the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Metagenomic insights into the metabolism of microbial communities that mediate iron and methane cycling in Lake Kinneret iron-rich methanic sediments
Michal Elul
Maxim Rubin-Blum
Zeev Ronen
Itay Bar-Or
Werner Eckert
Orit Sivan
Complex microbial communities facilitate iron and methane transformations in anoxic methanic sediments of freshwater lakes, such as Lake Kinneret (the Sea of Galilee, Israel). The phylogenetic and functional diversity of these consortia are not fully understood, and it is not clear which lineages perform iron reduction and anaerobic oxidation of methane (AOM). Here, we investigated microbial communities from both natural Lake Kinneret iron-rich methanic sediments (>20 cm depth) and iron-amended slurry incubations from this zone using metagenomics, focusing on functions associated with iron reduction and methane cycling. Analyses of the phylogenetic and functional diversity indicate that consortia of archaea (mainly Bathyarchaeia, Methanomicrobia, Thermoplasmata, and Thermococci) and bacteria (mainly Chloroflexi (Chloroflexota), Nitrospirae (Nitrospirota), and Proteobacteria) perform key metabolic reactions such as amino acid uptake and dissimilation, organic matter fermentation, and methanogenesis. The Deltaproteobacteria, especially Desulfuromondales (Desulfuromonadota), have the potential to transfer electrons extracellularly either to iron mineral particles or to microbial syntrophs, including methanogens. This is likely via transmembrane cytochromes, outer-membrane hexaheme c-type cytochrome (OmcS) in particular, or pilin monomers (PilA), all of which were attributed to this lineage. Bona fide anaerobic oxidizers of methane (ANME) and denitrifying methanotrophs Methylomirabilia (NC10) may mediate AOM in these methanogenic sediments; however we also consider the role of methanogens in active AOM or back flux of methanogenesis. Putative aerobes, such as methane-oxidizing bacteria Methylomonas and their methylotrophic syntrophs Methylotenera, are found among the anaerobic lineages in Lake Kinneret iron-amended slurries and are also involved in the oxidation of methane or its intermediates, as suggested previously. We propose a reaction model for the metabolic interactions in these sediments, linking the potential players that interact via intricate metabolic tradeoffs and direct electron transfer between species. Our results highlight the metabolic complexity of microbial communities in an energy-limited environment, where aerobe and anaerobe communities may co-exist and facilitate AOM as one strategy for survival.
- Article
(3006 KB) - Full-text XML
-
Supplement
(1216 KB) - BibTeX
- EndNote
Methane (CH4) is an effective greenhouse gas (Wuebbles and Hayhoe, 2002). The sources of methane to the atmosphere are of anthropogenic and natural origins, where natural methane is produced mainly biogenically (methanogenesis) as the final remineralization process in anoxic environments (Froelich et al., 1979). The natural sources of methane contribute up to 40 % of the global methane emissions (Saunois et al., 2020), almost entirely from freshwater systems (Bastviken et al., 2011; Zhu et al., 2020). In these freshwater environments, anaerobic oxidation of methane (AOM) consumes over 50 % of the produced methane (Segarra et al., 2015). These environments are usually low in sulfate, and thus other terminal electron acceptors such as metals, iron (Fe) in particular; nitrate; nitrite; and humic acids become available for this process, as explored in recent studies (Raghoebarsing et al., 2006; Ettwig et al., 2010; Adler et al., 2011; Haroon et al., 2013; Norði et al., 2013; Scheller et al., 2016; Bai et al., 2019). However, the diversity and metabolic potential of the microbial communities in natural anoxic ferruginous sediments are not fully understood (Vuillemin et al., 2018).
This study investigates the microbial communities in the sulfate-depleted, iron-rich methanic sediments of Lake Kinneret, a monomictic lake located in the north of Israel at mid-latitudes. The lake is thermally stratified from March until December. During the stratified period, due to the spring phytoplankton bloom decline, the hypolimnion of the lake becomes enriched with particulate organic carbon, and oxygen is gradually depleted (Eckert and Conrad, 2007). At the top 5 cm of the sediment, sulfate reduction is the main process throughout the year (Eckert and Conrad, 2007). At 5 cm below the sediment surface, methanogenesis becomes dominant (Adler et al., 2011). Methane production peaks at 7–12 cm in the sediment while sulfate is depleted (below 10 µM). At 20 cm depth, methane decrease is observed, and ferrous iron concentrations increase to about 200 µM (Adler et al., 2011; Bar-Or et al., 2015). Iron-dependent AOM (Fe-AOM) appears to play a role in methane removal from this deep methanogenic zone of the lake sediments, based on porewater depth profiles, rate modeling, microbial profiles, and incubation experiments on top cores and slurries (Adler et al., 2011; Sivan et al., 2011; Bar-Or et al., 2015, 2017).
