the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Comprehensive characterization of an aspen (Populus tremuloides) leaf litter sample that maintained ice nucleation activity for 48 years
Yalda Vasebi
Marco E. Mechan Llontop
Regina Hanlon
David G. Schmale III
Russell Schnell
Decaying vegetation was determined to be a potentially important source of atmospheric ice nucleation particles (INPs) in the early 1970s. The bacterium Pseudomonas syringae was the first microorganism with ice nucleation activity (INA) isolated from decaying leaf litter in 1974. However, the ice nucleation characteristics of P. syringae are not compatible with the characteristics of leaf litter-derived INPs since the latter were found to be sub-micron in size, while INA of P. syringae depends on much larger intact bacterial cells. Here we determined the cumulative ice nucleation spectrum and microbial community composition of the historic leaf litter sample 70-S-14 collected in 1970 that conserved INA for 48 years. The majority of the leaf litter-derived INPs were confirmed to be sub-micron in size and to be sensitive to boiling. Culture-independent microbial community analysis only identified Pseudomonas as potential INA. Culture-dependent analysis identified one P. syringae isolate, two isolates of the bacterial species Pantoea ananatis, and one fungal isolate of Mortierella alpina as having INA among 1170 bacterial colonies and 277 fungal isolates, respectively. Both Pa. ananatis and M. alpina are organisms that produce heat-sensitive sub-micron INPs. They are thus both likely sources of the INPs present in sample 70-S-14 and may represent important terrestrial sources of atmospheric INPs, a conclusion that is in line with other recent results obtained in regard to INPs from soil, precipitation, and the atmosphere.
- Article
(1387 KB) - Full-text XML
- BibTeX
- EndNote
Ice-nucleating particles (INPs) are necessary to initiate freezing of cloud droplets in mixed-phase clouds in order for precipitation to form. Therefore, the concentration of atmospheric INPs affects frequency and intensity of precipitation (Mülmenstädt et al., 2015). Identifying and characterizing INPs and their sources is thus the subject of intense research with important implications for modeling climate change (Morris et al., 2014; Coluzza et al., 2017).
In their search for sources of atmospheric INPs in the 1970s, Russell Schnell and Gabor Vali at the University of Wyoming found that decaying leaf litter contained unexpectedly high concentrations of INPs active at temperatures as warm as −1.3 ∘C (Schnell and Vali, 1972). These very active INPs were called leaf-derived nuclei (LDN). Because LDN were absent in green leaves and increased in concentration during leaf decay, it was hypothesized that they were of microbial origin. In fact, Pseudomonas syringae, the most active ice-nucleating microbe known to date, was later isolated from leaf litter (Maki et al., 1974). P. syringae is a genetically diverse species and includes plant pathogenic as well as non-pathogenic strains (Morris et al., 2013). Parallel to the discovery of P. syringae with INA in leaf litter, P. syringae with INA were also isolated from dried powdered corn leaves and identified as the culprit of frost damage in crop plants (Arny et al., 1976), and P. syringae with INA have been isolated from clouds, precipitation, and surface water (Morris et al., 2013). Because of its ubiquitous presence in compartments of the water cycle and other circumstantial evidence, P. syringae might contribute to the formation of precipitation in clouds (Morris et al., 2013). Other bacteria with INA have been identified in additional Pseudomonas species (Failor et al., 2017) and in additional genera within the Gammaproteobacteria: Erwinia (Phelps et al., 1986), Pantoea (Failor et al., 2017), a genus that includes several species classified in the past as members of the genus Erwinia, and Xanthomonas (Kim et al., 1987). A strain belonging to the genus Lysinibacillus was recently identified and characterized as the first ice-nucleating Gram-positive bacterium (Failor et al., 2017). Some fungal species have been found to have INA as well: Fusarium acuminatum and F. avenaceum (Pouleur et al., 1992), F. tricinctum and F. oxysporum (Richard et al., 1996), F. sporotrichioides (Huffman et al., 2013), Mortierella alpina (Fröhlich-Nowoisky et al., 2015), Isaria farinosa, and Acremonium implicatum (Huffman et al., 2013). An additional fungus with INA is the lichen fungus Rhizoplaca chrysoleuca (Kieft, 1988). Finally, pollen was added to this long list of biological INA (Diehl et al., 2001).
Microrganisms with INA produce different types of INA molecules. The Gammaproteobacteria produce a membrane-anchored INA protein that is inserted into the bacterial cell wall and is thus heat labile and cannot be separated from bacterial cells by filtration through membrane filters with a 0.22 µm pore size (Morris et al., 2004). However, in the case of Erwinia herbicola (now known as Pantoea ananatis), the INA protein can be released from cells as part of extra-cellular vesicles (EVs) (Phelps et al., 1986). Since EVs are submicron in size (tens to hundreds of nanometers), these vesicles can pass through a 0.22 µm filter.
All biological INPs outside of the Gammaproteobacteria pass through 0.22 µm filters. The INPs produced by the INA isolate of the genus Lysinibacillus are moreover very heat stable (remaining partially active even after an hour of boiling), are proteinase resistant, and do not pass through a 100 kDa protein filter with a pore size of approximately 50 nm. This suggests that they consist of relatively large non-proteinaceous secreted single molecules or stable aggregates of multiple smaller molecules (Failor et al., 2017). Fungal INPs are also mostly retained by 100 kDa filters, suggesting that they also consist of large secreted single molecules or of large aggregates of smaller molecules (Fröhlich-Nowoisky et al., 2015; O'Sullivan et al., 2015). Many fungal INPs are heat stable up to 60 ∘C but do not resist boiling and are variably susceptible to proteinases, and are thus believed to be mostly proteinaceous (Fröhlich-Nowoisky et al., 2015; O'Sullivan et al., 2015). Pollen INPs are mostly retained by 100 kDa filters, are heat and proteinase resistant (Pummer et al., 2012), and have been proposed to consist of polysaccharides of various chain lengths (Dreischmeier et al., 2017).
