisomes) could help to alleviate the limited amounts of nutrients in the restricted microenvironments provided by cloud droplets. The whole phenomenon resembles the rapid resumption of microbial activity in dry soils after rewetting by rain, which is known as the Birch effect and is described here for the first time for the atmosphere. This work provides unprecedented information on the modulations of an aeromicrobiome's functioning in relation to atmospheric conditions. In addition to contributing to the processing and fate of chemical compounds in the atmosphere, cloud-induced modulations of biological processes could have ecological repercussions by shaping airborne microbial diversity and their capacity to invade surface environments.
- Article
(5166 KB) - Full-text XML
-
Supplement
(3253 KB) - BibTeX
- EndNote
It is well established that biological material circulates in the atmosphere. This includes taxonomically and functionally diverse microorganisms, with frequent saprotrophs (Amato et al., 2007b; Tignat-Perrier et al., 2020), methylotrophs (Amato et al., 2007a), phototrophs (Dillon et al., 2020), and others that include obligatory or opportunist plant, animal, or human pathogens (Brown and Hovmøller, 2002). Once aerosolized as individual cells or fragments of biofilms, they can remain airborne for up to several days (Burrows et al., 2009) at concentrations typically ranging from ∼103 to (Amato et al., 2023; Šantl-Temkiv et al., 2022). Close to the ground, in the planetary boundary layer, the airborne microbial diversity reflects that of emitting surfaces and follows their spatial and temporal variations in relation to meteorological and (micro)climatic conditions (Bowers et al., 2011; Fierer et al., 2008; Gusareva et al., 2019; Prass et al., 2021; Tignat-Perrier et al., 2020). At high altitudes in the free troposphere, the plumes from multiple sources mix, which results in more evenly distributed assemblages (Péguilhan et al., 2021) at extremely low biomasses (Smith et al., 2018).
Viable microbial cells suspended in the air are exposed to conditions at the limits of life's capacities, including low water and nutrient availability, low temperatures, and high levels of UV radiation and oxidants (Šantl-Temkiv et al., 2022). Water availability in particular is among the most limiting factors in biological processes in nature (Stevenson et al., 2015).
Clouds are air masses where the relative humidity exceeds 100 %, resulting in the condensation of water vapor on the surfaces of aerosol particles, including microbial cells. This leads to the formation of droplets of a few micrometers in diameter (i.e., individual volumes of ), with a typical liquid water content of . Chemical compounds from the gas and particle phases dissolve into the aqueous phase, and complex chemical processes take place, with a notable influence on the composition of the air masses (Ervens et al., 2018; Herrmann et al., 2015; Lelieveld and Crutzen, 1990; Li et al., 2023). Microbiological processes can also, to some extent, participate in processing organic compounds and oxidants (Bianco et al., 2019; Khaled et al., 2021; Vaïtilingom et al., 2013). From the perspective of the microbiologist, cloud droplets can thus be considered short-lived aquatic microhabitats providing microorganisms with liquid water and a range of dissolved nutrients at nanomolar to micromolar concentrations (carboxylic acids, amino acids, ammonium, nitrate, and metals) (Deguillaume et al., 2014; Šantl-Temkiv et al., 2013). Bulk cloud water indeed proved during laboratory incubations to offer nutritional conditions compatible with microbial development, with impacts on the chemical composition (Amato et al., 2007a; Bianco et al., 2019; Sattler et al., 2001; Vaïtilingom et al., 2013). In addition, clouds, through precipitation, provide efficient access routes to the ground for microorganisms airborne at high altitude and thus contribute to aerial dissemination (Péguilhan et al., 2021; Woo and Yamamoto, 2020).
The highly diluted microbial biomass in the atmosphere, combined with a short residence time, makes any in situ assessment challenging, so how the functioning of living cells may be modulated during atmospheric transport remains largely unexplored. If conditions allow, airborne microorganisms can maintain or activate metabolic processes in response to environmental conditions (Amato et al., 2017; Hill et al., 2007; Klein et al., 2016; Šantl-Temkiv et al., 2018). For instance, bacteria (Sphingomonas aerolata) aerosolized in a simulation chamber increase ribosome numbers when exposed to volatile organic compounds (ethanol and acetic acid) and, thus, potentially, metabolic activity (Krumins et al., 2014). The data also suggest modulations of the energy metabolism of living bacteria in natural clouds in relation to oxidants (Wirgot et al., 2017).
So far, the current knowledge of microbial functioning in the atmosphere and clouds has thus been based almost exclusively on laboratory incubations of samples and isolated strains (Amato et al., 2007a; Bianco et al., 2019; Jousse et al., 2018; Vaïtilingom et al., 2013; Wirgot et al., 2019) or, at best, on experiments in atmospheric simulation chambers (Amato et al., 2015; Krumins et al., 2014), i.e., under conditions that do not fully reflect the in situ natural atmospheric conditions to which the cells are actually exposed. Metagenomics and, in particular, metatranscriptomics can provide instant snapshots of the biological processes taking place in a system. Over the last decade, the advent of high-throughput sequencing techniques has encouraged such approaches. These led to unprecedented insights into the functioning of microbiota in humans (Franzosa et al., 2014; Jorth et al., 2014), oceans (Salazar et al., 2019), rivers (Satinsky et al., 2014), soils (Rosado-Porto et al., 2022), and highly polluted environments (Chen et al., 2015). Clouds were explored once, revealing several biological processes that included responses to stresses, transport, and key catabolic and anabolic processes (Amato et al., 2019). By comparing cloud transcriptomes with other data available in the literature, this work highlighted functional peculiarities compared with surface biomes. Still, there is no information regarding possible specificities of microbial functioning in clouds compared with the clear, cloud-free atmosphere, which occupies most of the atmospheric volume.
Here, we postulate that clouds could act as atmospheric “oases”, i.e., specific masses providing water and nutrients to living organisms and allowing them to thrive within an otherwise vast and hostile atmospheric environment. By using an innovative combined non-targeted metagenomic and metatranscriptomic approach, we examine the functioning of airborne microbial cells in clouds as compared with a clear atmosphere, and we specify whether and which biological processes are indeed affected. Given that airborne particles, including bacteria, spend an average of 10 %–15 % of their atmospheric residence time in clouds (Ervens and Amato, 2020; Lelieveld and Crutzen, 1990), such oases would provide conditions of (temporary) habitats or “airborne ecosystems” and could therefore lead to enhanced survival, persistence, and dispersal of bacteria, similar to features of other dynamic environments. This study, based on unique and unprecedented datasets, provides valuable information regarding the active aeromicrobiome and its environmental drivers.
2.1 Sample collection
Samples were collected from the summit of the Puy de Dôme (PUY; 1465 m a.s.l.; 45.772° N, 2.9655° E; France; ∼400 km east of the Atlantic Ocean and ∼300 km north of the Mediterranean Sea), located in an area composed of deciduous forests and pastoral landscapes and exposed most of the time to air masses from the north and west (Deguillaume et al., 2014; Renard et al., 2020). This mountain station is part of the Cézeaux-Aulnat-Opme-Puy-de-Dôme (CO-PDD) instrumented platform network for atmospheric research (Baray et al., 2020). Meteorological variables are monitored, and the station is fully equipped for on-site sample processing and conditioning, including for microbiological and molecular analyses.
The main information pertaining to sample acquisition is summarized in Table 1. Totals of nine cloud and six clear-air events were sampled in 2019 and 2020 for periods of about 2 to 6 consecutive hours during the daytime. Under both conditions, two to four high-flow-rate impingers (HFRis; model DS6, Kärcher SAS, Bonneuil-sur-Marne, France) sampling with an airflow rate of 2 m3 min−1 were deployed in parallel. More details about these samplers and their applicability for collecting biological material are provided in Šantl-Temkiv et al. (2017). Nucleic acid analyses were carried out from samplers filled with filtered and autoclaved nucleic acid preservation (NAP) buffer solution (Camacho-Sanchez et al., 2013; Menke et al., 2017) as the collection liquid (1.7 L of 0.5X NAP for a clear atmosphere or 850 mL of 1X NAP for clouds, in order to account for the expected liquid evaporation or accumulation), following the procedures detailed in Péguilhan et al. (2023a, b), including decontamination and controls. The negative controls consisted of unexposed collection liquid and collection liquid exposed to the sampling tank for 10 min with the sampler off. These were taken immediately before sampling and processed in parallel to the samples. For the atmospheric samples, the volume of the collection liquid was checked by weighting every sampling hour and compensated if necessary with autoclaved ultrapure water. Samples and controls were processed immediately after sampling using the PUY station's microbiology facility and a laminar flow hood previously exposed to UV light for 15 min. The collection liquid from each individual sampler was filtered through 0.22 µm porosity mixed cellulose ester (MCE) filters (47 mm diameter; ref. 0421A00023; ClearLine®, Bernolsheim, France) using sterile Nalgene filtration units. The filters were rolled using sterile forceps and placed in 5 mL type-A bead tubes (ref. 740799.50; Macherey-Nagel, Hoerdt, France). A volume of 1200 µL MR1 lysis buffer (ref. 744351.125; Macherey-Nagel) was then added to each tube, and a bead-beating step of 10 min was performed using a Genie2 vortex set at maximum speed. Filters and lysates were finally stored at −80 °C in the bead tubes before further processing, as detailed in the next section. Meteorological variables during sampling were monitored by the PUY meteorological station, including temperature, relative humidity, liquid water content, wind speed, and direction (https://www.opgc.fr/data-center/public/data/copdd/pdd, last access: 20 February 2025). The planetary boundary layer height (BLH) was extracted from the ECMWF ERA5 global reanalysis (https://www.ecmwf.int/en/forecasts/datasets/reanalysis-datasets/era5, last access: 20 February 2025) (Hoffmann et al., 2019). The geographic origin of the sampled air masses was derived from 72 h backward trajectories computed using the Computing Atmospheric Trajectory (CAT) model (Baray et al., 2020), which used dynamical fields extracted from the ERA5 meteorological data archive with a spatial resolution of 0.125° for the present work. This tool was used to estimate percentages of air mass trajectory points in each of the eight direction sectors (Renard et al., 2020).
