the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Cryptic roles of tetrathionate in the sulfur cycle of marine sediments: microbial drivers and indicators
Subhrangshu Mandal
Sabyasachi Bhattacharya
Chayan Roy
Moidu Jameela Rameez
Jagannath Sarkar
Tarunendu Mapder
Svetlana Fernandes
Aditya Peketi
Aninda Mazumdar
To explore the potential role of tetrathionate in the sedimentary sulfur cycle, population ecology of microorganisms capable of metabolizing this polythionate was revealed at 15–30 cm resolution along two, ∼3 m long, cores collected from 530 and 580 m below the sea level, off India's west coast, within the oxygen minimum zone (OMZ) of the Arabian Sea. Metagenome analysis along the cores revealed widespread occurrence of genes involved in the formation, oxidation, and reduction of tetrathionate; high diversity and relative abundance were also detected for bacteria that are known to render these metabolisms in vitro. Results of slurry culture of the sediment samples in thiosulfate- or tetrathionate-containing microbial growth media, data obtained via pure-culture isolation, and finally metatranscriptome analyses corroborated the in situ functionality of the tetrathionate-forming, tetrathionate-oxidizing, and tetrathionate-reducing microorganisms. Ion chromatography of pore waters revealed the presence of up to 11.1 µM thiosulfate in the two cores, whereas tetrathionate remained undetected in spectroscopic assay based on its reaction with cyanide. While thiosulfate oxidation by chemolithotrophic bacteria prevalent in situ is the apparent source of tetrathionate in this ecosystem, high biochemical and geochemical reactivity of this polythionate could be instrumental in its cryptic status in the sulfur cycle. Potential abiotic origin of tetrathionate in the sediment horizon explored could neither be ruled out nor confirmed from the geochemical information available. On the other hand, tetrathionate potentially present in the system can be either oxidized to sulfate or reduced back to thiosulfate/sulfide via chemolithotrophic oxidation and respiration by native bacterial populations, respectively. Up to 2.01 mM sulfide present in the sediment cores may also reduce tetrathionate abiotically to thiosulfate and elemental sulfur. However, in the absence of measured data for O2 or other oxyanions having possibilities of serving as electron acceptors, the biogeochemical modalities of the oxidative half of the tetrathionate cycle remained unresolved.
- Article
(5686 KB) - Full-text XML
-
Supplement
(1653 KB) - BibTeX
- EndNote
Phylogenetically diverse microorganisms oxidize or reduce different sulfur species to meet their bioenergetic requirements and in doing so play profound roles in biogeochemical sulfur cycling in nature (Baumgartner et al., 2006; Ghosh and Dam, 2009; Wasmund et al., 2017). Within the marine realm, microbial processes of sulfur cycle are central to benthic biogeochemistry as they are linked to the in situ transformations, sequestrations, and fluxes of carbon, nitrogen, and iron. There have been extensive studies of the benthic–sedimentary sulfur cycle across the global ocean (Jørgensen, 1990; Jørgensen and Bak, 1991; Rudnicki et al., 2001; Tostevin et al., 2014), and the focus of such investigations has typically been on geomicrobial transformations of the two redox end-members sulfate and sulfide (Holmkvist et al., 2011; Jørgensen et al., 2019). Elemental sulfur and thiosulfate have also been envisaged as constituting key junctions in the network of sulfur species transformations in the marine sediments (Jørgensen, 1990; Jørgensen and Bak, 1991; Thamdrup et al., 1994). However, tetrathionate or other polythionates are rarely appreciated for their potential role(s) in marine sedimentary sulfur cycle, presumably because these sulfur species are not abundant in these environments.
Overall paucity of polythionates in natural environments is largely attributable to their high reactivity under biotic (Kanao et al., 2007; Ghosh and Dam, 2009; Boden et al., 2010; Pyne et al., 2017, 2018) as well as abiotic (Schippers et al., 1999; Schippers and Jørgensen, 2001) conditions. The cryptic nature of these sulfur species notwithstanding, several bacteria capable of producing and/or utilizing tetrathionate for bioenergetic purposes have been isolated from different terrestrial and aquatic (both fresh water and marine) habitats (Kaprálek, 1972; Oltmann and Stouthamer, 1975; Barrett and Clark, 1987; Price-Carter et al., 2001; Sorokin, 2003; Ghosh et al., 2005, 2006; Ghosh and Roy, 2007). At the same time, several enzymes catalyzing redox reactions involving tetrathionate have been characterized in taxonomically diverse microorganisms. For instance, thiosulfate dehydrogenase (TsdA), widespread in photo- or chemolithotrophic bacteria, is attributed to the formation of tetrathionate from thiosulfate (Hensen et al., 2006; Denkmann et al., 2012; Brito et al., 2015; Pyne et al., 2018; Rameez et al., 2019). In several other archaeal and bacterial chemolithotrophs, thiosulfate-to-tetrathionate conversion is mediated by thiosulfate:quinone oxidoreductase (TQO or DoxDA; see Muller et al., 2004; Rzhepishevska et al., 2007; Quatrini et al., 2009; Kikumoto et al., 2013). Chemolithotrophic oxidation of tetrathionate to sulfate, on the other hand, is rendered either (i) by the pyrroloquinoline quinone (PQQ)-binding tetrathionate hydrolase (TetH), as described in Acidithiobacillus species (De Jong et al.,1997; Kanao et al., 2007; Rzhepishevska et al., 2007; van Zyl et al., 2008; Kanao et al., 2013), or (ii) via coupling with glutathione (to form glutathione:sulfodisulfane and sulfite) by the action of another PQQ-binding protein called thiol dehydrotransferase (ThdT), followed by the oxidation of glutathione:sulfodisulfane via iterative actions of sulfate thiol esterase (SoxB) and c-type cytochrome containing sulfur dehydrogenase (SoxCD), as reported in Advenella kashmirensis (Pyne et al., 2017, 2018). On the reductive half, typical tetrathionate-reducing bacteria such as Salmonella, Citrobacter, and Proteus transform tetrathionate to thiosulfate by tetrathionate reductase (TtrABC) followed by the conversion of thiosulfate to sulfide by thiosulfate reductase (PhsAB and PsrA) (Oltmann and Stouthamer, 1975; Barrett and Clark, 1987; Hensel et al., 1999; Stoffels et al., 2011).
Here we use approaches of molecular microbiology to investigate the potential involvement of tetrathionate metabolism in the sulfur cycle of sediments underlying the perennial oxygen minimum zone (OMZ) of the Arabian Sea, off the west coast of India. Community structures and functions of tetrathionate-forming, tetrathionate-oxidizing, and tetrathionate-reducing microorganisms were revealed by metagenome analyses and slurry culture of sediment samples along two ∼3 m long cores collected from 530 and 580 m below the sea level (m b.s.l.), at sites having the GPS coordinates 16∘50.03′ N, 71∘59.50′ E and 16∘49.88′ N, 71∘58.55′ E respectively; sediment samples were also analyzed for the presence of tetrathionate and thiosulfate in their pore waters. In situ activity of tetrathionate metabolizers was corroborated by pure-culture isolation and metatranscriptome analysis for the deepest sample site, within the sulfate–methane transition zone, of one of the two cores. Correspondence was also explored between the de novo sequenced genomes of the isolates and the metatranscriptome or the metagenomes sequenced. The microbial ecology delineated in this way was considered in the context of the in situ geochemistry to infer implications for the sedimentary sulfur cycle.
2.1 Study site and sample collection and storage
During a comprehensive exploration of the sediment biogeochemistry of eastern Arabian Sea OMZ (ASOMZ), on board R/V Sindhu Sankalp (SSK42), a number of gravity cores were collected from water depths spanning 225 and 1275 m b.s.l., covering the entire thickness of the eastern ASOMZ (Fernandes et al., 2018). Of these, SSK42/5 and SSK42/6, on which the present study is based, were collected from 580 m b.s.l. (16∘49.88′ N, 71∘58.55′ E) and 530 m b.s.l. (16∘50.03′ N, 71∘59.50′ E) water depths, respectively (Fernandes et al., 2018), i.e., the approximate center of the vertical expanse of the ASOMZ off the west coast of India. Both the cores were ∼3 m long and 12 cm in diameter; their onboard sampling was carried out at 15 to 30 cm intervals, as described previously, under a constant shower of high-purity N2 to minimize exposure of the native microflora to aerial O2 and avoid aerial oxidation of the H2S, Fe2+, and other reduced chemical substances present in the sediments (Fernandes et al., 2018).
In order to protect the sediment samples from aerial oxidation, only one ∼30 cm long C-shaped part of PVC core liner was removed at a time, as shown in Fig. S1 in the Supplement. The 30 cm length exposed at a time for sampling was constantly and closely showered with high-purity N2 emitted from multiple nozzles fitted to multiple nitrogen generators. Immediately after the C-shaped longitudinal part of the PVC core liner was cut open, the top 1 cm of the exposed surface was scraped off along the core circumference, using sterile scalpels, to eliminate potential contaminations from the core liner's inner surface and/or seawaters through which the core had passed. Subsequently, to sample a particular sediment depth of the core for microbiological studies, an approximately 5 mm thick sediment slice (spanning equally on either side of the core-height marking) was scooped out with a sterile scalpel and put into a sterile polypropylene bottle. For every sediment depth investigated, two such slices or sample replicates were collected for duplicate metagenome analyses (these were designated as sample replicates 1 and 2; see Tables S1 and S2), a third one was taken for metatranscriptome analysis, while two more replicates were taken for culture-dependent investigations. All five replicates were collected in individual screw-capped bottles, and those meant for metatranscriptomics were treated immediately with RNAlater (Ambion Inc., USA). The headspace of each sample-containing bottle was flushed with high-purity N2, subsequent to which the bottles were sealed with Parafilm (Bemis Company Inc., Neenah, USA) and stored frozen or cool. Sample replicates meant for culture-independent and culture-dependent studies were stored at −20 and 4 ∘C, respectively. From the laboratory on board SSK42, en route to Bose Institute, and over subsequent preservation of samples, these temperatures were maintained.
For onboard extraction of pore waters, samples from a particular sediment depth were taken out by inserting sterile 50 mL cutoff syringes deep inside the core cross section, multiple times along the circumference on the exposed “C half”. The samples were immediately collected in sterile 50 mL centrifuge tubes. All these operations were carried out under focused streams of high-purity N2. The tubes were centrifuged at 4700×g for 15 min at 4 ∘C, and the supernatants collected were syringe-filtered through 0.22 µm cellulose acetate membranes. Aliquots for different chemical analyses were dispensed to individual glass vials containing sodium azide that arrests microbial activity; only the vials meant for precipitating dissolved sulfide (ΣHS−) from the aliquots (in the form of cadmium sulfide, CdS) contained cadmium nitrate [Cd(NO3)2] instead of sodium azide. All the vials were crimp sealed immediately after N2 flushing and stored at 4 ∘C until further analysis.
2.2 Analytical method
Sulfide and sulfate concentrations in the pore water samples were determined and reported previously by Fernandes et al. (2018). Concentration of dissolved thiosulfate in the pore water samples was determined by ion chromatography using an Eco IC (Metrohm AG, Herisau, Switzerland) equipped with a conductivity detector (Metrohm, IC detector 1.850.9010). Chemical suppression was used for this purpose, while separation was carried using a Metrosep A Supp5 – 250/4.0 anion exchange column (Metrohm AG). A mixed solution of 1.0 mM sodium bicarbonate and 3.2 mM sodium carbonate was used as the eluent; 100 mM sulfuric acid was used as the regenerant; flow rate was 0.7 mL min−1; and injection volume was 100 µL. Prior to analysis, pore water samples were diluted 1000-fold with deionized water (Siemens, <0.06 µS) and passed through 0.22 µm hydrophilic polyvinylidene fluoride membrane filters (Merck Life Science Private Limited, Bengaluru, India). Analytical-grade thiosulfate IC Standard (Sigma Aldrich, St. Louis, USA) was used to prepare the calibration curve for quantification of this anion. Three different concentrations of thiosulfate, 0.5, 5, and 20 µM, were measured for the construction of calibration curve by plotting peak height against concentration. Based on triplicate analysis of the standards, deviations from their actual concentrations were found to be less than 2.5 %.
Concentration of tetrathionate in the pore water samples was measured by cyanolytic method (Kelly and Wood, 1994), where tetrathionate reacts with cyanide to form thiocyanate according to the reaction , and the thiocyanate (SCN−) formed is quantified spectrophotometrically from the absorbance of ferric thiocyanate (ε=5030 M−1 cm−1, λ=460 nm), which is formed by reacting the thiocyanate ion with ferric nitrate. Differences in the reactivity of thionates with cyanide enable their discrimination and quantitative characterization within mixtures of such compounds. For instance, trithionate is stable at high pH and reacts with cyanide only at elevated temperatures; thiosulfate reacts with cyanide at room temperature, also only in the presence of copper(II) catalyst. In contrast, the higher polythionates (, where n=4 or more) react rapidly with cyanide at room temperature to form SCN−, , , and HCN. Furthermore, in aqueous systems where intermediate sulfur compounds are cycling and dissolved sulfide is present, cyanide may also react with zero-valent sulfur components of the colloidal fraction of particulate elemental sulfur, polysulfides (), and their protonated forms, albeit when administered under hot and slightly acidic conditions (Kamyshny, 2009), which is not the case in the cyanolytic procedure currently followed for tetrathionate estimation (Kelly and Wood, 1994). Here, 0.1 mL of the pore water sample was added to 0.8 mL of cyanolytic buffer (a mixture of 50 mL of 0.2 M NaH2PO4 and 39 mL of 0.2 M NaOH; pH 7.4), and the volume was made up to 2.0 mL. The mixture was then chilled on ice for 20 min, and 1 mL of 0.1 M pre-chilled potassium cyanide was added and mixed rapidly and incubated on ice for another 20 min. Finally, 0.6 mL of ferric nitrate reagent (30.3 g Fe(NO3)3⋅9H2O in 21.7 mL of 72 % perchloric acid, made up to 50 mL with distilled water) was added with continuous agitation and allowed to warm to room temperature until the white precipitate (if any) re-dissolved. Volume was made up to 5 mL, and optical density of the color due to ferric thiocyanate was measured on a spectrophotometer. The standard curve used for tetrathionate estimation was prepared using different concentrations (0.1, 0.5 1.0, 5.0, and 10.0 µM) of analytical grade tetrathionate (Sigma Aldrich, St. Louis, USA). Based on triplicate analysis of these standards, deviations from their actual concentrations were found to be less than 5 %.