Now that all these biological INPs have been identified, and at least partially characterized, we decided to go back to one of the original leaf litter samples, the aspen (Populus tremuloides) leaf litter sample 70-S-14, which had been collected by Russell Schnell in 1970 and whose characterization in regard to its INA was published in 1976 (Schnell and Vali, 1976). Intriguingly, sample 70-S-14 maintained a high concentration of INA until today, 48 years after collection, while being stored at room temperature. INPs in this sample had been found to be less than 0.1 µm in size (Schnell and Vali, 1976). Therefore, Pseudomonas species can be excluded as the main source of INPs in this sample. While the INPs in the sample could be of plant origin (Pummer et al., 2015), here we tested the specific hypothesis that the INPs in this leaf litter sample were produced by strains of Lysinibacillus or by other bacteria or fungi. To test this hypothesis, we performed a comprehensive characterization of sample 70-S-14 in regard to its content in INPs and the composition of its microbial community.
2.1 Description of leaf litter sample 70-S-14 and how it has been stored
Leaf litter sample 70-S-14 was collected in a grove of Populus tremuloides (aspen) about 1.5 km west of Penhold Airbase, Alberta, now known as Red Deer Regional Airport (52.1762∘ N, 113.8870∘ W) in the summer of 1970. Even though the dominant litter in the grove was from decaying aspen leaves that fell the previous fall, there were decaying leaves from shrubs and grasses that also were collected in the sample. To collect the litter, handfuls of litter were grasped in a 1 m square to fill a garbage bag. The litter was somewhat moist as it was collected in the shade of the trees. The grove is still there and litter samples from the same area have been collected many times over the intervening decades, most recently in June 2018. The litter still produces prodigious numbers of ice nuclei with freezing activity beginning at −4.5 to −5 ∘C in concentrations of 108 g−1 of leaf litter active at −10 ∘C.
Some months after collection, when the litter sample was dry, the twigs and other large debris were removed. Some of the cleaned sample was ground to a fine powder in a fluid energy mill and used for INA tests in the Colorado State University Cloud Chamber, where it proved to produce excellent INPs (Schnell and Vali, 1976, Fig. 3).
Sample 70-S-14 was stored at room temperature inside four different plastic bags and a paper bag from 1970 to 2018 at room temperature open to the air in Schnell's various offices and a portion mailed to Virginia Tech in 2017. At Virginia Tech, it was again stored at room temperature in a paper bag until processing.
2.2 Characterization of the cumulative ice nucleation spectrum of leaf litter sample 70-S-14
One gram of leaf litter was added to 100 mL double-distilled water (DDW) in a 250 mL flask under sterile conditions and stirred for 5 min. The leaf litter suspension was passed through 2.5 µm pore size Whatman cellulose filter paper (GE Healthcare, USA) to remove large leaf fragments. This primary suspension was then subject to 10 different treatments (see Fig. 1), and INA was tested after each treatment to determine the main characteristics of the INPs present in the leaf litter sample.
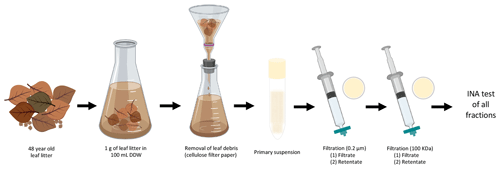
Figure 1Characterization of INA of aspen leaf litter 70-S-14. For each biological replicate (n=4), 1 g of aspen leaf litter was suspended in 100 mL of sterile double-stilled water (DDW). The primary suspension, the filtrates, and the filter retentates of the 0.22 µm and the 100 kDa filters were tested for INA. In parallel, a portion of the primary suspension and of both filtrates and of both filter retentates were boiled for 15 min and tested for INA as well.
In short, 10 mL of the primary suspension was passed through a 0.22 µm filter (Supor® 200 PES membrane Disc Filter, PALL, USA) to select for sub-micron INPs in the filtrate and for intact bacterial and fungal cells in the 0.22 µm filter retentate resuspended from the filter in the same volume (10 mL); 5 mL of the 0.22 µm filtrate was then passed through a 100 kDa protein filter with an approximate pore size of 50 nm (Macrosep Advance Centrifugal Device with 100 K MWCO, PALL, USA) for 10 min at 5000 rpm to separate sub 50 nm sized INPs in the 100 kDa filter filtrate from INPs larger than 50 nm in the 100 kDa filter retentate resuspended in the same volume (5 mL). Portions of the original suspension and of each filtrate and retentate were also boiled for 15 min to determine the heat sensitivity of each fraction.
Dilutions from 10−1 to 10−6 were made after each of the 10 treatments to obtain cumulative ice nucleation spectra. Thirty drops of 20 µL of each dilution were tested at −8, −10, and −12 ∘C, maintaining each temperature for 10 min on parafilm boats floating in a cryobath (Lauda Alpha RA24, LAUDA-Brinkmann, Delran, NJ, USA). The entire assay, including the 10 treatments, was repeated three times. The concentration of ice nuclei was calculated using the approach developed by Vali (1971) and described previously (Failor et al., 2017).
2.3 Culture-independent microbial community analysis
To determine the overall composition of the fungal and bacterial communities in the leaf litter sample, a culture-independent approach was used first. The primary leaf litter suspension described above was vacuum-filtered through a 0.22 µm pore-size filter membrane (Supor® 200 PES membrane Disc Filter, PALL, USA). DNA was extracted from the filter with the DNeasy PowerWater kit (Qiagen, USA) according to the manufacturer's protocol. DNA concentration and quality were evaluated by UV spectrophotometry (NanoDrop 1000, Thermo, USA) and visualized on a 1 % agarose gel.