Table 1Conditions of sample acquisition.
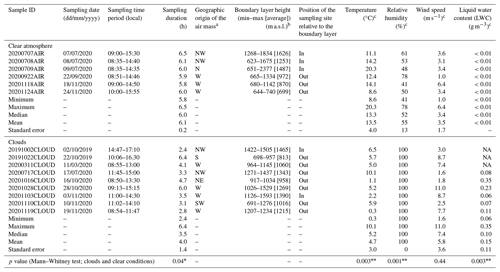
∗ Significant p value (<0.05). Highly significant p value (<0.01).
a Derived from the 72 h air mass backward trajectory, as detailed in the Supplement. b Data extracted from the ECMWF ERA5 model for the sampling period. c Average over the sampling period. NA: no data available.
2.2 Nucleic acid extraction and shotgun sequencing
For each sample, DNA and RNA were extracted in parallel from single MCE filters using a NucleoMag® DNA and RNA water kit (Macherey-Nagel, Hoerdt, France), following the protocols recommended by the manufacturer for filter membranes. All the facilities were previously treated with RNase-away spray solution (Thermo Scientific; Waltham, MA, USA). For DNA extraction, half of the lysate was processed (600 µL), and the final step consisted of the removal of RNA by adding 1:50 volume of RNase A (12 mg mL−1, stock solution from Macherey-Nagel). DNA was finally eluted into 50 µL of DNase-free H2O after 5 min of incubation at 56 °C and then quantified by fluorescence using the Quant-iT™ PicoGreen® dsDNA kit (Invitrogen; Thermo Fisher Scientific, Waltham, MA, USA). For RNA extraction, the remaining 600 µL of the lysate was processed, and the final step consisted of removal of DNA through the addition of 1:7 volume of reconstructed rDNase (as provided with kits from Macherey-Nagel). RNA was finally eluted into 30 µL of RNase-free H2O after 10 min of incubation at room temperature. Only trace amounts of DNA could be obtained from negative controls (7.3 ng of DNA on average and 11.4 ng at maximum), and these were, thus, not processed for sequencing. In contrast, the total amounts of DNA and RNA recovered from the environmental samples ranged from 42.6 to 838.7 ng and from 22.5 to 244.8 ng, respectively. The corresponding total DNA and RNA concentrations in the air volumes sampled as inferred from concentrations in the extracts ranged from 0.03 to 0.73 ng DNA m−3 and from 0.02 to 0.42 ng RNA m−3, respectively (Table S1 in the Supplement).
Individual DNA or RNA extracts from individual samplers from the same sampling event were pooled, and 30 µL was transferred to GenoScreen (Lille, France) for further processing of RNAs (quantification and reverse transcription to cDNAs) and shotgun sequencing of metagenomes (MGs) from DNAs and metatranscriptomes (MTs) from cDNAs on Illumina HiSeq (paired end reads of 150 bp). The first sample (20191022CLOUD) was sequenced deeply (∼200 M reads) and used to check the feasibility of the approach, adjust the sequencing depth, and elaborate on the bioinformatics workflows. The other samples were sequenced at a lower depth (40–60 M – million – reads per sample). Raw-sequencing MG and MT data are available as fastq.gz files through the European Nucleotide Archive at EMBL's European Bioinformatics Institute (EBI) under project accession PRJEB54740 and samples ERR9966616 to ERR9966643.
2.3 Bioinformatics and differential expression analyses
Raw MGs contained approximately 30 to 260 M reads (68.7 M on average), and raw MTs contained 65 M to 195 M reads (Tables S2 and S3). The bioinformatics workflow is detailed in the Supplement and summarized there in Fig. S1. Briefly, this consisted of (i) sequence preprocessing (quality control, trimming, etc.), (ii) taxonomic annotations of MGs and MTs (Kraken2 v2.1.1; Wood and Salzberg, 2014; PlusPF database), and (iii) construction of a gene catalog to serve as a unique reference for the study, as inspired by Salazar et al. (2019) (Fig. S2). This was elaborated on by (i) merging all the contigs from each individual MG, (ii) predicting genes (MetaGeneAnnotator v1.0.0; Noguchi et al., 2008), (iii) clustering in order to remove redundancy (CD-Hit v4.8.1; Li and Godzik, 2006), and (iv) annotating functions and taxonomies (using DIAMOND v2.0.8.0 in Buchfink et al., 2015, and the UniProtKB Swiss-Prot database from the UniProt Consortium, 2019). Finally, (v) non-rRNA (non-ribosome) reads in each MG and MT were mapped towards the annotated gene catalog. The log ratios of the number of reads associated with a gene, taxon, Enzyme Commission (E.C.) number, or Gene ontology (GO) term in an MT dataset to those of the corresponding MGs (abbreviated as RNA : DNA log ratios) were calculated using data normalized to the total counts. RNA : DNA ratios are commonly used as an appraisal of the relative level of metabolic activity, with higher ratios indicating potentially higher metabolic activity (Baldrian et al., 2012; Zhang et al., 2014). In addition, statistical differential expression analysis (DEA) was performed on the MT : MG mapping coverage ratio towards the gene catalog in order to detect overrepresented genes and functions as well as those significantly overrepresented in clouds compared with clear conditions or vice versa (MTX model v1.5.1; Zhang et al., 2021; see the Supplement for details). Metabolic pathways were reconstructed from the E.C. numbers obtained from UniProtKB identifiers using the KEGG database and resources (Kanehisa et al., 2023).
3.1 Microbial taxonomy in metagenomes and metatranscriptomes
The datasets include sequences from eukaryotes, bacteria, archaea, and viruses. Bacteria dominate in clear atmospheres (∼88 % and 71 %, on average, of the total number of reads in MGs and MTs, respectively), while eukaryotes (mainly fungi) prevail in clouds (∼51 % and 87 % on average) (Fig. S3), but both prokaryotic and eukaryotic diversity indices are statistically similar between cloud and clear-atmosphere samples (Kruskal–Wallis test; p>0.05) (Fig. S4(1)). Archaea (Euryarchaeota, Thaumarchaeota, and Crenarchaeota) and viruses (mainly bacteriophages) both contribute very low proportions of sequences (<0.1 %).
In bacteria, totals of 32 distinct phyla, 159 orders, and 1249 genera are identified in MGs (Data S1 in the Supplement). The dominant ones are Micrococcales, Corynebacteriales, Propionibacteriales (Actinobacteria), Pseudomonadales, Sphingomonadales, Burkholderiales, and Hyphomicrobiales (Proteobacteria) (Fig. 1a). The observed bacteria richness in the samples varies between 532 and 826 genera, the vast majority of which (963 out of 1249 genera; ∼77 %) are common to clouds and clear air (Fig. 1c). Shannon's diversity index ranges from ∼2.6 to ∼4.3, depending on the samples (Fig. S4).
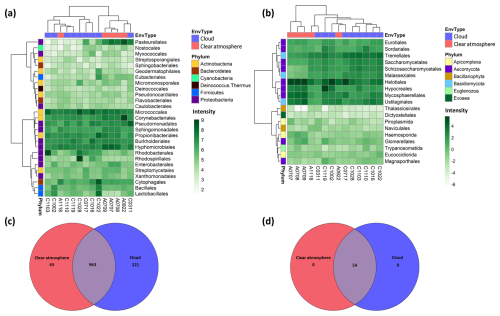
Figure 1Bacterial and eukaryotic diversity from metagenomes. (a, b) Distribution of the most abundant bacterial and eukaryotic orders in the metagenomes, together with the corresponding hierarchical clustering (Ward's method, “ward.D2”). The intensity scale depicts the centered log-ratio (clr) abundance. EnvType: environment type (the samples are identified as follows: “A” for clear atmosphere (air) or “C” for clouds, followed by the sampling date in the format “mmdd”). (c, d) Venn diagrams depicting the distribution of bacteria and eukaryotic genera between clouds and a clear atmosphere.
In eukaryotes, the identified richness is distributed between eight phyla, 21 orders, and 54 genera (Data S2). All are shared between cloudy and clear-atmosphere conditions (Fig. 1d). Fungal taxa largely predominate, with the most abundant ones affiliated with Helotiales, Hypocreales, and Mycosphaerellales in Ascomycota and Ustilaginales in Basidiomycota (Fig. 1b). Other unicellular eukaryotic phyla detected include Apicomplexa (parasites of Metazoa), Bacillariophyta (microalgae), Euglenozoa, Evosea, and Cercozoa (ameboids and flagellates). The observed eukaryotic richness in the samples varies between 41 and 43 genera, depending on the samples, with Shannon's index ranging from ∼1.1 to ∼2.4 (Fig. S4).
The taxa significantly overrepresented in MTs compared with MGs include 77 families of bacteria (198 genera) and 10 families of eukaryotes (27 genera) (Data S3). These are subsets of the total biodiversity seen in MGs, and they are not distinguishable between clouds and clear conditions (PCA: principal component analysis, Fig. S5). Bacterial taxa tend to exhibit a higher relative representation in MTs compared with MGs (termed the RNA : DNA ratio) and in clouds compared with a clear atmosphere, in contrast to eukaryotes (Figs. S6 and S7).
3.2 Functional aspects and differential expression analyses
The RNA : DNA concentration ratio is significantly higher in clouds than in a clear atmosphere (ranges 1.49–3.62 and 0.21–1.64; Mann–Whitney test, p value=0.004) (Table S1; Fig. 2a). No relationship with relative humidity is observed in a clear atmosphere.
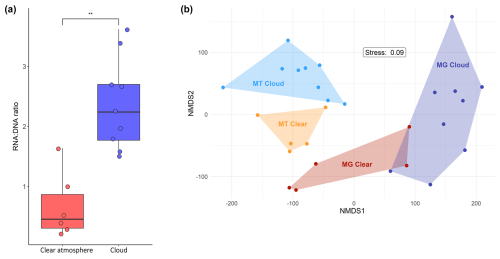
Figure 2(a) RNA : DNA concentration ratio in clouds and a clear atmosphere. The dots indicate individual samples. The boxplots display the medians and the 25th and 75th percentiles. The whiskers are the 1.5 interquartile ranges. ** indicates a significantly higher ratio in clouds (Mann–Whitney test, p value=0.004). (b) Nonmetric multidimensional scaling (NMDS) analysis based on the 21 046 functional gene entries detected in total, depicting clear distinctions between MGs and MTs and between MTs of cloudy and clear conditions.