Complex microbial consortia mediate the biogeochemical transformations in anoxic lake sediments (Vuillemin et al., 2018). These include methylotrophic and hydrogenotrophic methanogens from the Thermoplasmata, Methanomicrobia, Methanobacteria, and Bathyarchaeota clades, as well as potential sulfate or iron reducers, such as Deltaproteobacteria, Firmicutes, and Nitrospirae lineages (Vuillemin et al., 2018). Alongside the abovementioned lineages, Bar-Or et al. (2015) identified acetoclastic methanogens such as Methanothrix (formerly Methanosaeta, Methanosarcinales, recently reclassified as Methanothrichales by the Genome Taxonomy Database, GTDB; Parks et al., 2018), as well as the H2 and CO2 – using methanogenic Methanomicrobiales genera Methanolinea and Methanoregula in Lake Kinneret sediments. Very few bona fide anaerobic oxidizers of methane (ANME) were found, and the identity of microbes that perform Fe-AOM in these sediments was unknown. Desulfuromondales (Deltaproteobacteria/Desulfobacterota) and Thermodesulfovibrio (Nitrospirae/Nitrospirota) were suggested to couple catabolism of organic substances to the reduction of metals (Bar-Or et al., 2015). We note that deltaproteobacterial lineages have been recently reclassified based on the GTDB (e.g., Desulfuromondales were reclassified as Desulfuromonadota phylum; Parks et al., 2018). Hereafter, we use the “Deltaproteobacteria” terminology to describe the taxonomy of these lineages, as implemented in the Silva 132 database, as the new Genome Taxonomy Database classification has not been peer-reviewed and widely accepted at the time of preparation of this paper.
Here, we use metagenomic analyses to explore the phylogenetic diversity and the metabolic potential of the microbial communities in the deep (27–41 cm) iron-rich methanic part of Lake Kinneret sediments (LK-2017) and the slurry incubations from the Bar-Or et al. (2017) study. These incubations – including (a) 13CH4, (b) 13CH4 + hematite, or (c) 13CH4 + amorphous iron + molybdate (A.Fe(III) + MoO4) – produced substantial amounts of 13C-labeled dissolved inorganic carbon (DIC) over 470 d (80–450 ‰, Fig. S1 in the Supplement). In all the treatments, the diversity of bacteria and archaea was similar to that of the natural sediments. We supplemented these data with analyses of bacterial and archaeal diversity at the 16S rRNA gene level for a wider range of treatments analyzed by Bar-Or et al. (2017). We sought genetic evidence for the ability of the microorganisms to perform iron reduction and produce or oxidize methane and aimed to identify and classify genes that are necessary to catalyze reactions of the respective pathways. As in most natural sediments iron and manganese are present mainly as low-solubility oxide minerals at circumneutral pH (Norði et al., 2013; He et al., 2018) and microbial cells are impermeable to these solid minerals (Shi et al., 2007), we aimed to identify the strategies that may allow microorganisms to cope with this constraint and potentially perform metal-dependent AOM, including (1) direct transfer of electrons to the mineral at the cell surface via an electron carrier, such as cytochrome c (Shi et al., 2007); (2) bridging electrons to a mineral via a cellular appendage, such as conductive nanowires (Gralnick and Newman, 2007; Schwarz et al., 2007; Shi et al., 2016; Lovley and Walker, 2019); (3) indirect electron transfer via metal chelate (Paquete et al., 2014); and (4) indirect electron transfer by electron shuttling compounds, such as quinones or methanophenazines (Newman and Kolter, 2000; Wang and Newman, 2008; He et al., 2019).
2.1 Sample collection, DNA extraction, and sequencing
Lake Kinneret sediments were collected in 2013 from the center of the lake (station A, located at 42 m depth), and slurry incubations were set up under anaerobic conditions, as previously described (Bar-Or et al., 2017). Briefly, sediment from the sediment zone of 26–41 cm was mixed with porewater extracted from the parallel geochemical zone to create homogenized 1 : 5 sediment-to-porewater ratio slurries. The homogenized slurry was transferred under continuous N2 flushing in 40 mL portions into 60 mL bottles, crimped, and sealed. The sediment slurries were amended with 13C-labeled methane and, except the natural sample, treated with different iron oxide minerals. Each sample set was kept (I) without inhibitors, (II) with inhibition of sulfate reduction and sulfur disproportionation by sodium molybdate (Na2MoO4) addition, and (III) inhibition of methanogenesis by 2-bromoethanesulfonate (BES) addition. One of the sample sets was killed (autoclaved) as a control. The various treatments are summarized in the Supplement, Table S1. DNA was extracted from each incubation at the beginning and end of the experiment (after 470 d), and analysis of DNA 16S rRNA genes was performed for all of these incubations and the untreated sediments (Sect. S1 in the Supplement). Four metagenomic libraries (untreated sediments – at the initial time when the sediment arrived at the laboratory and before it was incubated (referred to as t0 hereafter) in 2013; incubations – +13CH4, 13CH4 + hematite, and 13CH4 + amorphous (A.) Fe(III) + MoO4) were prepared at the sequencing core facility at the University of Illinois at Chicago using the Nextera XT DNA library preparation kit (Illumina, USA). Our preliminary analyses of the microbial diversity (16S rRNA amplicons and metagenomics) at t0 in 2013 revealed contamination with common laboratory bacteria, such as Firmicutes and Bacilli (Figs. S2a and S4). To avoid the discovery of contaminant functions, we prepared a DNA library for an additional sample, collected from the same water and sediment depth in 2017 (LK-2017). A total of 12–28 million 2 × 150 bp paired-end reads per library were sequenced using Illumina NextSeq500.