For bacterial community analysis, the V4 hypervariable region of the 16S rRNA gene was amplified and sequenced using barcoded versions of the primers 799F (anti-chloroplast, 5′AACMGGATTAGATACCCKG3′) and 1115R (“universal”, 5′AGGGTTGCGCTCGTTG3′). For fungal community analysis, the ITS 2 region was amplified and sequenced using primers ITS9_F (GAACGCAGCRAAIIGYGA) and ITS4_R (TCCTCCGCTTATTGATATGC). All steps from PCR to paired-end (2×300 bp) amplicon sequencing on the Illumina MiSeq platform were performed at Molecular Research LP (MR DNA™, Shallowater, TX, USA).
Paired-end sequences were joined together into a single-sequence read. Quality trimming was performed and barcodes and primer sequences were depleted. Then, sequences shorter than 200 bp, sequences with ambiguous base calls, and sequences containing homopolymers longer than 6 bp were removed. High-quality sequences were processed using the Quantitative Insights into Microbial Ecology (QIIME) bioinformatic pipeline (Caporaso et al., 2010). Operational taxonomic units (OTUs) were assigned using an open-reference approach with a threshold of 97 % sequence similarity. OTU picking and taxonomy assignment were performed using UCLUST and the SILVA database (Quast et al., 2013). Fungal ITS paired-end sequences were processed as described above, but UNITE was used as the database instead (Abarenkov et al., 2010).
All OTUs assigned to mitochondria or chloroplasts and OTUs with fewer than five reads were excluded from further analysis. QIIME-generated output files were imported to R for data visualization using the Phyloseq 1.19.1 (McMurdie and Holmes, 2013) and ggplot 2 2.2.1 (Wickham, 2009) packages.
2.4 Culture-dependent bacterial community analysis
In parallel to the culture-independent approach, bacteria were cultured to determine the composition of the bacterial community present in the leaf litter sample using a culture-dependent approach. The undiluted primary leaf litter suspension described above and 10−1 and 10−2 dilutions were plated on R2A (Reasoner and Geldreich, 1985). Agar plates were incubated at 28 ∘C for 2–7 days. After incubation, the bacterial population was estimated by counting the bacterial colonies. This assay was performed three times. One hundred colonies were randomly selected for identification using PCR followed by Sanger sequencing of the V4 hypervariable region of the 16S rRNA gene as described previously (Failor et al., 2017).
2.5 INA testing of individual bacterial colonies
The suspensions and 10−1 and 10−2 dilutions described above were plated on R2A and LEM (Lysinibacillus enrichment medium). LEM is based on a medium originally developed for Lysinibacillus sphaericus (Russell et al., 1989) to enrich for Lysinibacillus strains similar to the Lysinibacillus strain with INA that we recently isolated (Failor et al., 2017). LEM contains, per liter, Na2HPO4, 5.57 g; KH2PO4, 2.4 g; (NH4)2SO4, 2.0 g; MgSO4⋅7H2O, 50 mg; MnCl2⋅4H2O, 4.0 mg; FeSO4⋅7H2O, 810 µg; CaCl2⋅2H2O, 1.5 mg; H2SO4, 0.3 µL; sodium acetate, 321.8 mg; thiamine 200 mg; and biotin, 20 µg. After incubation for up to 7 days, random bacterial colonies were resuspended in 140 µL of DDW. Five drops of 20 µL from each colony suspension were tested for INA at −8, −10, and −12 ∘C. Colonies for which at least one drop froze at any of the used temperatures were streaked onto new plates. Bacterial suspensions were made from the new plates for these colonies and five drops were tested for INA a second time. If still positive, a third test was performed similarly to the one described to determine the cumulative ice nucleation spectrum of leaf litter 70-S-14: 30 drops of 20 µL each were tested for each dilution of a 10−1 to 10−6 dilution series starting with a bacterial suspension of approximately 1×108 cfu mL−1. The same was done to determine the cumulative ice nucleation spectrum of Pa. ananatis BAV 3057. In this case, a bacterial suspension at a concentration of 3.2×108 cfu mL−1 was used as a starting suspension for the 10−1 to 10−6 dilution series and the bacterial starting suspension was subject to the same differential filtration and boiling combinations as the leaf litter.
2.6 INA testing of individual fungal colonies
Ten-fold serial dilutions were made from the primary suspension described above. One hundred microliters of each dilution was plated on potato dextrose agar (PDA) medium supplemented with either streptomycin (20 mg L−1) or lactic acid (0.1 %) to restrict bacterial growth. Plates were incubated for 7 days at room temperature. Single fungal colonies were transferred to new PDA plates supplemented with streptomycin (20 mg L−1) and kept at room temperature. For all fungal isolates, mycelium was scraped off the agar and resuspended in 1 mL of DDW, and three drops of 20 µL from each fungal suspension were tested for INA as described above. Fungal isolates for which at least one drop froze at any of the used temperatures were transferred to new PDA plates. To confirm INA in fungal isolates, new PDA plates were used to prepare fungal suspensions, and 30 drops were tested for a second time. For the cumulative ice nucleation spectrum of M. alpina strain LL118, 1 mg of mycelium was scraped off the PDA plates and suspended in 10 ml of DDW and processed as described in the protocol used for the cumulative ice nucleation spectrum of leaf litter 70-S-14.