2.3 Extraction of total DNA or RNA from sediment samples/pure-culture isolates
Total community DNA (metagenome) was extracted from the sediment samples using the PowerSoil DNA isolation kit (MoBio, Carlsbad, USA), as per the manufacturer's protocol. A microgram level of DNA was obtained from each batch of preparatory reaction that started with a 0.5 g sediment sample. Genomic DNA of pure-culture isolates was extracted using the HiPurA bacterial genomic DNA purification kit (Himedia Laboratories, Mumbai, India), following manufacturer's instructions. Quality of metagenomic and genomic DNA samples was checked by electrophoresis and considered to be of high quality when no degradation signs were apparent. DNA quantity was determined using the Qubit dsDNA HS assay kit (Thermo Fisher Scientific, Waltham, USA).
Total community RNA (metatranscriptome) was extracted from an RNAlater-treated sample replicate by using the RNA PowerSoil total RNA isolation kit (MoBio), as per manufacturer's protocol. Nanogram-level total RNA was obtained after pooling the products of 15 individual preparatory reactions, each carried out using the 2 g sediment sample. All the individual RNA preparations were subjected to DNase digestion by RNase-free DNase I (Thermo Fisher Scientific) and purified using the RNeasy MinElute cleanup kit (Qiagen, Hilden, Germany); their concentrations were measured using the Quant-iT RiboGreen RNA assay kit (Thermo Fisher Scientific). Integrity of RNA (RIN) within the individual preparations was determined on a TapeStation RNA ScreenTape electrophoretic system (Agilent Technologies, Santa Clara, USA), and only high-quality preparations having RIN value >7.0 were added to the RNA pool that were subsequently used for sequencing library construction.
2.4 Metagenome and genome sequencing
The duplicate set of metagenomes extracted for each sediment depth explored along SSK42/5 and SSK42/6 was shotgun sequenced individually on an Ion Proton sequencing platform (Thermo Fisher Scientific) using 200 nucleotide read chemistry, as described previously (Ghosh et al., 2015). Complete lists of sedimentary communities investigated along SSK42/5 and SSK42/6 are given in Tables S1 and S2, respectively, together with the accession numbers of the metagenomic sequence datasets.
A total of 1 µg DNA from each metagenome sample was taken for deep shotgun sequencing by the Ion Proton platform using 200 bp read chemistry on a PI V2 chip. Sequencing libraries were constructed using the Ion Plus fragment library kit (Thermo Fisher Scientific), following the manufacturer's Ion Plus gDNA library preparation user guide. The Proton library was generated using 1 µg of genomic DNA which was fragmented to approximately 200 base pairs by the Covaris S2 system (Covaris, Inc., Woburn, USA) and purified with 1.8X Agencourt Ampure XP beads (Beckman Coulter, Brea, USA). Fragmentation was followed by end-repair, blunt-end ligation of the Ion Xpress barcode and Ion P1 adaptor and nick translation.
Post-ligation, size selection was done using E-Gel Size-Select 2 % Agarose gels (Thermo Fisher Scientific) with 300 bp target size. Final polymerase chain reaction (PCR) was performed using Platinum PCR SuperMix High Fidelity and Library Amplification Primer Mix (Thermo Fisher Scientific), for five cycles of amplification. The resulting library was purified using 1.2X AMPure XP reagent (Beckman Coulter) and the concentration determined with Qubit dsDNA HS assay kit (Thermo Fisher Scientific); size distribution was done with the Agilent 2100 bioanalyzer high-sensitivity DNA kit (Agilent Technologies). Libraries were pooled in equimolar concentrations and used for template preparation.
Library templates for sequencing were prepared using OneTouch 2 protocols and reagents (Thermo Fisher Scientific). Library fragments were clonally amplified onto ion sphere particles (ISPs) through emulsion PCR and then enriched for template-positive ISPs. Proton emulsion PCR reactions utilized the Ion PI Template OT2 200 kit v3 (Thermo Fisher Scientific). Following recovery, enrichment was completed by selectively binding the ISPs containing amplified library fragments to streptavidin-coated magnetic beads, removing empty ISPs through washing steps and denaturing the library strands to allow for collection of the template-positive ISPs. For all reactions, these steps were accomplished using the ES module of the Ion OneTouch 2. The selected ISPs were loaded on the PI V2 chip and sequenced with the Ion PI 200 sequencing kit (Thermo Fisher Scientific) using the 500 flow (125 cycle) run format.
Whole genomes of isolated bacterial strains were sequenced on an Ion S5 platform (Thermo Fisher Scientific) using 400 nucleotide read chemistry on a 530 or 520 chip. Libraries were constructed by the Ion Xpress Plus fragment library kit (Thermo Fisher Scientific) using 100 ng genomic DNA from each isolate. In this procedure genomic DNA samples were fragmented using Ion Shear Plus reagents (Thermo Fisher Scientific). The fragmented libraries were purified using 1.8X Agencourt Ampure XP beads (Beckman Coulter) and subjected to barcode-adapter ligation and nick repair. Adapter-ligated and nick-repaired libraries were purified again by 1X Agencourt Ampure XP beads (Beckman Coulter).
Size selection of the libraries was done using E-Gel Size-Select 2 % Agarose gels (Thermo Fisher Scientific) with 480 bp target size. Final PCR was performed using Platinum SuperMix High Fidelity PCR system and Library Amplification Primer Mix (both from Thermo Fisher Scientific), for eight cycles of amplification. The resulting libraries were purified using 1X Agencourt AMPure XP reagent (Beckman Coulter). Concentrations of the purified libraries were determined with the Qubit dsDNA HS assay kit (Thermo Fisher Scientific). Libraries were then pooled in equimolar concentrations and used for template preparation.
The library template to be used for sequencing was prepared using Ion OneTouch 2 reagents (Thermo Fisher Scientific). Library fragments were clonally amplified onto ion sphere particles (ISPs) through emulsion PCR and then enriched for template-positive ISPs. Following emulsion PCR, enrichment was completed by selectively binding the ISP-containing amplified library fragments to streptavidin-coated magnetic beads, removing empty ISPs through washing steps and denaturing the library strands to allow for collection of the template-positive ISPs using the Ion OneTouch ES instrument (Thermo Fisher Scientific). The selected ISPs were loaded on a 530 or 520 chip, and sequencing was performed with the Ion S5 sequencing kit (Thermo Fisher Scientific) using the 850-flow run format.
2.5 Metatranscriptome (community mRNA) sequencing
The pooled total RNA preparations were selectively converted to a library of template molecules using the TruSeq Stranded mRNA and Total RNA kit (Illumina Inc., San Diego, USA). Depletion of rRNAs was carried out using the Ribo-Zero Gold system (Illumina Inc.), which is an integral part of the kit used for preparing the library. The rRNA-depleted RNA pool, which was expected to contain only the total mRNA, was fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were copied into first strand cDNAs using reverse transcriptase and random primers. This was followed by second strand cDNA synthesis using DNA Polymerase I and RNase H. cDNA fragments were then subjected to end repair, addition of single “A” bases, adaptor ligation, purification, and enrichment with PCR to create the final library, which was sequenced on a HiSeq4000 platform (Illumina Inc.) using paired-end, 2×150 nucleotide sequencing by synthesis read chemistry with dual indexing workflows. Furthermore, in order to extract and eliminate any rRNA read that may have remained in the raw metatranscriptomic sequence dataset, the 26 579 343 read pairs available in all were mapped onto the SILVA large subunit as well as small subunit rRNA gene sequence database (Quast et al., 2012), using the short read aligner Bowtie2 v.2.3.4.3 (Langmead and Salzberg, 2012) in default local (sensitive) alignment mode. This identified ∼0.3 % reads as ascribable to rRNAs, thereby leaving 26 496 769 read pairs in the final dataset used for downstream analyses.
2.6 De novo assembly and annotation of genomes, metagenomes, and metatranscriptome
High-quality reads (Phred score cutoff 20) from the duplicate metagenomic sequence datasets available for each sediment community were co-assembled using Megahit v1.2.x (Li et al., 2015) with the k-mer lengths of 21, 29, 39, 59, 79, 99, 119, and 141. Contigs of >100 bp length were searched using MetaGeneMark (Zhu et al., 2010) for genes encoding peptides having lengths of >30 amino acids. For the genomes of pure-culture isolates, high-quality reads (Phred score cutoff 20) were assembled using SPAdes 3.13.0 (Nurk et al., 2013), with k-mer lengths of 21, 33, 55, 77, 99, and 127 and minimum coverage cutoff of 35X. The whole genome sequences were deposited to the GenBank and annotated using Prokaryotic Genome Annotation Pipeline (PGAP located at https://www.ncbi.nlm.nih.gov/genome/annotation_prok/, last access: 22 April 2019) of the National Center for Biotechnology Information (NCBI), Bethesda, MD, USA. Total metagenomic or metatranscriptomic reads available for the different sediment communities were mapped onto the individual genome sequences or the manually curated gene catalogs obtained from the individual genomes, using Bowtie2 v.2.3.4.3 (Langmead and Salzberg, 2012) in default local (sensitive) alignment mode. The rRNA-sequence-free metatranscriptomic dataset was assembled using the Python script rnaspades.py, available within SPAdes 3.13.0 (Nurk et al., 2013), with default parameters. Genes encoding continuous stretches of minimum 30 amino acids were predicted in contigs longer than 100 bp using Prodigal v2.6.3 (Hyatt et al., 2010).
Gene catalogs obtained after de novo assembly of the individual metagenomes, or the solitary metatranscriptome, were functionally annotated by searching against the EggNOG v5.0 database (http://eggnog5.embl.de/download/eggnog_5.0/, last access: 7 April 2019) with EggNOG-mapper (Huerta-Cepas et al., 2016) (http://beta-eggnogdb.embl.de/{#} /app/emapper, last access: 7 April 2019) using HMMER algorithms. Putative protein sequence catalogs obtained for the individual genomes via PGAP annotation were re-annotated by searching against the EggNOG v5.0 database with EggNOG-mapper using the HMMER algorithm. Genes encoding proteins involved in tetrathionate formation, oxidation, and reduction were identified within the annotated genomes by their orthology numbers designated in the Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al., 2016).
2.7 Direct taxonomic annotation of raw metagenomic reads
Raw reads from the duplicate metagenomic sequence datasets of each sediment community were directly annotated for their taxonomic affiliation by searching the datasets individually against the NCBI non-redundant (nr) protein sequence database, using the Organism Abundance tool of MG-RAST 3.6 (Meyer et al., 2008). The two independent values obtained in this way for the relative abundances of genera within a community were averaged and used for comparisons between communities; percentage allocation of reads over various genera was taken as a direct measure of the relative abundance (prevalence) of those genera within the community (Tringe et al., 2005; Ghosh et al., 2015; Roy et al., 2020). Within MG-RAST, sequences were trimmed to contain no more than five successive bases with a Phred quality score <15. To classify reads using the Organism Abundance tool, the best-hit classification algorithm was followed, reporting only those BlastX search results which had minimum 45 nucleotides (15 amino acids) alignment, ≥60 % identity, and e value ; these cutoffs are stringent enough for genus-level classification of homologs of metabolically diverse genes, irrespective of their intrinsic levels of conservation.
2.8 Slurry culture of sediment samples
Ability of the different sedimentary microbial communities to form tetrathionate (from thiosulfate), oxidize tetrathionate (to sulfate), or reduce tetrathionate (to thiosulfate and/or sulfide) was tested via incubation of the samples in culture media supplemented with known concentrations of thiosulfate or tetrathionate. Within the slurry culture setups, in vitro rates of redox transformations of sulfur involving tetrathionate were determined and expressed as micromoles S per liter slurry per day. For each experiment testing the formation or oxidation of tetrathionate, 10 % (w∕v) sediment sample was suspended in artificial seawater (ASW) supplemented with thiosulfate (T) or tetrathionate (Tr), i.e., ASWT or ASWTr broth medium (Alam et al., 2013), respectively, to form a slurry; the culture flask was then incubated aerobically at 15 ∘C on a rotary shaker (150 rpm). For each anaerobic experiment testing tetrathionate reduction, 10 % (w∕v) sediment sample was suspended in tetrathionate-supplemented Rappaport Vassiliadis tetrathionate-supplemented (RVTr) medium (Vassiliadis, 1983) that was already made O2-free by addition of sodium thioglycolate. Addition of sediment samples to sterile O2-free RVTr media (contained in a screw-capped bottles) and subsequent incubation of the culture bottles were all carried out inside a Whitley H35 Hypoxystation (Don Whitley Scientific, West Yorkshire, UK) preset at 75 % humidity, 15 ∘C temperature, and 0 % partial pressure of O2, using the gas mixture ().
