the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Protists and collembolans alter microbial community composition, C dynamics and soil aggregation in simplified consumer–prey systems
Matthias C. Rillig
Andrea Carminati
Alexandre Jousset
Stefan Scheu
Microbes play an essential role in soil functioning including biogeochemical cycling and soil aggregate formation. Yet, a major challenge is to link microbes to higher trophic levels and assess consequences for soil functioning. Here, we aimed to assess how microbial consumers modify microbial community composition (PLFA markers), as well as C dynamics (microbial C use, SOC concentration and CO2 emission) and soil aggregation. We rebuilt two simplified soil consumer–prey systems: a bacterial-based system comprising amoebae (Acanthamoeba castellanii) feeding on a microbial community dominated by the free-living bacterium Pseudomonas fluorescens and a fungal-based system comprising collembolans (Heteromurus nitidus) grazing on a microbial community dominated by the saprotrophic fungus Chaetomium globosum. The amoeba A. castellanii did not affect microbial biomass and composition, but it enhanced the formation of soil aggregates and tended to reduce their stability. Presumably, the dominance of P. fluorescens, able to produce antibiotic toxins in response to the attack by A. castellanii, was the main cause of the unchanged microbial community composition, and the release of bacterial extracellular compounds, such as long-chained polymeric substances or proteases, in reaction to predation was responsible for the changes in soil aggregation as a side effect. In the fungal system, collembolans significantly modified microbial community composition via consumptive and non-consumptive effects including the transport of microbes on the body surface. As expected, fungal biomass promoted soil aggregation and was reduced in the presence of H. nitidus. Remarkably, we also found an unexpected contribution of changes in bacterial community composition to soil aggregation. In both the bacterial and fungal systems, bacterial and fungal communities mainly consumed C from soil organic matter (rather than the litter added). Increased fungal biomass was associated with an increased capture of C from added litter, and the presence of collembolans levelled off this effect. Neither amoebae nor collembolans altered SOC concentrations and CO2 production. Overall, the results demonstrated that trophic interactions are important for achieving a mechanistic understanding of biological contributions to soil aggregation and may occur without major changes in C dynamics and with or without changes in the composition of the microbial community.
- Article
(1196 KB) - Full-text XML
- BibTeX
- EndNote
Soil microbes are essential for soil functioning (Bardgett and Van der Putten, 2014). As major primary decomposers of organic matter, soil microbial communities play a crucial role in soil biogeochemical cycles (Bradford et al., 2013; Glassman et al., 2018). Microbes are key determinants of soil structure as well, notably soil aggregation (Lehmann et al., 2017). Soil aggregates are “soil-specific entities built from mineral and organic compounds with stronger bonds between building blocks than with neighbouring particles” (Yudina and Kuzyakov, 2019). Soil aggregate properties, in particular their stability, allow for the prediction of large-scale soil properties such as soil erosion (Barthes and Roose, 2002). The role of soil microbes on biogeochemical cycles and soil structure has been intensively studied in the past decades (Wieder et al., 2013; Lehmann et al., 2017; Malik et al., 2018). Yet, a major challenge is to link the soil microbial level to higher trophic levels (Grandy et al., 2016). To do so, it is required to investigate how soil microbial grazers and predators impact the composition of the soil microbial community (Thakur and Geisen, 2019) and assess consequences for soil functioning.
Soil microbes form highly diverse and interconnected communities (Delgado-Baquerizo et al., 2016; de Vries et al., 2018; Wagg et al., 2019), with bacteria and fungi as key players. In the last decade, increasing effort has been undertaken to decipher biotic interactions within microbial communities (Velicer and Vos, 2009; Schmidt et al., 2015; Deveau et al., 2018) and ultimately link microbial diversity and connectivity to soil biogeochemical cycles (Wagg et al., 2019). Bacteria and fungi interact in many ways, with numerous examples of competition and facilitation (Tiunov and Scheu, 2005; Velicer and Vos, 2009; Worrich et al., 2017; Bahram et al., 2018), often resulting in complementary actions favouring biogeochemical cycles (Deveau et al., 2018; Wagg et al., 2019). However, we know little about how the diversity and interactions within soil microbial communities drive soil structure. So far, general mechanistic underlying effects of bacteria and fungi on soil aggregation have been described using isolated microbial strains, generally pinpointing positive effects (Lehmann et al., 2017). Bacteria influence soil aggregation mainly through the production of extracellular polymeric substances (EPSs), which adsorb onto mineral surfaces, increase the viscosity of the soil solution and enhance soil particle cohesion (Chenu, 1993; Sandhya and Ali, 2014; Liu et al., 2013). Filamentous fungi promote soil aggregation in various ways including (i) the enmeshment of soil particles (Degens et al., 1996; Tisdall et al., 1997; Baldock, 2002); (ii) the secretion of polymeric substances (Chenu, 1989; Caesar-TonThat and Cochran, 2000; Daynes et al., 2012), enhancing mineral particle cohesion similar to bacterial EPSs; and (iii) the release of a variety of molecules, such as hydrophobins (Linder et al., 2005; Zheng et al., 2016) and phenolic acids (Caesar-Tonthat and Cochran, 2000), reducing wettability and preventing soil aggregates from collapsing when the soil is rewetted. However, effects vary between microbial strains. For example, good aggregators were reported for species of the bacterial genera Bacillus and Pseudomonas (Caesar-TonThat et al., 2007, 2014) and of the fungal genera Chaetomium, Mucor and Aspergillus (Swaby, 1949). Enhanced aggregative ability was related to higher lipid and protein content in bacterial EPSs (Liu et al., 2013) and to higher density of mycelial growth (Lehmann et al., 2020). Such developments with a microbial focus bring us closer to understanding how the diversity of microbes drive biogeochemical cycles and soil aggregation, but they fail to connect how these processes are modified in the presence of higher trophic levels.
Soil microbial communities have long been thought to be mainly structured by abiotic filters (Griffiths et al., 2011) and plant inputs (bottom-up regulation; Myers et al., 2001), whereas the importance of higher trophic levels has so far only been seen as a major but understudied factor (Thakur and Geisen, 2019) of crucial importance for C cycling (Crowther et al., 2015; Filser et al., 2016; Grandy et al., 2016; Morriën et al., 2017). Several soil organisms can consume bacteria and fungi, including protists, nematodes and collembolans. These consumers often modify the biomass and the composition of the microbial community (Rønn et al., 2002; Djigal et al., 2004; Rosenberg et al., 2009; Perez et al., 2013; Jiang et al., 2017; Geisen et al., 2018; Coulibaly et al., 2019). Such modifications are thought to result directly from consumptive effects, i.e. selective feeding, or indirectly from changes in the competition between microbes due to the consumption of certain microbial strains (Thakur and Geisen, 2019). However, other non-consumptive effects are also at stake (Bradford, 2016) as indicated by experimental evidence. For example, the release of N by protists and nematodes (Griffiths and Bardgett, 1997) and the excretion of urine or faeces by microarthropods (Cragg and Bardgett, 2001; Milcu et al., 2006), as well as their necromass (Coleman et al., 2002), modify the quality of organic matter processed by soil microorganisms, with expected effects on their biomass and composition. The comminution of plant debris (Lussenhop, 1992) as well as the transport of microbes on the body of soil microarthropods (Griffiths and Bardgett, 1997; Bardgett et al., 1998; Gormsen et al., 2004) is also assumed to contribute to non-consumptive effects of microarthropods on soil microbial communities. Further, consumers of soil microorganisms can modify microbial activity and physiology, such as the production of bacterial EPSs (Matz and Kjelleberg, 2005; Queck et al., 2006) and antibiotic compounds (Jousset and Bonkowski, 2010). Variations in enzymatic activity of wood-decomposing fungi in the presence of microbivores have also been reported (Crowther et al., 2015). Overall, the influence of trophic interactions on the structure and activity of soil microbial communities is an active research topic (Jiang et al., 2017; Thakur and Geisen, 2019; Lucas et al., 2020). By contrast, the integration of higher trophic levels into soil aggregation studies lags far behind, with only one study investigating how collembolans modify the effect of fungi on soil aggregation (Siddiky et al., 2012) and no studies targeting effects of protists or nematodes. However, given the importance of microbes for soil aggregation, modifications in microbial community composition or activity due to microbial consumers is expected to play a significant role.
Here, we aimed to investigate how microbial consumers modify microbial community composition and how this impacts C dynamics and soil aggregation. We focused on two simplified consumer–prey systems: a bacterial- and a fungal-based system, representing the two main, and often interconnected, channels of C fluxes in soil (de Vries and Caruso, 2016). The bacterial-based system comprised the free-living amoeba Acanthamoeba castellanii feeding on a microbial community dominated by bacteria (Pseudomonas fluorescens sensu lato), and the fungal-based system comprised the collembolan species Heteromurus nitidus grazing on a microbial community dominated by saprotrophic fungi (Chaetomium globosum). We conducted a microcosm experiment lasting for 6 weeks and assessed changes in microbial community composition, C dynamics and soil aggregation.
2.1 Experimental design and microcosm incubation
We reconstructed simplified bacterial-based and fungal-based prey–consumer systems in soil microcosms. The soil was autoclaved (2 h at 121 ∘C) to reduce and level off initial microbial abundance and diversity and was reinoculated with single common soil bacterial or saprotrophic fungal strains, i.e. P. fluorescens and C. globosum, to set up a bacterial- and a fungal-dominated system, respectively. The resulting microbial community was established from the introduced species as well as species that survived autoclaving and those introduced together with the consumers, notably the microbes used to culture the protists and collembolans in the laboratory (Escherichia coli and Saccharomyces cerevisiae, respectively). These microbial communities formed the prey of the consumers added. The consumers, i.e. the protist A. castellanii and collembolan H. nitidus, were added 2 weeks after the microbial inoculation to allow the microbial communities to establish. We accounted for the co-addition of microorganisms with the consumers to microcosms by adding E. coli and a microbial wash of the collembolans, respectively. In addition, we set up a zero control in which only autoclaved tap water was added (for full experimental design see Table 1). Overall, our experimental design allowed for the disentanglement of the effects of the addition of consumers on the microbial community due to trophic interactions (consumptive and non-consumptive effects) from those related to the co-addition of microbes with the consumers. Furthermore, we were able to assess how preceding steps of microbial community establishment influenced soil microbial community composition. We linked the changes in microbial community composition due to the preceding inoculation steps and the addition of consumers to C dynamics and soil aggregation.