2.2 Bioinformatics
For each library, taxonomic diversity was determined by mapping the reads to the Silva V132 database of the small subunit rRNA sequences using phyloFlash (Glöckner et al., 2017; Gruber-Vodicka et al., 2019), or by using a protein-level classifier, Kaiju (Menzel et al., 2016). The list of normalized taxa abundances, following removal of chloroplast and mitochondria sequences, was used as input for Figs. 1 and S4. Metagenomes were co-assembled from concatenated reads from four metagenomic libraries (LK-2017, +13CH4, 13CH4 + hematite, and 13CH4 + A.Fe(III) + MoO4) with Spades V3.12 (Bankevich et al., 2012; Nurk et al., 2013), following decontamination, quality filtering (QV = 10), and adapter trimming with the BBDuk tool from the BBMap suite (Bushnell B, http://sourceforge.net/projects/bbmap/, last access: 1 August 2020). Downstream analyses, including open reading frame (ORF) prediction, homology, and hidden Markov model (HMM)-based searches against taxonomic and functional databases; estimates of function abundance based on read coverage; and automatic binning were performed with the SqueezeMeta pipeline (Tamames and Puente-Sánchez, 2019). ORFs and Kyoto Encyclopedia of Genes and Genomes (KEGG) functions were quantified based on the mapping of metagenomic reads as counts per million (an equivalent of transcripts per million, TPM, in transcriptomics). To predict the general metabolic functions, we assigned KEGG functions to the 21 categories of the Functional Ontology Assignments for Metagenomes (FOAM) database (Prestat et al., 2014). Automatic binning using metabat2 (Kang et al., 2015) and maxbin (Wu et al., 2015), which was refined using DAStool (Sieber et al., 2018), and manual binning based on differential coverage and guanine–cytosine content with gbtools (Seah and Gruber-Vodicka, 2015) resulted in a limited number of high-quality metagenome-assembled genomes; therefore in this study we looked for specific functions in the metagenomes, followed by homology searches against the RefSeq (O'Leary et al., 2016) and GeneBank databases to evaluate taxonomy. Multiheme cytochromes (MHCs) were identified based on cytochrome c Pfam HMM (PF00034) (Boyd et al., 2019), which revealed only a limited number of sequences. Thus, we identified open reading frames that comprised more than three cytochrome c binding motif sites (CxxCH) (Leu et al., 2020). Putative transmembrane (first 60 amino acids <10, the expected number of amino acids in transmembrane helices >18) and secreted peptides (first 60 amino acids ≥10) were identified with TMHMM V2.0 (Moller et al., 2002). Putative transmembrane (first 60 amino acids <10, the expected number of amino acids in transmembrane helices >18) and secreted peptides (first 60 amino acids ≥10) were identified with TMHMM V2.0 (Moller et al., 2002). Secreted MHCs were also identified by SignalP v5.0, using the archaeal, gram-negative, and gram-positive options (Almagro Armenteros et al., 2019), and the list was curated based on both TMHMM and SignalP annotations. Other functions – including outer-membrane hexaheme c-type cytochrome (OmcS), pilin monomers (PilA), HdrA, HdrD, HdrE, FwdC/FmdC (K00202), Ftr (K00672), Mch (K01499), MtrA (K00577), Mer (K00320), Mtd (K00319), McrA (K00399), and Fpo subunits (K22158-K22170) – were identified based on their KEGG orthology, and their taxonomy was assigned based on BLAST searches against the RefSeq database (amino acid sequences are provided in Sect. S4, S.DB.6–8 in the Supplement). For conservative predictions of electroactive pilins, we selected PilA sequences with ≥9.8 % aromatic amino acids and ≤22-amino-acid aromatic-free gap using a Python script (https://github.com/GlassLabGT/Python-scripts, last access: 1 August 2020), as previously described (Bray et al., 2020).
3.1 Geochemical evidence for iron-coupled AOM in Lake Kinneret iron-rich methanic sediments
We explore here slurries amended with Lake Kinneret sediments from the deep methanic zone (26–41 cm). In this potentially ferruginous zone, sedimentary profiles show that the concentration of methane decreased from its maximum values of above 2 mM at around 10 cm depth to 500 µM at 40 cm depth, and that of dissolved ferrous iron increased (from 1–6 µM at the first 10 cm below the sediment surface to ∼ 60–100 µM, depending on sampling season). This, combined with an increase of δ13C of methane (from about −65 ‰ at 7 cm to −53 ‰ at 24 cm below the sediment surface) and a decrease of δ13C of total lipid compounds (from 27 ‰ at 23 cm to −31 ‰ at 27 cm below the sediment surface), suggests AOM in the deep sediment coupled to iron reduction (Adler et al., 2011; Sivan et al., 2011). The iron-coupled AOM process was supported by rate modeling and by microbial profiles (Adler et al., 2011; Sivan et al., 2011; Bar-Or et al., 2015, 2017). We note that alternative common electron acceptors were scarce: dissolved manganese oxides concentrations were ∼0.04 %, and nitrate and sulfate were below the detection limit (Sivan et al., 2011).