3.1 Aspen leaf litter 70-S-14 contains mostly INPs of submicron size that are heat-sensitive
Differential filtration using a 0.22 µm pore size filter and a 100 kDa filter (approximately 50 nm pore size) in combination with boiling each filtrate and each retentate allowed us to determine the concentration of total INPs in the leaf litter sample and their approximate size and heat sensitivity (Fig. 2). While there was no detectable INA at −6 ∘C and above, INA increased strongly when the temperature was lowered to −8 ∘C, at which temperature we detected 106 INPs per gram of leaf litter. The concentration of active INPs increased further to almost 107 g−1 at −10 and −12 ∘C. Based on the earlier tests of ice nucleation on less aged 70-S-14 by Schnell and Vali (1976), it appears that the sample of 70-S-14 lost between 1 and 1.5 ∘C of threshold ice nucleation activity and 2 orders of magnitude in total INP concentration active at −10 to −12 ∘C over the 48 years of storage.
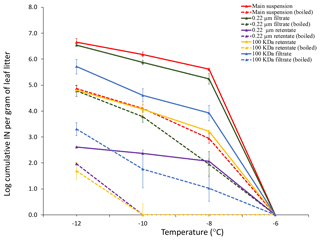
Figure 2Cumulative ice nucleation spectra of aspen leaf litter sample 70-S-14. Results are shown for all fractions shown in Fig. 1 based on droplet freezing assays at −6, −8, −10, and −12 ∘C. IN: ice nuclei.
Almost all INPs passed through the 0.22 µm filter, confirming the results obtained by Schnell in 1976 that leaf litter mostly contains sub-micron INPs (Schnell and Vali, 1976). The retentate of the 0.22 µm filter further confirmed this result since it had a very low concentration of INPs, approximately only 1∕100 to 1∕1000 of the 0.22 µm filtrate. Boiling the original unfiltered suspension and the 0.22 µm filtrate also reduced the INP concentration to 1∕100 to 1∕1000, revealing that the majority of INPs present in the leaf litter is heat sensitive. Further, passing the 0.22 µm filtrate through the 100 kDa filter reduced the concentration of INPs active at −8 ∘C approximately 100-fold, while the concentration of INPs active at −12 ∘C was reduced approximately 10-fold. This suggests that the majority of sub-micron INPs is larger than 50 nm and that a fraction of INPs is even smaller than 50 nm. Intriguingly, resuspensions of the INPs from the 100 kDa filter revealed a concentration of INPs in the 100 kDa filter retentate that was even lower than that in the 100 kDa filtrate. This could be due to the majority of INPs strongly binding to the filter. When the filtrate and the retentate of the 100 kDa filter were boiled, INPs active at −8 ∘C were almost completely abolished, further confirming the heat sensitivity of the majority of the INPs present in the 70-S-14 leaf litter sample. In summary, the majority of INPs in aspen leaf litter sample 70-S-14 consisted of heat-sensitive sub-micron-sized particles, with some being smaller than 50 nm and some being larger than 50 nm.
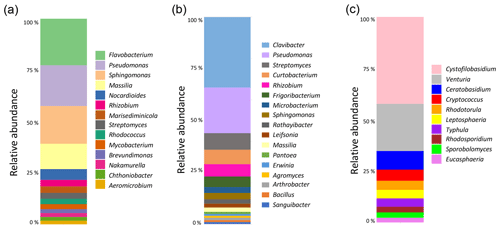
Figure 3Analysis of the composition of the microbial communities present in aspen leaf litter sample 70-S-14. (a) Results from the culture-independent analysis of bacterial diversity based on the V4 hypervariable region of the 16S rRNA gene (considering only classified reads); (b) results from sequencing the V4 hypervariable region of the 16S rRNA gene of 100 bacterial isolates cultured from sample 70-S-14; (c) results from the culture-independent analysis of fungal diversity based on the ITS region (considering only classified reads).
3.2 Microbial population analysis of the aspen leaf litter 70-S-14 sample reveals few known microbial taxa with INA
To identify the possible biological origin of the INPs present in the aspen leaf litter sample, the overall composition of the bacterial and fungal communities was determined. This was done using a culture-independent analysis by extracting DNA followed by amplification and sequencing of the bacterial V4 hypervariable region of the 16S rRNA gene and the fungal ITS region (Fig. 3).
The main bacterial phyla found in the leaf litter were Proteobacteria (49 %), Actinobacteria (34 %), and Bacteroidetes (16 %). Within the Proteobacteria, the genera Pseudomonas (13 %) and Sphingomonas (12 %) were the most common. Within the Actinobacteria, the most common taxon was an undescribed genus in the Microbacteriaceae family (22 %). Within the Bacteroidetes, Flavobacterium was the most common genus (14 %). Therefore, of all bacterial taxa known to include strains with INA, only the genus Pseudomonas was identified in this culture-independent approach. The fungal leaf litter community contained 42 % Basidiomycota, 38 % unidentified phyla, and 20 % Ascomycota. The most common genus was identified as Cystofilobasidium (14 %), followed by Venturia, Ceratobasidium, and unidentified genera in the Helotiales and Tremellomycetes. None of these genera is known to include INA strains.
For bacteria, we also performed a culture-dependent analysis by plating the primary suspension on R2A medium. Based on colony counts, the leaf litter contained a total of 3.13×105 colony-forming units (CFUs) per gram of leaf litter. One hundred random colonies were then selected for partially sequencing the 16S rRNA gene. This analysis revealed the presence of bacterial genera that are known to include strains with INA. In fact, not only were 22 % of colonies identified as Pseudomonas, but 1 % each were identified as Pantoea and Erwinia. However, the genus with the highest number of colonies (34 %), the Gram-positive bacterium Clavibacter, is not known to include any strains with INA.
3.3 INA testing of bacterial and fungal colonies reveals presence of INA strains belonging to the bacterial species P. syringae and Pa. ananatis strains and the fungal species M. alpina
To determine whether any culturable bacteria or fungi with INA were still present in the aspen leaf litter 48 years after collection, a total of 1170 bacterial colonies either grown on R2A (881 colonies, including the 100 colonies described above) or LEM (289 colonies) and 277 fungal isolates grown on PDA were tested for INA.