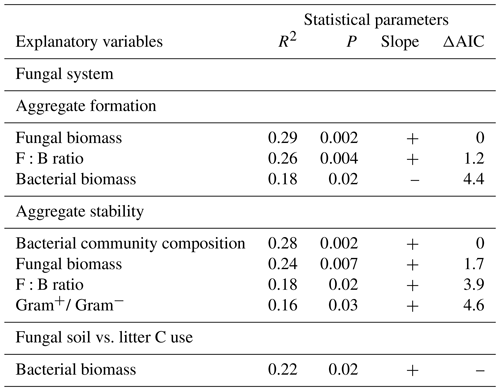
Table 1Details on the experimental design. X indicates which soil organisms have been added to the systems.
In the bacterial-based system, we first added P. fluorescens (2×109 cfu) and 2 weeks later A. castellanii (6×108 individuals) to half of the jars inoculated with P. fluorescens. In addition, the mucilage-producing Gram− bacterium E. coli (6×108 individuals; Danese et al., 2000), used to culture the amoebae, was added to the microcosms that did not receive amoebae (for full experimental design see Table 1). Bacteria and amoebae were added as suspension in autoclaved tap water. In the fungal-based system, we first added the saprotrophic fungus C. globosum (4 cm3 of colonized LB agar) and 2 weeks later the collembolan species H. nitidus (30 individuals per microcosm) to half of the jars inoculated with C. globosum. In addition, we added a microbial wash of collembolans (240 individuals) to the microcosms which did not receive the collembolans (for full experimental design see Table 1). The microbial wash as well as C. globosum was added as suspension in autoclaved tap water. H. nitidus was added using an entomological exhauster. The same amount of medium (autoclaved tap water with or within smashed LB agar) was added to the respective control treatments.
The bacterium P. fluorescens and the fungus C. globosum were chosen as they are abundant in soils (Dubuis et al., 2007; Domsch et al., 1993) and beneficially affect soil aggregation (Swaby, 1949; Caesar-TonThat et al., 2014; Tisdall et al., 2012). The amoeba species A. castellanii was selected as a representative and ubiquitous soil-dwelling amoeba able to consume a broad range of microorganisms and for its ability to produce proteases (Serrano-Luna et al., 2006; Weekers et al., 1995) able to perforate bacterial biofilms essential for soil aggregation. The amoeba species A. castellanii was shown to feed on our model strain P. fluorescens (Jousset et al., 2009) but prefers less toxic strains. We thus expected that the amoebae will feed not only on P. fluorescens but also on the other bacterial strains of the system (remaining microbial background and E. coli). The collembolan species H. nitidus was chosen because of its abundance in European temperate soils (Hopkin, 1997) and as it has been shown to preferentially feed on C. globosum (Pollierer et al., 2019); feeding on C. globosum it is able to survive and reproduce. H. nitidus also feeds on bacteria including P. fluorescens but cannot reproduce when feeding only on this bacterial species (Pollierer et al., 2019). In our system, we thus expected that H. nitidus would mainly feed on C. globosum but would also ingest bacteria present in the microcosms.
The soil used in the experiment comprised a mixture of sand (59.7 %) and agricultural soil (39.8 %), collected from a wheat agricultural experimental field of the University of Göttingen, managed under conventional tillage and located in the metropolitan area of Göttingen. The agricultural soil was crushed through a 1 mm sieve to destroy larger aggregates and then mixed with sand. The properties of the soil mixture were 6.0 % clay, 30.8 % silt, 13.7 % fine sand (63–200 µm), 41.4 % medium sand (200–630 µm), 8.1 % coarse sand (630–2000 µm), 4.5 % CaCO3 and 0.36 % organic carbon (analyses were conducted by LUFA Speyer, Germany, following the methods VDLUFA I, C2.2.1, 2012, and DIN ISO 10694:1996-08, 1996). A total of 120 g dry weight of soil was added per microcosm.
To analyse whether the use of soil C was influenced by consumer–prey interactions, we added a source of particulate organic matter, i.e. dried chopped litter (micrometre- to millimetre-sized pieces; 0.4 %) to the soil mixture. The litter was composed of a mixture of maize (85.3 %) and wheat (14.7 %) litter. Litter of maize as a C4 plant was chosen to allow tracing the incorporation of litter carbon into microorganisms. The organic carbon present in the agricultural soil was mainly derived from the residues of wheat as a C3 plant.
The microcosms consisted of glass jars (7.5 cm diameter and 10 cm high) containing the mixture of soil, sand and litter (120 g dry weight of soil mixture with litter) and were covered by non-sealed lids, allowing gas and water exchange but limiting potential contamination in our semi-sterile experimental design. Microcosms were incubated in darkness in a climate chamber at 20 ∘C for a total of 6 weeks (July–August 2017), with the first 2 weeks with microbial prey only. Soil water-holding capacity was adjusted weekly to 60 % of the maximum by adding autoclaved tap water under sterile conditions.
2.2 Microbial community composition
Changes in soil microbial abundance and composition were quantified by phospholipid fatty acid (PLFA) analysis. Lipids were extracted from fresh soil equivalent to 3.5 g dry weight. The soil was frozen at −20 ∘C after the experiment until further use according to the protocol of Buyer and Sasser (2012). PLFAs were measured and identified as described in Pollierer et al. (2015) using a gas chromatograph (GC; Clarus 500 with autosampler, PerkinElmer, USA). The mass (nmol g−1 dry soil) of all extracted and identified PLFAs was used as a measure of microbial biomass. The PLFA 18:2ω6,9 was used as a fungal biomarker, and eight PLFAs were used as bacterial biomarkers: i15:0, a15:0, i16:0; i17:0 (Gram+ bacteria), cy17:0, 18:1ω7 (Gram− bacteria), 16:1ω7 and 15:0 (general bacterial marker; Frostegård and Bååth, 1996; Contosta et al., 2015).
2.3 Microbial C use, CO2 emission and SOC concentration
The use of C originating from soil (organic C originally present in the wheat agricultural soil) vs. litter (added chopped litter mainly derived from maize; see below) by soil microbes was assessed by tracing the 13C signal of the litter and soil in the microbial PLFA markers. In particular, we investigated the 13C of the fungal PLFA marker and of the eight bacterial PLFA markers, which allowed us to trace the C source of fungal and bacterial communities. The 13C∕12C ratios of the PLFAs were measured using a trace gas chromatograph (GC; Thermo Finnigan, Bremen, Germany), equipped with a DB-5 and DB-1 column (30 and 15 m, both 0.25 µm i.d., Agilent), and coupled via a GP interface to a Delta Plus mass spectrometer (Thermo Finnigan, Bremen, Germany). The temperature program was run according to the following steps: 1 min at 80 ∘C, an increase to 170 ∘C at a rate of 10 ∘C min−1, an increase to 192 ∘C at a rate of 0.7 ∘C min−1, 4 min at 192 ∘C, an increase to 200 ∘C at a rate of 0.7 ∘C min−1, an increase to 210 ∘C at a rate of 1.5 ∘C min−1, a final increase to 300 ∘C at a rate of 10 ∘C min−1 and a final step at 300 ∘C for 10 min. Helium was used as a carrier gas for injections (250 ∘C). PLFAs were identified by comparison of their chromatographic retention times with those of standard mixtures composed of 37 different FAMEs (fatty acid methyl esters; Sigma-Aldrich, St Louis, USA) ranging from C11 to C24 and 26 BAMEs (bacterial fatty acid methyl esters; Sigma-Aldrich, St Louis, USA). Isotope ratios were expressed vs. the Vienna Pee Dee Belemnite standard (V-PDB) as δ13C [‰] = ((13C∕12C)sample∕ (13C∕12C)standard−1) × 1000. To compare 13C∕12C ratios of microbial PLFAs to the 13C∕12C ratios of C sources (organic soil carbon and added litter), we also measured the 13C∕12C ratios of the agricultural soil used in the microcosms and of the added litter using a Delta Plus mass spectrometer (Thermo Finnigan, Bremen, Germany). Aliquots of ca. 2 g of soil and litter samples were dried (70 ∘C, 24 h), milled (45 s, frequency 25 s−1; MM200, Retsch GmbH, Haan, Germany) and placed in a desiccator for 48 h. Aliquots of ca. 25 mg of soil and 0.7 mg of litter were analysed (eight replicates each). The difference between the average isotopic signature of the soil (δ13C ‰) and the litter (δ13C ‰) covered an amplitude of 13.45 ‰ and set the full range of isotopic variation (100 %) used to trace the C origin. Using these values as endpoints the proportion of C of soil origin in fungal and bacterial PLFA marker fatty acids (FAs; x) was calculated as follows: % Csoil(x)= [(δ13C] with δ13Cx the δ13C signature of the respective marker FA. For bacterial markers, we used average δ13C values weighted by the relative proportion of each bacterial marker considered. Using the mean δ13C value of litter in this calculation assumes that carbon from maize and wheat litter in the litter mixture was used indiscriminately, which we assume to be justified considering the short period of time when predominantly labile-litter compounds are used. Further, considering that the litter mixture comprised predominantly maize litter, differential incorporation of carbon from wheat is likely to affect the calculations only slightly. As local hotspots of soil can show lower δ13C signal than the average δ13C signature of homogenized soil, the calculation may result in an estimated percentage of C of soil origin higher than 100 %.
To monitor the activity of soil organisms (soil microbiota and soil animals), CO2 production was measured twice during the incubation period by titration with HCl of 2 mL KOH placed in the jar for 48 h (Fig. 4), following the protocol of Macfadyen (1970). Finally, to estimate whether the inoculation treatments modified the soil organic carbon (SOC) content over the incubation period, SOC concentration of aliquots of soil from the treatments was measured after the end of the incubation period (Fig. 3). Aliquots (ca. 500 mg) of dried (105 ∘C, 24 h) and milled (45 s, frequency 25 s−1; MM200, Retsch GmbH, Haan, Germany) soil samples were placed at 600 ∘C for 2 h to remove the organic carbon by combustion. Aliquots of 20 mg of milled soil, unburned and burned at 600 ∘C, were used to measure their carbon content (Vario EL, Elementar, Hanau, Germany), which represented the inorganic and organic carbon, respectively. The soil organic carbon content was obtained by the difference in the concentrations between the total organic carbon and the inorganic carbon.