The ASWT or ASWTr medium (both having pH 7.5) contained ASW supplemented with Na2S2O3⋅5H2O (10 mM) or K2S4O6 (5 mM), respectively (the two sulfur compounds were added to the media separately after filter sterilization) (Alam et al., 2013). ASW contained the following per liter distilled water: 25.1 g NaCl, 1 g (NH4)2SO4, 1.5 g MgSO4, 7H2O, 0.3 g CaCl2⋅2H2O, 0.2 g NaHCO3, 2.4 g Tris, 1 mL trace element solution, and 0.5 g K2HPO4 (added after autoclaving separately). A total of 1 L trace element solution (pH 6.0), in turn, contained 50 g EDTA, 22 g ZnSO4⋅7H2O, 5.06 g MnCl2, 4.99 g FeSO4, 1.1 g (NH4)6MoO26⋅4H2O, 1.57 g CuSO4 and 1.61 g CoCl2⋅6H2O. RVTr medium (pH 5.4) contained the following per liter of distilled water: 4.5 g soya peptone, 8.0 g NaCl, 0.4 g K2HPO4, 0.6 g KH2PO4, 29.0 g MnCl2, 0.036 g Malachite green, 10 mM K2S4O6, 0.5 g sodium thioglycolate (used as an O2 scavenger), and 0.1 mg resazurin (added to indicate the presence of any dissolved O2).
During RVTr preparation, inside a Whitley H35 Hypoxystation preset to 0 % partial pressure of O2, a pre-weighed amount of potassium tetrathionate salt was first dissolved in a premeasured volume of anoxic, deionized water (degassed for several hours inside the H35 Hypoxystation till the resazurin indicator added in the water became colorless). This anoxic tetrathionate solution was then added via filter sterilization to a separate pre-autoclaved O2-free solution that contained the rest of the RVTr components in an appropriate volume and had cooled down to room temperature within the Hypoxystation. Thioglycolate that was already there in the second solution had reacted irreversibly, during autoclaving, with the dissolved O2 present in the mixed-salt solution to form dithiodiglycolate. Post-autoclave cooling of this second solution within the Hypoxystation, therefore, did not breakdown the S–S bonds of dithiodiglycolate to regenerate the SH−-containing thioglycolate, so there was no possibility of thioglycolate attacking the incoming tetrathionate solution. Moreover, (i) at neutral pH, thiol-group-containing reducing agents do not attack tetrathionate under non-enzymatic (abiotic) conditions, or for that matter in the absence of specific inorganic catalysts (Pyne et al., 2018); (ii) zero-hour reading for all the slurry culture sets in the RVTr medium showed the intact presence of the 10 mM tetrathionate originally supplied in the medium; and (iii) abiotic control incubations involving autoclaved sediment samples showed that the 10 mM tetrathionate supplied to the RVTr medium was intact after prolonged incubation.
Concentrations of the various redox species of sulfur were estimated in the slurry cultures at every 6 h interval of incubation. End-point (final) concentrations of the different sulfur species were recorded when no further changes were detected in their concentrations over three consecutive estimations. Concentrations of thiosulfate, tetrathionate, and sulfate in the media were measured by iodometric titration, cyanolysis, and gravimetric sulfate precipitation, respectively, at different time intervals (Alam et al., 2013). Possible presence of dissolved sulfides was checked via precipitation as CdS by the addition of 2 M Cd(NO3)2, followed by spectroscopic estimation as described previously (Cline, 1969).
2.9 Enrichment, isolation, and characterization of bacterial strains
Isolation of sulfur-oxidizing chemolithotrophs from the 275 cm b.s.f. sediment sample of SSK42/6 was carried out in mineral salt–thiosulfate–yeast extract (MSTY), ASWT, and ASWTY media. While the ASWTY (pH 7.5) medium was a yeast-extract-supplemented (500 mg L−1) derivative of ASWT, MSTY (pH 7.0) contained modified basal and mineral salt (MS) solution supplemented with 20 mM Na2S2O3⋅5H2O and 500 mg L−1 yeast extract (Ghosh and Roy, 2006). MS, in turn, contained the following per liter distilled water: 1 g NH4Cl, 4 g K2HPO4, 1.5 g KH2PO4, 0.5 g MgSO4⋅7H2O, and 5.0 mL trace metal solution (Vishniac and Santer, 1957). Three portions of the 275 cm b.s.f. sediment sample of SSK42/6 were added (5 % w∕v) individually to MSTY, ASWT, and ASWTY broths and incubated aerobically at 15 ∘C until phenol red indicator present in the media turned yellow (apparently due to production of sulfuric acid from thiosulfate). Post yellowing, individual enrichment slurries were kept undisturbed for 1 h to allow sediment particles to settle down; 10 mL cell suspension from each flask was then centrifuged at 6000 g for 10 min, and the pellet was resuspended in milliliter of the corresponding medium, serially diluted, and spread onto agar plates of the corresponding medium and incubated at 15 ∘C. Morphologically distinct colonies were picked up and dilution-streaked until all colonies in individual plates looked similar; representative colonies from such pure-culture plates were taken as strains and maintained in their respective isolation medium. Only Methylophaga, though isolated in ASWT, was maintained in ASW supplemented with 0.3 % (v∕v) methanol (ASWM; this medium had a pH of 7.5) because its growth in ASWT waned after six straight sub-cultures. Chemolithotrophic abilities of the new isolates to oxidize thiosulfate to tetrathionate and/or tetrathionate to sulfate were tested in MSTY, MSTrY, ASWT, ASWTr, ASWTY, ASWTrY, ASWTM, and ASWTrM media. MSTrY (pH 7.0) contained MS solution supplemented with 10 mM K2S4O6 and 500 mg L−1 yeast extract. ASWTrY (pH 7.5) was a yeast-extract-supplemented (500 mg L−1) derivative of ASWTr. ASWTM (pH 7.5) and ASWTrM (pH 7.5) were thiosulfate-supplemented (10 mM Na2S2O3⋅5H2O) and tetrathionate-supplemented (10 mM K2S4O6) variants of ASWM, respectively. Concentrations of dissolved thiosulfate, tetrathionate, and sulfate in the spent culture media were measured as described above.
Tetrathionate-reducing bacterial strains were isolated from the 275 cm b.s.f. sediment sample of SSK42/6 in the RVTr medium (Vassiliadis, 1983) under strictly anaerobic conditions. A total of 2.5 g of sediment sample was added to 45 mL RVTr broth contained in a screw-capped bottle and prepared as described above. Sediment addition to the medium and subsequent incubation of the screw-capped bottles at 15 ∘C for 1 month were all carried out inside the Whitley H35 Hypoxystation preset to zero O2 as stated above. After 1 month, still inside the Hypoxystation, 1 mL of the sediment RVTr mixture was serially diluted and spread onto RVTr agar plates and incubated at 15 ∘C. After growth appeared in the RVTr agar plates they were taken out of the Hypoxystation; biomasses were serially dilution-streaked onto fresh plates and incubated aerobically until all colonies in the individual plates looked similar. Representative colonies from such pure-culture plates were taken and maintained aerobically in Luria broth medium. Tetrathionate-reducing abilities of the new isolates were tested by growing them anaerobically for 30 days in RVTr broth, inside the H35 Hypoxystation. Concentrations of dissolved thiosulfate, tetrathionate, and sulfide in the RVTr cultures were measured by the methods mentioned above.
Genomic DNA extracted from the individual isolates was used as template for PCR amplification of 16S rRNA genes with the Bacteria-specific universal primer pair 27f (fD1) and 1492r (rP2) (Weisburg et al., 1991). The 16S rRNA gene sequences were determined from the PCR products using a 3500xL genetic analyzer automated DNA sequencer (Thermo Fisher Scientific). The 16S rRNA gene sequence of each strain was compared against sequences available in the GenBank/EMBL/DDBJ databases, using BLASTN; strains were finally classified down to the lowest identifiable taxonomic category on the basis of their 16S rRNA gene sequence similarities with the closest validly published species having standing in nomenclature (http://www.bacterio.net/, last access: 10 March 2019; see also Euzéby, 1997; Parte, 2013).
3.1 Tetrathionate-forming, tetrathionate-oxidizing, and tetrathionate-reducing microorganisms and genes corresponding to such processes are abundant along SSK42/5 and SSK42/6
The duplicate metagenomic sequence datasets obtained for each of the 25 distinct sediment samples explored along SSK42/5 and SSK42/6 were co-assembled and annotated individually. A total of 23 out of the 25 contig collections obtained in this way were found to contain genes for tetrathionate formation (Table S3 in the Supplement), while all 25 encompassed genes for tetrathionate oxidation (Table S4). Furthermore, 24 out of the 25 contig collections contained genes for tetrathionate reduction (Table S5). The tetrathionate formation-related genes identified included those encoding for the different subunits of the thiosulfate dehydrogenases TsdA (Denkmann et al., 2012; Pyne et al., 2018) and DoxDA (Quatrini et al., 2009), which catalyze the oxidation of thiosulfate to tetrathionate in taxonomically diverse bacteria and archaea. While the genes identified for tetrathionate oxidation encoded the sulfate thiol esterase SoxB and the sulfur dehydrogenase SoxC (Lahiri et al., 2006; Pyne et al., 2018), those detected for tetrathionate reduction encoded subunits of tetrathionate (TtrABC) and thiosulfate reductases (PhsAB and PsrA) (Barrett and Clark, 1987; Stoffels et al., 2011).
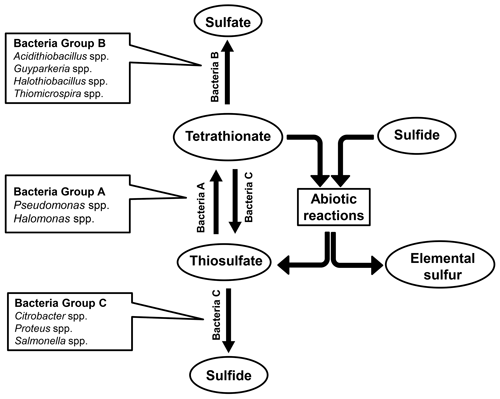
Concurrent with the above findings, direct taxonomic annotation of the raw (unassembled) metagenomic sequence datasets revealed that considerable proportions of the reads obtained for the individual sediment depths of SSK42/5 and SSK42/6 were ascribable to bacterial genera whose members are known to render tetrathionate formation, oxidation, or reduction. In that way, 1.3 %–4.36 % and 3 %–7.8 % of metagenomic reads obtained for the individual sample sites of SSK42/5 (Fig. 1a) and SSK42/6 (Fig. 1b) were ascribable to the genera Pseudomonas and Halomonas, the majority of marine strains of which are known to form tetrathionate from the oxidation of thiosulfate under aerobic or anaerobic conditions (Tuttle, 1980; Mason and Kelly, 1988; Sorokin et al., 1999, 2003). A total of 0.1 %–1.5 % and 0.4 %–6.4 % of metagenomic reads obtained for the individual sample sites of SSK42/5 (Fig. 1a) and SSK42/6 (Fig. 1b) were ascribable to the genera Acidithiobacillus, Guyparkeria, Halothiobacillus, and Thiomicrospira, all members of which oxidize tetrathionate chemolithotrophically (Hedrich and Johnson, 2013; Watsuji et al., 2016; Boden et al., 2017). A total of 0.1 %–0.3 % and 0.2 %–0.4 % of metagenomic reads obtained for the individual sample sites of SSK42/5 (Fig. 1a) and SSK42/6 (Fig. 1b) were ascribable to the genera Citrobacter, Proteus, and Salmonella, all members of which respire by reducing tetrathionate to thiosulfate and/or sulfide (Kaprálek, 1972; Barrett and Clark, 1987; Price-Carter et al., 2001; Stoffels et al., 2011).
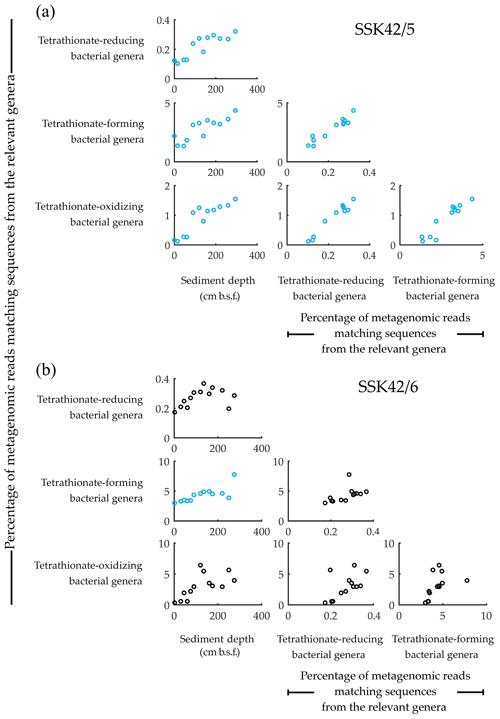
Figure 1Relative abundance of the tetrathionate-metabolizing bacterial groups plotted pair-wise against each other or against sediment depth, along (a) SSK42/5 and (b) SSK42/6. Relative abundances are expressed as the percentages of metagenomic reads matching protein-coding genomic sequences from the genera of tetrathionate-forming (Pseudomonas and Halomonas), tetrathionate-oxidizing (Acidithiobacillus, Guyparkeria, Halothiobacillus, and Thiomicrospira), and tetrathionate-reducing (Citrobacter, Proteus, and Salmonella) bacteria. Presence of these genera in the various sediment samples of SSK42/5 and SSK42/6 was corroborated via manual scrutiny of the amplified 16S rRNA gene-sequence-based diversity data reported previously for these cores (Fernandes et al., 2018) as well as by searching the individual metagenomic sequence datasets against the 16S rRNA gene sequence database of the Ribosomal Database Project (using BlastN with minimum alignment length 50 bp, minimum identity cutoff 90 %, and maximum e value cutoff 1e−5). Plots corroborated by Pearson correlation coefficient (CC) and/or Spearman rank correlation coefficient (RCC) values with P<0.05 are shown in blue. Whereas none of the plots were corroborated by negative CC or RCC values numerically ≥0.8 with P<0.05, those corroborated by positive/negative CC and/or RCC values numerically ≤0.8 are shown in black, irrespective of whether P is <0.05. All CC and RCC values pertaining to SSK42/5 and SSK42/6 are given in Tables S6 and S7, respectively.
