the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Co-occurrence of Fe and P stress in natural populations of the marine diazotroph Trichodesmium
Noelle A. Held
Eric A. Webb
Matthew M. McIlvin
David A. Hutchins
Natalie R. Cohen
Dawn M. Moran
Korinna Kunde
Maeve C. Lohan
Claire Mahaffey
E. Malcolm S. Woodward
Trichodesmium is a globally important marine microbe that provides fixed nitrogen (N) to otherwise N-limited ecosystems. In nature, nitrogen fixation is likely regulated by iron or phosphate availability, but the extent and interaction of these controls are unclear. From metaproteomics analyses using established protein biomarkers for nutrient stress, we found that iron–phosphate co-stress is the norm rather than the exception for Trichodesmium colonies in the North Atlantic Ocean. Counterintuitively, the nitrogenase enzyme was more abundant under co-stress as opposed to single nutrient stress. This is consistent with the idea that Trichodesmium has a specific physiological state during nutrient co-stress. Organic nitrogen uptake was observed and occurred simultaneously with nitrogen fixation. The quantification of the phosphate ABC transporter PstA combined with a cellular model of nutrient uptake suggested that Trichodesmium is generally confronted by the biophysical limits of membrane space and diffusion rates for iron and phosphate acquisition in the field. Colony formation may benefit nutrient acquisition from particulate and organic sources, alleviating these pressures. The results highlight that to predict the behavior of Trichodesmium, both Fe and P stress must be evaluated simultaneously.
- Article
(4404 KB) - Full-text XML
-
Supplement
(1906 KB) - BibTeX
- EndNote
The diazotrophic cyanobacterium Trichodesmium plays an important ecological and biogeochemical role in the tropical and subtropical oceans globally. By providing bioavailable nitrogen (N) to otherwise N-limited ecosystems, it supports basin-scale food webs, increasing primary productivity and carbon flux from the surface ocean (Capone et al., 1997; Carpenter and Romans, 1991; Coles et al., 2004; Deutsch et al., 2007; Sohm et al., 2011). Nitrogen fixation is energetically and nutritionally expensive, so it typically occurs when other sources of N are unavailable, i.e., in N-starved environments (Karl et al., 2002). However, nitrogen availability is not the sole control on nitrogen fixation, which must be balanced against the cell's overall nutritional status. Because it can access a theoretically unlimited supply of atmospheric N2, Trichodesmium often becomes phosphorus (P) limited (Frischkorn et al., 2018; Hynes et al., 2009; Orchard, 2010; Sañudo-Wilhelmy et al., 2001; Wu et al., 2000). It also has a tendency to experience iron (Fe) limitation because the nitrogenase enzyme is iron-demanding (Bergman et al., 2013; Chappell et al., 2012; Rouco et al., 2018; Sunda, 2012; Walworth et al., 2016a).
There is uncertainty about when and where Trichodesmium is Fe and P stressed and how this impacts nitrogen fixation in nature. Some reports suggest that Trichodesmium is primarily phosphate stressed in the North Atlantic and primarily Fe stressed in the Pacific, owing to relative Fe and P availability in these regions (Bergman et al., 2013; Chappell et al., 2012; Frischkorn et al., 2018; Hynes et al., 2009; Orchard, 2010; Sañudo-Wilhelmy et al., 2001). However, others have suggested that Fe and P can be co-limiting to Trichodesmium; one incubation study found two examples of Fe–P co-limitation in the field (Mills et al., 2004). Even less clear is how Fe and/or P stress impacts nitrogen fixation. For instance, despite the intuitive suggestion that nitrogen fixation is limited by Fe or P availability, laboratory evidence indicated that Trichodesmium is specifically adapted to co-limited conditions, with higher growth and N2-fixation rates under co-limitation than under single nutrient limitation (Garcia et al., 2015; Walworth et al., 2016a).
There are several protein biomarkers for Fe and P stress in Trichodesmium, many of which are periplasmic binding proteins involved in nutrient acquisition. For Fe, this includes the IdiA and IsiB proteins, and for phosphorus, specifically phosphate, it includes the PstS and SphX proteins (see Table S1 in the Supplement). In Trichodesmium, IdiA, an ABC transport protein, and IsiB, a flavodoxin, are expressed under Fe-limiting conditions, and both are conserved across species with high sequence identity (Chappell et al., 2012; Webb et al., 2007). Transcriptomic and proteomic studies have shown that they are more abundant during Fe stress, though there is low-level basal level expression (Chappell et al., 2012; Snow et al., 2015; Walworth et al., 2016a). In this dataset, IdiA and IsiB were both highly abundant and correlated to one another (Fig. S1 in the Supplement). IdiA is used as the molecular biomarker of Fe stress in the following discussion, but the same conclusions could be drawn from IsiB distributions. Like IdiA and IsiB, SphX and PstS are conserved across diverse Trichodesmium species (Chappell et al., 2012; Walworth et al., 2016a). SphX is abundant at the transcript and protein level under phosphate limitation (Orchard et al., 2009; Orchard, 2010). PstS, a homologous protein located a few genes downstream of SphX, responds less clearly to phosphate stress. In Trichodesmium, the reason may be that PstS is not preceded by a pho box, a regulatory DNA sequence which is necessary for P-based regulation (Orchard et al., 2009). Thus, in this study we focused on SphX as a marker of phosphate stress and IdiA as a marker of Fe stress.
Here, we present evidence based on field metaproteomes that Trichodesmium colonies were simultaneously Fe and P stressed, particularly in the tropical and subtropical Atlantic. While Fe–P stress has been suggested before, this study provides molecular evidence for co-stress in a broad geographical and temporal survey. This co-stress occurred across significant gradients in Fe and P concentration, suggesting nutrient stress was driven not only by biogeochemical gradients but also by Trichodesmium's response to nutrient depletion; we explore possible biophysical and biochemical mechanisms behind this. Fe and P stress were positively associated with nitrogen fixation and organic nitrogen uptake, suggesting that Trichodesmium's Fe, P, and N statuses are linked, perhaps via a regulatory network and/or due to the biophysical limits of nutrient uptake related to the intersection of membrane crowding and nutrient diffusion.
2.1 Sample acquisition
A total of 37 samples were examined in this study. Samples were acquired by the authors on various research expeditions, and most exist in biological duplicate or triplicate (Table S2). Trichodesmium colonies were handpicked from 200 or 130 µm surface plankton net tows, rinsed thrice in 0.2 µm filtered trace-metal-clean surface seawater into trace-metal-clean LDPE bottles, decanted onto 0.2–5 µm filters, and frozen until protein extraction. The samples were of mixed puff and tuff morphology, depending on the natural diversity present at the sampling location. The majority of samples considered in this study were taken in the early morning predawn hours. Details such as filter size, morphology, location, cruise, date, and time of sampling are provided in Table S2.
2.2 Protein extraction and digestion
Proteins were extracted by a detergent-based method following Saito et al. (2014) and Lu and Zhu (2005). To reduce protein loss and contamination, all tubes were ethanol rinsed and dried prior to use, and all water and organic solvents used were LC–MS grade. Sample filters were placed in a microfuge tube with 1–2 mL 1 % sodium dodecyl sulfate (SDS) extraction buffer (1 % SDS, 0.1 M Tris/HCL pH 7.5, 10 mM EDTA) and incubated for 10 min at 95 ∘C with shaking, then for 1 h at room temperature with shaking. The protein extract was decanted and clarified by centrifugation (14 100 g) at room temperature. The crude protein extracts were quantified with the colormetric BCA protein concentration assay using bovine serum albumin as a standard (Pierce catalog number 23225). Extracts were concentrated by 5 kD membrane centrifugation (Vivaspin spin columns, GE Healthcare). The protein extracts were purified by organic precipitation (0.5 mM HCl made in 50 % methanol and 50 % acetone) at −20 ∘C for at least 1 week, then collected by centrifugation at 14 100 g for 30 min at 4 ∘C, decanted, and dried by vacuum concentration for 10 min. The protein pellets were resuspended in a minimum amount of 1 % SDS extraction buffer and re-quantified by BCA protein concentration assay to assess extraction efficiency.
The proteins were embedded in a 500 µL final volume acrylamide gel, which was then cut up into 1 mm pieces to maximize surface area and rinsed in 50 : 50 acetonitrile / 25 mM ammonium bicarbonate overnight at room temperature. The next morning, the rinse solution was replaced and the rinse repeated for 1 h. Gels were dehydrated thrice in acetonitrile, dried by vacuum centrifugation, rehydrated in 10 mM dithiothreitol (DTT) in 25 mM ammonium bicarbonate, and then incubated for 1 h at 56 ∘C with shaking. Unabsorbed DTT solution was removed and the volume recorded, allowing the total gel volume to be calculated. Gels were washed in 25 mM ammonium bicarbonate, then incubated in 55 mM iodoacetamide for 1 h at room temperature in the dark. Gels were again dehydrated thrice in acetonitrile. Trypsin (Promega Gold) was added at a ratio of 1 : 20 µg total protein in 25 mM ammonium bicarbonate in a volume sufficient to barely cover the gel pieces. Proteins were digested overnight at 37 ∘C with shaking. Any unabsorbed solution was then removed to a new tube, and 50 µL of peptide extraction buffer (50 % acetonitrile, 5 % formic acid in water) was added and incubated for 20 min at room temperature. The supernatant was then decanted and combined with the unabsorbed solution, and the peptide extraction step was repeated. The resulting peptide mixture was concentrated by vacuum centrifugation to 1 µg µL−1 concentration based on the starting protein concentration. Finally, the peptides were clarified by centrifugation at room temperature, taking the top 90 % of the volume to reduce the carryover of gel debris.