2.4 Soil aggregate properties
Soil aggregate formation was assessed by soil dry sieving (six helicoidal movements; 30 cm amplitude) of air-dried (ca. 22 ∘C; 7 d) soil samples using the following sieves: 10, 5, 3 and 2 mm and 250 and 50 µm, resulting in seven diameter classes of aggregates. As soil was crushed through a 1 mm sieve during soil preparation for microcosm incubation, aggregates larger than 1 mm indicate enhanced cohesion acquired during incubation. The term “aggregate formation” is used to describe the quantity and size of aggregates obtained by dry sieving after incubation. Soil aggregate stability was measured following ISO/FDIS109030:2012(E) (2012) described in Le Bissonnais (1996) and Le Bissonnais and Arrouays (1997). Briefly, 8 g of dried (40 ∘C, 24 h) soil aggregates (3–5 mm) were gently rewetted by capillarity for 5 min on a buffer paper placed on a saturated sponge. Aggregates were then transferred into ethanol, and aggregates >50 µm were retrieved by sieving in ethanol. The aggregate fraction >50 µm was dried at 40 ∘C for 24 h and sieved using six sieves (2, 1 mm; 500, 200, 100, 50 µm), resulting in seven diameter classes of aggregates. The mean weight diameter (MWD) was calculated as the average diameter of aggregates weighted by the mass proportion of aggregates within each fraction. The MWD of the dry distribution of aggregates, indicating aggregate formation, is denoted by MWDdd. The MWD obtained after dispersion of aggregate by gently rewetting, indicating aggregate stability, is denoted by MWDas.
2.5 Data analyses
Differences between treatments in the concentration of microbial PLFA markers, microbial C use, CO2 emissions, SOC concentrations and degree of soil aggregation were inspected using generalized least-square (GLS) models, followed by ANOVA and post hoc Tukey tests. Differences in variance between treatments were accounted for in the GLS models. The effects of treatments on bacterial PLFA composition (eight specific markers) were investigated using non-metric multidimensional scaling (NMDS), followed by discriminant function analysis (DFA). Overall differences in bacterial PLFA composition within the bacterial and fungal system were first analysed by MANOVA. Pairwise differences between treatments were further inspected using Mahalanobis distances. Changes in soil bacterial community composition were captured by the scores (Axis 1) of principal component analyses (PCA) of the eight bacterial PLFA markers for further analyses. Axis 1 of the PCA on bacterial markers of the bacterial and fungal system explained 39.3 % and 37.6 % of total variability, respectively. Pairwise correlations among (i) descriptors of soil microbial community and (ii) soil aggregation and C dynamics were tested using Spearman's correlation coefficients (ρ). The best explanatory variables of soil aggregation, microbial C use, SOC concentrations and CO2 production were identified among the descriptors of the microbial community composition using simple linear generalized regression models (GLMs) and selected according to their R2 and ΔAIC (AIC is the Akaike information criterion). Only significant models are displayed. All statistical analyses were run separately for the bacterial and fungal systems. Data provided in the text represent means ± standard deviation. All statistical analyses were conducted in R version 3.6.1 (R Development Core Team, 2008).
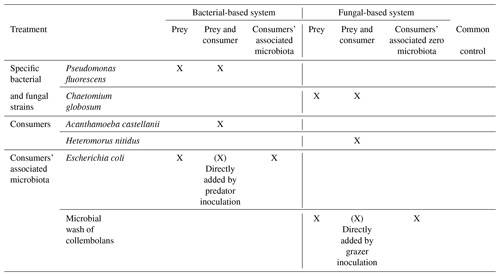
Figure 1Effect of bacterial and fungal consumer–prey inoculations on microbial biomass and composition. (a) Microbial biomass, (b) fungal biomass, (c) bacterial biomass, (d) fungal : bacterial PLFA ratio (F:B), proportion of (e) Gram+ bacteria and (f) Gram− bacteria, and (g) ratio between Gram+ and Gram− bacteria. Differences between treatments were analysed separately for the bacterial and fungal systems using GLS models followed by ANOVA and post hoc Tukey tests. Letters (blue lowercase for bacterial system and brown capital for fungal system) indicate significant differences between means according to Tukey tests. Grey background indicates control treatment of bacterial and fungal systems.
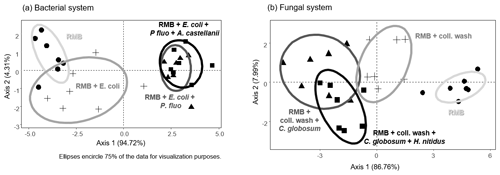
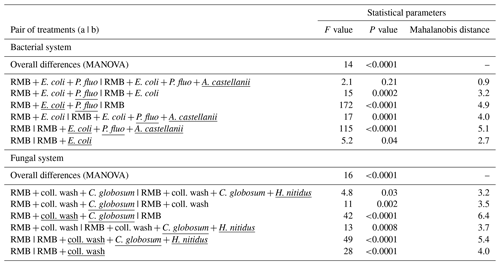
Figure 2Effect of bacterial and fungal consumer–prey inoculations on bacterial community composition. Discriminant function analysis of the bacterial PLFA markers in the (a) bacterial and (b) fungal systems. Overall differences in bacterial PLFA composition (eight markers) within the bacterial and fungal systems were analysed first by NMDS followed by MANOVA. Pairwise differences between means were further investigated using Mahalanobis distances. For details of the results of the statistical tests see Table 2. RMB: remaining microbial background; coll. wash: collembolan wash; P. fluo: P. fluorescens. Ellipses encircle 75 % of the data for visualization purposes.
3.1 Microbial community composition as affected by consumer–prey system
Overall, consumer–prey inoculation had little influence on soil microbial biomass but modified the composition of microbial communities (Fig. 1). In the bacterial system, the predator A. castellanii did not significantly affect the total microbial biomass, fungal-to-bacterial (F:B) PLFA ratio, bacterial community composition, and the proportions of Gram+ and Gram− bacteria (Figs. 1, 2; Table 2). In the fungal system, the grazer H. nitidus significantly decreased fungal biomass, as well as the F:B ratio, but did not induce changes in microbial and bacterial biomass or in the proportion of Gram+ and Gram− bacteria (Fig. 1). Despite this lack of changes in specific bacterial markers, the addition of collembolans influenced the overall composition of the bacterial community (pairwise comparison, NMDS, DFA, Mahalanobis distances; F=4.8, P=0.03; Fig. 2, Table 2). Initial steps of microbial community establishment also significantly affected microbial community composition. In the bacterial system, treatments where E. coli and P. fluorescens were added showed significantly higher proportions of Gram− bacteria and lower proportions of Gram+ bacteria (Fig. 1e–g). Total bacterial biomass was lowest in both of these treatments (Fig. 1c). Further, the addition of P. fluorescens significantly modified the bacterial community composition (pairwise comparison, NMDS, DFA, Mahalanobis distances; F=15.0, P=0.0002; Fig. 2, Table 2), whereas the addition of E. coli had no significant effect (pairwise comparison, NMDS, DFA, Mahalanobis distances; F=5.2, P=0.05; Fig. 2, Table 2). In the fungal system, the inoculation of the microbial wash did not affect fungal and bacterial biomass or the proportions of Gram+ and Gram− bacteria (Fig. 1). However, the microbial wash significantly modified the overall bacterial community composition (pairwise comparison, NMDS, DFA, Mahalanobis distances; F=28.0, P<0.0001; Fig. 2, Table 2). The inoculation of C. globosum significantly increased fungal biomass and decreased bacterial biomass, which led to an overall increase in the F:B ratio (Fig. 1b–d). Treatments where C. globosum was added showed a significantly lower proportion of Gram− bacteria, and the addition of C. globosum significantly modified the bacterial community composition (NMDS, DFA, Mahalanobis distances; F=11.0, P=0.002). Overall, the bacterial community composition was significantly modified by every step of the establishment of the prey–consumer fungal-based system as indicated by the significant differences between all pairs of treatments (Fig. 2, Table 2).
Table 2Effect of bacterial and fungal consumer–prey inoculations on bacterial community composition. Differences in the composition of bacterial PLFAs between treatments were analysed using non-metric multidimensional scaling (NMDS), followed by discriminant function analysis (DFA). Overall differences in terms of bacterial PLFA composition (eight markers) within the bacterial and fungal systems were first analysed by MANOVA. Pairwise differences between treatments were further investigated using Mahalanobis distances. Inoculation treatments are underlined. RMB: remaining microbial background; coll. wash: collembolan wash; P. fluo: P. fluorescens.
3.2 Microbial C use, CO2 emissions and SOC concentrations as affected by consumer–prey system
Based on 13C content in microbial PLFA markers, most of the bacterial (73±14 % across all treatments) and fungal (64±22 %) C originated from soil C (rather than litter C). In the bacterial systems these values did not vary significantly among treatments (Fig. 3). The addition of the amoeba A. castellanii thus did not modify the use of C sources. In the fungal system, the origin of fungal and bacterial C differed. In microcosms with fungi only, 52±11 % of fungal C originated from soil, whereas 72±4.3 % of bacterial C was of soil origin, indicating that fungi captured more litter C (48±11 %) than bacteria (28±4.3 %). These differences levelled off in the presence of collembolans (Fig. 3a), indicating that the addition of H. nitidus modified the use of C sources by soil microbes. Despite these differences between fungi and bacteria in their ability to capture C resources, no significant differences were observed in terms of CO2 emitted and SOC concentrations between treatments (Fig. 3b, c).