3.2 Synchronized population fluctuation of different tetrathionate-metabolizing types, along SSK42/5 and SSK42/6
Analyses based on the direct taxonomic annotation of the unassembled metagenomic data from discrete sediment depths of SSK42/5 revealed that the relative abundances of reads ascribed to the genera of tetrathionate-forming, tetrathionate-oxidizing, and tetrathionate-reducing bacteria fluctuate synchronously along the sediment surface to core-bottom trajectory (Fig. 1a). Corroboratively, pair-wise Pearson correlation coefficients (CCs) as well as Spearman rank correlation coefficients (RCCs) between the prevalence of the three metabolic types are also significantly high in SSK42/5 (Fig. 1a; Table S6), which indicate the existence of strong syntrophic interdependence between the three tetrathionate-metabolizing types in this sediment horizon. Relative abundances of metagenomic reads ascribed to the genera of tetrathionate-forming, tetrathionate-oxidizing, and tetrathionate-reducing bacteria also fluctuate more or less synchronously along SSK42/6, except the region between 250 and 275 cm b.s.f. (Fig. 1b), which is the sulfate–methane transition zone (SMTZ) of this sediment horizon (notably, SMTZ in SSK42/5 was below the 280 cm b.s.f. sediment depth explored in this core; see Fernandes et al., 2018, for the methane profiles of all the SSK42 cores). While lack of synchrony in the lower end of SSK42/6 apparently resulted in the lower correlation values obtained for this core (Table S7) compared to those obtained for SSK42/5, it seems quite plausible that the changes in geochemistry and community architecture associated with the shallowing of SMTZ in SSK42/6 impacted the population ecology of tetrathionate-metabolizing microorganisms in this region. Sedimentation rate, age–depth profile, and other geochemical features of the two cores separated by a distance of only 1 km are otherwise largely comparable (Bhattacharya et al., 2019). Consistent prevalence of reads ascribed to the thiosulfate-to-tetrathionate-converting bacterial genera Halomonas and Pseudomonas in the metagenomes extracted from the different sediment samples of SSK42/5 and SSK42/6 (Fig. 1a and b) indicated that tetrathionate could be bioavailable in the chemical milieu of these sediment horizons (notably, pure-culture strains belonging to these two genera were also isolated from the 275 cm b.s.f. sample of SSK42/6; see Sect. 3.3 below, and also Fig. 2). Apart from these two, several such genera were also found to be well represented in the metagenomes of SSK42/5 and SSK42/6, some members of which are known to produce tetrathionate as a free intermediate during the oxidation of thiosulfate to sulfate and release the same to the extracellular milieu (Tables S8 and S9). These organisms, affiliated with the genera Acidithiobacillus, Advenella, Halothiobacillus, Paracoccus, Pusillimonas, and Thiomicrospira, can increase tetrathionate availability in the ASOMZ sediments, even as they themselves are potential users of the tetrathionate produced (Ghosh et al., 2005; Hedrich and Johnson, 2013; Watsuji et al., 2016; Boden et al., 2017; Rameez et al., 2019). Tables S8 and S9 show the relevant references and the percentages of metagenomic reads that were found to be ascribed to these genera in the different sediment samples of SSK42/5 and SSK42/6, respectively.
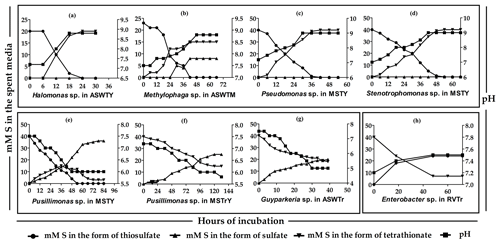
Figure 2Time course formation and transformations of tetrathionate by the
representative strains of the various species-level clusters isolated from
275 cm b.s.f. of SSK42/6. (a through d) Thiosulfate-to-tetrathionate conversion
by Halomonas sp. 15WGF, Methylophaga sp. SBPC3, Pseudomonas sp. SBBB, and Stenotrophomonas sp. SBPC3, respectively. (e, f)
Oxidation of thiosulfate to sulfate via tetrathionate and oxidation of
tetrathionate to sulfate by Pusillimonas ginsengisoli MTCC 12558, respectively. (g) Oxidation of
tetrathionate to sulfate by Guyparkeria sp. SB14A. (h) Reduction of tetrathionate to
thiosulfate by Enterobacter sp. RVSM5a. ![]() ,
, ![]() , and
, and ![]() indicate the
concentrations of sulfur (mM S) present in the medium at a given time point
of incubation in the form of thiosulfate, sulfate, and tetrathionate,
respectively.
indicate the
concentrations of sulfur (mM S) present in the medium at a given time point
of incubation in the form of thiosulfate, sulfate, and tetrathionate,
respectively. ![]() denotes the pH of a culture at a given time point of
incubation.
denotes the pH of a culture at a given time point of
incubation.
Tetrathionate can be oxidized in situ as a potential energy and electron source by members of the obligately chemolithotrophic genera Acidithiobacillus, Guyparkeria, Halothiobacillus, and Thiomicrospira that were detected via direct taxonomic annotation of the unassembled metagenomic data (Fig. 1a and b) and/or isolated as pure cultures from the 275 cm b.s.f. sample of SSK42/6 (Fig. 2). In addition, several such genera were detected (via direct annotation of metagenomic reads) along SSK42/5 and SSK42/6, some members of which are known to oxidize tetrathionate to sulfate. These organisms, affiliated with the genera Advenella, Bosea, Burkholderia, Campylobacter, Hydrogenovibrio, Pandoraea, Pusillimonas, Pseudaminobacter, Sulfurivirga, Thiohalorhabdus, and Thiobacillus, may contribute to further tetrathionate depletion from the sediments (Tables S10 and S11 show the relevant references and the metagenomic read percentages ascribed to these genera along SSK42/5 and SSK42/6, respectively).
Tetrathionate in the ASOMZ sediments can also be utilized as a respiratory substrate by bacteria such as Citrobacter, Proteus, and Salmonella, which were detected by direct annotation of metagenomic reads (Fig. 1a and b). In addition, strains of Enterobacter such as those isolated as pure cultures from 275 cm b.s.f. of SSK42/6 (Fig. 2) can add to the in situ reduction of tetrathionate to thiosulfate or sulfide. Furthermore, several such genera were also detected along SSK42/5 and SSK42/6 (via direct annotation of metagenomic reads), some members of which are known to respire via reduction of tetrathionate in the absence of O2; these included Alteromonas, Alcaligenes, Desulfotomaculum, Desulfovibrio, Edwardsiella, Morganella, Pasteurella, Providencia, Serratia, and Shewanella (Tables S12 and S13 show the relevant references and the metagenomic read percentages ascribed to these genera along SSK42/5 and SSK42/6, respectively).
3.3 The tetrathionate-forming and tetrathionate-oxidizing microorganisms of the ASOMZ sediments are alive and active in situ
Aerobic slurry culture of the sediment samples of SSK42/5 and SSK42/6 in thiosulfate-containing artificial seawater (ASWT) medium resulted in either the formation of tetrathionate with no further oxidation of this polythionate or the formation of tetrathionate followed by oxidation of the latter to sulfate, or no transformation at all. These findings, in conjunction with the results of pure-culture isolation, illustrated that the sulfur-chemolithotrophic microorganisms present in these sediment horizons are alive in situ and possess distinct pathways for oxidizing thiosulfate. For SSK42/5, ASWT incubation of the 0, 15, 90, and 160 cm b.s.f. samples resulted in the formation of tetrathionate as the sole and final product of thiosulfate oxidation, which happened in vitro at a rate of 6.45–17.72 µmol S per liter slurry per day (Fig. 3a, Table S14). In contrast, ASWT incubation of the 45, 60, and 295 cm b.s.f. samples of SSK42/5 resulted in the initial formation of tetrathionate from thiosulfate at a rate of 1.11–6.45 µmol S per liter slurry per day (notably, no sulfate was produced during this period of incubation); subsequently, the accumulated tetrathionate was converted to sulfate at a rate of 5.86–13.75 µmol S per liter slurry per day (Fig. 3a, Table S15). Microbial communities present in the remaining five sediment samples of SSK42/5 did not convert any thiosulfate out of the 20 mM S supplied (in the ASWT medium) to any higher oxidation state of sulfur. For SSK42/6, ASWT incubation of the 120, 175, and 275 cm b.s.f. samples resulted in the formation of tetrathionate as the sole and final product of thiosulfate oxidation, which happened in vitro at a rate of 17.2–29.71 µmol S per liter slurry per day (Fig. 3b, Table S16). In contrast, ASWT incubation of the 2, 30, and 45 cm b.s.f. samples of SSK42/6 resulted in the initial formation of tetrathionate from thiosulfate at a rate of 21.05–33.68 µmol S per liter slurry per day (notably, no sulfate was produced during this period of incubation); subsequently, the accumulated tetrathionate was converted to sulfate at a rate of 24–54 µmol S per liter slurry per day (Fig. 3b, Table S17). Microbial communities of the remaining seven sediment samples of SSK42/6 did not transform any part of the thiosulfate supplied to any higher oxidation state of sulfur. This could be due to a low number of metabolically active chemolithotrophic cells present in these samples. Notably, control incubation sets involving autoclaved sediment samples in ASWT showed no change in the thiosulfate concentration of the medium.
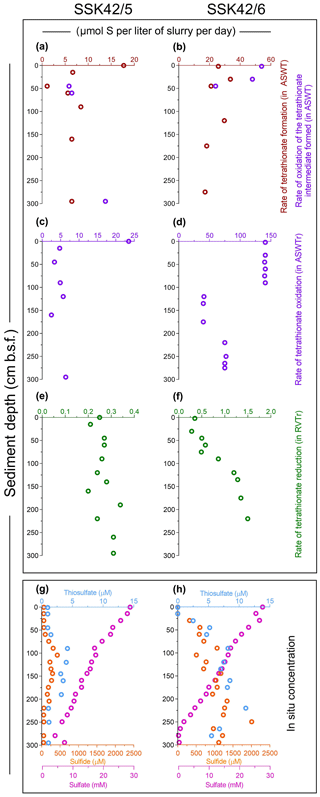
Figure 3Rates of formation and transformations of tetrathionate in slurry cultures of the sediment samples and concentrations of different sulfur species recorded along the two sediment cores. (a, b) In vitro rates of tetrathionate formation and its downstream oxidation (if any) in ASWT medium, for the different sedimentary communities of SSK42/5 and SSK42/6, respectively. (c, d) In vitro rates of tetrathionate oxidation in ASWTr medium, for the different sedimentary communities of SSK42/5 and SSK42/6, respectively. (e, f) In vitro rates of tetrathionate reduction in the RVTr medium, for the different sedimentary communities of SSK42/5 and SSK42/6, respectively. (g, h) Concentrations of sulfur species measured along SSK42/5 and SSK42/6, respectively. In (a) through (f), brown, violet, and green circles represent tetrathionate formation, oxidation, and reduction rates, respectively. In (g) and (h), orange, light blue, and purple circles represent the concentrations of sulfide, thiosulfate, and sulfate, respectively (sulfide and sulfate data were replotted from Fernandes et al., 2018).
Slurry culture of a number of sediment samples from SSK42/5 and SSK42/6 in ASWTr medium resulted in the oxidation of tetrathionate to sulfate. These data, together with the results of pure-culture isolation, indicated that the tetrathionate-oxidizing sulfur chemolithotrophs present in the different sediment samples were alive in situ. Of the individual microbial communities present in the different sediment samples of SSK42/5, those from 0, 15, 45, 90, 120, 160, and 295 cm b.s.f. oxidized tetrathionate at a rate of 2.5–23.5 µmol S per liter slurry per day (Fig. 3c, Table S18). Slurry culture of samples from the remaining five sediment depths of SSK42/5 did not result in any oxidation of tetrathionate, plausibly due to a low number of metabolically active chemolithotrophic cells present in these samples. For SSK42/6, ASWTr incubation of the 2, 30, 45, 60, 75, and 90 cm b.s.f. sediment samples resulted in the oxidation of tetrathionate to sulfate at an identical rate (approximately 140 µmol S per liter slurry per day). While ASWTr incubation of the samples from 120, 135, 160, and 175 cm b.s.f. resulted in tetrathionate oxidation at a common rate of approximately 40 µmol S per liter slurry per day, the same for the 220, 250, and 275 cm b.s.f. samples led to tetrathionate oxidation at a rate of approximately 75 µmol S per liter slurry per day (Fig. 3d, Table S19). Control incubations involving autoclaved sediment samples in ASWTr showed no oxidation of tetrathionate.
The in vitro rates of tetrathionate formation, oxidation (see above), and reduction (see Sect. 3.4 below) obtained by incubating the sediment samples in specific media and culture conditions are expected to have little or no correspondence with the potential in situ rates of such processes. Rather, the objective of these experiments was to check whether tetrathionate-metabolizing bacteria identified in this sediment system via culture-independent microbiological techniques were alive in situ (potential active state of the tetrathionate metabolizers was subsequently corroborated by pure-culture isolations and metatranscriptome analysis). In the slurry culture experiments it was peculiar to observe that the individual communities present within the sediment depths spanning 2–90, 120–175 cm b.s.f., or 220–275 cm b.s.f. of SSK42/6 exhibited mutually identical rates of tetrathionate oxidation in vitro, despite having dissimilar composition/abundance of chemolithotrophic taxa. This could be explained as follows. When a natural sample is incubated in selective culture media (such as ASWTr), certain specific microbial species present in the sample often outgrow all metabolic competitors by virtue of higher substrate affinity and culture-condition suitability. Consequently, the growth/substrate-utilization phenotype(s) manifested by such enriched consortia are actually contributed to by the selected few rather than the entire community of metabolic equivalents present in the sample (Roy et al., 2016). In light of this issue it seems plausible that distinct sets of chemolithotrophs more adept to growth in ASWTr medium are present across these sediment samples, and it was only their characteristic rates of tetrathionate oxidation which were manifested as the in vitro tetrathionate oxidation rates of the communities.