Independently of atmospheric conditions, many transcripts in the datasets are related to key biological functions and their regulation, including carbon, amino-acid, and protein processing; energy production; signaling; responses to stresses; and transport (Fig. 3; Fig. 4a, c, and e; Figs. S8 and S9). Of the total of 21 046 unique genes (UniProt IDs) in MGs constituting the reference catalog, 488 are found to be significantly more represented in MTs than in MGs (Data S4). These correspond to, at the GO term level, 419 biological processes, 284 molecular functions, and 140 cellular components overrepresented in transcriptomes (Data S5). Most of these (∼80 %) are affiliated with eukaryotes, in particular fungi, or with Gamma-Proteobacteria and Actinobacteria in bacteria (∼48 % and ∼25 %, respectively, of the bacteria transcripts) (Fig. S10).
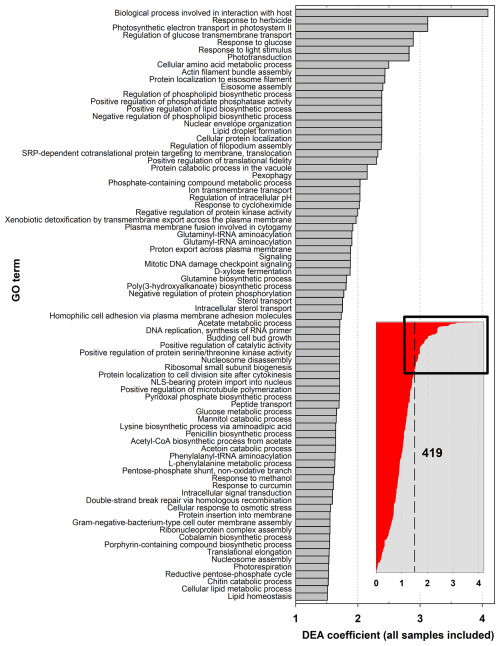
Figure 3Biological process GO terms associated with overrepresented transcripts in samples, as compared with their representation in metagenomes, from differential expression analysis (DEA). Only the 80 GO terms with DEA coefficients greater than 1.5, out of the 419 in total, are shown.
These genes and functions occur and are distributed differentially, depending on the presence of condensed water (Fig. 4b, d, and f; Figs. S8 and S9). Nonmetric multidimensional scaling (NMDS) indeed indicates distinct transcriptional patterns that depend on atmospheric conditions (Fig. 2b), and DEA specifies them (Fig. 5; Data S6 and S7): of the 488 genes significantly more represented in MTs than in MGs, 320 (∼66 %) are also significantly differentially represented between clouds and clear conditions, about two-thirds of them in clouds and contributed by eukaryotes (Fig. S11). In total, the differentially represented transcripts belong to 394 biological processes, 147 cellular components, and 279 molecular functions and correspond to 200 unique E.C. numbers (see the distribution of the subclasses in Table S4). Overall, the most diverse enzyme transcripts are NADH:ubiquinone oxidoreductase (E.C. 7.1.1.2, 11 distinct entries), RNA polymerase (E.C. 2.7.7.6), RNA helicase (E.C. 3.6.4.13), cytochrome-c oxidase (E.C. 7.1.1.9, six distinct entries each), and nonspecific serine and threonine protein kinase (E.C. 2.7.11.1, five distinct entries). Below we examine the similarities and specificities of microbial transcriptomes inside and outside clouds for different categories of functions and metabolisms based on the distribution of GOs and E.C. numbers (Figs. 5, 6, and S12–S15; Table S4; Data S6 and S7).
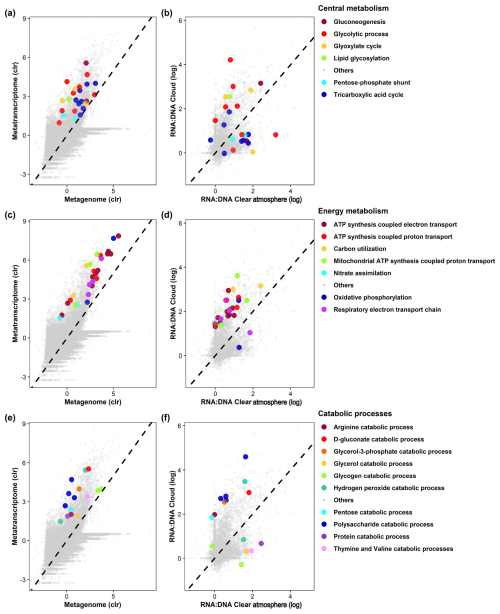
Figure 4GO term representation in metagenomes and metatranscriptomes (a, c, e) and relative representation (termed RNA : DNA) in metatranscriptomes in clouds and a clear atmosphere (b, d, f) for biological processes related to key metabolism (a, b), energy metabolism (c, d), and catabolic (e, f) processes. Other GO terms of interest are presented in Figs. S9 and S10.
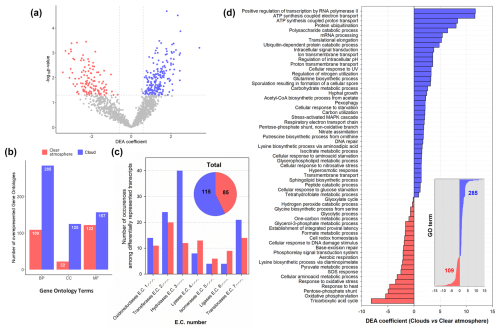
Figure 5Results of DEA of transcript overrepresentation in clouds (blue) compared with a clear atmosphere (red) at the gene (a), GO term (BP: biological process; CC: cellular component; MF: molecular function) (b), and E.C. number (c) levels. In panel (d), the DEA coefficients associated with selected biological process GO terms are shown (39 GO terms out of 285 in total for clouds and 22 out of 109 for a clear atmosphere).
3.2.1 Key, carbon, and energy metabolisms
Numerous transcripts related to translation and elongation factors in eukaryotes are overrepresented in clouds (Data S6), suggesting metabolic regulations and the production of new biomasses. Large numbers of transcripts relate to key functions of carbon and energy metabolisms in both clouds and clear-air samples (Figs. 4, S8, and S9): glycolysis and glucose-related metabolic processes (GO:0006096 and GO:0006006), glyoxylate and tricarboxylic acid (TCA) cycles (GO:0006097 and GO:0006099), and carbohydrate metabolisms (GO:0005975, GO:0006083, and GO:0019427). Consistently, transcript codes for the key enzymes of these pathways are abundant in both prokaryotes and eukaryotes, such as isocitrate dehydrogenase (IDH) (E.C. 1.1.1.42 and E.C. 1.1.1.41), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (E.C. 1.2.1.12), and aconitase (E.C. 4.2.1.3) (Figs. 6 and S12–S14).
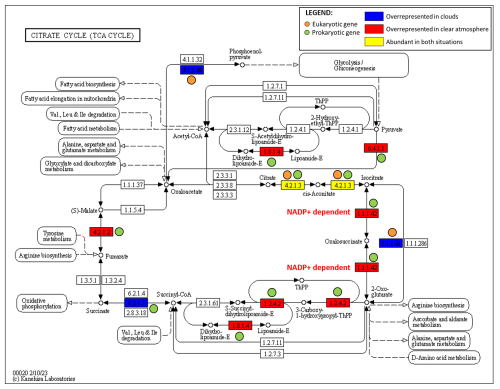
Figure 6The TCA cycle metabolic pathway, depicting overrepresented enzyme transcripts in clouds and/or a clear atmosphere by eukaryotes and/or prokaryotes (from UniProtKB identifiers and the KEGG database).
DEA indicates a strong overrepresentation in clouds of functional transcript codes for hydrolases (E.C. 3.-.-.-) and, to a lesser extent, translocases (E.C. 7.-.-.-), particularly those involved in the translocation of protons (E.C. 7.1.-.-). These relate to GOs, including carbon utilization (GO:0015976), polysaccharide catabolism (GO:0000272), and adenosine triphosphate (ATP) production-coupled electron and proton transport (GO:0042773 and GO:0015986) (Fig. 5). The pentose phosphate pathway (non-oxidative phase) (GO:0009051) also tends to be more represented in clouds than in a clear atmosphere, with an overrepresentation in particular of fructose–bisphosphate aldolase (fba) transcripts (E.C. 4.1.2.13) (Fig. S13). These observations concur with increased biochemical energy needs in clouds.
In turn, transcripts of lyases (E.C. 4.-.-.-) and ligases (E.C. 6.-.-.-) are overall more prevalent in a clear atmosphere than in clouds (Fig. 5; Table S4). In particular, the TCA cycle pathway (GO:0006099) appears upregulated by prokaryotes, with transcript coding for enzymes, including pyruvate carboxylase (pyc, pycA, and ylaP; E.C. 6.4.1.1), isocitrate dehydrogenase (nicotinamide adenine dinucleotide phosphate (NADP)-dependent) (IDH) (icd, Cgl0664, and cg0766; E.C. 1.1.1.42), fumarate hydratase (fumC; E.C. 4.2.1.2), and alpha-ketoglutarate dehydrogenase (sucA, odhA, and Oant; E.C. 1.2.4.2) (Fig. 6). Glycolytic processes and the glyoxylate cycle are overall barely affected by clouds based on DEA. Nevertheless, transcripts of enzymes involved in specific steps of these pathways are detected in higher proportions in clouds, including enolase (2-phospho-D-glycerate hydro-lyase; E.C. 4.2.1.11), phosphoenolpyruvate carboxykinase (E.C. 4.1.1.49), and acetyl-coenzyme A synthetase (E.C. 6.2.1.1) (Figs. S12 and S13).