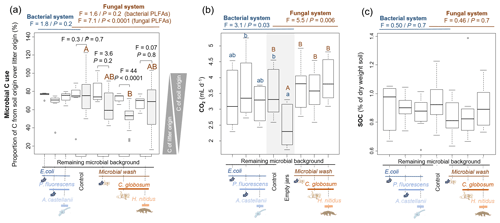
Figure 3Effect of bacterial and fungal consumer–prey inoculations on C dynamics. (a) Microbial C use, indicated by the relative contribution of C from soil over litter in bacterial and fungal PLFA markers. Grey boxes indicate fungal PLFAs. (b) CO2 production from soil microorganisms and animals; data are average emissions over 48 h after 4 and 6 weeks of incubation. Grey background indicates control treatment of both the bacterial and fungal system. (c) SOC concentrations. Differences between treatments were analysed separately for the bacterial and fungal systems using GLS models followed by ANOVA and post hoc Tukey tests. Letters (blue lowercase for bacterial system and brown capital for fungal system) indicate significant differences between means. In addition, in panel (a), for the four treatments of the fungal system, the differences in C origin in bacterial and fungal PLFAs were tested (pairwise comparisons).
3.3 Soil aggregation as affected by consumer–prey system
The 6-week incubation period in microcosms resulted in the formation of soil aggregates, regardless of the treatments (mean MWDdd across all treatments mm), compared to the initial soil conditions (MWD mm). The formed aggregates on average were unstable (mean MWDas across all treatments mm) and ranged from very unstable (min MWDas=0.31 mm) to moderately stable (max MWDas=1.04 mm), according to the classification of the international norm ISO/FDIS109030:2012(E)) (2012). The protist and collembolan consumers had contrasting effects on soil aggregate formation. Adding A. castellanii significantly increased the formation of soil aggregates, whereas H. nitidus significantly decreased it (Fig. 4a, b). Effects of consumers on soil aggregate stability were weaker but more consistent in the bacterial than in the fungal system. Both A. castellanii and H. nitidus tended to reduce the stability of soil aggregates (Fig. 4a, b). These effects were weak as no significant differences were observed by direct comparison of the treatments with P. fluorescens or C. globosum with and without their respective consumers. However, the significant increase in soil aggregate stability in response to the addition of P. fluorescens and C. globosum vanished when their associated consumers were added. Overall, this indicates that A. castellanii and H. nitidus reduce the positive effect of P. fluorescens and C. globosum on soil aggregate stability, although only a little. The initial inoculation of soil microorganisms also modified the formation and stability of soil aggregates. In particular, P. fluorescens increased the stability of the soil aggregates but did not modify their formation (Fig. 4a, b). C. globosum had a significant positive effect on both soil aggregate formation and stability (Fig. 4a, b). Neither the addition of E. coli nor the microbial wash significantly modified soil aggregation.
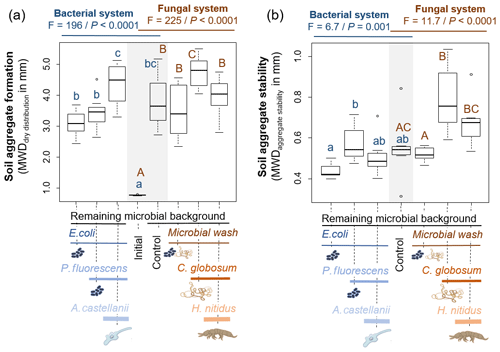
Figure 4Effect of bacterial and fungal consumer–prey inoculations on (a) soil aggregate formation and (b) soil aggregate stability. Differences between treatments were analysed separately for the bacterial and fungal systems using GLS models followed by ANOVA and post hoc Tukey tests. Letters (blue lowercase for bacterial system and brown capital for fungal system) indicate significant differences between means according to Tukey tests. Grey background indicates control treatments common to bacterial and fungal systems. P<0.001; P<0.01.
3.4 Relationships between microbial community composition, C dynamics and soil aggregation
Descriptors of the microbial community composition were poorly related among each other in the bacterial system, whereas in the fungal system they were more correlated (Fig. 5, Table A1 in Appendix). In the bacterial system, bacterial biomass positively correlated with the ratio of Gram+ to Gram− bacteria (ρ=0.58, P=0.0007). These two variables also were significantly related to the microbial community composition (as indicated by PCA scores, P<0.0001). Despite overall low levels of fungal biomass in the bacterial system, fungal biomass positively correlated with the F:B ratio (ρ=0.78, P<0.0001), which itself positively correlated with microbial biomass (ρ=0.39, P=0.03). In the fungal system, fungal biomass and the F:B ratio (themselves positively related, ρ=0.99, P<0.0001) were positively correlated with microbial biomass (P=0.003 and P=0.002, respectively). Fungal biomass and the F:B ratio also significantly correlated with the bacterial community composition (as indicated by PCA scores, P<0.0001). The composition of the bacterial community was significantly related to the Gram+ bacteria-to-Gram− bacteria ratio (ρ=0.71, P<0.0001), itself being related to fungal biomass (ρ=0.39, P=0.04), but not to the F:B ratio (ρ=0.29, P=0.13). Fungal biomass negatively correlated with bacterial biomass (, P=0.002).
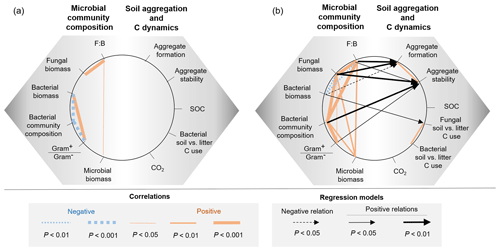
Figure 5Relationships between microbial community composition and soil aggregation and C dynamics in the (a) bacterial-based and (b) fungal-based systems. Coloured lines indicate significant correlations (Spearman) among descriptors of microbial community composition and soil functioning; for details of the statistical parameters see Tables A1 and A2. Black arrows indicate significant relationships between soil functioning variables and descriptors of the microbial community composition analysed by generalized linear models; for details of the statistical parameters see Table A3. Only significant relationships are displayed.
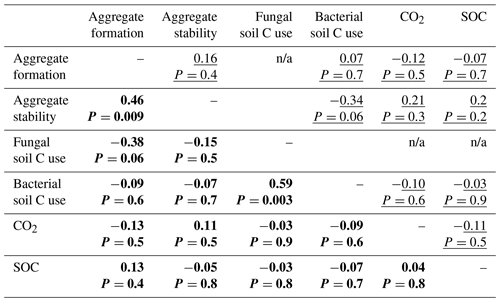
In both the bacterial and fungal system, soil aggregation, microbial C use, CO2 emissions and SOC concentrations were poorly correlated (Fig. 5, Table A2). In the bacterial system, no correlations were found between soil aggregate formation, soil aggregate stability, bacterial C use, CO2 emissions and SOC concentrations (Fig. 5a). In the fungal system, soil aggregate formation and stability were positively correlated (ρ=0.46, P=0.009). In addition, fungal C use positively correlated with bacterial C use (ρ=0.59, P=0.003), indicating that communities characterized by higher capture of litter C by fungi also presented higher incorporation of litter C into bacteria. SOC concentrations and CO2 emissions were not related to each other, nor were they related to microbial C use or soil aggregation.
Variations in soil aggregation and C dynamics were not explained by any of the descriptors of the microbial community in the bacterial system (Fig. 5, Table A3). In the fungal system, some microbial community parameters were related to soil aggregation and overall C use (single generalized linear models). In particular, soil aggregate formation and stability both positively correlated with fungal biomass (P=0.002 and 0.007, respectively; Table A3) and the F:B ratio (P=0.004 and 0.02, respectively; Table A3), which were strongly correlated variables (Table A2). Besides the dominant effect of fungal abundance (R2 and AIC), soil aggregate formation was negatively related to bacterial biomass (R2=0.18, P=0.02), but this was less strong according to R2 and the AIC (Fig. 5b, Table A3). Concerning soil aggregate stability, the effects of fungal abundance, i.e. fungal biomass and the F:B ratio, came second and third, respectively, following the effect of the bacterial community composition (R2 and AIC), which significantly correlated with soil aggregate stability (R2=0.28, P=0.002; Table A3). In addition, the Gram+ bacteria-to-Gram− bacteria ratio positively correlated with soil aggregate stability but less strongly (R2=0.16, P=0.03, higher AIC). Fungal C use positively correlated with bacterial biomass (R2=0.22, P=0.02) in the fungal system, indicating that fungi acquired more soil C when bacterial biomass was increased. The production of CO2 and the SOC concentration did not correlate with any microbial parameters in both the bacterial and fungal system.
Our results showed that simplified trophic interactions modified microbial community composition and soil aggregation but did not affect, or only slightly affected, C dynamics. Overall, the effects were stronger in the fungal-based system than in the bacterial-based system. In the latter, the inoculation of P. fluorescens as the dominant bacterial strain in large drove the changes in microbial community composition, whereas the addition of the amoeba predator A. castellanii did not induce further changes, presumably because P. fluorescens is a less preferred and toxic strain for A. castellanii. However, A. castellanii enhanced the formation of soil aggregates, presumably related to changes in the production of bacterial EPSs in response to the attack by A. castellanii. In the fungal-based system, conforming to our expectations, the inoculation of C. globosum increased fungal biomass and the addition of the grazer H. nitidus reduced it. These variations in fungal biomass were positively related to changes in soil aggregation, suggesting a detrimental effect of collembolans on soil aggregation. Surprisingly, the inoculation of C. globosum and H. nitidus resulted in significantly modified bacterial biomass and composition, and this was related to changes in soil aggregation. Finally, in the bacterial- and fungal-based systems, soil organic matter was the dominant C source, and inoculation steps only weakly modified the relative importance of soil vs. added chopped litter as the microbial C source. Notably, the inoculation treatments did not significantly affect SOC concentrations and CO2 emissions, suggesting that despite trophic interactions significantly modifying microbial communities and soil aggregation, this may not change soil C dynamics.
4.1 Bacteria-based prey–consumer system
The composition of the microbial community was mainly driven by the addition of P. fluorescens, which dominated the system and is less preferred toxic prey for A. castellanii (Jousset et al., 2009). As P. fluorescens is a Gram− bacterium, the increased proportion of Gram− bacteria in the system after its addition was expected. Interestingly, the addition of P. fluorescens also decreased the proportion of Gram+ bacteria as well as overall bacterial biomass. Thus, P. fluorescens presumably constrained the growth of the remaining microbial background community present in the system, notably by outcompeting Gram+ bacteria (Powers et al., 2015).