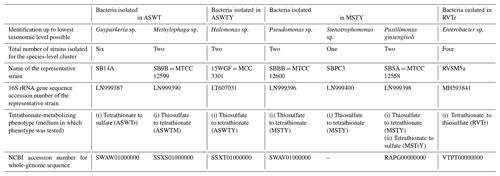
Whatever may be the actual tetrathionate formation/oxidation rates of the sedimentary communities in their natural habitat, growths and activities in the slurry culture experiments illustrated that the tetrathionate-forming and tetrathionate-oxidizing bacteria detected along SSK42/5 and SSK42/6 are alive in situ. In order to further verify whether these metabolic types were metabolically active in their native habitat, whole metatranscriptome of the 275 cm b.s.f. sediment sample of SSK42/6 was sequenced, and the paired end reads were assembled into contigs. The gene catalog obtained via annotation of the assembled contigs was found to encompass homologs of thiosulfate dehydrogenase (tsdA), which is involved in the conversion of thiosulfate to tetrathionate, and sulfate thiol esterase (soxB) and sulfur dehydrogenase (soxC), involved in tetrathionate oxidation (taxonomically, these homologs were ascribed to species of Gramella, Halothiobacillus, and Marinobacter; see Table S20). Furthermore, from 275 cm b.s.f. of SSK42/6, 15 such bacterial strains were isolated (Table 1) that could form tetrathionate from thiosulfate and/or oxidize tetrathionate to sulfate. The 16S rRNA gene-sequence-based taxonomic identification of the isolates clustered them under six species-level entities belonging to six distinct genera. Tetrathionate-forming and/or tetrathionate-oxidizing phenotypes of one representative strain each from the six species-level clusters are shown in Fig. 2. The isolates belonging to the genera Halomonas, Methylophaga, Pseudomonas, and Stenotrophomonas oxidized thiosulfate only up to tetrathionate (Fig. 2a–d); those belonging to Pusillimonas not only formed tetrathionate from thiosulfate but also oxidized tetrathionate to sulfate (Fig. 2e and f); the Guyparkeria isolates did not form tetrathionate from thiosulfate but oxidized tetrathionate to sulfate (Fig. 2g).
3.4 Active tetrathionate-reducing microorganisms in ASOMZ sediment
During anaerobic slurry culture in RVTr medium, microbial communities of all the sediment samples explored in SSK42/5 and SSK42/6 reduced tetrathionate to thiosulfate and/or sulfide at a rate of 0.5–1.5 µmol S per liter slurry per day (Fig. 3e and f, Tables S21 and S22); control sets involving autoclaved sediment samples exhibited no depletion of tetrathionate from the RVTr media. No tetrathionate reductase (ttrABC) or thiosulfate reductase (phsAB or psrA) was detected in the gene catalog obtained via assembly and annotation of the metatranscriptomic data from 275 cm b.s.f. of SSK42/6; but, the same catalog did contain many genes having the highest sequence identities with homologs belonging to the typical tetrathionate-reducer Salmonella. Furthermore, anaerobic enrichment of the 275 cm b.s.f. sediment samples of SSK42/6, followed by isolation of pure cultures, in RVTr medium yielded four such strains that reduced 30–32 mM S tetrathionate to the equivalent amount of thiosulfate over 72 h of anaerobic incubation in RVTr medium (Fig. 2h shows the tetrathionate-reduction kinetics of the representative strain RVSM5a). The 16S rRNA gene-sequence-based taxonomic identification of the four isolates clustered them under a single species-level entity belonging to the genus Enterobacter (Table 1).
3.5 The tetrathionate-metabolizing bacteria isolated from 275 cm b.s.f. of SSK42/6 are widespread across SSK42/5 and SSK42/6
Whole-genome sequencing and annotation were carried out for the three tetrathionate-forming isolates Halomonas sp. MCC 3301, Methylophaga sp. MTCC 12599, and Pseudomonas bauzanensis MTCC 12600; the two tetrathionate-oxidizing isolates Guyparkeria sp. SB14A and Pusillimonas ginsengisoli MTCC 12558; and the tetrathionate-reducing isolate Enterobacter sp. RVSM5a (see Table 1 and Table S23 for GenBank accession numbers and general features of the genomes, respectively). When metagenomic sequence data from the 25 distinct sediment samples of SSK42/5 and SSK42/6 were mapped separately onto each of these six genomes, significant percentages of the metagenomic read sets were found to match sequences from the individual genomes (Figs. 4 and 5, Table S23). In SSK42/5 and SSK42/6, 0.01 %–0.3 % and 0.02 %–19.05 % metagenomic reads from the individual sediment samples mapped onto the different genomes, respectively. Expectedly, the prevalence of reads matching sequences from the new isolates was relatively higher for the metagenomes of SSK42/6 (Fig. 5). Within this core it was highest for 275 cm b.s.f., i.e., the sample site from where all the strains were isolated (Table S23). These data corroborated the significant prevalence of the tetrathionate-metabolizing bacterial strains across the sediment horizons of SSK42/5 and SSK42/6.
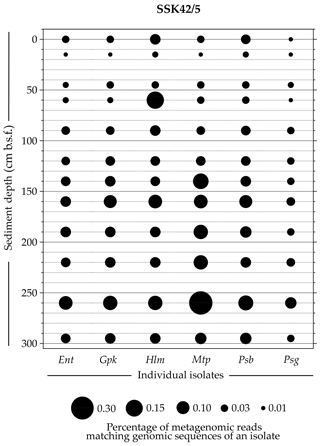
Figure 4Bubble plot showing the percentages of metagenomic reads from individual sediment samples of SSK42/5 that matched genomic sequences of the six tetrathionate-metabolizing bacterial isolates: (Ent) Enterobacter sp. RVSM5a, (Gpk) Guyparkeria sp. SB14A, (Hlm) Halomonas sp. MCC 3301, (Mtp) Methylophaga sp. MTCC 12599, (Psb) Pseudomonas bauzanensis MTCC 12600, and (Psg) Pusillimonas ginsengisoli MTCC 12558. Scales for sediment depth (plotted on y axis) and percentage of metagenomic reads from a given depth matching genomic sequences of an isolate (represented by bubble size) are both linear; the individual isolates are placed along the x axis.
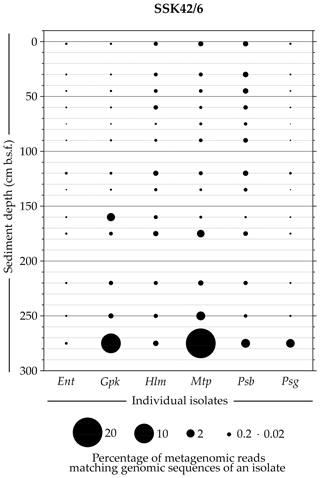
Figure 5Bubble plot showing the percentages of metagenomic reads from individual sediment samples of SSK42/6 that matched genomic sequences of the six tetrathionate-metabolizing bacterial isolates: (Ent) Enterobacter sp. RVSM5a, (Gpk) Guyparkeria sp. SB14A, (Hlm) Halomonas sp. MCC 3301, (Mtp) Methylophaga sp. MTCC 12599, (Psb) Pseudomonas bauzanensis MTCC 12600, and (Psg) Pusillimonas ginsengisoli MTCC 12558. Scales for sediment depth (plotted on y axis) and percentage of metagenomic reads from a given depth matching genomic sequences of an isolate (represented by bubble size) are both linear; the individual isolates are placed along the x axis.
3.6 Correspondence between genomic sequences of the tetrathionate-metabolizing isolates and metatranscriptomic sequences from their habitat
In order to check whether the newly isolated bacterial strains were metabolically active and growing in their natural habitat, the rRNA-sequence-free metatranscriptomic sequence dataset (i.e., all paired end mRNA reads) obtained from the 275 cm b.s.f. sediment sample of SSK42/6 was mapped onto six individual catalogs of tetrathionate-metabolizing and housekeeping genes curated from the genomes of the six different isolates. In this way, the metatranscriptomic sequence dataset was found to encompass reads matching the tsdA genes of the thiosulfate-to-tetrathionate-converting isolates Halomonas sp. MCC 3301 and Pseudomonas bauzanensis MTCC 12600 and the soxB and soxC genes of the tetrathionate-oxidizing isolates Guyparkeria sp. SB14A and Pusillimonas ginsengisoli MTCC 12558 (Table S24). In this context it is noteworthy that the annotated draft genome of the tetrathionate-forming isolate Methylophaga sp. MTCC 12599 did not contain any tsdA gene while that of the tetrathionate-reducing isolate Enterobacter sp. RVSM5a contained no ttrABC and phsABC genes; these could be attributable to either the incompleteness of the two genomes or the presence of novel thiosulfate-to-tetrathionate-converting and tetrathionate-reducing genes in the two bacteria respectively. On the other hand, mapping of the metatranscriptomic sequence dataset onto the individual isolates' gene sets concerning core metabolisms such as (a) genetic information processing (transcription, translation, DNA replication and repair), (b) environmental information processing (ABC transporters, phosphotransferase system, and bacterial secretion system), and (c) cell growth and division (cell cycle) resulted in the concordant matching of up to 1.1 % metatranscriptomic read pairs with the different sets. Table S24 shows the numbers of metatranscriptomic read pairs that matched concordantly with representative housekeeping genes of the six isolates.
3.7 Thiosulfate as the key source of tetrathionate in ASOMZ sediments
Findings of the present culture-independent and culture-dependent investigations showed that microbe-mediated oxidation of thiosulfate to tetrathionate (by members of the bacterial genera included under group A in Fig. 6) could be a key metabolic process in the sulfur cycle of ASOMZ sediments. While genes for the metabolic conversion of thiosulfate to tetrathionate were present across the two cores, so were live microorganisms which could accomplish this process. Furthermore, metatranscriptomic data highlighted the potentially functional (metabolically active) state of thiosulfate-to-tetrathionate-converting bacteria in situ. Tetrathionate formed in this way can have a number of fates: it can either be converted to sulfate (via chemolithotrophic oxidation by members of the genera included under Group B in Fig. 6) or reduced to thiosulfate and/or sulfide (by members of the genera included in Group C of Fig. 6). Whilst culture-independent as well as culture-dependent data supported the feasibility of these metabolic processes in situ, copious hydrogen sulfide present in the pore waters (Fernandes et al., 2018; also see Fig. 3g and h, Table S25) can potentially reduce tetrathionate to thiosulfate and elemental sulfur abiotically (Rowe et al., 2015). Corroborative to these possibilities, ion chromatographic analyses revealed up to 11.1 µM thiosulfate in the pore waters of all the sulfide-containing sediment samples of SSK42/5 and SSK42/6 (Fig. 3g and h, Table S25). These thiosulfate concentration values are consistent with those reported from a number of physicochemically similar sediment horizons across the global ocean (Troelsen and Jørgensen, 1982; Zopfi et al., 2004). Tandem absence of sulfide and thiosulfate in the upper 15 cm b.s.f. of SSK42/6 could be due to potentially high rates of chemolithotrophic conversion of sulfide/thiosulfate to sulfate in situ. Tetrathionate was not found to exist freely in the pore waters of SSK42/5 and SSK42/6; this is apparently attributable to the fact that its buildup to measurable quantities (1 µM for the methods used in this as well as previous studies; Podgorsek and Imhoff, 1999) could be debarred in natural environments due to high reactivity under the mediation of microbes (Kanao et al., 2007; Ghosh and Dam, 2009; Boden et al., 2010; Pyne et al., 2017, 2018) as well as naturally occurring chemical substances such as sulfide (Roy and Trudinger, 1970; Podgorsek and Imhoff, 1999; Schippers et al., 1999; Schippers and Jørgensen, 2001; Zopfi et al., 2004). From that perspective, the geomicrobiological information unearthed in this study illustrates the power of metaomics in discovering such cryptic interfaces between the chemosphere and the biosphere that are almost impossible to decipher from geochemical records alone.
4.1 Biogeochemical perspectives of tetrathionate metabolism in ASOMZ sediments
Concentrations and isotopic ratios of the various chemical constituents of sedimentary solid phases and pore fluids had long remained central to the deciphering of in situ biogeochemical pathways. Significant information on the carbon–sulfur cycles of modern as well as ancient, marine, and lacustrine sediments has been generated in this way. Currently, however, there is an increasing consensus that several questions in biogeochemistry – such as those concerning sulfur compound oxidation/disproportionation, relative importance of simple fatty acid catabolism and anaerobic methane oxidation in sedimentary sulfate reduction, and biogeochemical processes within sulfate–methane transition zones – cannot be answered from preserved geochemical records alone. In recent times a lot of advancement has taken place in our overall understanding of carbon–sulfur cycling in marine systems by virtue of data obtained from metagenomic, metatranscriptomic, and in situ as well as in vitro geomicrobiological experiments. Forensic-level detection power of these approaches in unearthing such cryptic biogeochemical processes that do not get manifested, or leave their imprints, as detectable geological records has been demonstrated in a number of recent papers (e.g., Canfield et al., 2010; Garcia-Robledo et al., 2017; Bhattacharya et al., 2019), which revealed such microbial community functions using metaomics approaches that were almost impossible to detect via geochemical analyses. So far as the present study is concerned, metagenome analysis along SSK42/5 and SSK42/6 revealed tetrathionate-metabolizing potential in bacterial communities present at different depths of the sediment horizons where tetrathionate is not easily detectable. At the same time, metatranscriptome analysis for the deepest sediment sample of SSK42/6 indicated that these tetrathionate-metabolizing communities are potentially active in situ.