2.3 Data acquisition
The global proteomes were analyzed by liquid chromatography–active modulation–liquid chromatography mass spectrometry (LCamLC-MS) using two orthogonal, in-line reverse-phase columns: a PLRP-S column (200 µm × 150 mm, 3 µm bead size, 300 Å pore size, NanoLCMS Solutions) and a C18 column packed in-house (100 m × 150 mm, 3 µm particle size, 120 Å pore size, C18 Reprosil-God, Dr. Maisch GmbH, packed in a New Objective PicoFrit column). The first dimension utilized an 8 h pH = 10 gradient (10 mM ammonium formate and 10 mM ammonium formate in 90 % acetonitrile) and was trapped every 30 min on alternating dual traps, then eluted at 500 nL min−1 onto the C18 column with a 30 min gradient (0.1 % formic acid and 0.1 % formic acid in 99.9 % acetonitrile); 10 µg of protein was injected per run directly onto the first column using a Thermo Dionex Ultimate3000 RSLCnano system (Waltham, MA), and an additional RSLCnano pump was used for the second dimension gradient. The samples were then analyzed on a Thermo Orbitrap Fusion mass spectrometer with a Thermo Flex ion source (Waltham, MA). MS1 scans were monitored between m∕z 380–1580, with a m∕z 1.6 MS2 isolation window (CID mode), 50 ms maximum injection time, and 5 s dynamic exclusion time.
2.4 Relative quantitation of peptides and proteins
Raw spectra were searched with the Sequest algorithm using a custom-built genomic database (Eng et al., 2008). The genomic database consisted of a publicly available Trichodesmium community metagenome available on the JGI IMG platform (IMG ID 2821474806), as well as the entire contents of the CyanoGEBA project genomes (Shih et al., 2013). Protein annotations were derived from the original metagenomes. SequestHT mass tolerances were set at ±10 ppm (parent) and ± 0.8 Dalton (fragment). Cysteine modification of +57.022 and methionine modification of +16 were included. Protein identifications were made with Peptide Prophet in Scaffold (Proteome Software) at the 95 % protein and peptide identification levels. Relative abundance was measured by averaging the precursor intensity (area under the MS1 peak) of the top three most abundant peptides in each protein, then by normalizing this value to total precursor ion intensity. Normalization and global false discovery rate (FDR) calculations, which were 0.1 % at the peptide level and 1.2 % at the protein level, were performed in Scaffold (Proteome Software). FDR was calculated by Scaffold using the probabilistic method by summing the assigned protein or peptide probabilities and dividing by the maximum probability (100 %) for each. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD016225 and can be accessed at https://doi.org/10.6019/PXD016225 (Perez-Riverol et al., 2019; Held and Saito, 2019). Statistical tests of relationships between proteins were conducted with the SciPy stats package (https://docs.scipy.org/doc/scipy/reference/stats.html, last access: December 2019) using linear Pearson tests when the relationship appeared to be linear and a Spearman rank order test when this was not the case.
2.5 Absolute quantitation of peptides
A small number of peptides were selected for absolute quantitation using a modified heterologous expression system. The peptides were ensured to be specific to Trichodesmium based on sequence identity compared to over 300 marine bacteria genomes, 3 metagenomes, and 956 specialized assemblies (see http://www.metatryp.whoi.edu, last access: December 2019) (Saito et al., 2015). A custom plasmid was designed that contained the Escherichia coli K12 optimized reverse translation sequences for peptides of interest separated by tryptic spacers (protein sequence: TPELFR). The peptides and transition ions included are provided in Table S7. To avoid repetition of the spacer nucleotide sequence, 12 different codons were utilized to encode the spacer. Six equine apomyoglobin and three peptides from the commercially available Pierce peptide retention time calibration mixture (product number 88320) were also included. The sequence was inserted into a pet(30a)+ plasmid using the BAMH1 5′ and XhoI 3′ restriction sites.
The plasmid was transformed into competent tuner(DE3)pLys E.coli cells and grown on kanamycin-amended LB agar plates to ensure plasmid incorporation. A single colony was used to inoculate a small amount of 15N-labeled, kanamycin-containing SOC media (Cambridge Isotope Laboratories) as a starter culture. These cells were grown overnight and then used to inoculate 10 mL of 15N-labeled, kanamycin-containing SOC media. Cells were grown to approximately OD600 0.6, then induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), incubated in the overexpression phase overnight at room temperature, and harvested by centrifugation.
Cells were lysed with BugBuster detergent with added benzonase nuclease. The extracts were centrifuged and a large pellet of insoluble cellular material remained. Because the plasmid protein was large, this pellet contained a large number of inclusion bodies containing nearly pure protein. The inclusion bodies were solubilized in 6 M urea at 4 ∘C overnight. The protein was reduced, alkylated, and trypsin digested in solution to generate a standard peptide mixture.
The standard mixture was calibrated to establish the exact concentration of the peptides. A known amount (10 fmol µL−1) of the commercially available Pierce standard peptide mixture (Catalog number 88320) and an apomyoglobin digest were spiked into the standard. The ratio of Pierce (isotopically labeled according to JPT standards) or apomyoglobin (light) to heavy standard peptide MS2 peak area was calculated and used to establish the final concentration of the standard peptide mixture (Fu et al., 2016; Milo, 2013). Multiple peptides were used for this calibration, and the standard deviation among them was approximately 10 %. Finally, the linearity of the peptide standard was tested by generating a dilution curve and ensuring that the concentration of each standard peptide versus MS2 peak area was linear between 0.001 and 20 fmol µL−1 concentration, using 10 µL injections consistent with experimental injection volumes.
The sample was prepared at 0.2 µg µL−1 concentration with 10 µL injected to give a total of 2 µg sample protein. The heavy labeled standard peptide mixture was spiked into each sample at a concentration of 10 fmol µL−1. The concentration of the light peptide was calculated as the ratio of the MS2 area of the light : heavy peptide multiplied by 10 µg µL−1. A correction was applied for protein recovery before and after purification, and the result was the absolute concentration of the peptide in femtomoles per microgram total protein.
The percent of the membrane occupied by the ABC transporter PstA was calculated by converting the absolute protein concentration to molecules per Trichodesmium cell, using average values for Trichodesmium cell volume (Hynes et al., 2012), carbon content per volume (Strathmann, 1967), protein content per gram carbon (Rouwenhorst et al., 1991), and the cross-sectional area of a calcium ATPase (Hudson and Morel, 1992) (see Table S5).
2.6 Self-organizing map analyses
Self-organizing maps were used to reduce the dimensionality of the data and explore relationships among co-varying proteins of interest. Only Trichodesmium proteins were considered. Analyses were conducted in Python 3.0, and fully reproducible code is available at https://github.com/naheld/self_organizing_map_tricho_metaP (last updated: December 2019).
The input data consisted of a table of protein names (rows) and samples (columns) such that the input vectors contained 2818 features. To eliminate effects of scaling, the data were unit normalized with the Scikit-learn preprocessing algorithm. The input vectors were used to initialize a 100-output-node (10×10) self-organizing map using the SOMPY Python library (https://github.com/sevamoo/SOMPY, last access: December 2019). The output nodes were then clustered using a k-means clustering algorithm (k=10) implemented in Scikit-learn. The input nodes (proteins) assigned to each map node were then retrieved, and the entire process was repeated 10 000 times. Proteins were considered in the same cluster if they appeared in the same cluster of output nodes more than 99.99 % of the time.
3.1 Proteome overview
This study presents 36 field metaproteomes of colonial Trichodesmium populations collected at 16 locations on four expeditions (Table S2). All but one location were in the subtropical and tropical Atlantic, and most samples were collected in the early morning hours to avoid changes occurring on the diel cycle (Fig. 1 and Table S2). The metaproteomes were analyzed with a two-dimensional LC-MS/MS workflow that provided deep coverage of the proteome. This resulted in 4478 protein identifications, of which 2944 were Trichodesmium proteins. The remaining proteins were from colony-associated epibionts, and they will be discussed in a future publication. Protein abundance is presented as precursor (MS1) intensity of the three most abundant peptides for each protein, normalized to total protein in the sample. Thus, changes in protein abundance were interpreted as changes in the fraction of the proteome devoted to the protein. The most abundant were GroEL, ribosomal, and phycobilisome proteins.