The addition of the amoeba A. castellanii overall weakly affected bacterial abundance and community composition. This lack of effect contrasts previous work reporting a strong decrease in bacterial biomass as well as significant and rapid changes in bacterial community composition in the presence of A. castellanii (Jousset et al., 2009; Rosenberg et al., 2009). One possible explanation for this discrepancy is the dominance of P. fluorescens and its ability to produce toxins in response to grazing by A. castellanii (Jousset and Bonkowski, 2010). In fact, P. fluorescens is not preferred prey for A. castellanii (Jousset et al., 2009). In our system, the amoebae thus presumably only moderately fed on P. fluorescens and exerted stronger pressure on the other less abundant bacterial strains, namely E. coli and the bacteria from the remaining microbial background. As a consequence, there may have been changes in the composition of non-toxic bacterial strains and in the proportions of Gram− and Gram+ bacteria, the latter usually being less preferred by protozoa due to their protective cell wall (Rønn et al., 2002; Murase et al., 2006). However, protected from protozoan predation, the dominance of P. fluorescens masked these changes. In addition, due to preferential feeding on non-toxic strains, one could have expected beneficial effects of amoebae on P. fluorescens, but no increase in Gram− bacteria occurred in treatments with A. castellanii. This lack of change may have resulted from several concomitant factors. First, P. fluorescens was dominating the bacterial community, leading to little release in the competitive pressure due to preying on the less abundant non-toxic bacteria. Further, low abundance of non-toxic bacterial strains in our system resulted in low nutrient release due to grazing by protists on these bacteria (Jousset et al., 2008). The potential benefit for the growth of P. fluorescens was thus presumably minor. Moreover, although P. fluorescens is non-preferred prey, it nonetheless is consumed by A. castellanii (Jousset and Bonkowski, 2010), and this may have counteracted positive effects on its growth. Finally, the activation of toxin production by P. fluorescens is costly (Zha et al., 2006), and the production of such defence molecules is usually associated with reduced investment in bacterial growth (Malik et al., 2020). A more detailed investigation of bacterial community composition using molecular tools may have allowed deeper insight into these processes and should be employed in future studies.
Soil aggregation, but not bacterial C use, SOC concentrations and CO2 production, was modified by the inoculation of the consumer–prey system. The bacterial community incorporated most of its C from soil (73.4±14.0 %) and this remained unchanged regardless of the inoculation treatment. More homogeneous distribution of soil organic matter, already processed and embedded into the soil matrix, compared to the added chopped litter, composed of relatively large pieces (micrometre to millimetre size) of non-processed plant debris, may explain this high incorporation (Malik et al., 2016; Tecon and Or, 2017). The unchanged proportion of C sources by bacteria when specific bacterial strains or the amoeba predator was added is consistent with the lack of effect of these inoculation steps on overall microbial biomass. The unchanged levels in CO2 emission and SOC concentrations as well were in line with the constant microbial biomass. What was more surprising is that despite microbial biomass and composition as well as C dynamics remaining unchanged, we observed modifications in soil aggregation after the amoeba predator was added. To the best of our knowledge, this is the first experimental evidence that protists affect soil aggregation and that significant effects of amoebae can occur without modifying the biomass and the composition of the microbial community. Possible explanations are expected changes in microbial activity. In our case, P. fluorescens was dominant and presumably weakly affected by A. castellanii in terms of biomass, but both may have changed their activity. In particular, P. fluoresencens responds to predation by A. castellanii through enhanced production of phenolic compounds with antibiotic properties (Jousset and Bonkowski, 2010). Enhanced mucilage production, with higher production of polysaccharides, is a common strategy used by bacteria as well in response to predation by protists, leading to higher bacterial survival and growth (Matz and Kjelleberg, 2005; Queck et al. 2006). Further, A. castellanii may have upregulated the production of proteases enabling the amoebae to perforate bacterial biofilms (Serrano-Luna et al., 2006). All these compounds released by the amoeba predator and its bacterial prey have a significant potential to induce changes in soil aggregation as a side effect. Polysaccharides (Chenu, 1989, 1993), proteins (Rillig, 2004; Liu et al., 2013; Erktan et al., 2017) and phenolic compounds (Yoshikawa et al., 2018) are all known for their important role in soil aggregation. It is plausible that an enhanced production of mucilage, notably polysaccharides, by P. fluorescens may be the underlying cause of the formation of larger soil aggregates after the addition of A. castellanii. Furthermore, the release of proteases by A. castellanii may have decreased the wettability of the mucilage and thereby have been responsible for the lack of associated significant increase in the stability of aggregates. Although our focus was not to quantify changes in extracellular compounds occurring in amoeba–bacteria interactions, our results suggest that further investigations in this direction may be promising and could explain the effects of protists on soil aggregation. Initial steps of the inoculation of the prey–consumer system also influenced soil aggregation. In the absence of the amoeba predator, the bacterial communities, activated by wet and dry cycles (Cosentino et al., 2006), promoted the formation of soil aggregates. This effect occurred regardless of the bacterial strains inoculated, suggesting a similar ability of these bacterial strains to increase soil particle cohesion. By contrast, the inoculation of P. fluorescens significantly stabilized soil aggregates, suggesting specific hydrophobic properties of its EPSs.
Overall, the effects of the amoeba A. castellanii on the microbial community were limited and did not induce changes in microbial biomass and composition or in C dynamics. However, despite this lack of effects, the addition of the amoeba predator significantly affected soil aggregation. Such changes in soil aggregation irrespective of changes in C dynamics and microbial composition suggest that protists preying on bacterial communities can significantly affect soil structure, without significantly affecting microbial growth and C cycling. Presumably, changes in the activity of microbiota, notably the release of extracellular compounds in response to predation by protists, are responsible for changes in soil aggregation as side effects.
4.2 Fungi-based prey–consumer system
The microbial community was influenced by every inoculation step in the fungal-based system, including by the addition of the collembolan grazers, and showed strong relations between the fungal and bacterial communities. As expected, fungal biomass was highest in the treatment with C. globosum, indicating that our inoculation was successful and triggered fungal development, which drove an overall increase in microbial biomass. Remarkably, the increase in fungal biomass was associated with a decrease in bacterial biomass as well as in Gram− bacteria, indicating competition between fungi and bacteria as is common in the top soil (Bahram et al., 2018). Adding collembolans reduced fungal biomass, presumably due to the consumption of fungal hyphae by H. nitidus, confirming that C. globosum is consumed by H. nitidus (Pollierer et al., 2019). In addition, as collembolans also feed on litter (Potapov et al., 2016), competition for litter resources between fungi and collembolans may have contributed to the reduced fungal biomass. Unexpectedly, bacterial biomass did not benefit from the decrease in fungal biomass due to the addition of H. nitidus. This might have been due to (i) the competitive pressure exerted by fungi remaining high in the presence of H. nitidus and/or (ii) the potential positive effects of reduced competition being compensated by H. nitidus also feeding on bacteria (Pollierer et al., 2019), even though probably at a low level. In fact, the composition of the bacterial community was significantly modified by collembolans. Presumably, these changes were due to selective feeding of H. nitidus on certain bacterial strains and/or through modifications in competitive interactions between certain bacterial strains and C. globosum (Lussenhop, 1992; Coulibaly et al., 2019; Thakur and Geisen, 2019). In addition, other non-consumptive effects may have occurred. For example, comminution of the added litter by collembolans (Coleman et al., 2002) may have contributed to changes in bacterial community composition (Scheu et al., 2005). Further, the release of specific organic compounds by living or dead collembolans, such as urine, faeces (Lussenhop, 1992) or chitin (Lucas et al., 2020), may also have modified the composition of the bacterial community. Notably, the addition of the collembolan wash also significantly modified the bacterial community composition. Although it has long been assumed that microorganisms from the body surface of microarthropods may affect soil microbiota (Lussenhop, 1992; Scheu et al., 2005), our results provide experimental evidence for it through the separate addition of a microbial wash. This effect highlights the importance of processes of community coalescence (Rillig et al., 2015) caused by the carriage of microorganisms on the body of the consumers. Far from anecdotic, these effects were as strong as those when the soil animals themselves were added, suggesting that future studies should integrate such non-consumptive effects as main drivers of the role of microarthropod grazers in soil microbial community composition.
Modifications in the composition of the microbial community were associated with changes in microbial C use and soil aggregation but not with changes in SOC concentrations and CO2 emissions. More C of litter origin was captured by fungi rather than by bacteria when fungal biomass was high. Superior ability of fungi compared to bacteria in capturing litter C has been observed previously in microcosm experiments (Malik et al., 2016) and is consistent with the network structure of fungal mycelia able to bridge air-filled pores (Otten et al., 2001) and thus able to reach distant resources such as pieces of litter widely distributed in the soil matrix. In our study, the increased use of litter C by fungi was associated by an increased use of litter C by bacteria. This positive correlation suggests that fungi facilitated the access of litter C to bacteria, confirming previous similar observations (Purahong et al., 2016; Gorka et al., 2019). This positive effect may have been due to the transfer of nutrients by fungal hyphae (Worrich et al., 2017) or increased mobility of bacteria along fungal hyphae (Simon et al., 2015; Worrich et al., 2016). Effects, however, were minor and did not affect the bacterial C source, which remained based mainly on soil C. Interestingly, all these changes in microbial biomass, composition and C use occurred without impacting the overall production of CO2 and SOC concentration. Potentially, concomitant opposite effects of increased microbial biomass and decreased microbial activity were responsible for the overall constant levels of soil respiration. Such compensatory effects have been proposed previously to be responsible for the absence of effects of higher-trophic-level consumers on soil respiration (Mikola and Setälä, 1998; Lucas et al., 2020).