3.2.2 Protein, amino-acid, and nitrogen metabolism
Many of the most abundant transcripts related to protein, amino-acid, and nitrogen metabolism are more prevalent in clouds than they are in a clear atmosphere (Figs. 4, 5, S8, and S9). These include post-translational protein modifications (ubiquitination, phosphorylation, glycosylation, and related processes: GO:0016567 and GO:0006511) known to participate in the regulation of enzymatic activities and several other cellular and metabolic processes (Yang et al., 2022). The overrepresentation in clouds of transcripts of the regulatory gene areA suggests that several nitrogen sources are targeted (Kudla et al., 1990), likely as a response to limited resources. Clouds are also associated with amino-acid starvation (GO:0034198).
Of the amino-acid-related transcripts and functions, only glutamine and lysine biosynthesis are overrepresented in clouds (Fig. 5). Catabolism is directed towards serine and arginine, in line with ornithine and putrescine production pathways within glutathione metabolism, which is a major pathway in the recycling of NADP+ and the regulation of oxidants (Fig. S15). Under clear conditions, processes of amino-acid biosynthesis prevail (arginine, leucine, threonine, lysine, beta-alanine, and glycine), potentially involving gaseous dinitrogen fixation (Dalton and Kramer, 2006).
In inorganic nitrogen metabolism, nitrate assimilation (GO:0042128) and the utilization of ammonium (GO:0042128) are both overrepresented in clouds. Both ions are abundant in atmospheric water (Péguilhan et al., 2021). The latter function is in agreement with the overrepresentation of L-glutamate:ammonia ligase transcripts (E.C. 6.3.1.2, GO:0004356, gln), which is an enzyme involved in the synthesis of glutamine and is one of the main regulators of cellular development and oxidative stress responses in fungi (Wang et al., 2022).
3.2.3 Transport, signaling, and response to stress
In clouds, transcripts linked to transmembrane transports of ions and protons (GO:0034220, GO:1902600, and GO:0055085) are among the most represented (Fig. S9). These participate in ATP synthesis (GO:0042773 and GO:0015986), regulation of substrate utilization (GO:0006808, GO:0015976, and GO:0009267), and homeostasis (GO:0045454) (Figs. 5 and 7).
In turn, oxidative stress (GO:0006979) and SOS response (GO:0009432) dominate in a clear atmosphere, whereas in cloud responses to osmotic and nitrosative stress (GO:0071470 and GO:0071500) the stress-activated MAPK cascade (GO:0051403), regulation of intracellular pH (GO:0051453), processes of starvation towards carbon and nitrogen (GO:0042149 and GO:0034198), and processes of autophagy and pexophagy (i.e., macropexophagy; GO:0051403) prevail (Figs. 5, 7, and S9). Such functional patterns can be interpreted as microbial responses to wetting. They indicate a probable sheltering effect of condensed water on oxidative stress, along with limited nutrient resources requiring metabolic adjustments.
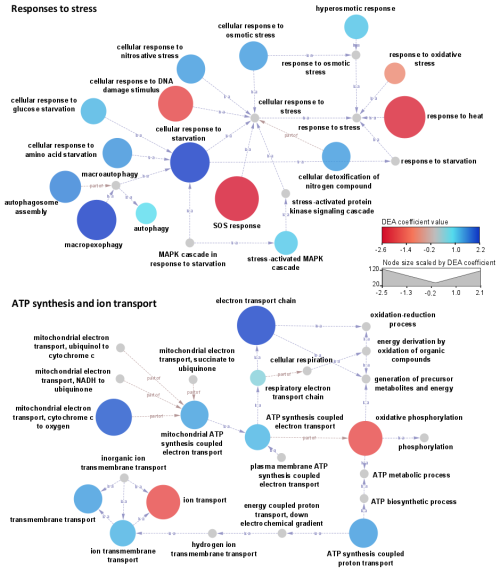
Figure 7Networks linking biological processes (GO terms) related to stress responses, together with ATP synthesis and ion transport. The color scale represents the associated DEA coefficients, with negative values (red shades) indicating a significant overrepresentation of related transcripts under clear conditions and positive values (blue shades) in clouds. Node size is scaled to the DEA coefficient's absolute value. The arrows indicate relationships between GOs (“is a” or “part of” as specified).
Here we report the most comprehensive dataset to date regarding aeromicrobiome functioning in the natural environment. For the first time, we demonstrate that multiple microbial functions are directly influenced by atmospheric conditions, specifically the presence of clouds. In previous work, using targeted and non-targeted transcriptomic approaches, we identified metabolically active bacteria and fungi in clouds (Amato et al., 2017) and then depicted their functioning through the analysis of transcriptomes (Amato et al., 2019). Here, we extend our knowledge by integrating a non-cloudy atmosphere in our analysis, thus replacing clouds in an atmospheric context, i.e., as volumes embedded within the clear atmosphere.
In terms of taxonomy, the airborne microbial assemblages in clouds are not distinguishable from those in a clear atmosphere, as we recently reported from amplicons for bacteria (Péguilhan et al., 2023a). They consist of diverse eukaryotes, bacteria, Archaea, and viruses, with Proteobacteria and Actinobacteria dominating in bacteria and Ascomycota and Basidiomycota dominating in eukaryotes, which is typical over continental vegetated areas (e.g., Bowers et al., 2013; Tignat-Perrier et al., 2020). Such similarity was to be expected given the large sizes of microbial aerosols and thus their ability to act as cloud condensation nuclei, considering that cell multiplication in clouds is unlikely to significantly affect the structure of microbial assemblages due to limited residence times (Ervens and Amato, 2020).
4.1 A Birch effect up in the sky
Higher concentrations of RNA with respect to DNA in clouds than in cloud-free air suggest higher levels of microbial metabolic activity (Baldrian et al., 2012; Salazar et al., 2019). This remains to be assessed quantitatively through more direct activity measurements. Accordingly, transcripts related to multiple biological processes are overrepresented in clouds compared with a clear atmosphere, such as energy metabolism, carbohydrate and polysaccharide catabolism, transcription, translation, transmembrane transports, and metabolic regulation mechanisms. Homeostasis regulation, starvation, and autophagy supplant the oxidative stress response and DNA repair functions (SOS response) that prevail under clear conditions. The switch in the functional gene expression observed in microorganisms between a clear atmosphere and clouds therefore suggests a phenomenon similar to the Birch effect, which occurs in dry soils in response to rewetting by rain, where the sudden influx of water triggers a burst of microbial activity (Griffiths and Birch, 1961; Unger et al., 2010). The Birch effect in soils typically lasts a few days, i.e., much longer than the lifetime of clouds and individual cloud droplets (Feingold et al., 1996). The metabolic modulations described here should therefore apply to any cloud, regardless of the time elapsed since its formation. As with soils – but necessarily to a much smaller extent because of the low biomass – the resurgence of microbial activity in clouds may lead to the release of gaseous biogenic compounds such as N2O through aerobic ammonium oxidation, i.e., nitrification (Jørgensen et al., 1998), which is also supported by the overrepresentation of ammonium utilization processes.
Dry–wet alternance in ecosystems contributes to shaping microbial assemblages, activity, and responsiveness to changing conditions. The lag time after which microorganisms start recovering in soil upon rewetting shortens after repeated dry–wet cycles, due to selection processes towards the most responsive ones (Zhou et al., 2016). Transcriptionally active taxa in our observations represent ∼20 % of the richness, and this could result from and attest to such selection processes during aerial transport. Typical atmospheric residence times of microbial cells are on the order of a few days (Burrows et al., 2009), during which they may undergo ∼10 water evaporation–condensation cycles before precipitating (Pruppacher and Jaenicke, 1995). While clouds can last for several hours (Dagan et al., 2018), individual cloud droplets can form and evaporate within a few minutes (Feingold et al., 1996). Such rapid fluctuations of water availability could favor the most responsive microorganisms, such as Proteobacteria and Actinobacteria, which include numerous generalists with high metabolic flexibility (Chen et al., 2021) and which in fact often dominate airborne microbial assemblages (Amato et al., 2017; Péguilhan et al., 2021; Šantl-Temkiv et al., 2022).
4.2 Responses to stress attest to multiple functional adjustments
Our data indicate that a clear atmosphere is dominated by responses to oxidative stress and DNA damage involving SOS responses, while clouds are characterized by osmotic stress, starvation, and autophagy. The functional patterns of aeromicrobiome stress responses are therefore very consistent with environmental conditions and help paint a more complete picture of the multiple aspects of the microbial journey in the high atmosphere.
In clouds, liquid water shelters cells from oxidants and radiations, but the rapid condensation and evaporation processes, long with the dissolution of solids and the solubilization of gases, generate large fluctuations of water activity (e.g., Koehler et al., 2006). Additionally, in the limited volumes provided by droplets, the nutrient requirements are likely not fully satisfied, and autophagy processes may contribute to alleviating the needs. Peroxisomes, organelles dedicated to the detoxification of oxidants in eukaryotes, are targeted in particular by autophagy (pexophagy), as during fungal spore germination. Such process could compromise survival if the cloud evaporates, but this may be a tradeoff with increased chances in the race for surface colonization if the cloud precipitates.
Here, clear air was collected at relative humidities between 41 % and 78 %, i.e., at the limits of compatibility with biological processes, around 0.6 aw (water activity) for the most tolerant organisms (i.e., 60 % pure water relative humidity) (Stevenson et al., 2015). At an aw value below 0.55, DNA becomes unstructured and metabolic regulations are no longer possible. Water limitation is a great challenge that many microorganisms have to face in their natural habitats. This affects cell turgor due to water efflux and slows down growth and metabolic activity (Chowdhury et al., 2011).
In order to manage the numerous environmental factors related to variations of water activity, such as temperature or osmotic pressure, microorganisms have developed a variety of strategies: modifications of the saturation level of lipids in membranes to adjust fluidity, synthesis and accumulation of intracellular-compatible solutes in order to prevent water efflux and maintain homeostasis (osmoprotectants and cryoprotectants, e.g., K+, sucrose, trehalose, and amino acids) (Poolman and Glaasker, 1998), and chaperones to protect molecular structures and membrane canal proteins such as aquaporins, in order to sustain water fluxes (Tong et al., 2019).