Regarding the biogeochemical feasibility of the three tetrathionate-metabolizing processes in the highly O2-constrained, sulfidic sediments of SSK42/5 and SSK42/6 (Fernandes et al., 2018), it is noteworthy that tetrathionate reduction is a mode of anaerobic respiration (Barrett and Clark, 1987; Hensel et al., 1999; Price-Carter et al., 2001), so it has no issue with lack of O2 in the chemical milieu; thiosulfate-to-tetrathionate conversion can also occur under both aerobic and anaerobic conditions (Sorokin et al., 1999). However, most of the sulfur chemolithotrophic bacteria known thus far, including some of those which form tetrathionate from thiosulfate and/or oxidize tetrathionate to sulfate, use O2 as the terminal electron acceptor (Ghosh and Dam, 2009; Wasmund et al., 2017; Patwardhan et al., 2018). Albeit it is peculiar that such microorganisms are alive and active in these apparently O2-depleted sediment horizons, aerobic respiration-related genes such as aa3- ∕ cbb3-type cytochrome-c oxidases (coxABCD ∕ ccoNOPQ) and cytochrome-bd ubiquinol oxidase (cydABX ∕ appX) are abundant in the metagenomes of all the sample sites explored in SSK42/5 and SSK42/6 (Bhattacharya et al., 2019). Furthermore, when the metatranscriptomic sequence dataset obtained for the 275 cm b.s.f. sediment sample of SSK42/6 was assembled and annotated, the resultant contigs were found to encompass several homologs corresponding to genes for aerobic respiration by aa3-type and cbb3-type cytochrome-c oxidases and cytochrome-bd ubiquinol oxidase (Table S26), together with those for other O2-requiring (oxidase enzyme catalyzed) biochemical reactions (Table S27). These data, together with the isolation of obligately aerobic strains from this sedimentary ecosystem (Bhattacharya et al., 2019), indicated that cryptic O2 source(s) supportive of aerobic metabolic processes are likely to be present in situ.
The nature and origin of the oxidants necessary to comprehensively realize the thiosulfate to tetrathionate, and tetrathionate to sulfate, oxidation potential of the ASOMZ sediment microflora remain unresolved. Whilst there are not many biochemical options for in situ O2 production, nor are there high chances of O2 influx to the system (Breuer et al., 2009; Cavan et al., 2017; Jessen et al., 2017), a plausible scenario in the form of cryptic aerobiosis by perchlorate-respiring microorganisms has been envisaged by Bhattacharya et al. (2019). In that study it has been postulated based on metagenomic, metatranscriptomic, and co-culture-based data that perchlorate respirers could be sustaining aerobic life in these sulfidic sediment systems via cryptic O2 supply–consumption partnership similar to the one reported for picocyanobacteria and nitrite oxidizers in the acutely hypoxic waters of the eastern tropical North and South Pacific (Garcia-Robledo et al., 2017). This said, in the absence of measured data for any potential oxidant, including O2 or other oxyanions having possibilities of serving as electron acceptors, only future investigations of biogeochemistry focused on possible pathways of cryptic aerobiosis can resolve the modalities of sulfur compound oxidation in these acutely O2 -scarce environments.
4.2 Trends of thiosulfate/sulfide concentration and prevalence of tetrathionate-metabolizing bacteria, along the sediment cores, implicate tetrathionate as a key intermediate of the sulfur cycle
Along both the sediment cores SSK42/5 and SSK42/6, relative abundance of tetrathionate-forming, tetrathionate-oxidizing, and tetrathionate-reducing bacteria increased with depth (Fig. 1a and b). This could be attributable to the corresponding overall increase in the concentration of thiosulfate (Fig. 3g and h), which plausibly is the key source of biogenic tetrathionate in the sediment. Sulfide concentration also increased with depth, consistent with the increase in thiosulfate concentration (Fig. 3g and h). Sulfide is not only a potential direct source of thiosulfate in marine sediments (Jørgensen, 1990), but it can also be a product of microbial tetrathionate reduction (Barrett and Clark, 1987; Price-Carter et al., 2001). Furthermore, sulfide, when present in sediment cores, can chemically (abiotically) reduce tetrathionate to thiosulfate and elemental sulfur (Roy and Trudinger, 1970; Rowe et al., 2015). In view of these dependencies, depth trends of sulfide concentration are expected to show a certain degree of correlation with trends of thiosulfate concentration.
In both the sediment cores, relative abundance of tetrathionate-oxidizing bacteria increased with depth even as sulfate concentration decreased along the same trajectory (Figs. 1a, b, 3g and h). This indicated that the amounts of sulfate produced from potential tetrathionate oxidation at individual sediment depths were far less than the amounts of sulfate that were reduced to sulfide in situ. Furthermore, in this context, it is noteworthy that in neither of the two cores did rates of in vitro tetrathionate formation or tetrathionate oxidation (in slurry culture experiments) exhibit any parity with the trends of fluctuation observed for the relative abundance of tetrathionate-forming or tetrathionate-oxidizing bacteria (Figs. 1a, b, 3a–d). This could be reflective of the fact that the substrate-utilization rates manifested in the slurry culture experiments actually resulted from the activities of the few chemolithotrophic species/strains that were potentially enriched in the specific media used in these experiments and not that of the whole community present in situ. In contrast, however, the rate of in vitro tetrathionate reduction along both the sediment cores exhibited overall parity with the trends of fluctuation observed for the relative abundance of tetrathionate-reducing bacteria (both parameters showing overall increase with sediment depth; Figs. 1a, b, 3e and f). This suggests that tetrathionate reduction rates observed in the slurry cultures of the sediment samples were attributable to all the tetrathionate-reducing species/strains present in situ, which in turn indicates a general ability of tetrathionate reducers to grow in RVTr medium.
4.3 Potential abiotic drivers of tetrathionate formation in ASOMZ sediments
A number of other studies, based on geochemical experiments and preserved records, have also revealed the occurrence, and complex transformations, of tetrathionate and other intermediate sulfur species in ecologically diverse (but mostly non-sulfidic) environments, including marine sediments. Bak et al. (1993) measured tetrathionate, trithionate, and thiosulfate in diverse natural samples, while Zopfi et al. (2004) used different techniques of analytical geochemistry to track the abiotic as well as biotic (microbe-mediated) transformations of sulfur cycle intermediates, including tetrathionate, in the sediments of the Black Sea and North Sea. Findlay and Kamyshny (2017) envisaged the potential fates and transformation rates of intermediate sulfur species in lacustrine water columns and sediments by introducing and tracking 35S-labeled sulfur compounds in the samples. Furthermore, both Zopfi et al. (2004) and Findlay and Kamyshny (2017), in concurrence with previous reports (Schippers and Jørgensen, 2001), envisaged the in situ oxidation of pyrite (FeS2) by MnO2 as an abiotic source of tetrathionate in the sediments. Such microbe-independent processes are also not improbable in the marine sediments explored in this study as (i) Fe (9232–17234 ppm), Mn (71–172 ppm), and pyrite (0.05 wt %–1.09 wt %) were all detected in the solid phase of SSK42/5 and SSK42/6 samples; (ii) genes for Mn(II)-to-Mn(IV) oxidation were identified in the assembled metagenomes of all the sediment samples explored; (iii) sequences corresponding to manganese oxidase (cotA) and other accessory proteins involved in Mn(II)-to-Mn(IV) oxidation were there in the assembled metatranscriptome analyzed; and (iv) a high percentage of reads ascribed to MnO-depositing bacteria such as Aeromonas, Citrobacter, Enterobacter, Gallionella, Hyphomicrobium, Leptothrix, and Proteus (that reduce Mn+4 to Mn+2 for anaerobic respiration; Ghiorse, 1984), and MnO-to-MnO2-converting bacteria such as Arthrobacter, Oceanospirillum, and Vibrio (Tebo et al., 2005; Sujith and Bharathi, 2011), were detected in all the metagenomes sequenced. Notably, however, manganese concentrations detected in the sediment samples of SSK42/5 and SSK42/6 are orders of magnitude lower than the threshold (>0.2 % (w∕w)) reported previously for FeS2 dissolution (Schippers and Jørgensen, 2001). Thus, to confirm MnO2–FeS2 interaction as a potent source of tetrathionate in these sediment horizons, future studies of geochemistry yielding fine-resolution depth trends for pyrite and MnO2 contents of the sediments are necessary alongside comprehensive data for pore water metal ion concentrations and pyrite-specific (tracer) slurry incubations.
4.4 Biogeochemical underpinnings of the cryptic status of tetrathionate in the sulfur cycle
Previous investigations based on the top few centimeters of Baltic Sea sediments had reported microbial production of tetrathionate and highlighted its role in the sulfur cycle (Podgorsek and Imhoff, 1999). Similar to the present scenario, in that study of sediments collected from coastal locations off Hiddensee (Germany), thiosulfate and tetrathionate were undetectable in the 7–8 mM sulfate-containing and 4–20 µM sulfide-containing native pore water samples tested along a 6 cm b.s.f. sediment profile (detection limits for thiosulfate and tetrathionate, in the methods employed by Podgorsek and Imhoff (1999), were 0.5 and 1.0 µM, respectively). But when an approximately 15 m2 area of the same Baltic Sea location was covered with a plastic sheet for 5 months to artificially construct a stable anoxic condition, up to 5 µM tetrathionate was found to accumulate within the 6 cm b.s.f. sediment profile explored, concomitant with the buildup of 80–280 µM thiosulfate and 320–1200 µM sulfide and depletion of sulfate to a concentration of 0.4 mM within the first 4 cm b.s.f. In the above findings of Podgorsek and Imhoff (1999) it was peculiar that tetrathionate was absent in the native Baltic Sea (off Hiddensee coast) sediment samples containing 4–20 µM sulfide but accumulated to a concentration of 5 µM under induced anoxia involving 320–1200 µM sulfide. Concurrently, in the other Baltic Sea study site (Gotland Basin) explored by Podgorsek and Imhoff (1999), tetrathionate accumulation of up to 5 nmol cm−3 sediment was recorded, during a period of annual anoxia, concomitant with increased concentrations of sulfide, elemental sulfur, and thiosulfate. To explain these data, Podgorsek and Imhoff (1999) invoked the cyclic sequence of reactions that was proposed and demonstrated previously (Roy and Trudinger, 1970; Hansen, 1974; Sorokin et al., 1996) and which involved bacterial oxidation of thiosulfate to tetrathionate on the one hand and chemical reaction between sulfide and tetrathionate, forming elemental sulfur and thiosulfate, on the other. While it was added that accumulation of tetrathionate to high concentrations is debarred in euxinic marine environments owing to its high chemical reactivity with sulfide, and that even low concentrations of thiosulfate and tetrathionate acting as a catalytic couple were sufficient to promote large-scale net oxidation of sulfide to elemental sulfur (Podgorsek and Imhoff, 1999), questions remained as to how tetrathionate accumulation in the Baltic Sea sediments occurred only amidst heightened sulfide buildup. In the context of this apparent biogeochemical paradox, findings of the current study illuminate the centrality of microbe-mediated oxidation/reduction mechanisms (over and above the abiotic role of sulfide) in keeping the tetrathionate pool cryptic (below easily detectable quantities) within the sediment systems.
Sulfur cycling is a crucial component of sediment biogeochemistry within the marine realm. Apart from controlling in situ sulfide–sulfate balance, microbe-mediated processes of the sulfur cycle work in conjunction with those of the carbon and nitrogen cycles to remineralize organic matter sequestered in the seabed and also influence metal deposition/mobilization. Of the various redox species of sulfur, tetrathionate is seldom considered to be an important intermediate of sulfur cycling in marine sediments. The present study of the eastern Arabian Sea sediment system used microbiological data to reveal tetrathionate as a potential key intermediate of the sulfur cycle and identify the biochemical pathways possibly involved in its formation and transformations in situ. Albeit the microbiological drivers and indicators underlying the potential status of tetrathionate as a key junction of sedimentary sulfur cycle were revealed here in the context of an oxygen minimum zone, there was no apparent reason to presume that such processes do not have their equivalents in other geomicrobiologically distinct sediment horizons of the marine realm. The absence of measured data for key in situ geochemical parameters disallowed any direct inference regarding the abiotic mechanisms potentially involved in the formation and transformation of tetrathionate in this sediment system. However, circumstantial evidence was there which could be indicative of pyrite (via abiotic reaction with MnO2) being a source of tetrathionate in the system. Consistent with previous reports from other analogous sediment systems, tetrathionate was not detectable in the sulfidic ASOMZ sediments explored in the current study. In the context of this cryptic nature of tetrathionate in the sulfur cycle, our data revealed the key role of microbial redox metabolisms in preempting the accumulation of this highly reactive polythionate in sediment pore fluids, over and above the known abiotic mechanisms of tetrathionate scavenging by in situ sulfide. However, in the absence of any measured data for O2 or other oxyanions having possibilities of serving as electron acceptors, the biogeochemical modalities of the oxidative half of the tetrathionate cycle remain subject to future investigations of biogeochemistry focused on possible pathways of cryptic aerobiosis. Overall, appreciation of the scope and significance of our molecular and classical microbiology-based forecast of tetrathionate being a central intermediate of the marine sedimentary sulfur cycle remains subject to further biogeochemical substantiation, which among other things should explore the plausible presence of tetrathionate in the system in real time.
All data analysis codes used in this study are in the published domain and have been appropriately cited in the text.
All nucleotide sequence data have been deposited in NCBI Sequence Read Archive (SRA) or GenBank under the BioProject accession number PRJNA309469: (i) the whole metagenome shotgun sequence datasets have the Run accession numbers SRR3646127 through SRR3646132, SRR3646144, SRR3646145, SRR3646147, SRR3646148, SRR3646150 through SRR3646153, SRR3646155 through SRR3646158, and SRR3646160 through SRR3646165, and (ii) the metatranscriptome sequence dataset has the Run accession number SRR7991972. (iii) the whole-genome sequences have the GenBank accession numbers SWAW01000000, SSXS01000000, RAPG00000000.1., SSXT01000000, SWAV01000000, and VTPT00000000.