We amended the slurries with isotopically labeled 13CH4, 13CH4 + hematite, and 13CH4 + amorphous iron + molybdate for 470 d. Methane concentration increased with time (by up to ∼650 µM in the treatment with hematite addition, up to ∼1000 µM enrichment in the natural treatment, and up to ∼300 µM in the treatment with amorphous iron + molybdate), reflecting net methanogenesis. In these incubations, we also observed a marked enrichment of labeled carbon in the DIC pool after 10 months of incubation (up to 250 ‰ enrichment in the treatment with hematite addition, up to 80 ‰ enrichment in the natural treatment, and up to 450 ‰ enrichment in the treatment with amorphous iron + molybdate, Fig. S1), indicating conversion of methane to DIC. Ferrous iron concentrations increased by ∼ 20–50 µM following iron oxide amendments (with and without molybdate addition), indicating that iron was reduced. The BES amendments resulted in the highest increase in ferrous iron concentrations (∼ 50–110 µM), most likely due to the abiotic reaction of BES with iron minerals. The evidence for iron reduction, together with the fact that δ13CDIC values increased by 250–450 ‰ in the different iron-amended treatments but not in methane-only additions (only up to 80 ‰, Fig. S1), suggests iron-coupled AOM. Sulfate did not seem to play a role in the AOM, as the addition of molybdate, sulfate reduction, and disproportionation antagonist did not inhibit methane turnover (Fig. S1). The addition of BES to specific slurries inhibited the production of δ13CDIC, indicating the essential role of methanogens in the AOM activity (Fig. S1).
3.2 Diverse microbial consortia mediate biogeochemical cycles in Lake Kinneret sediments
Diverse microbial consortia inhabit Lake Kinneret iron-rich methanic sediments (>20 cm depth, Fig. 1). In these sediments, Bacteria outnumber archaea based on mapping of the metagenomic reads either to the Silva (V132) database of the 16S rRNA gene sequences (73–76 % and 24–27 % reads mapped to bacterial and archaeal sequences, respectively; Sect. S4, S.DB.1) or to the MAR (MARine) database of prokaryotic genomes (81–85 % and 15–19 % reads mapped to bacterial and archaeal sequences, respectively; Sect. S4, S.DB.2). We explored here mainly the microbial community in the deep sediment incubations (>20 cm) amended with 13CH4 alone, 13CH4 with hematite, or 13CH4 with amorphous iron oxides plus molybdate using metagenome analyses (Fig. 1). This diversity of microbes resembled that described previously for Lake Kinneret sedimentary profiles with amplicon sequencing of the 16S rRNA gene (Bar-Or et al., 2015), as well as that determined with either amplicon sequencing or metagenomics in ferruginous lakes across the globe (e.g., Vuillemin et al., 2018; Kadnikov et al., 2019). According to the metagenome analyses, the variation in the diversity of microbial communities in the natural samples and the incubations was small, possibly because iron is not a limiting electron acceptor throughout the sampling interval in the sediments (Sivan et al., 2011). Amplicon sequencing of the bacterial and archaeal 16S rRNA genes in a wider range of treatments (Figs. S2 and S3), which included additions of various iron minerals as well as amendments of molybdate (inhibitor of sulfate reduction) and BES (inhibitor of methanogenesis), also revealed minor changes in the phylogenetic diversity of the microbial consortia.
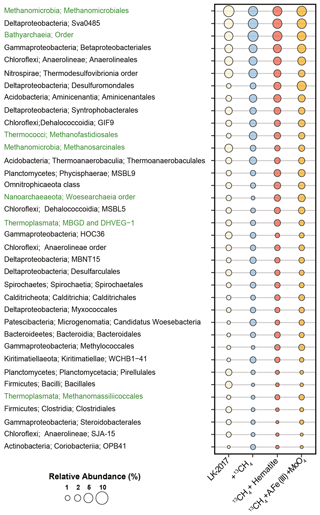
Figure 1Relative abundance of bacteria (black) and archaea (green) at the order level based on mapping of metagenomic reads to the Silva 132 database of the small subunit rRNA sequences. Lineages <1 %, which account together for 28–32 % of the microbial community, were removed from the display. LK 2017 refers to the natural sediments at depth 26 cm at t0.
Anaerolineales (Chloroflexi), Thermodesulfobrionia (Nitrospirae), and the deltaproteobacterial Sva0485 clade were among the most dominant bacterial lineages in these samples (3–6 % read abundance, Fig. 1). While all of these lineages may carry out dissimilatory sulfate reduction (Vuillemin et al., 2018), genetic evidence suggests that bacteria from the Sva0485 clade, which was recently named Candidatus Acidulodesulfobacterales, have the potential to reduce iron (Tan et al., 2019). Sva0485 was suggested recently to be involved in iron reduction also in the methanic zone of marine sediments (Vigderovich et al., 2019). Both these studies revealed a correlation between the distribution of this lineage and ferrous iron concentrations in sediment porewater. Interestingly, Ca. Acidulodesulfobacterales appear to thrive mainly under acidic conditions, while Thermodesulfobrionia was suggested to be either neutrophilic or alkaliphilic (Sekiguchi et al., 2008; Frank et al., 2016). Thus, the co-occurrence of these taxa hints that microenvironments with distinct physicochemical conditions may be present in Lake Kinneret sediments.