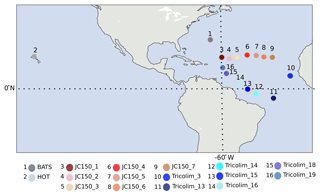
Figure 1Sampling locations. Red/pink colors indicate JC150 stations, blue colors indicate Tricolim stations, dark gray indicates the Bermuda Atlantic Time-series Study (BATS), and light gray indicates Hawaii Ocean Time-series (HOT). Most samples exist in duplicate or triplicate; see Table S2 for detailed information.
A self-organizing map analysis identified groups of proteins with similar profiles, i.e., proteins whose abundances changed cohesively, suggesting proteins that may be regulated similarly (Reddy et al., 2016). This revealed the central importance of nitrogen fixation to Trichodesmium. The nitrogenase proteins were among the most abundant in the proteome and were located in clusters 1 and 2 (Fig. 2 and Table S3). Also in these clusters were nitrogen metabolism proteins including glutamine synthetase, glutamine hydrolyzing guanosine monophosphate (GMP) synthase, and glutamate racemase. This is consistent with previous reports finding that N assimilation is synchronized with nitrogen fixation (Carpenter et al., 1992).
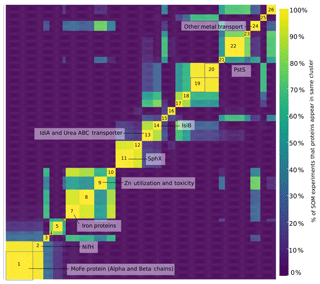
Figure 2Heatmap displaying results of self-organizing map (SOM) analysis. Each protein was mapped to a self-organizing map grid, and the grids subsequently clustered by a k-means clustering algorithm. The process was repeated 10 000 times, and the results are displayed here as a heatmap with warm colors representing proteins that appear in the same cluster. The color bar indicates the percent of SOM experiments in which two proteins appear in the same cluster. Only the top 500 most abundant proteins are displayed. Dark yellow indicates proteins that appear in the same cluster 99.99 % of the time. Clusters 1 and 2 contain nitrogen fixation, carbon fixation, and nitrogen assimilation proteins, as well as the regulatory systems NtcA and P-II. The cluster assignments for the proteins are available in Table S4.
Nitrogen fixation was closely linked to carbon fixation. Many photosystem proteins clustered with the nitrogenase proteins, including phycobilisome proteins, photosystem proteins, and the citric acid cycle protein 2-oxoglutarate dehydrogenase. This clustering indicated the possibility of direct regulatory links between C and N fixation. The nitrogen regulators P-II and NtcA were also present in this cluster and may mediate this association. In non-nitrogen fixing cyanobacteria, the high abundance of the nitrogen regulators NtcA and P-II is suggestive of nitrogen stress (Flores and Herrero, 2005; Saito et al., 2014). In diazotrophs, the role of these regulators is unclear because they do not respond to nitrogen compounds such as ammonia as they do in other cyanobacteria (Forchhammer and de Marsac, 1994). Here, the clustering of NtcA and P-II with C and N fixation proteins suggested that they play a role in balancing these processes in field populations, though the details of this role have yet to be elucidated.
In addition to identifying links between C and N fixation, the metaproteomes demonstrated that field populations of Trichodesmium invest heavily in macro- and micronutrient acquisition. There were clusters of proteins involved in trace metal acquisition and management, including Fe, zinc, and metal transport proteins, with the latter including proteins likely involved in Ni and Mo uptake (protein IDs TCCM_0270.00000020 and TCCM_0481.00000160). We also noted clusters of proteins involved in phosphate acquisition. Importantly, SphX and PstS appear in separate clusters, highlighting differential regulation of these functionally similar proteins.
3.2 Trichodesmium is simultaneously iron and phosphate stressed throughout the North Atlantic
A surprising emergent observation from the Trichodesmium metaproteomes was the co-occurrence of the iron (IdiA) and phosphate (SphX) stress biomarkers across the samples. The ubiquitous and highly abundant presence of these proteins relative to total protein implied that co-stress may be the norm rather than the exception for Trichodesmium colonies in the field, particularly in the North Atlantic. Even though low-level basal expression of IdiA and SphX has been observed, it was clear that the colonies were devoting a large fraction of their cellular resources to Fe and P uptake, respectively (see Tables S8 and S9) (Webb et al., 2001, 2007; Chappell and Webb, 2010; Orchard, 2010; Snow et al., 2015; Walworth et al., 2016a; Frischkorn et al., 2018). This, combined with the responsiveness of IdiA and SphX to nutrient availability in Trichodesmium filaments in the laboratory, indicated that co-stress was occurring.
Interestingly, biomarker abundance was not necessarily correlated to nutrient concentrations in the surface ocean, suggesting that the colonies were experiencing stress despite variation in nutrient availability (Fig. 3c–d). SphX abundance varied up to 7.5-fold and was negatively associated with dissolved phosphate concentrations, though analytical differences across the field expeditions may have forced this relationship (Fig. S2). Oceanographically, SphX was most abundant in the P-depleted, summer-stratified North Atlantic gyre (JC150 expedition) compared with winter waters near the Amazon river plume (Tricolim expedition) or at station ALOHA, where phosphate concentrations were greater (Hynes et al., 2009; Sañudo-Wilhelmy et al., 2001; Wu et al., 2000). IdiA varied up to 8-fold, but there was no observable relationship with Fe concentrations at the surface. Instead, IdiA may be responsive to other factors such as the varying iron requirements of the populations/species examined. It should be highlighted that in this study only Trichodesmium colonies were considered, so factors such as colony size may have affected iron availability and biomarker expression. Additionally, because the surface ocean iron inventory was low, transient dust inputs such as from the Sahara desert could dramatically impact iron availability on the short timescales of particle deposition, sinking, solubilization, and iron uptake. The timescale of these processes relative to changes in biomarker abundance is not well understood (Kunde et al., 2019). Carefully calibrated laboratory datasets relating IdiA and SphX abundance to nutrient-limited growth rates of Trichodesmium in both the filamentous and colonial forms would facilitate quantitative interpretation of these data.
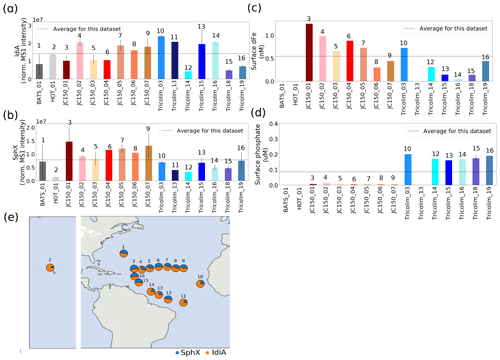
Figure 3Relative abundance of iron stress protein IdiA (a) and phosphate stress protein SphX (b). IdiA and SphX were among the most abundant proteins in the entire dataset. Error bars are 1 standard deviation from the mean. Dashed lines represent average values across the dataset. Protein abundances were normalized such that the total MS1 peak area across the entire proteome was the same for each sample. (c, d) Concentrations of dissolved Fe and dissolved phosphate nutrients. (e) Relative abundance of IdiA (orange) and SphX (blue) overlaid on the sampling locations.
3.3 The intersection of Fe, P, and N stress
The metaproteomes enabled the relationship between Fe and P stress and overall cellular metabolism to be explored. Nitrogenase protein abundance was positively correlated with both IdiA and SphX and was in fact highest at the intersection of high Fe and P stress (Fig. 4). This observation contrasts with the current paradigm that Trichodesmium downregulates nitrogen fixation when it is Fe or P stressed (Frischkorn et al., 2018; Rouco et al., 2018; Bergman et al., 2013; Shi et al., 2007). Instead, it is consistent with the idea that the nutritional demands of nitrogen fixation could drive the organism to Fe and P stress, thereby initiating an increase in Fe and P acquisition proteins including IdiA and SphX. This pattern may be specific to Trichodesmium colonies, which may aggregate during stress. This indicates that the cell's Fe, P, and N statuses are linked, perhaps involving one or more regulatory networks, which are particularly common in marine bacteria (Fig. 5) (Held et al., 2019). This network may regulate a specific physiological adaptation to nutrient co-stress. For instance, Fe and P co-limited Trichodesmium cells may reduce their cell size to optimize their surface area : volume quotient for nutrient uptake. However, a putative cell-size biomarker, Tery_1090, while abundant in co-limited cells in culture, was not identified in these metaproteomes despite bioinformatic efforts to target it, likely because it is a low-abundance protein (Walworth et al., 2016a, b).