The positive effect of C. globosum on soil aggregation is consistent with previous studies highlighting positive effects of fungi, in particular saprotrophic ones, on soil aggregation (Caesar-TonThat and Cochran, 2000; Daynes et al., 2012; Lehmann et al., 2017), especially when fungi dominate the microbial community (McMalla et al., 1958). Collembolans reduced the positive effect of C. globosum on soil aggregation. As hypothesized, the reduction in fungal biomass by collembolans was presumably responsible for the negative effect of H. nitidus on soil aggregation in our system. More intriguing changes in bacterial community composition related to inoculation steps also contributed to variations in soil aggregate stability. Previous studies reported positive effects of interactions between fungi and bacteria on soil aggregation (Aspiras et al., 1971) as well as differential effects of Gram+ and Gram− bacteria (Mu'minah et al., 2015). Our results further suggest that changes in the bacterial community composition contribute to a similar extent as fungal biomass to changes in soil aggregate stability. Our design does not allow the disentanglement of the individual effects of fungal biomass and bacterial community composition, but we suggest that a combination of both was responsible for the observed overall effects on soil aggregate stability. Altogether, we showed that collembolans modified the microbial community composition, presumably through both consumptive and non-consumptive effects, which impacted soil aggregation.
We demonstrated that simplified consumer–prey interactions influence soil microbial community composition, microbial C use and soil aggregation but not CO2 emissions and SOC concentrations. Remarkably, effects on soil aggregation were not related to SOC or CO2 emissions, suggesting that subtle but significant effects related to trophic interactions can occur at constant levels of SOC and microbial respiration. In the bacterial-based system, changes in soil aggregation occurred without any modification in microbial biomass and community composition. This lack of change presumably resulted from the dominance of P. fluorescens, toxic and non-preferred prey for A. castellanii. We conjecture that changes in bacterial physiology, such as increased EPS production and/or the release of secondary metabolites in response to predation, are the main cause of the formation of larger aggregates. In the fungal-based system, the collembolan species H. nitidus significantly modified the microbial community via decreasing the biomass of C. globosum and changing bacterial community composition. Notably, we found that the effects of collembolans on the soil microbial community did not only result from consumptive effects, suggesting that the transport of soil microorganisms on the body surface of collembolans is of major importance for the effect of higher trophic levels on soil microbial communities. As expected, fungal biomass was a main factor promoting soil aggregation and was negatively modulated in the presence of the fungal grazer H. nitidus. Remarkably, we also found an unexpected contribution of changes in bacterial community composition to soil aggregation. We conclude that the effects of consumer–prey interactions on soil aggregation can be either positive or negative and were in our case stronger on the formation of soil aggregates than on their stabilization. Our results highlight that non-trivial changes in microbial community composition and/or activity caused by microbial consumers impact soil aggregation as side effects. Integration of more complex trophic interactions is needed to assess how the mechanisms highlighted in our simplified systems are embedded into more complex processes in natural systems.
Table A1Correlation matrix among descriptors of microbial community. Spearman's correlation coefficients (ρ) for the bacterial-based system (underlined) and the fungal-based system (in bold).
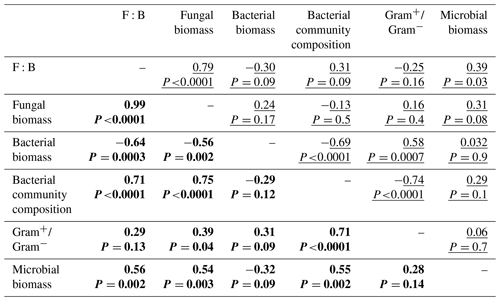
Table A2Correlation matrix among variables describing soil aggregation and overall C use. Spearman's correlation coefficients (ρ) for the bacterial-based system (underlined) and the fungal-based system (in bold). The term n/a indicates not applicable.
Data are available from the Zenodo public repository: https://doi.org/10.5281/zenodo.4005651 (Erktan et al., 2020).
AE and SS designed the study. MR, AJ and AC provided advice on the experimental design. AE conducted the microcosms experiment. AJ provided the culture of Acanthamoebae castellanii. AE analysed the PLFA data. AE ran the statistical analyses and wrote the first draft of the manuscript, and all authors contributed substantially to revisions.
The authors declare that they have no conflict of interest.
We are thankful to Theodora Volovei for her help in the set-up of the microcosm experiment and more generally for her technical help and advice. We thank Guido Humpert for conducting the PLFA extraction and measurement, Zhilei Gao for preparing the A. castellanii culture, and Leonie Schardt for conducting the measurement of soil organic carbon concentrations. We thank Jens Dyckmans for measuring the δ13C isotopic signal of PLFA samples.
This research has been supported by the European Commission – Horizon 2020 research and innovation program, Marie Skłodowska-Curie actions (grant no. 750249).
This open-access publication was funded
by the University of Göttingen.
This paper was edited by Jianming Xu and reviewed by three anonymous referees.
Aspiras, R. B., Allen, O. N., Harris, R. F., and Chesters, G.:Aggregate stabilization by filamentous microorganisms, Soil Sci., 112, 282–284, 1971.
Bahram, M., Hildebrand, F., Forslund, S. K., Anderson, J. L., Soudzilovskaia, N. A., Bodegom, P. M., Bengtsson-Palme, J., Anslan, S., Coelho, L. P., Herend, H., Huerta-Cepas, J., Medama, M. H., Maltz, M. R., Mundra, S., Olsson, P. A., Pent, M., Polme, S., Sunagawa, S., Ryberg, M., Tedersoo, L., and Bork, P.: Structure and function of the global topsoil microbiome, Nature, 560, 233–237, 2018.
Baldock, J. A.: Interactions of organic materials and microorganisms with minerals in the stabilization of soil structure, in: Interactions Between Soil Particles and Microorganisms, edited by: Huang, P. M., Bollag J.-M., and Senesi N., John Wiley & Sons, New York, NY, 85–131, 2002.
Bardgett, R. D. and Van Der Putten, W. H.: Belowground biodiversity and ecosystem functioning, Nature, 515, 505–511, 2014.
Bardgett, R. D., Keiller, S., Cook, R., and Gilburn, A. S.: Dynamic interactions between soil animals and microorganisms in upland grassland soils amended with sheep dung: a microcosm experiment, Soil Biol. Biochem., 30, 531–539, 1998.
Barthes, B. and Roose, E.: Aggregate stability as an indicator of soil susceptibility to runoff and erosion; validation at several levels, Catena, 47, 133–149, 2002.
Bradford, M. A.: Re-visioning soil food webs, Soil Biol. Biochem., 102, 1–3, 2016.
Bradford, M. A., Keiser, A. D., Davies, C. A., Mersmann, C. A., and Strickland, M. S.: Empirical evidence that soil carbon formation from plant inputs is positively related to microbial growth, Biogeochemistry, 113, 271–281, 2013.
Buyer, J. S. and Sasser, M.: High throughput phospholipid fatty acid analysis of soils, Appl. Soil Ecol., 61, 127–130, 2012.
Caesar-TonThat, T. C. and Cochran, V. L.: Soil aggregate stabilization by a saprophytic lignin-decomposing basidiomycete fungus I. Microbiological aspects, Biol. Fert. Soils, 32, 374–380, 2000.
Caesar-TonThat, T. C., Caesar, A. J., Gaskin, J. F., Sainju, U. M., and Busscher, W. J.: Taxonomic diversity of predominant culturable bacteria associated with microaggregates from two different agroecosystems and their ability to aggregate soil in vitro, Appl. Soil Ecol., 36, 10–21, 2007.
Caesar-TonThat, T. C., Stevens, W. B., Sainju, U. M., Caesar, A. J., West, M., and Gaskin, J. F.: Soil-Aggregating Bacterial Community as Affected by Irrigation, Tillage, and Cropping System in the Northern Great Plains, Soil Sci., 179, 11–20, 2014.
Chenu, C.: Influence of a fungal polysaccharide, scleroglucan, on clay microstructures, Soil Biol. Biochem., 21, 299–305, 1989.
Chenu, C.: Clay- or sand-polysaccharide associations as models for the interface between micro-organisms and soil: water related properties and microstructure, in: Proceedings of the International Workshop on Methods of Research on Soil Structure/Soil Biota Interrelationships, Elsevier, Wageningen, the Netherlands, 24–28 November 1991, 143–156, 1993.
Coleman, D., Fu, S. L., Hendrix, P., and Crossley, D.: Soil foodwebs in agroecosystems: impacts of herbivory and tillage management, Eur. J. Soil Biol., 38, 21–28, 2002.
Contosta, A. R., Frey, S. D., and Cooper, A. B.: Soil microbial communities vary as much over time as with chronic warming and nitrogen additions, Soil Biol. Biochem., 88, 19–24, 2015.
Cosentino, D., Chenu, C., and Le Bissonnais, Y.: Aggregate stability and microbial community dynamics under drying–wetting cycles in a silt loam soil, Soil Biol. Biochem., 38, 2053–2062, 2006.
Coulibaly, S. F. M., Winck, B. R., Akpa-Vinceslas, M., Mignot, L., Legras, M., Forey, E., and Chauvat, M.: Functional Assemblages of Collembola Determine Soil Microbial Communities and Associated Functions, Front. Environ. Sci., 7, 52, https://doi.org/10.3389/fenvs.2019.00052, 2019.
Cragg, R. G. and Bardgett, R. D.: How changes in soil faunal diversity and composition within a trophic group influence decomposition processes, Soil Biol. Biochem., 33, 2073–2081, 2001.
Crowther, T. W., Thomas, S. M., Maynard, D. S., Baldrian, P., Covey, K., Frey, S. D., van Diepen, L. T. A., and Bradford, M. A.: Biotic interactions mediate soil microbial feedbacks to climate change, P. Natl. Acad. Sci. USA, 112, 7033–7038, 2015.
Danese, P. N., Pratt, L. A., and Kolter, R.: Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture, J. Bacteriol., 182, 3593–3596, 2000.
Daynes, C. N., Zhang, N., Saleeba, J. A., and McGee, P. A.: Soil aggregates formed in vitro by saprotrophic Trichocomaceae have transient water-stability, Soil Biol. Biochem., 48, 151–161, 2012.
de Vries, F. T. and Caruso, T.: Eating from the same plate? Revisiting the role of labile carbon inputs in the soil food web, Soil Biol. Biochem., 102, 4–9, 2016.
de Vries, F. T., Griffiths, R. I., Bailey, M., Craig, H., Girlanda, M., Gweon, H. S., Hallin, S., Kaisermann, A., Keith, A., Kretzschmar, M., Lemanceau, P., Lumini, E., Mason, K. E., Olivier, A., Ostle, N., Prosser, J. I., Thin, C., Thomson, B., and Bardgett, R.: Soil bacterial networks are less stable under drought than fungal networks, Nat. Commun., 9, 3033, https://doi.org/10.1038/s41467-018-05516-7, 2018.