4.3 Airborne fungal spores initiate germination in clouds
In agreement with the overrepresentation of transcripts, it is likely that fungal spores initiate germination in clouds. These include translation initiation and elongation factors affiliated with several taxa of fungi (elF4E and eEF3) (van Leeuwen et al., 2013; Li et al., 2022; Osherov and May 2001), chitin deacetylase (Leroch et al., 2013), and other regulatory protein genes such as areA (Kudla et al., 1990). Fungal spores are propagules designed for (aerial) dispersion (Brown and Hovmøller, 2002), which germinate (i.e., initiate growth) when they reach favorable conditions for water availability. During germination, functions of cell protection give way to anabolic processes within minutes (van Leeuwen et al., 2013; Leroch et al., 2013). Cellular growth and respiration are promoted by the sudden availability of water, which is associated with the release and solubilization of readily bioavailable organic compounds. While dormant, functions of cell protection prevail in spores against osmotic stress, heat, and oxidants. In the presence of water, mitogen-activated protein kinase (MAPK) cascade signaling pathways mediate the activation of key metabolic functions of energy production and biosynthesis (van Leeuwen et al., 2013).
Autophagy processes in particular can participate in the early steps of appressorium synthesis in parasitic and symbiotic fungi (Veses et al., 2008), which is a structure designed to invade host cells. Although we did not consider time series, our observations strongly concur with such a sequence, and it is reasonable to assert that airborne fungal spore germination occurs in clouds.
4.4 Utilization of nutrients and interactions with chemistry
Microbial activity is driven by the balance between water availability and accessibility to substrates (Skopp et al., 1990). While not evaluable here, the amounts of water retained by efflorescent aerosols below water vapor saturation may be sufficient for sustaining microbial activity, down to very low values of relative humidity (Cruz and Pandis, 2000; Ervens et al., 2024). In clouds, i.e., above saturation levels, the large amounts of available water make it even conceivable that bacterial multiplication occurs. Bulk cloud water indeed contains enough nutrients to sustain microbial growth, including carboxylic acids, aldehydes, sugars, amino acids, ammonium, and nitrates (Amato et al., 2007a; Bianco et al., 2016, 2018, 2019; Deguillaume et al., 2014; Renard et al., 2022), and the level of microbial activity at 0 °C was shown to be compatible with this (Sattler et al., 2001). Field observations indicate that fog has a higher biomass than a clear atmosphere (Fuzzi et al., 1997; Saikh and Das, 2023), while estimations suggest that microbial mass may double during a cloud's lifetime (Ervens and Amato, 2020). The fact that, statistically, only 1 out of ∼10 000 droplets contains a microbial cell in aerially suspended water, as opposed to bulk water, potentially causes very efficient and rapid depletion of nutrients in these low biotic volumes (Khaled et al., 2021) ( for 20 µm diameter droplets, i.e., a cell concentration of at least ∼109 cells per milliliter in biotic droplets), which exposes cells to starvation and may limit metabolic processes (Gray et al., 2019).
The overrepresentation of transcripts related to carbon, ammonium, and nitrate utilization in clouds confirms that carbon and nitrogen biological processing occurs. Despite the low biomass, the impact of microbial activity on organic carbon chemistry has been assessed as potentially significant, depending on the volatility and solubility of the compounds (Ervens and Amato, 2020; Khaled et al., 2021; Nuñez López et al., 2024). The impacts on nitrogen species have barely been examined (Hill et al., 2007; Jaber et al., 2021). Estimates indicate potential biodegradation of amino acids, but there is as yet no information on inorganic compounds such as ammonium and nitrate, which are among the most abundant ions in cloud water (Deguillaume et al., 2014).
The recovery of active metabolic processes in airborne microorganisms prior to their deposition could facilitate surface or host invasion if clouds precipitate. In turn, in the likely event of the cloud evaporating instead of precipitating, triggering germination and sacrificing essential cellular structures while conditions may soon become inhospitable could compromise future chances of survival, including further dispersion. It thus remains to be seen whether these metabolic regulations offer an ecological advantage for microorganisms in such transient environments as clouds, where they have no chance of establishing themselves due to limited residence times, or whether they may influence survival in the atmosphere and invasion processes upon deposition.
Metatranscriptomic approaches are undoubtedly among the most powerful methods for examining microbial functioning. Still, they remain limited by multiple constraints inherently associated with them. For instance, they do not allow attribution of biological processes to individual cells or to specific taxa with certainty. They are also limited by current functional genomic knowledge and existing databases, in particular in natural environments like the atmosphere, where rare taxa often represent an important fraction of the richness (Péguilhan et al., 2023b). In the atmosphere, microorganisms have a short residence time of a few days, and air volumes contain mixed populations of cells of different origins and atmospheric ages. Analyses such as metatranscriptomics, based on RNA, assume a high turnover of these molecules in cells, in particular mRNA, and are therefore considered to reflect quasi-instantaneous cell activity. Nevertheless, rRNA can persist for several days at low temperatures (Schostag et al., 2020) and may therefore be the result of recent past activity. These “residual” signatures of activity, along with the high level of mixing, can blur our vision of the actual situation in such a highly dynamic environment as the atmosphere.
Transcriptomes attest to potential cellular activity, but they do not provide quantitative information on microbial activity in terms of fluxes of elements or energy. Quantitative measurements of microbial activity therefore remain necessary for confirming the “atmospheric Birch effect” caused by clouds. The transitions from clear to cloudy conditions in particular remain to be examined for evaluating the temporal responsiveness of airborne microbial assemblages to cloud formation and evaporation. While this is potentially achievable in an atmospheric simulation chamber, assessing microbial activity in naturally aerially suspended biological microorganisms remains highly challenging, if not impossible (yet). The development of methods able to detect and quantify microbial metabolic activity in air-suspended cells and at high frequency therefore appears to be a prerequisite.
Our study focused in particular on the potential impact of clouds on microbial functioning, and this relies on samples collected at a single site, using unique sampling methods in order to avoid introducing site effects and methodological bias. We were able to show qualitatively that there are differences in microbial gene expressions in samples collected in cloud-free and cloudy air masses. We used “water availability” as a proxy to distinguish between the two air mass types. However, cloudy air masses also differ from those outside clouds in a multitude of other environmental factors, which are expected to play roles in aeromicrobiome functioning, and they still need to be evaluated (Amato et al., 2023). Such variables include temperature, solar radiation, and chemical composition, and they are linked not only to clouds but also altitude, location, day–night cycles, and season. The synergy, temporal dynamics, and arrangement of these variables (shocks, cloud cycles, freezing events, and combination of chemicals) could also contribute to shaping aeromicrobiomes in even more complex ways.
The raw-sequencing MG and MT data are available as fastq.gz files from the European Nucleotide Archive at EBI, under project accession PRJEB54740 (samples ERR9966616 to ERR9966643) (https://www.ebi.ac.uk/ena/browser/view/PRJEB54740, Péguilhan, 2025).
The supplement related to this article is available online at https://doi.org/10.5194/bg-22-1257-2025-supplement.
Conceptualization: PA. Methodology: RP, FR, FE, BB, and EN. Investigation: RP and PA. Visualization: RP, MJ, and PA. Funding acquisition: PA and BE. Supervision: PA. Writing – original draft: RP and PA. Writing – review and editing: RP, PA, FR, MJ, FE, EN, BB, and BE.
The contact author has declared that none of the authors has any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
We thank the OPGC facility for running the PUY atmospheric station, sharing their meteorological data, and helping logistically during the sampling. We are also grateful to Nadia Goué at the computational center of Clermont Auvergne University (Mésocentre) and the Auvergne Bioinformatique network for providing computational power and excellent support with software deployment in the Galaxy environment.
This research has been supported by the Agence Nationale de la Recherche (grant-no. ANR-17-MPGA-0013).
This paper was edited by Paul Stoy and reviewed by two anonymous referees.
Amato, P., Demeer, F., Melaouhi, A., Fontanella, S., Martin-Biesse, A.-S., Sancelme, M., Laj, P., and Delort, A.-M.: A fate for organic acids, formaldehyde and methanol in cloud water: their biotransformation by micro-organisms, Atmos. Chem. Phys., 7, 4159–4169, https://doi.org/10.5194/acp-7-4159-2007, 2007a.
Amato, P., Parazols, M., Sancelme, M., Laj, P., Mailhot, G., and Delort, A.-M.: Microorganisms isolated from the water phase of tropospheric clouds at the Puy de Dôme: major groups and growth abilities at low temperatures, FEMS Microbiol. Ecol., 59, 242–254, https://doi.org/10.1111/j.1574-6941.2006.00199.x, 2007b.
Amato, P., Joly, M., Schaupp, C., Attard, E., Möhler, O., Morris, C. E., Brunet, Y., and Delort, A.-M.: Survival and ice nucleation activity of bacteria as aerosols in a cloud simulation chamber, Atmos. Chem. Phys., 15, 6455–6465, https://doi.org/10.5194/acp-15-6455-2015, 2015.
Amato, P., Joly, M., Besaury, L., Oudart, A., Taib, N., Moné, A. I., Deguillaume, L., Delort, A.-M., and Debroas, D.: Active microorganisms thrive among extremely diverse communities in cloud water, PloS one, 12, e0182869, https://doi.org/10.1371/journal.pone.0182869, 2017.
Amato, P., Besaury, L., Joly, M., Penaud, B., Deguillaume, L., and Delort, A.-M.: Metatranscriptomic exploration of microbial functioning in clouds, Sci. Rep.-UK, 9, 4383, https://doi.org/10.1038/s41598-019-41032-4, 2019.
Amato, P., Mathonat, F., Nuñez Lopez, L., Péguilhan, R., Bourhane, Z., Rossi, F., Vyskocil, J., Joly, M., and Ervens, B.: The aeromicrobiome: the selective and dynamic outer-layer of the Earth's microbiome, Front. Microbiol., 14, 1186847, https://doi.org/10.3389/fmicb.2023.1186847, 2023.
Baldrian, P., Kolařík, M., Stursová, M., Kopecký, J., Valášková, V., Větrovský, T., Zifčáková, L., Snajdr, J., Rídl, J., Vlček, C., and Voříšková, J.: Active and total microbial communities in forest soil are largely different and highly stratified during decomposition, ISME J., 6, 248–258, https://doi.org/10.1038/ismej.2011.95, 2012.