Methanomicrobiales (5–7 % read abundance) and Bathyarchaeia (4–7 % read abundance) were the most dominant archaeal lineages in the sediment (Fig. 1). The vast majority of the Methanomicrobiales sequences belonged to Methanoregulaceae genera Methanoregula and Methanolinea, which are known to obtain energy by CO2 reduction to methane, with H2 or formate as electron donors (Bräuer et al., 2015; Imachi and Sakai, 2016). Bathyarchaeia may occur in large numbers in lake sediments (e.g., Vuillemin et al., 2018; Kadnikov et al., 2019; Zhang et al., 2019). The role of these generalists – which are capable of using various carbon and energy sources, often fueling their metabolism through acetogenesis and methanogenesis – is not well understood (Evans et al., 2015; He et al., 2016; Yu et al., 2018; Zhou et al., 2018). Other notable archaeal lineages included the acetoclastic Methanothrix (1–3 % read abundance), which are often found in anoxic lake sediments (Smith and Ingram-Smith, 2007; Schwarz et al., 2007; Carr et al., 2018) as well as in oxygenated soil, as recently discovered for Methanothrix paradoxum (Angle et al., 2017). Their role in the anaerobic degradation of alkanes as syntrophs of Anaerolineaceae has been recently proposed based on enrichment culturing (Liang et al., 2015), as well as their ability to oxidize methane (Valenzuela et al., 2017, 2019). We also identified the putative obligate H2-dependent methylotrophic methanogen lineages Methanofastidiosales (Thermococci, 1–3 % read abundance) and Methanomassiliicoccales (Thermoplasmata, 0.4–1 % read abundance), as well as the putative degraders of detrital proteins Marine Benthic Group D (Thermoplasmata, 1–2 % read abundance) (Lazar et al., 2017; Evans et al., 2019). Less than 1 % of the total reads were mapped to sequences of the anaerobic methanotrophs, such as ANME-1 (0.3–0.8 %), as well as those of the nitrite-reducing methane oxidizers Methylomirabilales (NC10, 0.3–0.6 %, Sect. S4, S.DB.1).
Some type I Methylococcales methanotrophs were found (0.4–1.8 %) in Lake Kinneret sediments. This finding is supported by the quantitative polymerase chain reaction analyses of the pmoA gene (Bar-Or et al., 2017); our analyses of bacteria diversity at the 16S rRNA gene level (Fig. S2); and the 13C-labeled methane carbon incorporation in phospholipid-derived fatty acids that are typical of type I methanotrophs (Bar-Or et al., 2017), suggesting that methane metabolism was active in these bacteria. Methylotrophic Methylotenera (recently reclassified as Burkholderiales, Betaproteobacteriales in Silva 132), which were shown to co-occur with type I methanotrophs under nearly hypoxic conditions (Beck et al., 2013; Cao et al., 2019), were also found (0–1 %, Sect. S4, S.DB.1). The mechanisms behind the increase and elevated activity of the presumably aerobic methanotrophs in the anoxic sediments have not been fully understood, although this phenomenon appears to be widespread (Bar-Or et al., 2017; Martinez-Cruz et al., 2018).
3.3 The general metabolic potential of microbial communities
3.3.1 General metabolic potential
Based on the mapping of the metagenomic reads to ORFs to which KEGG orthology was assigned, several metabolic pathways were found to be dominant in Lake Kinneret iron-rich methanic sediments (Fig. 2). Overall, all four samples had a similar metabolic repertoire. The vast majority of mapped reads were attributed to amino acid utilization and biosynthesis, suggesting that the turnover of organic nitrogen plays an important role in fueling these microbial communities. Indeed, the ORFs that encode the five components of the branched-chain amino acid transport system (KEGG IDs KO1995–KO1999) were among the top 30 most abundant KEGG functions (out of a total of 9058 KEGG IDs that were assigned to the metagenomic ORFs, Sect. S4, S.DB.3). Circa 97 % of ORFs that encoded these components were assigned to bacteria (mainly Deltaproteobacteria and Chloroflexi, 46 and 6 % of reads mapped, respectively). The main intermediates of amino acid catabolism are fatty acids (Park et al., 2014; Narihiro et al., 2016; Aepfler et al., 2019; Scully and Orlygsson, 2019). Indeed, the KEGG IDs that are associated with the β-oxidation of fatty acids, including long-chain acyl-CoA synthetase (K01897), aldehyde : ferredoxin oxidoreductase (K03738), and acetyl-CoA C acetyltransferase (K00626) were among the top 10 most abundant functions, based on read mapping (acyl-CoA dehydrogenase K00249 was among the top 50 most abundant functions, Sect. S4, S.DB.3). Under anoxic conditions, the catabolism of many amino acids and their fatty acid derivatives is not thermodynamically favorable; thus this process is often coupled to syntrophic hydrogenotrophic methanogenesis or sulfate reduction (Sieber et al., 2012; Scully and Orlygsson, 2019; Ziels et al., 2019). In Lake Kinneret deep sediments (>20 cm), the abundant hydrogenotrophic Methanomicrobiales methanogens are the likely syntrophic scavengers of hydrogen. The hydrogen concentration in the deep methanic horizon is ∼20 µM g−1 sediment (Adler, 2015). Given that sulfate is below the detection limit there (<10 µM; Adler et al., 2011; Sivan et al., 2011), hydrogen scavenging may also be coupled to metal reduction, most likely by deltaproteobacterial lineages, some of which may be syntrophic (e.g., Syntrophobacterales). Through syntrophy, amino acids can be converted to acetate and propionate (Narihiro et al., 2016), which further fuel other organisms, including, among others, acetoclastic methanogens.
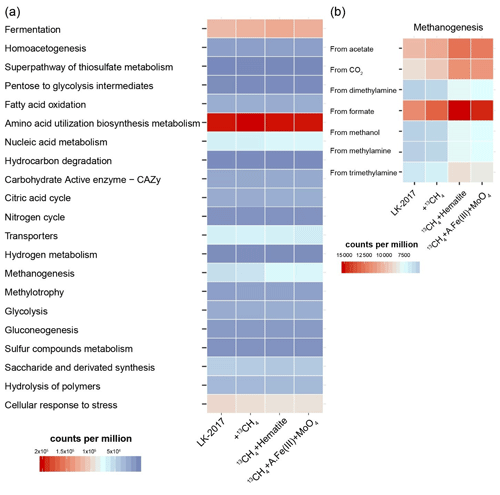
Figure 2The metabolic potential of microbial communities in Lake Kinneret sediments and slurry incubations. Reads were mapped to metagenomic ORFs to which KEGG functions were assigned. KEGG functions were organized and grouped into FOAM (Functional Ontology Assignments for Metagenomes) categories. The relative abundance of each FOAM category is presented in counts per million. (a) General pathways. (b) Zooming into the pathway of methanogenesis. A.Fe(III) + MoO4: amorphous iron and molybdate.