Degens, B. P., Sparling, G. P., and Abbott, L. K.: Increasing the length of hyphae in a sandy soil increases the amount of water-stable aggregates, Appl. Soil. Ecol., 3, 149–159, 1996.
Delgado-Baquerizo, M., Maestre, F. T., Reich, P. B., Jeffries, T. C., Gaitan, J. J., Encinar, D., Berdugo, M., Campbell, C. D., and Singh, B. K.: Microbial diversity drives multifunctionality in terrestrial ecosystems, Nat. Commun., 7, 10541, https://doi.org/10.1038/ncomms10541, 2016.
Deveau, A., Bonito, G., Uehling, J., Paoletti, M., Becker, M., Bindschedler, S., Hacquard, S., Hervé, V., Labbé, J., Lastovetsky, O. A., Mieszkin, S., Millet, L. J., Vajna, B., Junier, P., Bonfante, P., Krom, B. P., Olsson, S., van Elsas, J. D., and Wick, L. Y.: Bacterial–fungal interactions: ecology, mechanisms and challenges, FEMS Microbiol. Rev., 42, 335–352, 2018.
DIN ISO 10694:1996-08: Bodenbeschaffenheit – Bestimmung von organischem Kohlenstoff und Gesamtkohlenstoff nach trockener Verbrennung (Elementaranalyse; ISO 10694:1995), Beuth publishing DIN, Germany, 1996.
Djigal D., Brauman A., Diop T. A., Chotte J. L., and Villenave C.: Influence of bacterial-feeding nematodes (Cephalobidae) on soil microbial communities during maize growth, Soil Biol. Biochem., 36, 323–331, 2004.
Domsch, K. H., Gams, W., and Anderson, T.-H.: Compendium of soil fungi, Vols. I and II, 2nd edn., Academic Press, Oxford, 1993.
Dubuis, C., Keel, C., and Haas, D.: Dialogues of root-colonizing biocontrol pseudomonads, Eur. J. Plant Pathol., 119, 311–328, 2007.
Erktan, A., Balmot, J., Mérino-Martin, L., Monnier, Y., Pailler, F., Coq, S., Abiven, S., Le Bissonnais, Y., and Stokes, A.: Immediate and long-term effect of tannins in the stabilization of Mediterranean soil aggregates, Soil Biol. Biochem., 105, 197–205, 2017.
Erktan, A., Rillig, C. M., Carminati, A., Jousset, A., and Scheu, S.: Dataset_2020_Biogeosciences_Protists and collembolans alter microbial community composition, C dynamics and soil aggregation in simplified consumer – prey systems, Zenodo, https://doi.org/10.5281/zenodo.4005651, 2020.
Filser, J., Faber, J. H., Tiunov, A. V., Brussaard, L., Frouz, J., De Deyn, G., Uvarov, A. V., Berg, M. P., Lavelle, P., Loreau, M., Wall, D. H., Querner, P., Eijsackers, H., and Jiménez, J. J.: Soil fauna: key to new carbon models, SOIL, 2, 565–582, https://doi.org/10.5194/soil-2-565-2016, 2016.
Frostegård, Å. and Bååth, E.: The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil, Biol. Fert. Soils, 22, 59–65, 1996.
Geisen, S., Mitchell, E. A., Adl, S., Bonkowski, M., Dunthorn, M., Ekelund, F., Fernández, L. D., Jousset, A., Krashevska, A., Singer, D., Spiegel, F. W., Walochnik, J., and Lara, E.: Soil protists: a fertile frontier in soil biology research, FEMS Microbiol. Rev., 42, 293–323, 2018.
Glassman, S. I., Weihe, C., Li, J., Albright, M. B., Looby, C. I., Martiny, A. C., Treseder, K. K., Allison, S. D., and Martiny, J. B. H.: Decomposition responses to climate depend on microbial community composition, P Natl. Acad. Sci. USA, 115, 11994–11999, 2018.
Gorka, S., Dietrich, M., Mayerhofer, W., Gabriel, R., Wiesenbauer, J., Martin, V., Zheng, Q., Imai, B., Prommer, J., Weidinger, M., Schweiger, P., Eichorst, S. A., Wagner, M., Richter, A., Schintlmeister, A., Woebken, D., Kaiser, C.: Rapid transfer of plant photosynthates to soil bacteria via ectomycorrhizal hyphae and its interaction with nitrogen availability. Front. Microbiol., 10, 168, https://doi.org/10.3389/fmicb.2019.00168, 2019.
Gormsen, D., Olsson, P. I. A., and Hedlund, K.: The influence of collembolans and earthworms on AM fungal mycelium, Appl. Soil Ecol., 27, 211–220, 2004.
Grandy, A. S., Wieder, W. R., Wickings, K., and Kyker-Snowman, E.: Beyond microbes: Are fauna the next frontier in soil biogeochemical models?, Soil Biol. Biochem., 102, 40–44, 2016.
Griffiths, B. S. and Bardgett, R. D.: Interactions between microbe-feeding invertebrates and soil microorganisms, in: Modern soil microbiology, edited by: Van Elsas, J. D., Trevors, J. T., and Wellington, E. M. H, Editions M. Dekker, New York, 165–182, 1997.
Griffiths, R. I., Thomson, B. C., James, P., Bell, T., Bailey, M., and Whiteley, A. S.: The bacterial biogeography of British soils, Environ. Microbiol., 13, 1642–1654, 2011.
Hopkin, S. P. (Ed.): Biology of the springtails - Insecta: Collembola, Oxford University Press, Oxford, UK, 1997.
ISO/FDIS109030:2012(E): Soil quality – Measurement of the stability of soil aggregates subjected to the action of water, International Organization for Standardization, France, 2012.
Jiang, Y., Liu, M., Zhang, J., Chen, Y., Chen, X., Chen, L., Li, H., Zhang, X.-X., and Sun, B.: Nematode grazing promotes bacterial community dynamics in soil at the aggregate level, ISME J., 11, 2705–2717, 2017.
Jousset, A. and Bonkowski, M.: The model predator Acanthamoeba castellanii induces the production of 2, 4, DAPG by the biocontrol strain Pseudomonas fluorescens Q2-87, Soil Biol. Biochem., 42, 1647–1649, 2010.
Jousset, A., Scheu, S., and Bonkowski, M.: Secondary metabolite production facilitates establishment of rhizobacteria by reducing both protozoan predation and the competitive effects of indigenous bacteria, Funct. Ecol., 22, 714–719, 2008.
Jousset, A., Rochat, L., Péchy-Tarr, M., Keel, C., Scheu, S., and Bonkowski, M: Predators promote defence of rhizosphere bacterial populations by selective feeding on non-toxic cheaters, ISME J., 3, 666–674, 2009.
Le Bissonnais, Y. L.: Aggregate stability and assessment of soil crustability and erodibility: I. Theory and methodology, Eur. J. Soil Sci., 47, 425–437, 1996.
Le Bissonnais, Y. and Arrouays, D. : Aggregate stability and assessment of soil crustability and erodibility: II. Application to humic loamy soils with various organic carbon contents, Eur. J. Soil Sci., 48, 39–48, 1997.
Lehmann, A., Zheng, W., and Rillig, M. C.: Soil biota contributions to soil aggregation, Nat. Ecol. Evol., 1, 1828–1835, 2017.
Lehmann, A., Zheng, W., Ryo, M., Soutschek, K., Roy, J., Rongstock, R., Maaß, S., and Rillig, M. C.: Fungal Traits Important for Soil Aggregation, Front. Microbiol., 10, 2904, https://doi.org/10.3389/fmicb.2019.02904, 2020.
Linder, M. B., Szilvay, G. R., Nakari-Setälä, T., and Penttilä, M. E.: Hydrophobins: the protein-amphiphiles of filamentous fungi, FEMS Microbiol. Rev., 29, 877–896, 2005.
Liu, X., Eusterhues, K., Thieme, J., Ciobota, V., Höschen, C., Mueller, C. W., Küsel, K., Kögel-Knaber, I., Rösch, P., Popp, J., and Totsche, K.: STXM and NanoSIMS Investigations on EPS Fractions before and after Adsorption to Goethite, Environ. Sci. Technol., 47, 3158–3166, 2013.
Lucas, J. M., McBride, S. G., and Strickland, M. S.: Trophic level mediates soil microbial community composition and function, Soil Biol. Biochem., 143, 107756, https://doi.org/10.1016/j.soilbio.2020.107756, 2020.
Lussenhop, J.: Mechanisms of microarthropod–microbial interactions in soil, Adv. Ecol. Res., 23, 1–33, 1992.
Macfadyen, A.: Simple methods for measuring and maintaining the proportion of carbon dioxide in air, for use in ecological studies of soil respiration, Soil Biol. Biochem., 2, 9–18, 1970.
Malik, A. A., Chowdhury, S., Schlager, V., Oliver, A., Puissant, J., Vazquez, P. G. M., Jehmlich, N., von Bergen, M., Griffiths, R. I, and Gleixner, G.: Soil Fungal : Bacterial Ratios Are Linked to Altered Carbon Cycling, Front. Microbiol., 7, 1247, https://doi.org/10.3389/fmicb.2016.01247, 2016.
Malik, A. A., Puissant, J., Buckeridge, K. M., Goodall, T., Jehmlich, N., Chowdhury, S., Gweon, H. S., Peyton, J. M., mason, K. E., van Agtmaal, M., Blaud, A., Clark, I. M., Whitaker, J., Pywell, R. F., Ostle, N., Gleixner, G., and Griffiths, R. I.: Land use driven change in soil pH affects microbial carbon cycling processes, Nat. Commun., 9, 1–10, 2018.
Malik, A. A., Martiny, J. B., Brodie, E. L., Martiny, A. C., Treseder, K. K., and Allison, S. D.: Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change, ISME J., 14, 1–9, 2020.
Matz, C. and Kjelleberg, S.: Off the hook–how bacteria survive protozoan grazing, Trends Microbiol., 13, 302–307, 2005.