Only three bacterial colonies were found to have stable INA in all tests starting at −8 ∘C or above. The strains were identified by sequencing the hypervariable V4 region of the 16S rRNA gene. Two strains were identified as members of the genus Pantoea: one strain had 99 % DNA identity over 935 nt with P. ananatis strain Ta030 (NCBI accession number MH973238), and the other strain had 99 % DNA identity over 922 nt with P. ananatis strain 12WE (NCBI accession number MH010898.1). The third strain was identified as a member of the genus Pseudomonas since it had 99 % DNA identity over 794 nt with P. syringae pv. syringae strain CFBP4215 (NCBI accession number LT962480).
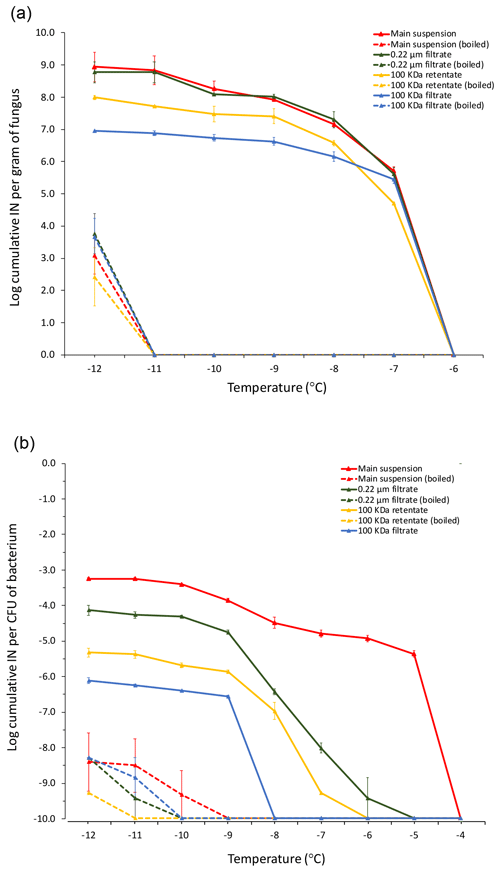
Figure 4Cumulative ice nucleation spectra of M. alpina strain LL118 (a) isolated from leaf litter sample 70-S-14 and of Pa. ananatis BAV 3057 (b) isolated from rain, but over 99 % identical in its 16S rRNA sequence to the two Pa. ananatis isolates from sample 70-S-14. The analyzed fractions derived from differential filtration and boiling are the same as shown in Fig. 1; besides that, instead of leaf litter, suspensions of M. alpina mycelium and of a Pa. ananatis bacterial suspension were used as the starting material. IN: ice nuclei; CFU: colony-forming units.
Among the fungal isolates, only one was found to have stable INA at −8 ∘C. This isolate, LL118, was identified as a member of the species M. alpina based on ITS sequencing since it had 99 % identity to M. alpina isolate F08ID36 with NCBI accession number KJ469836.1 (Fröhlich-Nowoisky et al., 2015).
Unfortunately, the P. syringae and the two Pa. ananatis strains isolated from the leaf litter were lost and could not be further characterized. However, we characterized the cumulative ice nucleation spectrum of M. alpina LL118 and of Pa. ananatis strain BAV 3057, which was previously isolated from rain (Failor et al., 2017) and that had over 99 % identity in its 16S rRNA sequence to one of the two Pa. ananatis strains from leaf litter. Because of the genetic similarity of Pa. ananatis BAV 3057 to the two Pa. ananatis strains isolated from the leaf litter, these strains can be expected to have very similar cumulative ice nucleation spectra. The number of INPs produced by M. alpina LL118 per gram of fungal mycelium active at −7 ∘C (the temperature at which drops of the fungal suspension started freezing) was approximately 1×106. The number of INPs produced by Pa. ananatis BAV 3057 at −5 ∘C (the temperature at which drops of the bacterial suspension started freezing) was approximately per CFU. Figure 4 shows how the INPs produced by both M. alpina LL118 and Pa. ananatis BAV 3057 were mostly heat sensitive. M. alpina INPs were all sub-micron in size, and the majority could be resuspended from the 100 kDa filter and were thus larger than 50 nm. Only approximately 1∕10 of M. alpina INPs passed through the 100 kDa filter and were thus smaller than 50 nm. Pa. ananatis INPs active at −5 ∘C were mostly larger than 0.22 µm, but 1∕10th of the Pa. ananatis INPs active at −9 ∘C and below were sub-micron in size. Similarly to M. alpina, more of the sub-micron INPs could be resuspended from the 100 kDa filter (and were thus larger than 50 nm) compared to the INPs that passed through the filter (and were thus smaller than 50 nm).
Aspen leaf litter sample 70-S-14 (Schnell and Vali, 1976) collected in 1970 has maintained remarkable INA over 48 years. When its characterization was first published in 1976, P. syringae was the only known organism with INA, and microbial community analysis was not possible. Therefore, with the discovery of many bacterial, fungal, and plant INA organisms since then and today's ease in determining the composition of microbial communities, this historic sample represented a great opportunity for re-evaluation of its INA and identification of the INA organisms possibly still present and alive in this sample 48 years later.
The cumulative INP spectra of 70-S-14 obtained after a combination of filtration and boiling (Fig. 2) clearly confirmed the result obtained in 1976 by Russel Schnell; i.e., the INPs in this sample are submicron in size. Moreover, the INPs were found to be mostly heat sensitive. Since INPs produced by Lysinibacillus are heat resistant (Failor et al., 2017), this result excludes our initial hypothesis that recently discovered INA strains of the genus Lysinibacillus might have contributed to the INA of 70-S-14. Also, heat-resistant INPs produced by pollen can be excluded (Pummer et al., 2012).