Baray, J.-L., Deguillaume, L., Colomb, A., Sellegri, K., Freney, E., Rose, C., Van Baelen, J., Pichon, J.-M., Picard, D., Fréville, P., Bouvier, L., Ribeiro, M., Amato, P., Banson, S., Bianco, A., Borbon, A., Bourcier, L., Bras, Y., Brigante, M., Cacault, P., Chauvigné, A., Charbouillot, T., Chaumerliac, N., Delort, A.-M., Delmotte, M., Dupuy, R., Farah, A., Febvre, G., Flossmann, A., Gourbeyre, C., Hervier, C., Hervo, M., Huret, N., Joly, M., Kazan, V., Lopez, M., Mailhot, G., Marinoni, A., Masson, O., Montoux, N., Parazols, M., Peyrin, F., Pointin, Y., Ramonet, M., Rocco, M., Sancelme, M., Sauvage, S., Schmidt, M., Tison, E., Vaïtilingom, M., Villani, P., Wang, M., Yver-Kwok, C., and Laj, P.: Cézeaux-Aulnat-Opme-Puy De Dôme: a multi-site for the long-term survey of the tropospheric composition and climate change, Atmos. Meas. Tech., 13, 3413–3445, https://doi.org/10.5194/amt-13-3413-2020, 2020.
Bianco, A., Voyard, G., Deguillaume, L., Mailhot, G., and Brigante, M.: Improving the characterization of dissolved organic carbon in cloud water: Amino acids and their impact on the oxidant capacity, Sci. Rep.-UK, 6, 37420, https://doi.org/10.1038/srep37420, 2016.
Bianco, A., Deguillaume, L., Vaïtilingom, M., Nicol, E., Baray, J.-L., Chaumerliac, N., and Bridoux, M.: Molecular characterization of cloud water Samples collected at the Puy de Dôme (France) by Fourier Transform Ion Cyclotron Resonance Mass Spectrometry, Environ. Sci. Technol., 52, 10275–10285, https://doi.org/10.1021/acs.est.8b01964, 2018.
Bianco, A., Deguillaume, L., Chaumerliac, N., Vaïtilingom, M., Wang, M., Delort, A.-M., and Bridoux, M. C.: Effect of endogenous microbiota on the molecular composition of cloud water: a study by Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR MS), Sci. Rep.-UK, 9, 7663, https://doi.org/10.1038/s41598-019-44149-8, 2019.
Bowers, R. M., McLetchie, S., Knight, R., and Fierer, N.: Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments, ISME J., 5, 601–612, https://doi.org/10.1038/ismej.2010.167, 2011.
Bowers, R. M., Clements, N., Emerson, J. B., Wiedinmyer, C., Hannigan, M. P., and Fierer, N.: Seasonal variability in bacterial and fungal diversity of the near-surface atmosphere, Environ. Sci. Technol., 47, 12097–12106, https://doi.org/10.1021/es402970s, 2013.
Brown, J. K. M. and Hovmøller, M. S.: Aerial Dispersal of pathogens on the global and continental scales and its impact on plant disease, Science, 297, 537–541, https://doi.org/10.1126/science.1072678, 2002.
Buchfink, B., Xie, C., and Huson, D. H.: Fast and sensitive protein alignment using DIAMOND, Nat. Methods, 12, 59–60, https://doi.org/10.1038/nmeth.3176, 2015.
Burrows, S. M., Butler, T., Jöckel, P., Tost, H., Kerkweg, A., Pöschl, U., and Lawrence, M. G.: Bacteria in the global atmosphere – Part 2: Modeling of emissions and transport between different ecosystems, Atmos. Chem. Phys., 9, 9281–9297, https://doi.org/10.5194/acp-9-9281-2009, 2009.
Camacho-Sanchez, M., Burraco, P., Gomez-Mestre, I., and Leonard, J. A.: Preservation of RNA and DNA from mammal samples under field conditions, Mol. Ecol. Resour., 13, 663–673, https://doi.org/10.1111/1755-0998.12108, 2013.
Chen, L., Hu, M., Huang, L., Hua, Z., Kuang, J., Li, S., and Shu, W.: Comparative metagenomic and metatranscriptomic analyses of microbial communities in acid mine drainage, ISME J., 9, 1579–1592, https://doi.org/10.1038/ismej.2014.245, 2015.
Chen, Y.-J., Leung, P. M., Wood, J. L., Bay, S. K., Hugenholtz, P., Kessler, A. J., Shelley, G., Waite, D. W., Franks, A. E., Cook, P. L. M., and Greening, C.: Metabolic flexibility allows bacterial habitat generalists to become dominant in a frequently disturbed ecosystem, ISME J., 15, 2986–3004, https://doi.org/10.1038/s41396-021-00988-w, 2021.
Chowdhury, N., Marschner, P., and Burns, R.: Response of microbial activity and community structure to decreasing soil osmotic and matric potential, Plant Soil, 344, 241–254, https://doi.org/10.1007/s11104-011-0743-9, 2011.
Cruz, C. N. and Pandis, S. N.: Deliquescence and hygroscopic growth of mixed inorganic–organic atmospheric aerosol, Environ. Sci. Technol., 34, 4313–4319, https://doi.org/10.1021/es9907109, 2000.
Dagan, G., Koren, I., Altaratz, O., and Lehahn, Y.: Shallow convective cloud field lifetime as a key factor for evaluating aerosol effects, iScience, 10, 192–202, https://doi.org/10.1016/j.isci.2018.11.032, 2018.
Dalton, D. A. and Kramer, S.: Nitrogen-fixing bacteria in non-legumes, in: Plant-Associated Bacteria, edited by: Gnanamanickam, S. S., Springer Netherlands, Dordrecht, 105–130, https://doi.org/10.1007/978-1-4020-4538-7_3, 2006.
Deguillaume, L., Charbouillot, T., Joly, M., Vaïtilingom, M., Parazols, M., Marinoni, A., Amato, P., Delort, A.-M., Vinatier, V., Flossmann, A., Chaumerliac, N., Pichon, J. M., Houdier, S., Laj, P., Sellegri, K., Colomb, A., Brigante, M., and Mailhot, G.: Classification of clouds sampled at the puy de Dôme (France) based on 10 yr of monitoring of their physicochemical properties, Atmos. Chem. Phys., 14, 1485–1506, https://doi.org/10.5194/acp-14-1485-2014, 2014.
Dillon, K. P., Correa, F., Judon, C., Sancelme, M., Fennell, D. E., Delort, A.-M., and Amato, P.: Cyanobacteria and algae in clouds and rain in the area of puy de Dôme, Central France, Appl. Environ. Microbiol., https://doi.org/10.1128/AEM.01850-20, 2020.
Ervens, B. and Amato, P.: The global impact of bacterial processes on carbon mass, Atmos. Chem. Phys., 20, 1777–1794, https://doi.org/10.5194/acp-20-1777-2020, 2020.
Ervens, B., Sorooshian, A., Aldhaif, A. M., Shingler, T., Crosbie, E., Ziemba, L., Campuzano-Jost, P., Jimenez, J. L., and Wisthaler, A.: Is there an aerosol signature of chemical cloud processing?, Atmos. Chem. Phys., 18, 16099–16119, https://doi.org/10.5194/acp-18-16099-2018, 2018.
Ervens, B., Amato, P., Aregahegn, K., Joly, M., Khaled, A., Labed-Veydert, T., Mathonat, F., Nuñez López, L., Péguilhan, R., and Zhang, M.: Ideas and perspectives: Microorganisms in the air through the lenses of atmospheric chemistry and microphysics, EGUsphere [preprint], https://doi.org/10.5194/egusphere-2024-2377, 2024.
Feingold, G., Cotton, W. R., Stevens, B., and Frisch, A. S.: The Relationship between Drop In-Cloud Residence Time and Drizzle Production in Numerically Simulated Stratocumulus Clouds, J. Atmos. Sci., 53, 1108–1122, https://doi.org/10.1175/1520-0469(1996)053<1108:TRBDIC>2.0.CO;2, 1996.
Fierer, N., Liu, Z., Rodríguez-Hernández, M., Knight, R., Henn, M., and Hernandez, M. T.: Short-term temporal variability in airborne bacterial and fungal populations, Appl. Environ. Microbiol., 74, 200–207, https://doi.org/10.1128/AEM.01467-07, 2008.
Franzosa, E. A., Morgan, X. C., Segata, N., Waldron, L., Reyes, J., Earl, A. M., Giannoukos, G., Boylan, M. R., Ciulla, D., Gevers, D., Izard, J., Garrett, W. S., Chan, A. T., and Huttenhower, C.: Relating the metatranscriptome and metagenome of the human gut, P. Natl. Acad. Sci. USA, 111, E2329–E2338, https://doi.org/10.1073/pnas.1319284111, 2014.
Fuzzi, S., Mandrioli, P., and Perfetto, A.: Fog droplets – an atmospheric source of secondary biological aerosol particles, Atmos. Environ., 31, 287–290, https://doi.org/10.1016/1352-2310(96)00160-4, 1997.
Gray, D. A., Dugar, G., Gamba, P., Strahl, H., Jonker, M. J., and Hamoen, L. W.: Extreme slow growth as alternative strategy to survive deep starvation in bacteria, Nat. Commun., 10, 890, https://doi.org/10.1038/s41467-019-08719-8, 2019.
Griffiths, E. and Birch, H. F.: Microbiological Changes in Freshly Moistened Soil, Nature, 189, 424–424, https://doi.org/10.1038/189424a0, 1961.