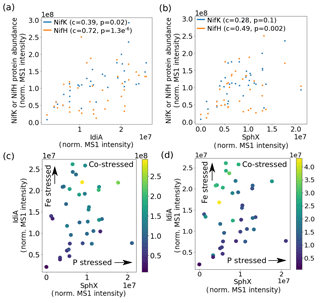
Figure 4Nitrogenase abundance is highest at the intersection of high iron and phosphate stress. (a) IdiA and (b) SphX abundance is positively related to nitrogenase MoFe (NifK) and Fe (NifH) protein abundance (c represents the Spearman rank-order correlation coefficient and p represents the Spearman p value). Effects of combined iron and phosphate stress biomarkers on nitrogenase abundance. Marker colors represent abundance of NifK (c) and NifH (d).

Figure 5The metaproteomes suggest that there is a currently unknown regulatory link between cellular Fe, P, and N regulation. Fur is the ferric uptake regulator, PhoRB is the phosphate two-component sensory system, Tery_1090 is the putative cell-size regulator, and P-II and NtcA are nitrogen regulatory proteins.
Nitrogen fixation is not the only way that Trichodesmium can acquire fixed N (Dyhrman et al., 2006; Küpper et al., 2008; Mills et al., 2004; Sañudo-Wilhelmy et al., 2001). In culture, Trichodesmium can be grown on multiple nitrogen sources including urea; in fact, it has been reported that nitrogen fixation provides less than 20 % of the fixed N demand of cells, and a revised nitrogen fixation model suggests that Trichodesmium takes up fixed N in the field (McGillicuddy, 2014; Mulholland and Capone, 1999). In this dataset, a urea ABC transporter was abundant, indicating that urea could be an important source of fixed nitrogen to colonies (Fig. 6a). The transporter is unambiguously attributed to Trichodesmium rather than a member of the epibiont community. Of course, this does not rule out the possibility that urea or other organic nitrogen sources such as trimethylamine (TMA) are also utilized by epibionts, although no such epibiont transporters were identified in the metaproteomes.
Typically, elevated urea concentration decreases or eliminates nitrogen fixation in colonies (Ohki et al., 1991). However, in laboratory studies urea exposure must be unrealistically high for this to occur, often over 20 µM (Ohki et al., 1991; Wang et al., 2000). In the field, urea utilization and nitrogen fixation seem to occur simultaneously, with a urea uptake protein positively correlated to nitrogenase abundance (Fig. 6b). Urea and other organic nitrogen sources such as trimethylamine (TMA) could be sources of nitrogen for Trichodesmium, and the relationship to nitrogenase abundance may indicate a general N stress signature driving both organic nitrogen uptake and nitrogen fixation (Walworth et al., 2018). Alternatively, urea uptake could be a colony-specific behavior, since colonies were sampled here as opposed to laboratory cultures that typically grow as single filaments. For instance, urea could be used for recycling fixed N within the colony, or there could be heterogeneity in nitrogen fixation, with some cells taking up organic nitrogen and others fixing it. These unexpected observations of co-occurring nitrogen fixation and organic nitrogen transport show the value of exploratory metaproteomics, which does not require targeting a specific protein based on a prior hypothesis.
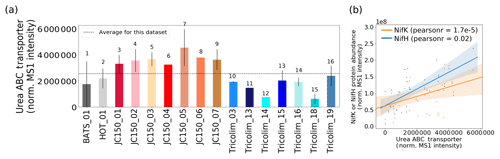
Figure 6(a) Relative abundance of the Trichodesmium urea ABC transporter. (b) The abundance of the urea ABC transporter is positively correlated with NifH and NifK abundance. Pearson linear correlation coefficients (r values) are provided (p value for NifK = , NifH = 0.02). Shaded bars indicate 95 % confidence intervals.
3.4 Mechanisms of simultaneous iron and phosphate stress – membrane crowding
ABC transporters are multiunit, transmembrane protein complexes that use ATP to shuttle substrates across membranes. Specific ABC transporters are required for both iron and phosphate uptake (Chappell et al., 2012; Orchard et al., 2009). Nutrient transport rates can be modulated by changing the number of uptake proteins installed on the cell membrane or the efficiency of these uptake proteins. One way to increase efficiency is through the expression of assisting proteins such as IdiA and SphX, which bind Fe and P respectively in the periplasm and shuttle the elements to their respective membrane transport complexes (Hudson and Morel, 1992). The high abundance of proteins involved in ABC transport suggested that nutrient transport rates could limit the amount of Fe and P Trichodesmium can acquire. Thus, we explored whether membrane crowding, i.e., lack of membrane space, can constrain the nutrient acquisition of Trichodesmium.
To investigate this, we quantified the absolute concentration of the phosphate ABC transporter PstA, which interacts with the phosphate stress biomarkers SphX and PstS. This analysis is distinct from the above global metaproteomes, which allowed patterns to be identified but did not allow for absolute quantitation. The analysis was performed for three Tricolim and six JC150 stations in a way similar to an isotope dilution experiment, in which isotopically labeled standards (in this case for each peptide) are used to control for analytical biases. Briefly, 15N-labeled peptide standards were prepared and spiked into the samples prior to parallel reaction monitoring (PRM) LC-MS/MS analysis. The concentration of the peptide in femtomoles per microgram total protein was calculated using the ratio of product ion intensities for the heavy (spike) and light (sample) peptide and converted to PstA molecules per cell (Table 1 and see also Table S5). The peptide used for quantitation of PstA is specific to Trichodesmium species. Based on these calculations, on average 19 % to 36 % of the membrane was occupied by the PstA transporter. In one population (JC150 expedition, Station 7), up to 83 % of the membrane was occupied by PstA alone. While these are first estimates, it is clear that the majority of Trichodesmium cells devoted a large fraction of their membrane surface area to phosphate uptake.
Table 1Quantification of the ABC transporter PstA and estimation of membrane space occupied.
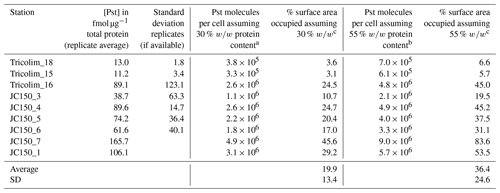
a Calculated using Trichodesmium cell volume of 3000 µm3 (Berman-Frank et al., 2001), cell volume to carbon conversion (Strathman, 1967), protein content of a cyanobacterium 30 % w∕w (López et al., 2010), carbon to total protein conversion 0.53 g C g−1 total protein (Rouwenhorst et al., 1991). b Calculated as in footnote a but with protein content of a cyanobacterium 55 % w∕w (López et al., 2010). c Calculated using cross-sectional area of a Ca ATPase of 0.0000167 µm2 (Hudson and Morel, 1992).
To examine whether membrane crowding can indeed cause nutrient stress or limitation, we developed a model of cellular nutrient uptake in Trichodesmium. The model identifies the concentration at which free Fe or phosphate limits the growth of Trichodesmium cells. This is distinct from nutrient stress, which changes the cell's physiological state but does not necessarily impact growth. In the model, nutrient limitation occurs when the daily cellular requirement is greater than the uptake rate, a function of the cell's growth rate and elemental quota. Following the example of Hudson and Morel (1992), the model assumes that the intake of nutrients once bound to the ABC transporter protein is instantaneous, i.e., that nutrient uptake is limited by the formation of the metal-transporter complex at the cell surface. This is an idealized scenario, because if intake is the slow step, for instance in a high-affinity transport system, the uptake rate will be slower and nutrient limitation exacerbated (discussed below).
We considered two types of nutrient limitation in the model (Table S10). First, we considered a diffusion-limited case in which the rate of uptake is determined by the diffusion of the nutrient to the cell's boundary layer ( [nutrient], where μ represents the cell growth rate, Q represents the cell nutrient quota, and kD represents the diffusion rate constant, dependent on the surface area and diffusion coefficient of the nutrient in seawater). Based on empirical evidence provided by Hudson and Morel (1992), limitation occurs when the cell quota is greater than two-thirds the diffusive-limited flux because beyond this, depletion of the nutrient in the boundary layer occurs. In the second case, membrane crowding limitation, the rate of uptake is determined by the rate of transporter-metal complex formation ( [transport protein] [nutrient], where kf represents the rate of ligand-nutrient complex formation). Here, up to 50 % of the membrane can be occupied by the transport protein following the example of Hudson and Morel (1992). This is within the range of the above estimates of membrane occupation by the phosphate transporter PstA. The model uses conservative estimates for diffusion coefficients, cell quotas, growth rates, and membrane space occupation to identify the lowest concentration threshold for nutrient limitation; as a result it is likely that Trichodesmium becomes limited at higher nutrient concentrations than the model suggests. At this time, the model can only consider labile dissolved Fe and inorganic phosphate, though Trichodesmium can also acquire particulate iron, organic phosphorus, phosphite, and phosphonates (Dyhrman et al., 2006; Frischkorn et al., 2018; Polyviou et al., 2015; Poorvin et al., 2004; Rubin et al., 2011).