The result is more in line with secreted INPs produced by strains of the species Erwinia herbicola (Phelps et al., 1986), of which most strains were later assigned to the species Pantoea ananatis (Walterson and Stavrinides, 2015), and with INPs secreted by fungi in the genus Fusarium (O'Sullivan et al., 2015) and in the species M. alpina (Fröhlich-Nowoisky et al., 2015). In fact, Erwinia/Pantoea cells secrete some of the heat-sensitive INA protein as part of sub-micron EVs (Phelps et al., 1986), and F. avenaceum and M. alpina both secrete sub-micron INPs that are heat sensitive and probably proteinaceous in nature (Fröhlich-Nowoisky et al., 2015; O'Sullivan et al., 2015). However, culture-independent analysis of 70-S-14 did not identify any of these organisms, although a high number of bacterial 16S rRNA fragments (131 796 sequencing reads) and a high number of fungal ITS fragments (153 546 sequencing reads) were sequenced. This suggests that these known INA organisms are very minor constituents of the overall microbial communities present in this leaf litter sample.
Interestingly though, sequencing a 16S rRNA fragment of a relatively small number (100) of randomly selected bacterial colonies cultured from 70-S-14 did reveal the presence of Pantoea/Erwinia (two colonies). This may be due to enrichment of this genus by the employed culture media. Even more importantly, two Pa. ananatis isolates and one M. alpina isolate with INA were identified among the 1170 bacterial colonies and the 277 fungal isolates, respectively. The cumulative ice nucleation spectra of M. alpina and Pa. ananatis fit the overall ice nucleation spectrum of 70-S-14 (with the majority of INPs being heat sensitive and in large part sub-micron in size) and are thus likely sources of the INPs present in 70-S-14.
Only one Pseudomonas strain with INA was isolated from 70-S-14. However, the typical ice nucleation spectrum of Pseudomonas species does not fit the leaf litter ice nucleation spectrum since most Pseudomonas species do not shed the INA protein as part of EVs, but INA of Pseudomonas species is generally associated with intact bacterial cells that do not pass through 0.22 µm pores (Failor et al., 2017; Maki et al., 1974; Morris et al., 2004).
Although Mortierella was the only fungal genus with known INA identified in 70-S-14, it is possible that other fungal strains are present (and culturable) at very low frequency in this leaf litter sample since the number of tested fungal isolates was relatively low (277 isolates). Therefore, we cannot exclude that testing additional fungal isolates could reveal the presence of additional fungal genera with INA in 70-S-14. However, it is also possible that some INPs still present in the leaf litter sample 48 years after collection were originally produced by bacteria or fungi that are not culturable anymore and whose DNA has been degraded. In fact, it is interesting that the number of INPs produced per gram of pure culture of the isolated M. alpina strain is approximately only 100-fold larger than the number of INPs produced per gram of leaf litter. Considering that the total mass of fungi and bacteria with INA in the leaf litter probably constitutes only a very minor fraction of the total leaf mass in the leaf litter sample suggests that organisms that produce a very high number of INPs must have been present in the leaf litter sample at some point. It is also possible that M. alpina, Pa. ananatis, and other INA organisms produced a much higher number of INPs when they originally grew in the decaying leaves compared to the number of INPs that they produced when grown on nutrient-rich agar plates in our lab before INA testing. In fact, low nutrient availability has been shown to increase production of INPs by various INA bacteria (Failor et al., 2017).
One important question in regard to the present study is whether the presence of viable Pa. ananatis and M. alpina isolates in a leaf litter sample that has maintained INA for 48 years strengthens the evidence for a role of these organisms as a source of atmospheric INPs. Several lines of evidence support a positive answer to this question. In fact, 106 strains among 593 strains found to have stable INA among 33 134 strains isolated from precipitation and tested for INA in our previous work (Failor et al., 2017) were identified as Pantoea or Erwinia as well. Also, Du and colleagues recently found submicron INPs in precipitation (Du et al., 2017). Although Pantoea/Erwinia and M. alpina and other known organisms with INA could not be identified in the analyzed precipitation samples using a culture-independent approach (Du et al., 2017), these organisms could still be present since we could not identify them in our culture-independent leaf litter analysis either, but we still found them by culturing. Finally, Conen and Yakutin recently identified a large fraction of heat-sensitive submicron INPs in soils from various continents (Conen and Yakutin, 2018). Since these INPs were only inactivated through boiling but not by incubation at 60 ∘C, they are more likely of fungal origin than a product of Erwinia/Pantoea. In conclusion, we think that combining the results from these recent studies with our new finding that 70-S-14 still contains viable Pa. ananatis and M. alpina with INA supports a role of these organisms as important sources of atmospheric INPs. Importantly, finding that heat-sensitive sub-micron INPs are still active after 48 years in leaf litter suggests that leaf litter might represent an important reservoir of atmospheric INPs. The relative importance of leaf litter compared to live plants and soil as a contributor to the atmospheric pool of INPs is thus an important question that warrants further investigation.
What we could not do in the present study and what we could not do in our previous study of bacterial sources of INPs in precipitation (Failor et al., 2017) was to directly determine the presence of different genes coding for different biological INPs in metagenomic sequences. In fact, while direct culture-independent metagenomic sequencing of environmental samples is possible today (Behzad et al., 2015), the limitation is that the only gene known so far to encode a biological molecule with INA is the INA gene of the Gammaproteobacteria, including Pseudomonas species, Xanthomonas species, and Pantoea/Erwinia species (Edwards et al., 1994). Identifying the genetic basis of biological INPs produced by additional bacteria and by fungi would instead allow determination of the presence of all these various INA genes in environmental samples, such as soil, plants, leaf litter, precipitation, and even clouds. Comparison of presence and abundance of various INA genes between samples could in turn help infer the migration of microbes with INA among environments and their relative contribution to atmospheric INPs.