The Supplement includes an MS Word file named “1_Supplementary Information” and an MS Excel file named “2_Supplementary Dataset”. The supplement related to this article is available online at: https://doi.org/10.5194/bg-17-4611-2020-supplement.
WG conceived the study, designed the experiments, interpreted the results, and wrote the paper. SM, in conjunction with SB, anchored the whole microbiological work, performed the experiments, analyzed the data, and contributed to paper writing. AM led the entire SSK42 mission and all geochemical investigations therein. CR, MJR, JS, and TM performed microbiological experiments and data analyses. SF and AP performed geochemical experiments. All authors read and vetted the manuscript.
The authors declare that they have no conflict of interest.
We thank the director of the CSIR-National Institute of Oceanography for facilitating the geochemical studies and the research cruise SSK42 for acquisition of sediment cores. All the support received from the CSIR-NIO Ship Cell members and the crew members of SSK42 is gratefully acknowledged. Sabyasachi Bhattacharya received a fellowship from Bose Institute. Subhrangshu Mandal received a fellowship from the Department of Science and Technology, GoI. Moidu Jameela Rameez and Chayan Roy received fellowship from the University Grants Commission, GoI. Svetlana Fernandes received a fellowship from the Council of Scientific and Industrial Research, GoI.
Financial support for conducting the microbiological studies was provided by the Bose Institute (via annual intramural faculty grants) and the Earth System Science Organization, Ministry of Earth Sciences (MoES), Government of India (via grant number MoES/36/00IS/Extra/19/2013). MoES also funded the research cruise SSK42 (via GAP2303).
This paper was edited by Tina Treude and reviewed by three anonymous referees.
Alam, M., Pyne, P., Mazumdar, A., Peketi A., and Ghosh, W.: Kinetic enrichment of 34S during proteobacterial thiosulfate oxidation and the conserved role of SoxB in S-S bond breaking, Appl. Environ. Microbiol., 79, 4455–4464, https://doi.org/10.1128/AEM.00956-13, 2013.
Bak, F., Schuhmann, A., and Jansen, K. H.: Determination of tetrathionate and thiosulfate in natural samples and microbial cultures by a new, fast and sensitive ion chromatographic technique, FEMS Microbiol. Ecol., 12, 257–264, https://doi.org/10.1111/j.1574-6941.1993.tb00038.x, 1993.
Barrett, E. L. and Clark, M. A.: Tetrathionate reduction and production of hydrogen sulfide from thiosulfate, Microbiol. Rev., 51, 192–205, 1987.
Baumgartner, L. K., Reid R. P., Dupraz, C., Decho, A. W., Buckley, D. H., Spear, J. R., Przekop, K. M., and Visscher, P. T.: Sulfate reducing bacteria in microbial mats: changing paradigms, new discoveries, Sediment. Geol., 185, 131–145, https://doi.org/10.1016/j.sedgeo.2005.12.008, 2006.
Bhattacharya, S., Roy, C., Mandal, S., Rameez, M. J., Sarkar, J., Fernandes, S., Mapder, T., Alam, M., Roy, R., Mondal, N., Pyne, P., Halder, P. K., Peketi, A., Chakraborty, R., Mazumdar, A., and Ghosh, W.: Metabolically-active obligate aerobes in anoxic (sulfidic) marine sediments, bioRxiv, 728287, https://doi.org/10.1101/728287, 2019.
Boden, R., Kelly, D. P., Murrell, J. C., and Schäfer, H.: Oxidation of dimethylsulfide to tetrathionate by Methylophaga thiooxidans sp. nov.: a new link in the sulfur cycle, Environ. Microbiol., 12, 2688–2699, https://doi.org/10.1111/j.1462-2920.2010.02238.x, 2010.
Boden, R., Scott, K. M., Williams, J., Russel, S., Antonen, K, Rae, A. W., and Hutt, L. P.: An evaluation of Thiomicrospira, Hydrogenovibrio and Thioalkalimicrobium: reclassification of four species of Thiomicrospira to each Thiomicrorhabdus gen. nov. and Hydrogenovibrio, and reclassification of all four species of Thioalkalimicrobium to Thiomicrospira, Int. J. Syst. Evol. Microbiol., 67, 1140–1151, https://doi.org/10.1099/ijsem.0.001855, 2017.
Breuer, E. R., Law, G. T., Woulds, C., Cowie, G. L., Shimmield, G. B., Peppe, O., Schwartz, M., and McKinlay, S.: Sedimentary oxygen consumption and microdistribution at sites across the Arabian Sea oxygen minimum zone (Pakistan margin), Deep-Sea Res. Pt. II, 56, 296–304, https://doi.org/10.1016/j.dsr2.2008.06.010, 2009.
Brito, J. A., Denkmann K., Pereira, I. A., Archer, M., and Dahl, C.: Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum structural and functional insights into thiosulfate oxidation, J. Biol. Chem., 290, 9222–9238, https://doi.org/10.1074/jbc.M114.623397, 2015.
Canfield, D. E., Stewart, F. J., Thamdrup, B., De Brabandere, L., Dalsgaard, T., Delong, E. F., Revsbech, N. P., and Ulloa, O.: A cryptic sulfur cycle in oxygen-minimum–zone waters off the Chilean coast, Science, 330, 375–1378, https://doi.org/10.1126/science.1196889, 2010.
Cavan, E.L., Trimmer, M., Shelley, F., and Sanders, R.: Remineralization of particulate organic carbon in an ocean oxygen minimum zone, Nat. Commun., 8, 1–9, https://doi.org/10.1038/ncomms14847, 2017.
Cline, J. D.: Spectrophotometric determination of hydrogen sulfide in natural waters, Limnol. Oceanogr., 14, 454–458, 1969.
De Jong, G. A. H., Hazeu, W., Bos, P., and Kuenen, G.: Polythionate degradation by tetrathionate hydrolase of Thiobacillus ferrooxidans, Microbiology, 143, 499–504. https://doi.org/10.1099/00221287-143-2-499, 1997.
Denkmann, K., Grein, F., Zigann, R., Siemen, A., Bergmann, J., van Helmont, S., Nicolai, A., Pereira, I. A., and Dahl, C.: Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome, Environ. Microbiol., 14, 2673–2688, https://doi.org/10.1111/j.1462-2920.2012.02820.x, 2012.
Euzéby, J. P.: List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet, Int. J. Syst. Evol. Microbiol., 47, 590–592, https://doi.org/10.1099/00207713-47-2-590, 1997.
Fernandes, S., Mazumdar, A., Bhattacharya, S., Peketi, A., Mapder, T., Roy, R., Carvalho, M. A., Roy, C., Mahalakshmi, P., Silva, R., Rao, P. S., Banik, S., and Ghosh, W.: Enhanced carbon-sulfur cycling in the sediments of Arabian Sea oxygen minimum zone center, Sci. Rep., 8, 8665, https://doi.org/10.1038/s41598-018-27002-2, 2018.
Findlay, A. J. and Kamyshny, A.: Turnover rates of intermediate sulfur species (S, S0, S2O, S4O, SO) in anoxic freshwater and sediments, Front. Microbiol., 8, 2551, https://doi.org/10.3389/fmicb.2017.02551, 2017.
Garcia-Robledo, E., Padilla, C. C., Aldunate, M., Stewart, F. J., Ulloa, O., Paulmier, A., Gregori, G., and Revsbech, N. P.: Cryptic oxygen cycling in anoxic marine zones, P. Natl. Acad. Sci. USA, 114, 8319–8324, https://doi.org/10.1073/pnas.1619844114, 2017.
Ghiorse, W. C.: Biology of iron-and manganese-depositing bacteria, Annu. Rev. Microbiol., 38, 515–550, https://doi.org/10.1146/annurev.mi.38.100184.002503, 1984.
Ghosh, W. and Dam, B.: Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea, FEMS Microbiol. Rev., 33, 999–1043, https://doi.org/10.1111/j.1574-6976.2009.00187.x, 2009, 2009.
Ghosh, W. and Roy, P.: Mesorhizobium thiogangeticum sp. nov., a novel sulfur-oxidizing chemolithoautotroph from rhizosphere soil of an Indian tropical leguminous plant, Int. J. Syst. Evol. Microbiol., 56, 91–97, https://doi.org/10.1099/ijs.0.63967-0, 2006.
Ghosh, W. and Roy, P.: Chemolithoautotrophic oxidation of thiosulfate, tetrathionate and thiocyanate by a novel rhizobacterium belonging to the genus Paracoccus, FEMS Microbiol. Lett., 270, 124–131, https://doi.org/10.1111/j.1574-6968.2007.00670.x, 2007.
Ghosh, W., Bagchi, A., Mandal, S., Dam, B., and Roy, P.: Tetrathiobacter kashmirensis gen. nov., sp. nov., a novel mesophilic, neutrophilic, tetrathionate-oxidizing, facultatively chemolithotrophic betaproteobacterium isolated from soil from a temperate orchard in Jammu and Kashmir, India, Int. J. Syst. Evol. Microbiol., 55, 1779–1787, https://doi.org/10.1099/ijs.0.63595-0, 2005.
Ghosh, W., Mandal, S., and Roy, P.: Paracoccus bengalensis sp. nov., a novel sulfur-oxidizing chemolithoautotroph from the rhizospheric soil of an Indian tropical leguminous plant, Syst. Appl. Microbiol., 29, 396–403, https://doi.org/10.1016/j.syapm.2005.10.004, 2006.
Ghosh, W., Roy, C., Roy, R., Nilawe, P., Mukherjee, A., Haldar, P. K., Chauhan, N. K., Bhattacharya, S., Agarwal, A., George, A., and Pyne, P.: Resilience and receptivity worked in tandem to sustain a geothermal mat community amidst erratic environmental conditions, Sci. Rep., 5, 12179, https://doi.org/10.1038/srep12179, 2015.
Hansen, T. A.: Sulfide als Electronendonor voor Rhodospirillaceae, Proefschrift, University of Groningen, 59–60, 1974.
Hedrich, S. and Johnson, D. B.: Acidithiobacillus ferridurans sp. nov., an acidophilic iron-, sulfur- and hydrogen-metabolizing chemolithotrophic gammaproteobacterium, Int. J. Syst. Evol. Microbiol., 63, 4018–4025, https://doi.org/10.1099/ijs.0.049759-0, 2013.
Hensel, M., Hinsley, A. P., Nikolaus, T., Sawers, G., and Berks, B. C.:The genetic basis of tetrathionate respiration in Salmonella typhimurium, Mol. Microbiol., 32, 275–287, https://doi.org/10.1046/j.1365-2958.1999.01345.x, 1999.
Hensen, D., Sperling, D., Trü per, H. G., Brune, D. C., and Dahl, C.: Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum, Mol. Microbiol., 62, 794–810, https://doi.org/10.1111/j.1365-2958.2006.05408.x, 2006.
Holmkvist, L., Ferdelman, T. G., and Jørgensen, B. B.: A cryptic sulfur cycle driven by iron in the methane zone of marine sediment (Aarhus Bay, Denmark), Geochim. Cosmochim. Ac., 75, 3581–3599, https://doi.org/10.1016/j.gca.2011.03.033, 2011.
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., Rattei, T., Mende, D. R., Sunagawa, S., Kuhn, M., and Jensen, L. J.: eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences, Nucl. Acids Res., 44, D286–D293, https://doi.org/10.1093/nar/gkv1248, 2016.
Hyatt, D., Chen, G. L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J.: Prodigal: prokaryotic gene recognition and translation initiation site identification, BMC Bioinformatics, 11, 119, https://doi.org/10.1186/1471-2105-11-119, 2010.
Jessen, G. L., Lichtschlag, A., Ramette, A., Pantoja, S., Rossel, P. E., Schubert, C. J., Struck, U., and Boetius, A.: Hypoxia causes preservation of labile organic matter and changes seafloor microbial community composition (Black Sea), Sci. Adv., 3, e1601897, https://doi.10.1126/sciadv.1601897, 2017.
Jørgensen, B. B.: A thiosulfate shunt in the sulfur cycle of marine sediments, Science, 249, 152–154, https://doi.org/10.1126/science.249.4965.152, 1990.
Jørgensen, B. B. and Bak, F.: Pathways and microbiology of thiosulfate transformations and sulfate reduction in a marine sediment (Kattegat, Denmark), Appl. Environ. Microbiol., 57, 847–856, 1991.
Jørgensen, B. B., Findlay, A. J., and Pellerin, A.: The Biogeochemical Sulfur Cycle of Marine Sediments, Front. Microbiol., 10, 849, https://doi.org/10.3389/fmicb.2019.00849, 2019.
Kamyshny Jr., A.: Improved cyanolysis protocol for detection of zero-valent sulfur in natural aquatic systems, Limnol. Oceanogr.-Method., 7, 442–448, https://doi.org/10.4319/lom.2009.7.442, 2009.
Kanao, T., Kamimura, K., and Sugio, T.: Identification of a gene encoding a tetrathionate hydrolase in Acidithiobacillus ferrooxidans, J. Biotechnol., 132, 16–22, https://doi.org/10.1016/j.jbiotec.2007.08.030, 2007.
Kanao, T., Kosaka, M., Yoshida, K., Nakayama, H., Tamada,T., Kuroki, R., Yamada, H., Takada, J., and Kamimura, K.: Crystallization and preliminary X-ray diffraction analysis of tetrathionate hydrolase from Acidithiobacillus ferrooxidans, Acta. Cryst. F., 69, 692–694, https://doi.org/10.1107/S1744309113013419, 2013.