Notably, the best-represented KEGG function was the heterodisulfide reductase subunit A (HdrA, K03388, Sect. S4, S.DB.3). Heterodisulfide reductases play an important role in the energy metabolism of anaerobic bacteria and archaea (such as sulfate reducers and methanogens), driving flavin-based electron bifurcation from various electron donors (Pereira et al., 2011; Ramos et al., 2015; Wagner et al., 2017; Buckel and Thauer, 2018). The HdrA protein itself carries the flavin adenine dinucleotide that is needed for bifurcation (Wagner et al., 2017). In our samples, read mapping suggests that the major taxonomic groups that carry the hdrA genes include Deltaproteobacteria (13–26 %), Chloroflexi (14–21 %), Bathyarchaeia (6–10 %), and Methanomicrobiales (3–4 %) as well as other bacteria and archaea (grouped under “below 3 %”, Fig. S5). These results highlight the presence of bifurcation-driven metabolic processes in Lake Kinneret sediments.
In agreement with previous metagenomic assessments of metabolic pathways in microbes from anoxic lake sediments (Vuillemin et al., 2018), fermentation and methanogenesis account for a substantial part of the metabolic repertoire (Fig. 2a). KEGG IDs that are associated with the fermentative metabolism – such as formate dehydrogenase (K00123), 2-oxoglutarate ferredoxin oxidoreductase (K00174-5), and acetolactate synthase (K01652) – were among the functions with the highest metagenomic coverage (Sect. S4, S.DB.3). Functional analysis of the metagenome suggests that carbon dioxide, formate, acetate, and methylated compounds can fuel methanogenesis (Fig. 2b). These findings are in line with the fact that archaeal lineages known to be capable of using the respective pathways were present (Fig. 1).
3.3.2 Metabolic potential for AOM
In the incubations, methane oxidation was indicated based on the transformation of 13C-labeled methane to 13C DIC and was suggested to involve methanogens due to the inhibition of this transformation by BES (inhibitor of the methyl coenzyme M reductase (McrA)). Based on the taxonomic assignment of genes that encode the enzymes in anaerobic methane metabolism pathways, archaeal lineages from Lake Kinneret iron-rich methanic sediments – such as Methanomicrobiales, Methanosarcinales, and Methanomassiliicoccales (66, 26, and 4 % of reads mapped to K00399 McrA-encoding ORFs, respectively) – are capable of performing the methane transformations (Fig. 3). As described above, all of these lineages are methanogens that convert inorganic carbon, acetate, and methylated compounds to methane under most environmental conditions. No mcrA sequences were assigned to Bathyarchaeia (Fig. 3), in agreement with previous observations of the rare occurrence of the mcrA gene in this lineage (Evans et al., 2015; Lazar et al., 2015; He et al., 2016; Maus et al., 2018). Thus, Bathyarchaeia are most likely not involved directly in methanogenesis or methanotrophy.
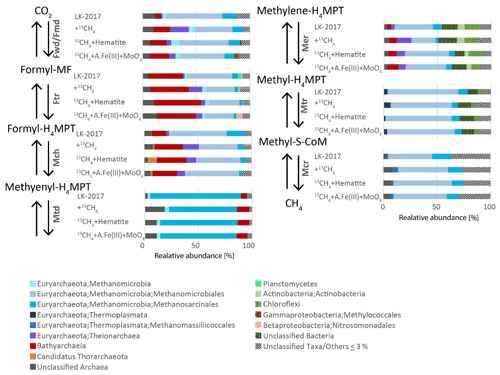
Figure 3Phylogenetic diversity of the seven core genes of methanogenesis from CO2. Phylogenetic assignments are based on BLAST mapping against the RefSeq database. Taxonomic classifications at the highest level possible (up to the order level) are shown. A.Fe(III) + MoO4: amorphous iron and molybdate.