All data is is available upon request from the corresponding author.
YV and MEML performed the experiments and analyzed the data with the help and advice of RH for fungal isolation and culturing. DGS contributed expertise during the interpretation of the data. RS provided the leaf litter and suggested the overall research project. BAV, with the help of YV and MEML, developed the experimental plan and wrote the manuscript. All authors read and edited the manuscript. YV and MEML contributed equally to this work.
The authors declare that they have no conflict of interest.
This research was supported by the National Science Foundation under grant IOS-1754721. Funding to Boris A. Vinatzer and David G. Schmale III was also provided in part by the Virginia Agricultural Experiment Station and the Hatch Program of the National Institute of Food and Agriculture, US Department of Agriculture.
This paper was edited by Kees Jan van Groenigen and reviewed by Cindy Morris and one anonymous referee.
Abarenkov, K., Henrik Nilsson, R., Larsson, K.-H., Alexander, I. J., Eberhardt, U., Erland, S., Høiland, K., Kjøller, R., Larsson, E., Pennanen, T., Sen, R., Taylor, A. F. S., Tedersoo, L., Ursing, B. M., Vrålstad, T., Liimatainen, K., Peintner, U., and Kõljalg, U.: The UNITE database for molecular identification of fungi – recent updates and future perspectives, New Phytol., 186, 281–285, https://doi.org/10.1111/j.1469-8137.2009.03160.x, 2010.
Arny, D. C., Lindow, S. E., and Upper, C. D.: Frost sensitivity of Zea mays increased by application of Pseudomonas syringae, Nature, 262, 282–284, https://doi.org/10.1038/262282a0, 1976.
Behzad, H., Gojobori, T., and Mineta, K.: Challenges and Opportunities of Airborne Metagenomics, Genome Biol. Evol., 7, 1216–1226, https://doi.org/10.1093/gbe/evv064, 2015.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., Fierer, N., Peña, A. G., Goodrich, J. K., Gordon, J. I., Huttley, G. A., Kelley, S. T., Knights, D., Koenig, J. E., Ley, R. E., Lozupone, C. A., McDonald, D., Muegge, B. D., Pirrung, M., Reeder, J., Sevinsky, J. R., Turnbaugh, P. J., Walters, W. A., Widmann, J., Yatsunenko, T., Zaneveld, J., and Knight, R.: QIIME allows analysis of high-throughput community sequencing data, Nat. Method., 7, 335–336, https://doi.org/10.1038/nmeth.f.303, 2010.
Coluzza, I., Creamean, J., Rossi, M., Wex, H., Alpert, P., Bianco, V., Boose, Y., Dellago, C., Felgitsch, L., Fröhlich-Nowoisky, J., Herrmann, H., Jungblut, S., Kanji, Z., Menzl, G., Moffett, B., Moritz, C., Mutzel, A., Pöschl, U., Schauperl, M., Scheel, J., Stopelli, E., Stratmann, F., Grothe, H., and Schmale, D.: Perspectives on the Future of Ice Nucleation Research: Research Needs and Unanswered Questions Identified from Two International Workshops, Atmosphere, 8, 138, https://doi.org/10.3390/ijerph13010019, 2017.
Conen, F. and Yakutin, M. V.: Soils rich in biological ice-nucleating particles abound in ice-nucleating macromolecules likely produced by fungi, Biogeosciences, 15, 4381–4385, https://doi.org/10.5194/bg-15-4381-2018, 2018.
Diehl, K., Quick, C., Matthias-Maser, S., Mitra, S. K., and Jaenicke, R.: The ice nucleating ability of pollen: Part I: Laboratory studies in deposition and condensation freezing modes, Atmos. Res., 58, 75–87, https://doi.org/10.1016/S0169-8095(01)00091-6, 2001.
Dreischmeier, K., Budke, C., Wiehemeier, L., Kottke, T., and Koop, T.: Boreal pollen contain ice-nucleating as well as ice-binding “antifreeze” polysaccharides, Sci. Rep., 7, 41890, https://doi.org/10.1038/srep41890, 2017.
Du, R., Du, P., Lu, Z., Ren, W., Liang, Z., Qin, S., Li, Z., Wang, Y., and Fu, P.: Evidence for a missing source of efficient ice nuclei, Sci. Rep., 7, 39673, https://doi.org/10.1038/srep39673, 2017.
Edwards, A. R., Van den Bussche, R. A., Wichman, H. A., and Orser, C. S.: Unusual pattern of bacterial ice nucleation gene evolution, Mol. Biol. Evol., 11, 911–920, https://doi.org/10.1093/oxfordjournals.molbev.a040172, 1994.
Failor, K. C., Schmale, D. G., 3rd, Vinatzer III, B. A., and Monteil, C. L.: Ice nucleation active bacteria in precipitation are genetically diverse and nucleate ice by employing different mechanisms, ISME J., 11, 2740–2753, https://doi.org/10.1038/ismej.2017.124, 2017.
Fröhlich-Nowoisky, J., Hill, T. C. J., Pummer, B. G., Yordanova, P., Franc, G. D., and Pöschl, U.: Ice nucleation activity in the widespread soil fungus Mortierella alpina, Biogeosciences, 12, 1057–1071, https://doi.org/10.5194/bg-12-1057-2015, 2015.