Gusareva, E. S., Acerbi, E., Lau, K. J. X., Luhung, I., Premkrishnan, B. N. V., Kolundžija, S., Purbojati, R. W., Wong, A., Houghton, J. N. I., Miller, D., Gaultier, N. E., Heinle, C. E., Clare, M. E., Vettath, V. K., Kee, C., Lim, S. B. Y., Chénard, C., Phung, W. J., Kushwaha, K. K., Nee, A. P., Putra, A., Panicker, D., Yanqing, K., Hwee, Y. Z., Lohar, S. R., Kuwata, M., Kim, H. L., Yang, L., Uchida, A., Drautz-Moses, D. I., Junqueira, A. C. M., and Schuster, S. C.: Microbial communities in the tropical air ecosystem follow a precise diel cycle, P. Natl. Acad. Sci. USA, 116, 23299–23308, https://doi.org/10.1073/pnas.1908493116, 2019.
Herrmann, H., Schaefer, T., Tilgner, A., Styler, S. A., Weller, C., Teich, M., and Otto, T.: Tropospheric Aqueous-Phase Chemistry: Kinetics, mechanisms, and its coupling to a changing gas phase, Chem. Rev., 115, 4259–4334, https://doi.org/10.1021/cr500447k, 2015.
Hill, K. A., Shepson, P. B., Galbavy, E. S., Anastasio, C., Kourtev, P. S., Konopka, A., and Stirm, B. H.: Processing of atmospheric nitrogen by clouds above a forest environment, J. Geophys. Res., 112, D11301, https://doi.org/10.1029/2006JD008002, 2007.
Hoffmann, L., Günther, G., Li, D., Stein, O., Wu, X., Griessbach, S., Heng, Y., Konopka, P., Müller, R., Vogel, B., and Wright, J. S.: From ERA-Interim to ERA5: the considerable impact of ECMWF's next-generation reanalysis on Lagrangian transport simulations, Atmos. Chem. Phys., 19, 3097–3124, https://doi.org/10.5194/acp-19-3097-2019, 2019.
Jaber, S., Joly, M., Brissy, M., Leremboure, M., Khaled, A., Ervens, B., and Delort, A.-M.: Biotic and abiotic transformation of amino acids in cloud water: experimental studies and atmospheric implications, Biogeosciences, 18, 1067–1080, https://doi.org/10.5194/bg-18-1067-2021, 2021.
Jørgensen, R. N., Jørgensen, B. J., and Nielsen, N. E.: N2O emission immediately after rainfall in a dry stubble field, Soil. Biol. Biochem., 30, 545–546, https://doi.org/10.1016/S0038-0717(97)00144-2, 1998.
Jorth, P., Turner, K. H., Gumus, P., Nizam, N., Buduneli, N., and Whiteley, M.: Metatranscriptomics of the human oral microbiome during health and disease, mBio, 5, e01012-14, https://doi.org/10.1128/mBio.01012-14, 2014.
Jousse, C., Dalle, C., Canet, I., Lagrée, M., Traïkia, M., Lyan, B., Mendes, C., Sancelme, M., Amato, P., and Delort, A.-M.: Metabolomic study of the response to cold shock in a strain of Pseudomonas syringae isolated from cloud water, Metabolomics, 14, 11, https://doi.org/10.1007/s11306-017-1295-7, 2018.
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M., and Ishiguro-Watanabe, M.: KEGG for taxonomy-based analysis of pathways and genomes, Nucleic Acids Res., 51, D587–D592, https://doi.org/10.1093/nar/gkac963, 2023.
Khaled, A., Zhang, M., Amato, P., Delort, A.-M., and Ervens, B.: Biodegradation by bacteria in clouds: an underestimated sink for some organics in the atmospheric multiphase system, Atmos. Chem. Phys., 21, 3123–3141, https://doi.org/10.5194/acp-21-3123-2021, 2021.
Klein, A. M., Bohannan, B. J. M., Jaffe, D. A., Levin, D. A., and Green, J. L.: Molecular evidence for metabolically active bacteria in the atmosphere, Front. Microbiol., 772, https://doi.org/10.3389/fmicb.2016.00772, 2016.
Koehler, K. A., Kreidenweis, S. M., DeMott, P. J., Prenni, A. J., Carrico, C. M., Ervens, B., and Feingold, G.: Water activity and activation diameters from hygroscopicity data – Part II: Application to organic species, Atmos. Chem. Phys., 6, 795–809, https://doi.org/10.5194/acp-6-795-2006, 2006.
Krumins, V., Mainelis, G., Kerkhof, L. J., and Fennell, D. E.: Substrate-dependent rRNA production in an airborne bacterium, Environ. Sci. Tech. Lett., 1, 376–381, https://doi.org/10.1021/ez500245y, 2014.
Kudla, B., Caddick, M. X., Langdon, T., Martinez-Rossi, N. M., Bennett, C. F., Sibley, S., Davies, R. W., and Arst Jr., H. N.: The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger, EMBO J., 9, 1355, https://doi.org/10.1002/j.1460-2075.1990.tb08250.x, 1990.
Lelieveld, J. and Crutzen, P. J.: Influences of cloud photochemical processes on tropospheric ozone, Nature, 343, 227–233, https://doi.org/10.1038/343227a0, 1990.
Leroch, M., Kleber, A., Silva, E., Coenen, T., Koppenhöfer, D., Shmaryahu, A., Valenzuela, P. D. T., and Hahn, M.: Transcriptome profiling of Botrytis cinerea conidial germination reveals upregulation of infection-related genes during the prepenetration stage, Eukaryot. Cell., 12, 614–626, https://doi.org/10.1128/ec.00295-12, 2013.
Li, C., Jia, S., Rajput, S. A., Qi, D., and Wang, S.: Transcriptional Stages of Conidia Germination and Associated Genes in Aspergillus flavus: An Essential Role for Redox Genes, Toxins, 14, 560, https://doi.org/10.3390/toxins14080560, 2022.
Li, K., Guo, Y., Nizkorodov, S. A., Rudich, Y., Angelaki, M., Wang, X., An, T., Perrier, S., and George, C.: Spontaneous dark formation of OH radicals at the interface of aqueous atmospheric droplets, P. Natl. Acad. Sci. USA, 120, e2220228120, https://doi.org/10.1073/pnas.2220228120, 2023.
Li, W. and Godzik, A.: Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences, Bioinformatics, 22, 1658–1659, https://doi.org/10.1093/bioinformatics/btl158, 2006.
Menke, S., Gillingham, M. A. F., Wilhelm, K., and Sommer, S.: Home-made cost effective preservation buffer is a better alternative to commercial preservation methods for microbiome research, Front. Microbiol., 8, 102, https://doi.org/10.3389/fmicb.2017.00102, 2017.
Noguchi, H., Taniguchi, T., and Itoh, T.: MetaGeneAnnotator: Detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes, DNA Res., 15, 387–396, https://doi.org/10.1093/dnares/dsn027, 2008.
Nuñez López, L., Amato, P., and Ervens, B.: Bacteria in clouds biodegrade atmospheric formic and acetic acids, Atmos. Chem. Phys., 24, 5181–5198, https://doi.org/10.5194/acp-24-5181-2024, 2024.
Osherov, N. and May, G. S.: The molecular mechanisms of conidial germination, FEMS Microbiol. Lett., 199, 153–160, https://doi.org/10.1111/j.1574-6968.2001.tb10667.x, 2001.
Péguilhan, R.: Metatranscriptomic analysis of cloud and aerosol samples at puy de Dôme, France, ENA [data set], https://www.ebi.ac.uk/ena/browser/view/PRJEB54740 (last access: 4 March 2025), 2025.
Péguilhan, R., Besaury, L., Rossi, F., Enault, F., Baray, J.-L., Deguillaume, L., and Amato, P.: Rainfalls sprinkle cloud bacterial diversity while scavenging biomass, FEMS Microbiol. Ecol., 97, fiab144, https://doi.org/10.1093/femsec/fiab144, 2021.
Péguilhan, R., Rossi, F., Rué, O., Joly, M., and Amato, P.: Comparative analysis of bacterial diversity in clouds and aerosols, Atmos. Environ., 298, 119635, https://doi.org/10.1016/j.atmosenv.2023.119635, 2023a.
Péguilhan, R., Rossi, F., Rué, O., Joly, M., and Amato, P.: Experimental and methodological framework for the assessment of nucleic acids in airborne microorganisms, bioRxiv [preprint], https://doi.org/10.1101/2023.10.10.561683, 2023b.
Poolman, B. and Glaasker, E.: Regulation of compatible solute accumulation in bacteria, Mol. Microbiol., 29, 397–407, https://doi.org/10.1046/j.1365-2958.1998.00875.x, 1998.
Prass, M., Andreae, M. O., de Araùjo, A. C., Artaxo, P., Ditas, F., Elbert, W., Förster, J.-D., Franco, M. A., Hrabe de Angelis, I., Kesselmeier, J., Klimach, T., Kremper, L. A., Thines, E., Walter, D., Weber, J., Weber, B., Fuchs, B. M., Pöschl, U., and Pöhlker, C.: Bioaerosols in the Amazon rain forest: temporal variations and vertical profiles of Eukarya, Bacteria, and Archaea, Biogeosciences, 18, 4873–4887, https://doi.org/10.5194/bg-18-4873-2021, 2021.
Pruppacher, H. R. and Jaenicke, R.: The processing of water vapor and aerosols by atmospheric clouds, a global estimate, Atmos. Res., 38, 283–295, https://doi.org/10.1016/0169-8095(94)00098-X, 1995.
Renard, P., Bianco, A., Baray, J.-L., Bridoux, M., Delort, A.-M., and Deguillaume, L.: Classification of Clouds Sampled at the Puy de Dôme Station (France) Based on Chemical Measurements and Air Mass History Matrices, Atmosphere-Basel, 11, 732, https://doi.org/10.3390/atmos11070732, 2020.
Renard, P., Brissy, M., Rossi, F., Leremboure, M., Jaber, S., Baray, J.-L., Bianco, A., Delort, A.-M., and Deguillaume, L.: Free amino acid quantification in cloud water at the Puy de Dôme station (France), Atmos. Chem. Phys., 22, 2467–2486, https://doi.org/10.5194/acp-22-2467-2022, 2022.
Rosado-Porto, D., Ratering, S., Moser, G., Deppe, M., Müller, C., and Schnell, S.: Soil metatranscriptome demonstrates a shift in C, N, and S metabolisms of a grassland ecosystem in response to elevated atmospheric CO2, Front. Microbiol., 13, 937021, https://doi.org/10.3389/fmicb.2022.937021, 2022.