AOM by reversal of methanogenesis is unlikely in obligate hydrogenotrophic methanogens Methanomicrobiales (Rotaru et al., 2014), and it has not been shown also in methylotrophic methanogens Candidatus Methanofastidiosa and Methanomassiliicoccales (Lang et al., 2015; Nobu et al., 2016; Yan and Ferry, 2018). More likely candidates to perform this AOM in Lake Kinneret sediments are ANME-1, which has been shown to perform this process and exist in our deep sediment, and Methanothrix. Methanothrix is closely related to Methanosarcinales (now classified as order Methanothrichales in class Methanosarcinia) and was suggested to perform AOM (Valenzuela et al., 2017, 2019). Several studies suggest that reverse methanogenesis may occur in Methanosarcinales species, based on (1) observation of trace methane oxidation (Zehnder and Brock, 1979; Moran et al., 2005), (2) catalyzation of reverse methanogenesis upon insertion of an mcr gene clone of ANME-1 (Soo et al., 2016), and (3) phylogenetic affiliation of bona fide ANME such as Methanoperedenaceae (ANME-2d) and ANME-2a with this clade (Cai et al., 2018; Yan et al., 2018). Methanothrix was shown to receive electrons from Geobacter or other organotrophs such as Sphaerochaeta during acetoclastic growth (Rotaru et al., 2014; Zhao et al., 2020), suggesting that its cell surface may be conductive. Moreover, the electron transfer enables this strict acetoclastic methanogen to use these electrons and reduce CO2 to methane, a metabolic capability that was unknown in these organisms previously. In line with the abovementioned study, our metagenomic analyses revealed that all the seven enzymes needed to perform both forward and reverse methanogenesis (Fmd/Fwd, Ftr, Mch, Mtd, Mer, Mtr, and Mcr) were assigned to Methanosarcinales (Fig. 3). We note that, since we measured net methanogenesis, the observed 13C-DIC enrichment can represent the back flux of the close-to-equilibrium methanogenesis. In principle, any methanogen can produce DIC via the back-flux reactions, yet we cannot estimate the isotopic fingerprint and the magnitude of this process in this set of incubations. Thermodynamically, both scenarios (back flux or active Fe-AOM) are plausible, and the bona fide Fe-AOM is feasible and competitive with organoclastic iron reduction (ΔG of about −200 kJ mol−1 for both processes (Table S2).
3.4 Genomic evidence for the microbial iron reduction in Lake Kinneret sediment
We asked whether direct extracellular electron transfer, either to a mineral or interspecies, could potentially play a role in Lake Kinneret methane oxidation, in case it is active Fe-AOM, and which taxa may be involved in this process. Direct interspecies electron transfer (DIET) between electrogenic microbes, such as Desulfuromondales, and their partners, such as Methanosarcinales methanogens, requires the presence of electrically conductive pili (e-pili) on the deltaproteobacterial partner, while in both DIET and Fe (metal)-AOM the archaeal partner/methane oxidizer needs to produce either secreted or membranal conductive entities (Rotaru et al., 2014; McGlynn et al., 2015; Holmes et al., 2018; Yan et al., 2018; Walker et al., 2020; Leu et al., 2020). It was previously suggested that thermophilic AOM coupled to sulfate reduction is conducted via DIET between ANME-1 and sulfate-reducing bacteria (Wegener et al., 2015); thus we examined if DIET might also contribute to Fe-AOM in Lake Kinneret.
Our metagenomic results suggest that Lake Kinneret microbiota may transfer electrons directly to extracellular minerals, either through multiheme c-type cytochromes or microbial nanowires, according to mechanisms described in previous reviews (Lovley, 2011; Shi et al., 2016). We investigated the trans-membranal MHCs that are anchored in either the bacterial membrane or archaeal S layer, as well as secreted MHCs. The putative transmembrane and secreted MHCs were encoded by 66 and 592 ORFs per metagenome, respectively. In both MHC types, most of the ORFs were classified as deltaproteobacterial (Fig. 4, 36–52 % in transmembrane MHCs and 29–35 % in secreted MHCs). This is not surprising, as deltaproteobacterial lineages are known to reduce particulate metals through extracellular electron transfer (EET), and to conduct DIET (Leang et al., 2003; Reguera et al., 2005; Lovley, 2011; Adhikari et al., 2016). Other MHC sequences were associated with Nitrospirae, Chloroflexi, and Acidobacteria (both secreted and trans-membranal) and with Actinobacteria (secreted MHCs, Fig. 4). Very few or no archaeal sequences were detected in both types of MHC (0–1.5 %).
In Deltaproteobacteria (Desulfuromonadota) such as Geobacter and Synthrophus, the protein nanowires are assembled from PilA monomers (Lovley and Walker, 2019; Walker et al., 2020). The role of c-type cytochrome OmcS nanowires in DIET has been recently suggested and debated (Filman et al., 2019; Wang et al., 2019; Lovley and Walker, 2019). We surveyed the metagenome for the presence of both PilA- and OmcS-encoding ORFs, most of which were indeed classified as deltaproteobacterial sequences by BLAST against the NCBI nr/nt database (Fig. 4c, d). The overall abundance of the secreted MHCs, trans-membranal MHCs, PilA, and OmcS ORFs was 364–493, 35–45, 4–9, and 4–9 counts per million reads mapped, respectively. Our findings confirm that the phylogenetic diversity of microbes is capable of nanowire-mediated DIET extents beyond deltaproteobacterial lineages (Bray et al., 2020), as strict searches attributed pilA-like sequences not only to Desulfobacterota (Deltaproteobacteria) but also to Thermodesulfovibrionales, Burkholderiales, Gemmatimonadales, Aminicenantales, and WOR-3 and Firmicutes (Fig. 4d). The identified OmcS ORFs were predominantly classified as deltaproteobacterial (36–55 %), and some were assigned as Actinobacteria (0–23 %, Fig. 4c). The vast majority of classified deltaproteobacterial OmcS hits were assigned as Desulfuromondales at the order level (representing 36–55 % overall and 66–100 % out of Deltaproteobacteria). The evidence for the presence of bacteria that transfer electrons via nanowires not only implies DIET but also indicates the potential of sediment microbiota to conduct EET using the particulate metals (Lovley, 2011; Liu et al., 2019b), and thus mediate iron and manganese reduction.