Milcu, A., Partsch, S., Langel, R., and Scheu, S.: The response of decomposers (earthworms, springtails and microorganisms) to variations in species and functional group diversity of plants, Oikos, 112, 513–524, 2006.
Mikola, J. and Setälä, H.: No evidence of trophic cascades in an experimental microbial- based soil food web, Ecology, 79, 153–164, 1998.
Morriën, E., Hannula, S. E., Snoek, L. B., Helmsing, N. R., Zweers, H., De Hollander, M., Soto, R. L., Bouffaud, M.-L., Buée, M., Dimmers, W., Duyts, H., Geisen, S., Girlanda, M., Griffiths, R. I., Jørgensen, H.-B., Jensen, J., Plassart, P., Redecker, D., Schmelz, R. M., Schmidt, O., Thomson, B. C., Tisserant, E., Uroz, S., Winding, A., Bailey, M. J., Bonkowski, M., Faber, J. H., Martin, F., Lemanceau, P., de Boer, W., van Veen, J. A., and van der Putten, W. H.: Soil networks become more connected and take up more carbon as nature restoration progresses, Nat. Commun., 8, 1–10, 2017.
Mu'minah, B., Hazarin Subair, F., and Baso, D.: Isolation and screening of exoplysaccharide producing bacteri (EPS) from potato rhizosphere dor soil aggregation, Int. J. Curr. Microbiol. App. Sci., 4, 341–349, 2015.
Murase, J., Noll, M., and Frenzel, P.: Impact of protists on the activity and structure of the bacterial community in a rice field soil, Appl. Environ. Microb., 72, 5436–5444, 2006.
Myers, R. T., Zak, D. R., White, D. C., and Peacock, A.: Landscape-level patterns of microbial community composition and substrate use in upland forest ecosystems, Soil Sci. Soc. Am. J., 65, 359–367, 2001.
Otten, W., Hall, D., Harris, K., Ritz, K., Young, I. M., and Gilligan, C. A.: Soil physics, fungal epidemiology and the spread of Rhizoctonia solani, New Phytol., 151, 459–468, 2001.
Perez, G., Decaëns, T., Dujardin, G., Akpa-Vinceslas, M., Langlois, E., and Chauvat, M.: Response of collembolan assemblages to plant species successional gradient, Pedobiologia, 56, 169–177, 2013.
Pollierer, M. M., O. Ferlian, and S. Scheu.: Temporal dynamics and variation with forest type of phospholipid fatty acids in litter and soil of temperate forests across regions, Soil Biol. Biochem., 91, 248–257, 2015.
Pollierer, M. M., Larsen, T., Potapov, A., Brückner, A., Heethoff, M., Dyckmans, J., and Scheu, S.: Compound-specific isotope analysis of amino acids as a new tool to uncover trophic chains in soil food webs, Ecol. Monogr., 89, e01384, https://doi.org/10.1002/ecm.1384, 2019.
Potapov, A. A., Semenina, E. E., Korotkevich, A. Y., Kuznetsova, N. A., and Tiunov, A. V.: Connecting taxonomy and ecology: Trophic niches of collembolans as related to taxonomic identity and life forms, Soil Biol. Biochem., 101, 20–31, 2016.
Powers, M. J., Sanabria-Valentín, E., Bowers, A. A., and Shank, E. A.: Inhibition of cell differentiation in Bacillus subtilis by Pseudomonas protegens, J. Bacteriol., 197, 2129–2138, 2015.
Purahong, W., Wubet, T., Lentendu, G., Schloter, M., Pecyna, M. J., Kapturska, D., Hofrichter, M., Krüger, D., and Buscot, F.: Life in leaf litter: novel insights into community dynamics of bacteria and fungi during litter decomposition, Mol. Ecol., 25, 4059–4074, 2016.
Queck, S. Y., Weitere, M., Moreno, A. M., Rice, S. A., and Kjelleberg, S.: The role of quorum sensing mediated developmental traits in the resistance of Serratia marcescens biofilms against protozoan grazing, Environ. Microbiol., 8, 1017–1025, 2006.
R Development Core Team: R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria, ISBN 3-900051-07-0, available at: http://www.R-project.org (last access: 10 August 2020), 2008.
Rillig, M. C.: Arbuscular mycorrhizae, glomalin, and soil aggregation, Can. J. Soil Sci., 84, 355–363, 2004.
Rillig, M. C., Antonovics, J., Caruso, T., Lehmann, A., Powell, J. R., Veresoglou, S. D., and Verbruggen, E.: Interchange of entire communities: microbial community coalescence, Trends Ecol. Evol., 30, 470–476, 2015.
Rønn, R., McCaig, A. E., Griffiths, B. S., and Prosser, J. I.: Impact of protozoan grazing on bacterial community structure in soil microcosms, Appl. Environ. Microb., 68, 6094–6105, 2002.
Rosenberg, K., Bertaux, J., Krome, K., Hartmann, A., Scheu, S., and Bonkowski, M.: Soil amoebae rapidly change bacterial community composition in the rhizosphere of Arabidopsis thaliana, ISME J., 3, 675–684, 2009.
Sandhya, V. and Ali, S.: Exopolysaccharide production by drought tolerant Bacillus spp. and effect on soil aggregation under drought stress, J. Microbiol. Biotech. Food Sci., 4, 51–57, 2014.
Scheu, S., Ruess, L., and Bonkowski, M.: Interactions between microorganisms and soil micro-and mesofauna, in: Microorganisms in soils: roles in genesis and functions, edited by: Varma, A. and Buscot, F., Springer, Berlin, Heidelberg, Germany, 253–275, 2005.
Schmidt, R., Cordovez, V., De Boer, W., Raaijmakers, J., and Garbeva, P.: Volatile affairs in microbial interactions, ISME J., 9, 2329–2335, 2015.
Serrano-Luna, J. D. J., Cervantes-Sandoval, I., Calderón, J., Navarro-García, F., Tsutsumi, V., and Shibayama, M.: Protease activities of Acanthamoeba polyphaga and Acanthamoeba castellanii, Can. J. Microbiol., 52, 16–23, 2006.
Siddiky, M. R. K., Schaller, J., Caruso, T., and Rillig, M. C.: Arbuscular mycorrhizal fungi and collembola non-additively increase soil aggregation, Soil Biol. Biochem., 47, 93–99, 2012.
Simon, A., Bindschedler, S., Job, D., Wick, L. Y., Filippidou, S., Kooli, W. M., Verrecchia, E. P., and Junier, P.: Exploiting the fungal highway: development of a novel tool for the in situ isolation of bacteria migrating along fungal mycelium, FEMS Microbiol. Ecol., 91, fiv116, https://doi.org/10.1093/femsec/fiv116, 2015.
Swaby, R. J.: The relationship between micro-organisms and soil aggregation, J. Gen. Microbiol., 3, 236–254, 1949.
Tecon, R. and Or, D.: Biophysical processes supporting the diversity of microbial life in soil, FEMS Microbiol. Rev., 41, 599–623, 2017.
Thakur, M. P. and Geisen, S.: Trophic regulations of the soil microbiome, Trends Microbiol., 27, 771–780, 2019.
Tisdall, J. M., Smith, S. E., and Rengasamy, P.: Aggregation of soil by fungal hyphae, Soil Res., 35, 55–60, 1997.
Tisdall, J. M., Nelson, S. E., Wilkinson, K. G., Smith, S. E., and McKenzie, B. M.: Stabilisation of soil against wind erosion by six saprotrophic fungi, Soil Biol. Biochem., 50, 134–141, 2012.
Tiunov, A. V. and Scheu, S.: Facilitative interactions rather than resource partitioning drive diversity-functioning relationships in laboratory fungal communities, Ecol. Lett., 8, 618–625, 2005.
VDLUFA I, C 2.2.1: Texturanalyse des Feinbodens – Kombination von Nasssiebung und Pipettmethode nach KÖHN, I, VDLUFA-Verlag, Darmstadt, Germany, 2012.
Velicer, G. J. and Vos, M.: Sociobiology of the myxobacteria, Annu. Rev. Microbiol., 63, 599–623, 2009.
Wagg, C., Schlaeppi, K., Banerjee, S., Kuramae, E. E., and van der Heijden, M. G.: Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning, Nat. Commun., 10, 1–10, 2019.
Weekers, P. H., Engelberts, A. M., and Vogels, G. D.: Bacteriolytic activities of the free-living soil amoebae, Acanthamoeba castellanii, Acanthamoeba polyphaga and Hartmannella vermiformis, A. Van Leeuw. J. Microb, 68, 237–243, 1995.
Wieder, W. R., Bonan, G. B., and Allison, S. D.: Global soil carbon projections are improved by modeling microbial processes, Nat. Clim. Change., 3, 909–912, 2013.
Worrich, A., König, S., Miltner, A., Banitz, T., Centler, F., Frank, K., Thullner, M., Harms, H., Kästner, M., and Wick, L. Y.: Mycelium-like networks increase bacterial dispersal, growth, and biodegradation in a model ecosystem at various water potentials, Appl. Environ. Microb., 82, 2902–2908, 2016.
Worrich, A., Stryhanyuk, H., Musat, N., König, S., Banitz, T., Centler, F., Frank, K., Thullner, M., Harms, H., Richnow, H.-H., Miltner, A., Kästner, M., and Wick, L. Y.: Mycelium-mediated transfer of water and nutrients stimulates bacterial activity in dry and oligotrophic environments, Nat. Commun., 8, 1–9, 2017.
Yoshikawa, S., Kuroda, Y., Ueno, H., Kajiura, M., and Ae, N.: Effect of phenolic acids on the formation and stabilization of soil aggregates, Soil Sci. Plant Nutr., 64, 323–334, 2018.
Yudina, A. and Kuzyakov, Y.: Saving the face of soil aggregates, Global Change Biol., 25, 3574–3577, 2019.
Zha, W., Rubin-Pitel, S. B., and Zhao, H.: Characterization of the substrate specificity of PhlD, a type III polyketide synthase from Pseudomonas fluorescens, J. Biol. Chem., 281, 32036–32047, 2006.
Zheng, W. S., Morris, E. K., Lehmann, A., and Rillig, M. C.: Interplay of soil water repellency, soil aggregation and organic carbon. A meta-analysis, Geoderma, 283, 39–47, 2016.