We first considered a spherical cell, where the surface area : volume quotient decreases as cell radius increases (Fig. 7). As the cell grows in size, higher nutrient concentrations are required to sustain growth. This is consistent with the general understanding that larger microbial cells with lower surface area : volume quotient are less competitive in nutrient uptake (Chisholm, 1992; Hudson and Morel, 1992). For a given surface area : volume quotient, the mechanism driving nutrient limitation is whichever model (diffusion or membrane crowding) results in a higher minimum nutrient concentration below which limitation occurs. For a spherical cell, Fe limitation is driven by diffusion when the cell is large and the surface area : volume quotient is low (Fig. 7a). However, when cells are smaller and the surface area : volume quotient is high, membrane crowding drives nutrient limitation, meaning that the number of ligands – and not diffusion from the surrounding environment – is the primary control on nutrient uptake. For phosphate, diffusion is almost always the driver of nutrient limitation owing to the higher rate of transporter-nutrient complex formation (kf) for phosphate, which causes very fast membrane transport rates and relieves membrane-crowding pressures across all cell sizes (Fig. 7b) (Froelich et al., 1982).
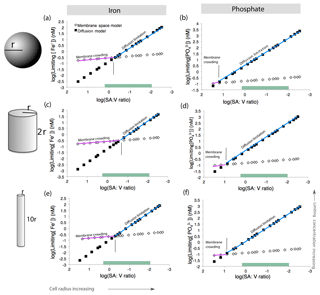
Figure 7Model calculations for membrane space and diffusion-based nutrient limitation reveal that membrane crowding could drive Trichodesmium to iron or phosphate stress, particularly when cells are small. Two cell morphologies (sphere and cylinder) were modeled for both iron and phosphate limitation. Calculations are detailed in Table S10. As the cell radius increases and the surface area : volume quotient decreases, the limiting concentration increases. This is concurrent with the current understanding that the low surface area : volume quotient of large cells leads to limitation. Green bars represent common SA : V quotients for T. theibautii (Hynes et al., 2012) (a–b). Membrane crowding (purple) occurs if the limiting nutrient concentration is greater than in the diffusion limitation model (blue). Membrane crowding is more significant for cylindrical cells in particular (c–d); altering the length of the cylinder minimally affects the model (e–f).
While this model may be directly applicable to some N2-fixing cyanobacteria such as Groups B and C, which have roughly spherical cells, Trichodesmium cells are not spheres but rather roughly cylindrical (Hynes et al., 2012). Thus, we repeated the model calculations for cylinders with varying radii (r) and heights (2r or 10r) based on previous estimates of Trichodesmium cell sizes (Bergman et al., 2013; Hynes et al., 2012). Cylinders have lower surface area : volume quotient than spheres of similar sizes. In addition, the rate constant (kD) for diffusion, which is a function of cell geometry, is greater. This increases the slope of the diffusion limitation line such that membrane crowding is important across a greater range of cell sizes (Fig. 7c–d). Trichodesmium cell sizes vary in nature; for instance, the cylinder height can be elongated, improving the surface area : volume quotient. However, the impact of cell elongation to radius r and height 10r on both diffusion limitation and membrane crowding is subtle (Fig. 7e–f). Furthermore, though not explicitly considered here, cylindrical cells living in filaments would have a reduced surface area available for nutrient uptake. Thus, we conclude that in certain scenarios, lack of membrane space could hypothetically limit Fe and perhaps P acquisition by Trichodesmium, particularly when the cells live in filaments or colonies as occurs in nature.
A key assumption of the model is that uptake rates are instantaneous. In the above calculations, we use the dissociation kinetics of Fe from water and phosphate from common seawater cations as the best case (i.e., fastest possible) kinetic scenario for nutrient acquisition. The model does not account for delays caused by internalization kinetics, which would exacerbate nutrient limitation. The involvement of the periplasmic binding proteins IdiA and SphX suggests that uptake is not simultaneous; their participation is likely associated with a kinetic rate of binding and dissociation from the periplasmic proteins in addition to any rate of ABC transport. Additionally, the model does not consider nutrient speciation, which could affect internalization rates, particularly for Fe (Hudson and Morel, 1992).
Membrane crowding could produce real cellular challenges, leading to the observation of Fe and P co-stress across the field populations examined. The above model explicitly allows 50 % of the cell surface area to be occupied by any one type of transporter, consistent with our estimate of cell surface area occupied by the PstA transporter. If 50 % of the membrane is occupied by phosphate transporters and another 50 % for Fe transporters, this would leave no room for other essential membrane proteins and even the membrane lipids themselves. The problem is further exacerbated if the cell installs transporters for nitrogen compounds such as urea, as the metaproteomes suggest. Thus, installation of transporters for any one nutrient must be balanced against transporters for other nutrients. This interpretation is inconsistent with Liebig's law of nutrient limitation, which assumes that nutrients are independent (Liebig, 1855; Saito et al., 2008). In an oligotrophic environment, membrane crowding could explicitly link cellular Fe, P, and N uptake statuses, driving the cell to be co-stressed for multiple nutrients.
3.5 Advantages of the colonial form for nutrient acquisition
Living in a colony has specific advantages and disadvantages for a Trichodesmium cell. Colonies may be able to access nutrient sources that single cells or filaments could not feasibly use. For instance, Trichodesmium colonies have a remarkable ability to entrain dust particles and can move these particles into the center of said colony (Basu et al., 2019; Basu and Shaked, 2018; Poorvin et al., 2004; Rubin et al., 2011). In this study, which focused on Trichodesmium colonies, the chemotaxis response regulator CheY was very abundant, particularly in populations sampled near the Amazon and Orinoco river plumes. CheY was positively correlated with the Fe stress biomarker IdiA but not with the phosphate stress biomarker SphX, suggesting that chemotactic movement is involved in the entrainment of trace metals including from particulate sources (Fig. 8).
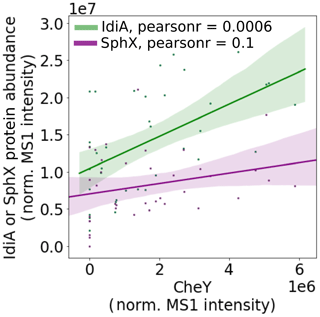
Figure 8CheY is positively correlated with the iron stress biomarker IdiA but has a weaker association with the phosphate stress biomarker SphX. This suggests that it might be involved in iron acquisition, for instance by helping colonies to move dust particles to the colony center. Pearson linear correlation coefficients (r values) are provided (p value for IdiA = , SphX = 0.1). Shaded bars indicate 95 % confidence intervals.
The metaproteomes and nutrient uptake model presented in this paper support the growing understanding that Trichodesmium must be able to access particulate and organic matter. Living in a colony can be advantageous because such substrates can be concentrated, improving the viability of extracellular nutrient uptake systems. Trichodesmium's epibiont community produces siderophores, which assist in Fe uptake, particularly from particulate sources (Chappell and Webb, 2010; Lee et al., 2018). Siderophore production is energetically and nutritionally expensive, so it is most advantageous when resource concentrations are high and loss is low, as would occur in the center of a colony (Leventhal et al., 2019). Colonies may similarly enjoy advantages for phosphate acquisition, particularly when the excreted enzyme alkaline phosphatase is utilized to access organic sources (Frischkorn et al., 2018; Orchard, 2010; Orcutt et al., 2013; Yamaguchi et al., 2016; Yentsch et al., 1970). Additionally, the concentration of cells in a colony means that the products of nitrogen fixation, including urea, can be recycled and are less likely to be lost to the environment.
A key hallmark of Trichodesmium colony formation is the production of mucus, which can capture particulate matter and concentrate it within the colony (Eichner et al., 2019). In addition to particle entrainment, the mucus layer can benefit cells by inhibiting oxygen diffusion, facilitating epibiont associations, regulating buoyancy, defending against grazers, and helping to “stick” trichomes together (Eichner et al., 2019; Lee et al., 2018; Sheridan, 2002). However, these benefits come at a cost because the mucus layer hinders diffusion to the cell surface (Fig. 9), reducing contact with the surrounding seawater. Despite this, the benefits of colony formation seem to outweigh the costs since Trichodesmium forms colonies in the field, particularly under stress (Bergman et al., 2013; Capone et al., 1997; Hynes et al., 2012).
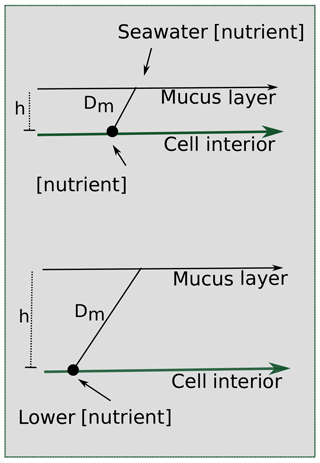
Figure 9Scheme for the effect of a mucus layer on nutrient diffusion. h represents the height of the mucus membrane and Dm represents the diffusion coefficient of the mucus. Assuming some diffusion constant for the nutrient through the mucus and the same starting seawater nutrient concentration, a thicker layer of mucus surrounding a cell in a colony would result in a lower concentration of nutrients experienced at the cell surface.