Saikh, S. R. and Das, S. K.: Fog-Induced Alteration in Airborne Microbial Community: a Study over Central Indo-Gangetic Plain in India, Appl. Environ. Microb., 89, e01367–22, https://doi.org/10.1128/aem.01367-22, 2023.
Salazar, G., Paoli, L., Alberti, A., Huerta-Cepas, J., Ruscheweyh, H.-J., Cuenca, M., Field, C. M., Coelho, L. P., Cruaud, C., Engelen, S., Gregory, A. C., Labadie, K., Marec, C., Pelletier, E., Royo-Llonch, M., Roux, S., Sánchez, P., Uehara, H., Zayed, A. A., Zeller, G., Carmichael, M., Dimier, C., Ferland, J., Kandels, S., Picheral, M., Pisarev, S., Poulain, J., Acinas, S. G., Babin, M., Bork, P., Boss, E., Bowler, C., Cochrane, G., de Vargas, C., Follows, M., Gorsky, G., Grimsley, N., Guidi, L., Hingamp, P., Iudicone, D., Jaillon, O., Kandels-Lewis, S., Karp-Boss, L., Karsenti, E., Not, F., Ogata, H., Pesant, S., Poulton, N., Raes, J., Sardet, C., Speich, S., Stemmann, L., Sullivan, M. B., Sunagawa, S., Wincker, P., Acinas, S. G., Babin, M., Bork, P., Bowler, C., de Vargas, C., Guidi, L., Hingamp, P., Iudicone, D., Karp-Boss, L., Karsenti, E., Ogata, H., Pesant, S., Speich, S., Sullivan, M. B., Wincker, P., and Sunagawa, S.: Gene expression changes and community turnover differentially shape the global ocean metatranscriptome, Cell, 179, 1068–1083.e21, https://doi.org/10.1016/j.cell.2019.10.014, 2019.
Šantl-Temkiv, T., Finster, K., Dittmar, T., Hansen, B. M., Thyrhaug, R., Nielsen, N. W., and Karlson, U. G.: Hailstones: A window into the microbial and chemical inventory of a storm cloud, PloS one, 8, e53550, 2013.
Šantl-Temkiv, T., Amato, P., Gosewinkel, U., Thyrhaug, R., Charton, A., Chicot, B., Finster, K., Bratbak, G., and Löndahl, J.: High-flow-rate impinger for the study of concentration, viability, metabolic activity, and ice-nucleation activity of airborne bacteria, Environ. Sci. Technol., https://doi.org/10.1021/acs.est.7b01480, 2017.
Šantl-Temkiv, T., Gosewinkel, U., Starnawski, P., Lever, M., and Finster, K.: Aeolian dispersal of bacteria in southwest Greenland: their sources, abundance, diversity and physiological states, FEMS Microbiol. Ecol., 94, fiy031, https://doi.org/10.1093/femsec/fiy031, 2018.
Šantl-Temkiv, T., Amato, P., Casamayor, E. O., Lee, P. K. H., and Pointing, S. B.: Microbial ecology of the atmosphere, FEMS Microbiol. Rev., 46, fuac009, https://doi.org/10.1093/femsre/fuac009, 2022.
Satinsky, B. M., Zielinski, B. L., Doherty, M., Smith, C. B., Sharma, S., Paul, J. H., Crump, B. C., and Moran, M. A.: The Amazon continuum dataset: quantitative metagenomic and metatranscriptomic inventories of the Amazon River plume, June 2010, Microbiome, 2, 17, https://doi.org/10.1186/2049-2618-2-17, 2014.
Sattler, B., Puxbaum, H., and Psenner, R.: Bacterial growth in supercooled cloud droplets, Geophys. Res. Lett., 28, 239–242, 2001.
Schostag, M. D., Albers, C. N., Jacobsen, C. S., and Priemé, A.: Low Turnover of Soil Bacterial rRNA at Low Temperatures, Front Microbiol., 11, 962, https://doi.org/10.3389/fmicb.2020.00962, 2020.
Skopp, J., Jawson, M. D., and Doran, J. W.: Steady-State Aerobic Microbial Activity as a Function of Soil Water Content, Soil Sci. Soc. Am. J., 54, 1619–1625, https://doi.org/10.2136/sssaj1990.03615995005400060018x, 1990.
Smith, D. J., Ravichandar, J. D., Jain, S., Griffin, D. W., Yu, H., Tan, Q., Thissen, J., Lusby, T., Nicoll, P., Shedler, S., Martinez, P., Osorio, A., Lechniak, J., Choi, S., Sabino, K., Iverson, K., Chan, L., Jaing, C., and McGrath, J.: Airborne bacteria in Earth's lower stratosphere resemble taxa detected in the troposphere: results from a new NASA aircraft bioaerosol collector (ABC), Front Microbiol., 9, 1752, https://doi.org/10.3389/fmicb.2018.01752, 2018.
Stevenson, A., Cray, J. A., Williams, J. P., Santos, R., Sahay, R., Neuenkirchen, N., McClure, C. D., Grant, I. R., Houghton, J. D., Quinn, J. P., Timson, D. J., Patil, S. V., Singhal, R. S., Antón, J., Dijksterhuis, J., Hocking, A. D., Lievens, B., Rangel, D. E. N., Voytek, M. A., Gunde-Cimerman, N., Oren, A., Timmis, K. N., McGenity, T. J., and Hallsworth, J. E.: Is there a common water-activity limit for the three domains of life?, ISME J., 9, 1333–1351, https://doi.org/10.1038/ismej.2014.219, 2015.
The UniProt Consortium: UniProt: a worldwide hub of protein knowledge, Nucl. Acid. Res., 47, D506–D515, https://doi.org/10.1093/nar/gky1049, 2019.
Tignat-Perrier, R., Dommergue, A., Thollot, A., Magand, O., Amato, P., Joly, M., Sellegri, K., Vogel, T. M., and Larose, C.: Seasonal shift in airborne microbial communities, Sci. Total Environ., 716, 137129, https://doi.org/10.1016/j.scitotenv.2020.137129, 2020.
Tong, H., Hu, Q., Zhu, L., and Dong, X.: Prokaryotic Aquaporins, Cells, 8, 1316, https://doi.org/10.3390/cells8111316, 2019.
Unger, S., Máguas, C., Pereira, J. S., David, T. S., and Werner, C.: The influence of precipitation pulses on soil respiration – Assessing the “Birch effect” by stable carbon isotopes, Soil. Biol. Biochem., 42, 1800–1810, https://doi.org/10.1016/j.soilbio.2010.06.019, 2010.
Vaïtilingom, M., Deguillaume, L., Vinatier, V., Sancelme, M., Amato, P., Chaumerliac, N., and Delort, A.-M.: Potential impact of microbial activity on the oxidant capacity and organic carbon budget in clouds, P. Natl. Acad. Sci. USA, 110, 559–564, https://doi.org/10.1073/pnas.1205743110, 2013.
van Leeuwen, M. R., Krijgsheld, P., Bleichrodt, R., Menke, H., Stam, H., Stark, J., Wösten, H. A. B., and Dijksterhuis, J.: Germination of conidia of Aspergillus niger is accompanied by major changes in RNA profiles, Stud. Mycol., 74, 59–70, https://doi.org/10.3114/sim0009, 2013.
Veses, V., Richards, A., and Gow, N. A.: Vacuoles and fungal biology, Curr. Opin. Microbiol., 11, 503–510, https://doi.org/10.1016/j.mib.2008.09.017, 2008.
Wang, S., Lin, R., Tumukunde, E., Zeng, W., Bao, Q., Wang, S., and Wang, Y.: Glutamine synthetase contributes to the regulation of growth, conidiation, sclerotia development, and resistance to oxidative stress in the fungus Aspergillus flavus, Toxins-Basel, 14, 822, https://doi.org/10.3390/toxins14120822, 2022.
Wirgot, N., Vinatier, V., Deguillaume, L., Sancelme, M., and Delort, A.-M.: H2O2 modulates the energetic metabolism of the cloud microbiome, Atmos. Chem. Phys., 17, 14841–14851, https://doi.org/10.5194/acp-17-14841-2017, 2017.
Wirgot, N., Lagrée, M., Traïkia, M., Besaury, L., Amato, P., Canet, I., Sancelme, M., Jousse, C., Diémé, B., Lyan, B., and Delort, A.-M.: Metabolic modulations of Pseudomonas graminis in response to H2O2 in cloud water, Sci. Rep.-UK, 9, 1–14, https://doi.org/10.1038/s41598-019-49319-2, 2019.
Woo, C. and Yamamoto, N.: Falling bacterial communities from the atmosphere, Environ. Microbiome, 15, 22, https://doi.org/10.1186/s40793-020-00369-4, 2020.
Wood, D. E. and Salzberg, S. L.: Kraken: ultrafast metagenomic sequence classification using exact alignments, Genome Biol., 15, R46, https://doi.org/10.1186/gb-2014-15-3-r46, 2014.
Yang, K., Tian, J., and Keller, N. P.: Post-translational modifications drive secondary metabolite biosynthesis in Aspergillus: a review, Environ. Microbiol., 24, 2857–2881, https://doi.org/10.1111/1462-2920.16034, 2022.
Zhang, Y., Zhao, Z., Dai, M., Jiao, N., and Herndl, G. J.: Drivers shaping the diversity and biogeography of total and active bacterial communities in the South China Sea, Mol. Ecol., 23, 2260–2274, https://doi.org/10.1111/mec.12739, 2014.
Zhang, Y., Thompson, K. N., Huttenhower, C., and Franzosa, E. A.: Statistical approaches for differential expression analysis in metatranscriptomics, Bioinformatics, 37, i34–i41, https://doi.org/10.1093/bioinformatics/btab327, 2021.
Zhou, X., Fornara, D., Ikenaga, M., Akagi, I., Zhang, R., and Jia, Z.: The resilience of microbial community under drying and rewetting cycles of three forest soils, Front Microbiol., 7, 1101, https://doi.org/10.3389/fmicb.2016.01101, 2016.