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M.: KEGG as a reference resource for gene and protein annotation, Nucl. Acids Res., 44, 457–462, https://doi.org/10.1093/nar/gkv1070, 2016.
Kaprálek, F.: The physiological role of tetrathionate respiration in growing Citrobacter, Microbiology, 71, 133–139, https://doi.org/10.1099/00221287-71-1-133, 1972.
Kelly, D. P. and Wood, A. P.: Synthesis and determination of thiosulfate and polythionates, Methods. Enzymol., 243, 475–501, https://doi.org/10.1016/0076-6879(94)43037-3, 1994.
Kikumoto, M., Nogami, S., Kanao, T., Takada, J., and Kamimura, K.: Tetrathionate-forming thiosulfate dehydrogenase from the acidophilic, chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans, Appl. Environ. Microbiol., 79, 113–120, https://doi.org/10.1128/AEM.02251-12, 2013.
Lahiri, C., Mandal, S., Ghosh, W., Dam, B., and Roy, P.: A novel gene cluster soxSRT is essential for the chemolithotrophic oxidation of thiosulfate and tetrathionate by Pseudaminobacter salicylatoxidans KCT001, Curr. Microbiol., 52, 267–273, https://doi.org/10.1007/s00284-005-0176-x, 2006.
Langmead, B. and Salzberg, S. L.: Fast gapped-read alignment with Bowtie 2, Nat. Methods., 9, 357–359, https://doi.org/10.1038/nmeth.1923, 2012.
Li, D., Liu, C. M., Luo, R., Sadakane, K., and Lam, T. W.: MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph, Bioinformatics., 31, 1674–1676, https://doi.org/10.1093/bioinformatics/btv033, 2015.
Mason, J. and Kelly, D. P.: Thiosulfate oxidation by obligately heterotrophic bacteria, Microb. Ecol., 15, 123–134, https://doi.org/10.1007/BF02011707, 1988.
Meyer, F., Paarmann, D., D'Souza, M., Olson, R., Glass, E. M., Kubal, M., Paczian, T., Rodriguez, A., Stevens, R., Wilke, A., and Wilkening, J.: The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes, BMC Bioinformatics, 9, 386, https://doi.org/10.1186/1471-2105-9-386, 2008.
Müller, F. H., Bandeiras, T. M., Urich, T., Teixeira, M., Gomes, C. M. and Kletzin, A.: Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate: quinone Oxidoreductase, Mol. Microbiol., 53, 1147–1160, https://doi.org/10.1111/j.1365-2958.2004.04193.x, 2004.
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A. A., Korobeynikov, A., Lapidus, A., Prjibelski, A. D., Pyshkin, A., Sirotkin, A., Sirotkin, Y., and Stepanauskas, R.: Assembling single-cell genomes and mini-metagenomes from chimeric MDA products, J. Comput. Biol., 20, 714–737, https://doi.org/10.1089/cmb.2013.0084, 2013.
Oltmann, L. F. and Stouthamer, A. H.: Reduction of tetrathionate, trithionate and thiosulphate, and oxidation of sulphide in Proteus mirabilis, Arch. Microbiol., 105, 135–142, https://doi.org/10.1007/BF00447128, 1975.
Parte, A. C.: LPSN – list of prokaryotic names with standing in nomenclature, Nucl. Acids Res., 42, 613–616, https://doi.org/10.1093/nar/gkt1111, 2013.
Patwardhan, S., Foustoukos, D. I., Giovannelli, D., Yucel, M., and Vetriani, C.: Ecological succession of sulfur-oxidizing Epsilon-and Gammaproteobacteria during colonization of a shallow-water gas vent, Front. Microbiol., 9, 2970, https://doi.org/10.3389/fmicb.2018.02970, 2018.
Podgorsek, L. and Imhoff, J. F.: Tetrathionate production by sulfur oxidizing bacteria and the role of tetrathionate in the sulfur cycle of Baltic Sea sediments, Aquat. Microb. Ecol., 17, 255–265, https://doi.org/10.3354/ame017255, 1999.
Price-Carter, M., Tingey, J., Bobik, T. A., and Roth, J. R.: The Alternative Electron Acceptor Tetrathionate Supports B12 Dependent Anaerobic Growth of Salmonella enteric Serovar Typhimurium on Ethanolamine or 1,2-Propanediol, J. Bacteriol., 183, 2463–2475, https://doi.org/10.1128/JB.183.8.2463-2475.2001, 2001.
Pyne, P., Alam, M., and Ghosh, W.: A novel soxO gene, encoding a glutathione disulfide reductase, is essential for tetrathionate oxidation in Advenella kashmirensis, Microbiol. Res., 205, 1–7, https://doi.org/10.1016/j.micres.2017.08.002, 2017.
Pyne, P., Alam, M., Rameez, M. J., Mandal, S., Sar, A., Mondal, N., Debnath, U., Mathew, B., Misra, A. K., Mandal, A. K., and Ghosh, W.: Homologs from sulfur oxidation (Sox) and methanol dehydrogenation (Xox) enzyme systems collaborate to give rise to a novel pathway of chemolithotrophic tetrathionate oxidation, Mol. Microbiol., 109, 169–191, https://doi.org/10.1111/mmi.13972, 2018.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glöckner, F. O.: The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucl. Acids Res., 41, 590–596, https://doi.org/10.1093/nar/gks1219, 2012.
Quatrini, R., Appia-Ayme, C., Denis, Y., Jedlicki, E., Holmes, D. S., and Bonnefoy, V.: Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans, BMC Genomics, 10, 394, https://doi.org/10.1186/1471-2164-10-394, 2009.
Rameez, M. J., Pyne, P., Mandal, S., Chatterjee, S., Alam, M., Bhattacharya, S., Mondal, N., Sarkar, J., and Ghosh, W.: Two pathways for thiosulfate oxidation in the alphaproteobacterial chemolithotroph Paracoccus thiocyanatus SST, Microbiol. Res., 230, 126345, https://doi.org/10.1016/j.micres.2019.126345, 2019.
Rowe Annette, R., Chellamuthu, P., Lam, B., Okamoto, A., and Nealson, K. H.: Marine sediments microbes capable of electrode oxidation as a surrogate for lithotrophic insoluble substrate metabolism, Front. Microbiol., 5, 784, https://doi.org/10.3389/fmicb.2014.00784, 2015.
Roy, A. B. and Trudinger, P. A.: The Biochemistry of Inorganic Compounds of Sulfur, Cambridge University Press, Cambridge, 7–41, 1970.
Roy, C., Alam, M., Mandal, S., Haldar, P.K., Bhattacharya, S., Mukherjee, T., Roy, R., Rameez, M. J., Misra, A. K., Chakraborty, R. and Nanda, A. K., Mukhopadhyay, S. K., and Ghosh, W.: Global association between thermophilicity and vancomycin susceptibility in bacteria, Front. Microbiol., 7, 412, https://doi.org/10.3389/fmicb.2016.00412, 2016.
Roy, C., Rameez, M. J., Haldar, P. K., Peketi, A., Mondal, N., Bakshi, U., Mapder, T., Pyne, P., Fernandes, S., Bhattacharya, S., Roy, R., Mandal, S., O'Neill, W. K., Mazumdar, A., Mukhopadhyay, S. K., Mukherjee, A., Chakraborty, R., Hallsworth, J. E., and Ghosh, W.: Microbiome and ecology of a hot spring-microbialite system on the Trans-Himalayan Plateau, Sci. Rep., 10, 5917, https://doi.org/10.1038/s41598-020-62797-z, 2020.
Rudnicki, M. D., Elderfield, H., and Spiro, B.: Fractionation of sulfur isotopes during bacterial sulfate reduction in deep ocean sediments at elevated temperatures, Geochim. Cosmochim. Ac., 65, 777–789, https://doi.org/10.1016/S0016-7037(00)00579-2, 2001.
Rzhepishevska, O. I., Valdé s, J., Marcinkeviciene, L., Gallardo, C. A., Meskys, R., Bonnefoy, V., Holmes, D. S., and Dopson, M.: Regulation of a novel Acidithiobacillus caldus gene cluster involved in metabolism of reduced inorganic sulfur compounds, Appl. Environ. Microbiol., 73, 7367–7372, https://doi.org/10.1128/AEM.01497-07, 2007.
Schippers, A. and Jørgensen, B. B.: Oxidation of pyrite and iron sulfide by manganese dioxide in marine sediments, Geochim. Cosmochim. Ac., 65, 915–922, https://doi.org/10.1016/S0016-7037(00)00589-5, 2001.
Schippers, A., Rohwerder, T., and Sand, W.: Intermediary sulfur compounds in pyrite oxidation: implications for bioleaching and biodepyritization of coal, Appl. Microbiol. Biotechnol., 52, 104–110, https://doi.org/10.1007/s002530051495, 1999.
Sorokin, D.: Oxidation of inorganic sulfur compounds by obligatory organotrophic bacteria, Mikrobiologiia, 72, 725–739, 2003.
Sorokin, D. Y., Robertson, L. A., and Kuenen, J. G.: Sulfur cycling in Catenococcus thiocyclus, FEMS Mlcrobiol. Ecol., 19, 117–125, https://doi.org/10.1111/j.1574-6941.1996.tb00204.x, 1996.
Sorokin, D. Y., Teske, A., Robertson, L. A., and Kuenen, J. G.: Anaerobic oxidation of thiosulfate to tetrathionate by obligately heterotrophic bacteria, belonging to the Pseudomonas stutzeri group, FEMS Microbiol. Ecol., 30, 113–123, https://doi.org/10.1111/j.1574-6941.1999.tb00640.x, 1999.
Stoffels, L., Krehenbrink, M., Berks, B. C., and Unden, G.: Thiosulfate Reduction in Salmonella enterica Is driven by the proton motive force, Appl. Environ. Microbiol., 194, 475–485. https://doi.org/10.1128/JB.06014-11, 2011.
Sujith, P. P. and Bharathi, P. L.: Manganese oxidation by bacteria: biogeochemical aspects, Molecular Biomineralization, 52, 49–76, 2011.
Tebo, B. M., Johnson, H. A., McCarthy, J. K., and Templeton, A. S.: Geomicrobiology of manganese (II) oxidation, Trends Microbiol., 13, 421–428, https://doi.org/10.1016/j.tim.2005.07.009, 2005.
Thamdrup, B., Finster, K., Fossing, H., Hansen, J. W., and Jørgensen, B. B.: Thiosulfate and sulfite distributions in porewater of marine sediments related to manganese, iron, and sulfur geochemistry, Geochim. Cosmochim. Ac., 58, 67–73, 1994.
Tostevin, R., Turchyn, A. V., Farquhar, J., Johnston, D. T., Eldridge, D. L., Bishop, J. K., and McIlvin, M.: Multiple sulfur isotope constraints on the modern sulfur cycle, Earth Planet. Sc. Lett., 396, 14–21, https://doi.org/10.1016/j.epsl.2014.03.057, 2014.
Tringe, S. G., von Mering, C., Kobayashi, A., Salamov, A. A., Chen, K., Chang, H. W., Podar, M., Short, J. M., Mathur, E. J., Detter, J. C., Bork, P., Hugenholtz, P., and Rubin, E. M.: Comparative metagenomics of microbial communities, Science, 308, 554–557, https://doi.org/10.1126/science.1107851, 2005.
Troelsen, H. and Jørgensen, B. B.: Seasonal dynamics of elemental sulfur in two coastal sediments, Estuar. Coast Shelf S., 15, 255–266, https://doi.org/10.1016/0272-7714(82)90062-2, 1982.
Tuttle, J. H.: Thiosulfate oxidation and tetrathionate reduction by intact cells of marine pseudomonad strain 16B, Appl. Environ. Microbiol., 39, 1159–1166, 1980.
van Zyl, L. J., van Munster, J. M., and Rawlings, D. E.: Construction of arsB and tetH mutants of the sulfur-oxidizing bacterium Acidithiobacillus caldus by marker exchange, Appl. Environ. Microbiol., 74, 5686–5694, https://doi.org/10.1128/AEM.01235-08, 2008.
Vassiliadis, P.: The Rappaport-Vassiliadis (RV) enrichment medium for the isolation of salmonellas: An overview, J. Appl. Bacteriol., 54, 69–76, https://doi.org/10.1111/j.1365-2672.1983.tb01302.x, 1983.
Vishniac, W. and Santer, M.: The Thiobacilli, Bacteriol. Rev., 21, 195–213, 1957.
Wasmund, K., Muß mann, M., and Loy, A.: The life sulfuric: microbial ecology of sulfur cycling in marine sediments, Environ. Microbiol. Rep., 9, 323–344, https://doi.org/10.1111/1758-2229.12538, 2017.
Watsuji, T. O., Had, E., Miyazaki, M., Ichimura, M., and Takai, K.: Thiomicrospira hydrogeniphila sp. nov., an aerobic, hydrogen-and sulfur-oxidizing chemolithoautotroph isolated from a seawater tank containing a block of beef tallow, Int. J. Syst. Evol. Microbiol., 66, 3688–3693, https://doi.org/10.1099/ijsem.0.001250, 2016.
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J.: 16S ribosomal DNA amplification for phylogenetic study, J. Bacteriol., 173, 697–703, https://doi.org/10.1128/jb.173.2.697-703.1991, 1991.
Zhu, W., Lomsadze, A., and Borodovsky, M.: Ab initio gene identification in metagenomic sequences, Nucl. Acids Res., 38, e132, https://doi.org/10.1093/nar/gkq275, 2010.
Zopfi, J., Ferdelman, T. G., and Fossing, H.: Distribution and fate of sulfur intermediates – sulfite, tetrathionate, thiosulfate, and elemental sulfur – in marine sediments, in: Sulfur Biogeochemistry – Past and Present, edited by: Amend, J. P., Edwards, K. J., and Lyons, T. W., Geological Society of America Special Paper 379, Boulder, Colorado, 97–116, 2004.