Trichodesmium's colonial lifestyle likely produces challenges for dissolved Fe and P acquisition, which must be compensated for by the production of multiple nutrient transport systems, such as for particulate iron and organic phosphorous, at a considerable cost. While laboratory studies have largely focused on single nutrient stresses in free filaments, these metaproteomic observations and the accompanying nutrient uptake model demonstrate that Fe and P co-stress may be the norm rather than the exception for colonies in the North Atlantic Ocean. This means that the emphasis on single limiting nutrients in culture studies and biological models may not capture the complexities of Trichodesmium's physiology in situ. Thus, biogeochemical models should consider incorporating Fe and P co-stress conditions. Specifically, in this study and in others there is evidence that nitrogen fixation is optimal under co-limited or co-stressed conditions, implying that an input of either Fe or P could counterintuitively decrease N2-driven new production (Garcia et al., 2015; Walworth et al., 2016a).
These data demonstrate that Trichodesmium cells are confronted by the biophysical limits of membrane space and diffusion rates for their Fe, P, and possibly urea acquisition systems. This means that there is little room available for systems that interact with other resources such as light, CO2, Ni, and other trace metals, providing a mechanism by which nutrient stress could compromise the acquisition of other supplies. The cell membrane could be a key link allowing Trichodesmium to optimize its physiology in response to multiple environmental stimuli. This is particularly important in an ocean where nutrient availability is sporadic and unpredictable. Future studies should aim to characterize the specific regulatory systems, chemical species and phases (i.e., dissolved versus particulate nutrient sources), and symbiotic interactions that underlie Trichodesmium's unique behavior and lifestyle.
All new data are provided in the Supplement and are also available at the Biological and Chemical Oceanography Data Management Office (BCO-DMO) with the following DOIs: https://doi.org/10.26008/1912/bco-dmo.787093.1 (Held and Saito, 2020a), https://doi.org/10.26008/1912/bco-dmo.787078.1 (Held and Saito, 2020b), https://doi.org/10.26008/1912/bco-dmo.787147.1 (Held and Saito, 2020c), https://doi.org/10.26008/1912/bco-dmo.787168.1 (Held and Saito, 2020d), and https://doi.org/10.26008/1912/bco-dmo.787181.1 (Held and Saito, 2020e). The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD016225 and https://doi.org/10.6019/PXD016225 (Held and Saito, 2019).
Supplementary information is provided in a separate file (Figs. S1 and S2, Tables S1, S2, S5, S6, S7, S8, S9) with Tables S3, S4, and S10 provided separately due to their large sizes. The supplement related to this article is available online at: https://doi.org/10.5194/bg-17-2537-2020-supplement.
NAH and MAS conceptualized the study. DAH and EAW led the Tricolim expedition. CM and MCL led the JC150 expedition. NRC, EMSW, and KK measured nutrient distributions on the Tricolim and JC150 expeditions. DMM and MMM helped with proteomic analyses. NAH prepared the paper with contributions from all co-authors.
The authors declare that they have no conflict of interest.
We acknowledge Elena Cerdan Garcia, Asa Conover, Joanna Harley, Despo Polyviou, and Petroc Shelley for assistance with sampling and nutrient measurements while at sea, in addition to the entire crew of the JC150 and Tricolim expeditions. We thank Ben Van Mooy for insightful discussions regarding this work. This work was supported by a National Science Foundation Graduate Research Fellowship under grant 1122274 (Noelle A. Held), Gordon and Betty Moore Foundation grant number 3782 (Mak A. Saito), and National Science Foundation grants OCE-1657755 (Mak A. Saito), EarthCube-1639714 (Mak A. Saito), OCE-1657757 (David A. Hutchins), and OCE-1851222 (David A. Hutchins). We also acknowledge funding from the UK Natural Environment Research Council (NERC) grants awarded to CM (NE/N001079/1) and ML (NE/N001125/1). NRC was supported by grant 544236 from the Simons Foundation.
This research has been supported by the National Science Foundation (Division of Graduate Education (grant nos. 1122274), Division of Ocean Sciences (grant nos. 1657755, 1657757, and 1851222), Directorate for Geosciences (grant no. 1639714)), the Gordon and Betty Moore Foundation (grant no. 3782), and the Natural Environment Research Council (NERC) (grant nos. NE/N001079/1 and NE/N001125/1).
This paper was edited by Koji Suzuki and reviewed by two anonymous referees.
Basu, S. and Shaked, Y.: Mineral iron utilization by natural and cultured Trichodesmium and associated bacteria, Limnol. Oceanogr., 63, 2307–2320, https://doi.org/10.1002/lno.10939, 2018.
Basu, S., Gledhill, M., de Beer, D., Matondkar, S. G. P., and Shaked, Y.: Colonies of marine cyanobacteria Trichodesmium interact with associated bacteria to acquire iron from dust, Communications Biology, 2, 1–8, https://doi.org/10.1038/s42003-019-0534-z, 2019.
Bergman, B., Sandh, G., Lin, S., Larsson, J., and Carpenter, E. J.: Trichodesmium – a widespread marine cyanobacterium with unusual nitrogen fixation properties, FEMS Microbiol. Rev., 37, 286–302, https://doi.org/10.1111/j.1574-6976.2012.00352.x, 2013.
Berman-Frank, I., Cullen, J. T., Shaked, Y., Sherrell, R. M., and Falkowski, P. G.: Iron availability, cellular iron quotas, and nitrogen fixation in Trichodesmium, Limnol. Oceanogr., 46, 1249–1260, https://doi.org/10.4319/lo.2001.46.6.1249, 2001.
Capone, D. G., Zehr, J. P., Paerl, H. W., Bergman, B., and Carpenter, E. J.: Trichodesmium, a globally significant marine cyanobacterium, Science, 276, 1221–1229, https://doi.org/10.1126/science.276.5316.1221, 1997.
Carpenter, E. J. and Romans, K.: Major Role of the Cyanobacterium Trichodesmium in Nutrient Cycling in the North Atlantic Ocean, Science, 254, 1989–1992, 1991.
Carpenter, E. J., Bergman, B., Dawson, R., Siddiqui, P. J., Söderbäck, E., and Capone, D. G.: Glutamine synthetase and nitrogen cycling in colonies of the marine diazotrophic cyanobacteria Trichodesmium spp., Appl. Environ. Microb., 58, 3122–3129, 1992.
Chappell, P. D. and Webb, E. A.: A molecular assessment of the iron stress response in the two phylogenetic clades of Trichodesmium, Environ. Microbiol., 12, 13–27, https://doi.org/10.1111/j.1462-2920.2009.02026.x, 2010.
Chappell, P. D., Moffett, J. W., Hynes, A. M., and Webb, E. A.: Molecular evidence of iron limitation and availability in the global diazotroph Trichodesmium, ISME J., 6, 1728–1739, https://doi.org/10.1038/ismej.2012.13, 2012.
Chisholm, S. W.: Phytoplankton Size, in: Primary Productivity and Biogeochemical Cycles in the Sea, edited by: Falkowski, P. G., Woodhead, A. D., and Vivirito, K., Springer, Boston, USA, 213–237, https://doi.org/10.1007/978-1-4899-0762-2_12, 1992.
Coles, V. J., Hood, R. R., Pascual, M., and Capone, D. G.: Modeling the impact of Trichodesmium and nitrogen fixation in the Atlantic ocean, J. Geophys. Res.-Oceans, 109, 1–17, https://doi.org/10.1029/2002JC001754, 2004.
Deutsch, C., Sarmiento, J. L., Sigman, D. M., Gruber, N., and Dunne, J. P.: Spatial coupling of nitrogen inputs and losses in the ocean, Nature, 445, 163–167, https://doi.org/10.1038/nature05392, 2007.
Dyhrman, S. T., Chappell, P. D., Haley, S. T., Moffett, J. W., Orchard, E. D., Waterbury, J. B., and Webb, E. A.: Phosphonate utilization by the globally important marine diazotroph Trichodesmium, Nature, 439, 68–71, https://doi.org/10.1038/nature04203, 2006.
Eichner, M., Thoms, S., Rost, B., Mohr, W., Ahmerkamp, S., Ploug, H., Kuypers, M. M. M., and de Beer, D.: N2 fixation in free-floating filaments of Trichodesmium is higher than in transiently suboxic colony microenvironments, New Phytol., 222, 852–863, https://doi.org/10.1111/nph.15621, 2019.
Eng, J. K., Fischer, B., Grossmann, J., and MacCoss, M. J.: A fast SEQUEST cross correlation algorithm, Journal of Proteome Research, 7(10), 4598–4602, https://doi.org/10.1021/pr800420s, 2008.
Flores, E. and Herrero, A.: Nitrogen assimilation and nitrogen control in cyanobacteria, Biochem. Soc. T., 33, 164–167, https://doi.org/10.1042/BST0330164, 2005.
Forchhammer, K. and de Marsac, N. T.: The P(II) protein in the cyanobacterium Synechococcus sp. strain PCC 7942 is modified by serine phosphorylation and signals the cellular N-status, J. Bacteriol., 176, 84–91, 1994.