Huffman, J. A., Prenni, A. J., DeMott, P. J., Pöhlker, C., Mason, R. H., Robinson, N. H., Fröhlich-Nowoisky, J., Tobo, Y., Després, V. R., Garcia, E., Gochis, D. J., Harris, E., Müller-Germann, I., Ruzene, C., Schmer, B., Sinha, B., Day, D. A., Andreae, M. O., Jimenez, J. L., Gallagher, M., Kreidenweis, S. M., Bertram, A. K., and Pöschl, U.: High concentrations of biological aerosol particles and ice nuclei during and after rain, Atmos. Chem. Phys., 13, 6151–6164, https://doi.org/10.5194/acp-13-6151-2013, 2013.
Kieft, T. L.: Ice Nucleation Activity in Lichens, Appl. Environ. Microbiol., 54, 1678–1681, 1988.
Kim, H. K., Orser, C., Lindow, S. E., and Sands, D. C.: Xanthomonas campestris pv. translucens strains active in ice nucleation, Plant Dis., 71, 994–997, 1987.
Maki, L. R., Galyan, E. L., Chang-Chien, M.-M., and Caldwell, D. R.: Ice Nucleation Induced by Pseudomonas syringae, Appl. Microbiol., 28, 456–459, 1974.
McMurdie, P. J. and Holmes, S.: phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data, PLOS ONE, 8, e61217, https://doi.org/10.1371/journal.pone.0061217, 2013.
Morris, C. E., Georgakopoulos, D. G., and Sands, D. C.: Ice nucleation active bacteria and their potential role in precipitation, J. Phys. IV France, 121, 87–103, 2004.
Morris, C. E., Monteil, C. L., and Berge, O.: The Life History of Pseudomonas syringae: Linking Agriculture to Earth System Processes, Annu. Rev. Phytopathol., 51, 85–104, https://doi.org/10.1146/annurev-phyto-082712-102402, 2013.
Morris, C. E., Conen, F., Alex Huffman, J., Phillips, V., Pöschl, U., and Sands, D. C.: Bioprecipitation: a feedback cycle linking Earth history, ecosystem dynamics and land use through biological ice nucleators in the atmosphere, Glob. Change Biol., 20, 341–351, https://doi.org/10.1111/gcb.12447, 2014.
Mülmenstädt, J., Sourdeval, O., Delanoë, J., and Quaas, J.: Frequency of occurrence of rain from liquid-, mixed-, and ice-phase clouds derived from A-Train satellite retrievals, Geophys. Res. Lett., 42, 6502–6509, https://doi.org/10.1002/2015GL064604, 2015.
O'Sullivan, D., Murray, B. J., Ross, J. F., Whale, T. F., Price, H. C., Atkinson, J. D., Umo, N. S., and Webb, M. E.: The relevance of nanoscale biological fragments for ice nucleation in clouds, Sci. Rep., 5, 8082, https://doi.org/10.1038/srep08082, 2015.
Phelps, P., Giddings, T. H., Prochoda, M., and Fall, R.: Release of cell-free ice nuclei by Erwinia herbicola, J. Bacteriol., 167, 496–502, 1986.
Pouleur, S., Richard, C., Martin, J.-G., and Antoun, H.: Ice Nucleation Activity in Fusarium acuminatum and Fusarium avenaceum, Appl. Environ. Microbiol., 58, 2960–2964, 1992.
Pummer, B. G., Bauer, H., Bernardi, J., Bleicher, S., and Grothe, H.: Suspendable macromolecules are responsible for ice nucleation activity of birch and conifer pollen, Atmos. Chem. Phys., 12, 2541–2550, https://doi.org/10.5194/acp-12-2541-2012, 2012.
Pummer, B. G., Budke, C., Augustin-Bauditz, S., Niedermeier, D., Felgitsch, L., Kampf, C. J., Huber, R. G., Liedl, K. R., Loerting, T., Moschen, T., Schauperl, M., Tollinger, M., Morris, C. E., Wex, H., Grothe, H., Pöschl, U., Koop, T., and Fröhlich-Nowoisky, J.: Ice nucleation by water-soluble macromolecules, Atmos. Chem. Phys., 15, 4077–4091, https://doi.org/10.5194/acp-15-4077-2015, 2015.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glöckner, F. O.: The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucl. Acids Res., 41, D590–D596, https://doi.org/10.1093/nar/gks1219, 2013.
Reasoner, D. J. and Geldreich, E. E.: A new medium for the enumeration and subculture of bacteria from potable water, Appl. Environ. Microbiol., 49, 1–7, 1985.
Richard, C., Martin, J. G., and Pouleur, S.: Ice nucleation activity identified in some phytopathogenic Fusarium species, Phytoprotection, 77, 83–92, https://doi.org/10.7202/706104ar, 1996.
Russell, B. L., Jelley, S. A., and Yousten, A. A.: Carbohydrate metabolism in the mosquito pathogen Bacillus sphaericus 2362, Appl. Environ. Microbiol., 55, 294–297, 1989.
Schnell, R. C. and Vali, G.: Atmospheric Ice Nuclei from Decomposing Vegetation, Nature, 236, 163–165, https://doi.org/10.1038/236163a0, 1972.
Schnell, R. C. and Vali, G.: Biogenic Ice Nuclei: Part I. Terrestrial and Marine Sources, J. Atmos. Sci., 33, 1554–1564, 1976.
Vali, G.: Quantitative Evaluation of Experimental Results an the Heterogeneous Freezing Nucleation of Supercooled Liquids, J. Atmos. Sci., 28, 402–409, 1971.
Walterson, A. M. and Stavrinides, J.: Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae, FEMS Microbiol. Rev., 39, 968–984, https://doi.org/10.1093/femsre/fuv027, 2015.
Wickham, H.: ggplt2, Use R!, Springer, New York, 213 pp., 2009.