Frischkorn, K. R., Krupke, A., Guieu, C., Louis, J., Rouco, M., Salazar Estrada, A. E., Van Mooy, B. A. S., and Dyhrman, S. T.: Trichodesmium physiological ecology and phosphate reduction in the western tropical South Pacific, Biogeosciences, 15, 5761–5778, https://doi.org/10.5194/bg-15-5761-2018, 2018.
Froelich, P. N., Bender, M. L., Luedtke, N. A., Heath, G. R., and DeVries, T.: The Marine Phosphorus Cycle, Am. J. Sci., 282, 464–511, 1982.
Fu, Q., Chen, Z., Zhang, S., Parker, S. J., Fu, Z., Tin, A., Liu, X., and Van Eyk, J. E.: Multiple and Selective Reaction Monitoring Using Triple Quadrupole Mass Spectrometer: Preclinical Large Cohort Analysis, in: Quantitative Proteomics by Mass Spectrometry, edited by: Sechi, S., Springer, New York, USA, 249–264, https://doi.org/10.1007/978-1-4939-3524-6_15, 2016.
Garcia, N. S., Fu, F., Sedwick, P. N., and Hutchins, D. A.: Iron deficiency increases growth and nitrogen-fixation rates of phosphorus-deficient marine cyanobacteria, ISME J., 9, 238–245, https://doi.org/10.1038/ismej.2014.104, 2015.
Held, N. A. and Saito, M. A.: Trichodesmium field metaproteomes, PRIDE, https://doi.org/10.6019/PXD016225, 2019.
Held, N. A. and Saito, M. A.: Trichodesmium sample provenance from samples collected in North Atlantic surface waters, station BATS, and station ALOHA between 2000 and 2018, Biological and Chemical Oceanography Data Management Office (BCO-DMO), Dataset version 2020-01-10, https://doi.org/10.26008/1912/bco-dmo.787093.1, 2020a.
Held, N. A. and Saito, M. A.: Metaproteomes of Trichodesmium from samples collected in North Atlantic surface waters, station BATS, and station ALOHA between 2000 and 2018, Biological and Chemical Oceanography Data Management Office (BCO-DMO), Dataset version 2020-01-10, https://doi.org/10.26008/1912/bco-dmo.787078.1, 2020b.
Held, N. A. and Saito, M. A.: Net tow metaproteome of Trichodesmium species mapped to a Trichodesmium metagenome plus cyanoGEBA species genomes in units of normalized protein spectral counts from samples collected in the Atlantic and Pacific Ocean between 2000 and 2018, Biological and Chemical Oceanography Data Management Office (BCO-DMO), Dataset version 2020-04-30, https://doi.org/10.26008/1912/bco-dmo.787147.1, 2020c.
Held, N. A. and Saito, M. A.: Net tow metaproteoome of Trichodesmium species mapped to a Trichodesmium metagenome plus cyanoGEBA species genomes in units of normalized peptide spectral counts from samples collected in the Atlantic and Pacific Ocean between 2000 and 2018, Biological and Chemical Oceanography Data Management Office (BCO-DMO), Dataset version 2020-04-30, https://doi.org/10.26008/1912/bco-dmo.787168.1, 2020d.
Held, N. A. and Saito, M. A.: FASTA file of sequences in Trichodesmium field metaproteomes mapped to a Trichodesmium metagenome plus cyanoGEBA species genomes from samples collected in the Atlantic and Pacific Ocean between 2000 and 2018, Biological and Chemical Oceanography Data Management Office (BCO-DMO), Dataset version 2020-01-14, https://doi.org/10.26008/1912/bco-dmo.787181.1, 2020e.
Held, N. A., McIlvin, M. R., Moran, D. M., Laub, M. T., and Saito, M. A.: Unique Patterns and Biogeochemical Relevance of Two-Component Sensing in Marine Bacteria, MSystems, 4, e00317-18, https://doi.org/10.1128/mSystems.00317-18, 2019.
Hudson, R. J. and Morel, F. M. M.: Trace metal transport by marine microorganisms: implications of metal coordination kinetics, Deep-Sea Res. Pt. I, 40, 129–150, 1992.
Hynes, A. M., Chappell, P. D., Dyhrman, S. T., Doney, S. C., and Webb, E. A.: Cross-basin comparison of phosphorus stress and nitrogen fixation in Trichodesmium, Limnol. Oceanogr., 54, 1438–1448, https://doi.org/10.4319/lo.2009.54.5.1438, 2009.
Hynes, A. M., Webb, E. A., Doney, S. C., and Waterbury, J. B.: Comparison of cultured Trichodesmium (Cyanophyceae) with species characterized from the field, J. Phycol., 48, 196–210, https://doi.org/10.1111/j.1529-8817.2011.01096.x, 2012.
Karl, D., Michaels, A., Bergman, B., Capone, D., Carpenter, E., Letelier, R., Lipschultz, F., Paerl, H., Sigman, D., and Stal, L.: Dinitrogen fixation in the world's oceans, Biogeochemistry, 57–58, 47–98, https://doi.org/10.1023/A:1015798105851, 2002.
Kunde, K., Wyatt, N. J., González-Santana, D., Tagliabue, A., Mahaffey, C., and Lohan, M. C.: Iron Distribution in the Subtropical North Atlantic: The Pivotal Role of Colloidal Iron, Global Biogeochem. Cy., 33, 1532–1547, https://doi.org/10.1029/2019GB006326, 2019.
Küpper, H., Šetlík, I., Seibert, S., Prášil, O., Šetlikova, E., Strittmatter, M., Levitan, O., Lohscheider, J., Adamska, I., and Berman-Frank, I.: Iron limitation in the marine cyanobacterium Trichodesmium reveals new insights into regulation of photosynthesis and nitrogen fixation, New Phytol., 179, 784–798, 2008.
Lee, M. D., Webb, E. A., Walworth, N. G., Fu, F. X., Held, N. A., Saito, M. A., and Hutchins, D. A.: Transcriptional activities of the microbial consortium living with the marine nitrogenfixing cyanobacterium Trichodesmium reveal potential roles in community-level nitrogen cycling, Appl. Environ. Microb., 84, e02026-17 https://doi.org/10.1128/AEM.02026-17, 2018.
Leventhal, G. E., Ackermann, M., and Schiessl, K. T.: Why microbes secrete molecules to modify their environment: The case of iron-chelating siderophores, J. R. Soc. Interface, 16, 20180674, https://doi.org/10.1098/rsif.2018.0674, 2019.
Liebig, J. V.: Principles of agricultural chemistry with special reference to the late researches made in England, Dowden, Hutchinson, and Ross, London, UK, 1855.
López, C. V. G., García, M. d. C., Fernández, F. G. A., Bustos, C. S., Chisti, Y., and Sevilla, J. M. F.: Protein measurements of microalgal and cyanobacterial biomass, Bioresource Technol., 101, 7587–7591, https://doi.org/10.1016/j.biortech.2010.04.077, 2010.
Lu, X. and Zhu, H.: Tube-Gel Digestion: A Novel Proteomic Approach for High Throughput Analysis of Membrane Proteins, Mol. Cell. Proteomics, 4, 1948–1958, https://doi.org/10.1074/mcp.M500138-MCP200, 2005.
McGillicuddy, D. J.: Do Trichodesmium spp. populations in the North Atlantic export most of the nitrogen they fix?, Global Biogeochem. Cy., 28, 103–114, https://doi.org/10.1002/2013GB004652, 2014.
Mills, M. M., Ridame, C., Davey, M., La Roche, J., and Geider, R. J.: Iron and phosphorus co-limit nitrogen fixation in the eastern tropical North Atlantic, Nature, 429, 292–294, https://doi.org/10.1038/nature02550, 2004
Milo, R.: What is the total number of protein molecules per cell volume? A call to rethink some published values, BioEssays, 35, 1050–1055, https://doi.org/10.1002/bies.201300066, 2013.
Mulholland, M. R. and Capone, D. G.: Nitrogen utilization and metabolism relative to patterns of N2 fixation in populations of Trichodesmium from the North Atlantic Ocean and Caribbean Sea, Mar. Ecol.-Prog. Ser., 188, 33–49, 1999.
Ohki, K., Zehr, J. P., Falkowski, P. G., and Fujita, Y.: Regulation of nitrogen-fixation by different nitrogen sources in the marine non-heterocystous cyanobacterium Trichodesmium sp. NIBB1067, Arch. Microbiol., 156, 335–337, https://doi.org/10.1007/BF00248706, 1991.
Orchard, E. D.: Phosphorus physiology of the marine Cyanobacterium Trichodesmium, Massachusetts Institute of Technology, 130, 81–122, https://doi.org/10.1575/1912/3366, 2010.
Orchard, E. D., Webb, E. A., and Dyhrman, S. T.: Molecular analysis of the phosphorus starvation response in Trichodesmium spp., Environ. Microbiol., 11, 2400–2411, https://doi.org/10.1111/j.1462-2920.2009.01968.x, 2009.
Orcutt, K. M., Gundersen, K., and Ammerman, J. W.: Intense ectoenzyme activities associated with Trichodesmium colonies in the Sargasso Sea, Mar. Ecol.-Prog. Ser., 478, 101–113, https://doi.org/10.3354/meps10153, 2013.
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J. Inuganti, A., Griss, J., Mayer G., Eisenacher, M., Perez, E., Uszkoriet, J., Pfeuffer, J., Sachsenberg, T., Yilmaz, S., Tiwary, S., Cox, J., Audain, E., Walzer, M., Januczak, A. F., Ternent, T., Brazma, A., and Vizcaíno, J. A.: The PRIDE database and related tools and resources in 2019: Improving support for quantification data, Nucleic Acids Res., 47, D442–D450, https://doi.org/10.1093/nar/gky1106, 2019.
Polyviou, D., Hitchcock, A., Baylay, A. J., Moore, C. M., and Bibby, T. S.: Phosphite utilization by the globally important marine diazotroph Trichodesmium, Env. Microbiol. Rep., 7, 824–830, https://doi.org/10.1111/1758-2229.12308, 2015.
Poorvin, L., Rinta-kanto, J. M., Hutchins, D. A., and Wilhelm, S. W.: Viral release of iron and its bioavailability to marine plankton, Limnol. Oceanogr., 49, 1734–1741, 2004.
Reddy, R. J., Gajadhar, A. S., Swenson, E. J., Rothenberg, D. A., Curran, T. G., and White, F. M.: Early signaling dynamics of the epidermal growth factor receptor, P. Natl. Acad. Sci. USA, 113, 201521288, https://doi.org/10.1073/pnas.1521288113, 2016.
Rouco, M., Frischkorn, K. R., Haley, S. T., and Alexander, H.: Transcriptional patterns identify resource controls on the diazotroph Trichodesmium in the Atlantic and Pacific oceans, ISME J., 1486–1495, https://doi.org/10.1038/s41396-018-0087-z, 2018.
Rouwenhorst, R. J., Frank Jzn, J., Scheffers, W. A., and van Dijken, J. P.: Determination of protein concentration by total organic carbon analysis, J. Biochem. Bioph. Meth., 22, 119–128, https://doi.org/10.1016/0165-022X(91)90024-Q, 1991.
Rubin, M., Berman-Frank, I., and Shaked, Y.: Dust-and mineral-iron utilization by the marine dinitrogen-fixer Trichodesmium, Nat. Geosci., 4, 529–534, https://doi.org/10.1038/ngeo1181, 2011.
Saito, M. A., Goepfert, T. J., and Ritt, J. T.: Some thoughts on the concept of colimitation: Three definitions and the importance of bioavailability, Limnol. Oceanogr., 53, 276–290, https://doi.org/10.4319/lo.2008.53.1.0276, 2008.
Saito, M. A., McIlvin, M. R., Moran, D. M., Goepfert, T. J., DiTullio, G. R., Post, A. F., and Lamborg, C. H.: Multiple nutrient stresses at intersecting Pacific Ocean biomes detected by protein biomarkers, Science, 345, 1173–1177, https://doi.org/10.1126/science.1256450, 2014.
Saito, M. A., Dorsk, A., Post, A. F., McIlvin, M. R., Rappé, M. S., DiTullio, G. R., and Moran, D. M.: Needles in the blue sea: Sub-species specificity in targeted protein biomarker analyses within the vast oceanic microbial metaproteome, Proteomics, 15, 3521–3531, https://doi.org/10.1002/pmic.201400630, 2015.
Sañudo-Wilhelmy, S. A., Kustka, A. B., Gobler, C. J., Hutchins, D. A., Yang, M., Lwiza, K., Burns, J., Capone, D. G., Raven, J. A., and Carpenter, E. J.: Phosphorus limitation of nitrogen fixation by Trichodesmium in the central Atlantic Ocean, Nature, 411, 66–69, https://doi.org/10.1038/35075041, 2001.
Sheridan, C. C.: The microbial and metazoan community associated with colonies of Trichodesmium spp.: a quantitative survey, J. Plankton Res., 24, 913–922, https://doi.org/10.1093/plankt/24.9.913, 2002.
Shi, T., Sun, Y., and Falkowski, P. G.: Effects of iron limitation on the expression of metabolic genes in the marine cyanobacterium Trichodesmium erythraeum IMS101, Environ. Microbiol., 9, 2945–2956, 2007.
Shih, P. M., Wu, D., Latifi, A., Axen, S. D., Fewer, D. P., Talla, E., Calteau, A., Cai, F., Tandeau de Marsac, N., Rippka, R., Herdman, M., Sivonen, K., Coursin, T., Laurent, T., Goodwin, L., Nolan, M., Davenport, K. W., Han, C. S., Rubin, E. M., Eisen, J. A., Woyke, T., Gugger, M., and Kerfeld, C. A.: Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing, P. Natl. Acad. Sci. USA, 110, 1053–1058, https://doi.org/10.1073/pnas.1217107110, 2013.
Snow, J. T., Polyviou, D., Skipp, P., Chrismas, N. A. M., Hitchcock, A., Geider, R., Moore, C. M., and Bibby, T. S.: Quantifying integrated proteomic responses to iron stress in the globally important marine diazotroph Trichodesmium, PLoS ONE, 10, 1–24, https://doi.org/10.1371/journal.pone.0142626, 2015.
Sohm, J. A., Webb, E. A., and Capone, D. G.: Emerging patterns of marine nitrogen fixation, Nat. Rev. Microbiol., 9, 499–508, https://doi.org/10.1038/nrmicro2594, 2011.
Strathmann, R. R.: Estimating Organic Carbon Content of Phytoplankton from Cell Volume or Plasma Volume, Limnol. Oceanogr., 12, 411–418, 1967.
Sunda, W. G.: Feedback interactions between trace metal nutrients and phytoplankton in the ocean, Front. Microbiol., 3, 1–22, https://doi.org/10.3389/fmicb.2012.00204, 2012.
Walworth, N. G., Fu, F.-X., Webb, E. A., Saito, M. A., Moran, D., Mcllvin, M. R., Lee, M. D., and Hutchins, D. A.: Mechanisms of increased Trichodesmium fitness under iron and phosphorus co-limitation in the present and future ocean, Nat. Commun., 7, 1–11, https://doi.org/10.1038/ncomms12081, 2016a.
Walworth, N. G., Lee, M. D., Fu, F.-X., Hutchins, D. A., and Webb, E. A.: Molecular and physiological evidence of genetic assimilation to high CO2 in the marine nitrogen fixer Trichodesmium, P. Natl. Acad. Sci. USA, 3, 201605202, https://doi.org/10.1073/pnas.1605202113, 2016b.
Walworth, N. G., Fu, F. X., Lee, M. D., Cai, X., Saito, M. A., Webb, E. A., and Hutchins, D. A.: Nutrient-colimited Trichodesmium as a nitrogen source or sink in a future ocean, Appl. Environ. Microb., 84, 1–14, https://doi.org/10.1128/AEM.02137-17, 2018.
Wang, Q., Li, H., and Post, A. F.: Nitrate assimilation genes of the marine diazotrophic, filamentous cyanobacterium Trichodesmium sp. strain WH9601, J. Bacteriol., 182, 1764–1767, https://doi.org/10.1128/JB.182.6.1764-1767.2000, 2000.
Webb, E. A., Moffett, J. W., and Waterbury, J. B.: Iron Stress in Open-Ocean Cyanobacteria (Synechoccoccus, Trichodesmium, and Crocosphaera spp.): Identification of the IdiA Protein, Appl. Environ. Microb., 67, 5444–5452, https://doi.org/10.1128/AEM.67.12.5444-5452.2001, 2001.
Webb, E. A., Jakuba, R. W., Moffett, J. W., and Dyhrman, S. T.: Molecular assessment of phosphorus and iron physiology in Trichodesmium populations from the western Central and western South Atlantic, Limnol. Oceanogr., 52, 2221–2232, https://doi.org/10.4319/lo.2007.52.5.2221, 2007.
Wu, J. F., Sunda, W., Boyle, E. A., and Karl, D. M.: Phosphate depletion in the western North Atlantic Ocean, Science, 289, 759–762, https://doi.org/10.1126/Science.289.5480.759, 2000.
Yamaguchi, T., Furuya, K., Sato, M., and Takahashi, K.: Phosphate release due to excess alkaline phosphatase activity in Trichodesmium erythraeum, Plankton Benthos Res., 11, 29–36, https://doi.org/10.3800/pbr.11.29, 2016.
Yentsch, C. M., Yentsch, C. S., and Perras, J. P.: Alkaline phosphatase activity in the tropical marine blue-green alga Trichodesmium, Limnol. Oceanogr., 17, 72–774, https://doi.org/10.4319/lo.1972.17.5.0772, 1972.