Some methanogens, such as Methanosarcina barkeri and Methanothrix sp., do not possess outer-surface MHCs, yet they are capable of DIET-based syntrophy (Rotaru et al., 2014; Holmes et al., 2018; Yee and Rotaru, 2020). Tubular sheaths that are made of major sheath protein (MspA) may enable some related archaea, including Methanothrix thermophila and Methanospirillum hungatei, to conduct electron transfer (Dueholm et al., 2015; Christensen et al., 2018; Liu et al., 2019a). The protein sheet of Methanothrix shoeghenii was described as concentrating metal ions like iron, copper, nickel, and zinc (Patel et al., 1986). It was thus proposed to give them an advantage in retrieving electrons from extracellular mineral-rich environments (Yee and Rotaru, 2020), such as our study site. BLAST searches using Methanothrix thermophila and Methanospirillum hungatei MspA queries (ABK14853.1 and WP_011449234.1) in Lake Kinneret metagenomes resulted only in poor (11.9–27.5 %) hits of the WP_011449234.1 query, all of which were annotated as a hypothetical Methanoregulaceae protein. Thus, no known Methanosarcinales MspA proteins were detected, and it is still unclear if and how these archaea conduct DIET.
Of the ANME playing a role in AOM, ANME-2d was recently proposed to perform MHC-mediated metal-dependent AOM (McGlynn et al., 2015; Ettwig et al., 2016; Fu et al., 2016; Scheller et al., 2016; Cai et al., 2018). Thus, we used the previously published ANME-2d MHC sequences (Sect. S4, S.DB.4) as a query for performing a BLAST search against the metagenomes. This analysis resulted in zero MHC BLAST hits, corresponding to the fact that only ANME-1 were found among ANME (as 16S rRNA gene sequences and low-quality bins), while ANME-2d were absent. Other Methanosarcinales lineages such as Methanosarcina acetivorans, were also suggested to conduct metal-dependent AOM (Cai et al., 2018; Yan et al., 2018; Leu et al., 2020). In Methanosarcinales, the dimeric membrane-bound HdrDE complex catalyzes the oxidation of CoM and CoB to CoMS–SCoB, and electron shuttling within the membranes is mediated by methanophenazines, which appear to be important in the reduction of extracellular iron by this lineage (Bond and Lovley, 2002; Sivan et al., 2016; Bar-Or et al., 2017; Yan et al., 2018; Holmes et al., 2019). While the taxonomic assignments of ORFs that encoded the HdrD subunit (K08264) were diverse (34–39 % Deltaproteobacteria, 9–14 % Chloroflexi, 2–4 % Bathyarchaeia, and 1.5–3 % Methanosarcinales; Fig. S5), 77–97 % of all the ORFs that encoded the HdrE subunit (K08265) belonged to Methanosarcinales genus Methanothrix (Fig. S5). However, these sequences were scarce (3 : 1000 hdrE to hdrA based on read mapping). We also detected all the possible genes involved in the formation of the membrane-bound coenzyme F420 : methanophenazine dehydrogenase complex Fpo (fpoABCDHIJKLMNO) which couple reduction of F420H2 with methanophenazine oxidation and proton translocation (Welte and Deppenmeier, 2014; Evans et al., 2019; Holmes et al., 2019), and the majority of Fpo-encoding ORFs were classified as Methanothrix (Sect. S4, S.DB.5). As we were unable to identify Methanosarcinales MHCs or proteins that encode other conductive features such as the tubular sheaths, the question regarding their involvement in Lake Kinneret metal-dependent AOM remains open.
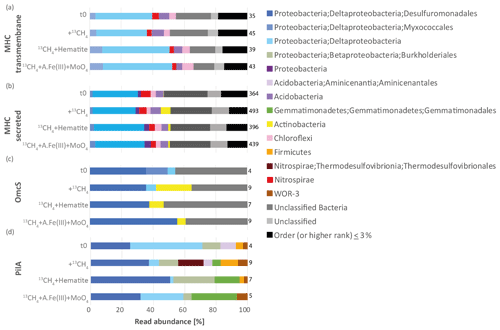
Figure 4Taxonomic affiliation of open reading frames that are needed for extracellular electron transfer, and their relative abundances based on the read mapping. (a) Transmembrane multiheme c-type cytochromes (MHCs), (b) secreted MHC, (c) outer-membrane hexaheme c-type cytochrome (OmcS), and (d) fimbrial protein PilA. Numbers of total reads per million reads mapped to a gene according to the treatments are listed on the right side of each bar plot. A.Fe(III) + MoO4: amorphous iron and molybdate. We note that the taxonomy of several lineages here is based on the NCBI classification and differs from that of the Genome Taxonomy Database (GTDB). In particular, Deltaproteobacteria have been reclassified in the GTDB; thus the Desulfuromondales lineages described here belong to the phylum Desulfobacterota and class Desulfuromonadia, and Myxococcales to the phylum Myxococcota. The phylum Nitrospirae is reassigned as Nitrospirota, and the order Betaproteobacteriales as Burkholderiales in Gammaproteobacteria.
Metagenomic analyses of natural deep iron-rich sediments of Lake Kinneret and slurry incubations from this zone suggest that, similarly to other freshwater systems (e.g., Vuillemin et al., 2018), a community of bacteria and archaea drives the mineralization of organic matter in the deep part of Lake Kinneret sediments, through degradation of amino and fatty acids, as well as hydrogenotrophic, acetoclastic, and methylotrophic methanogenesis (Fig. 5).
We present here a reaction model of possible methane and iron cycling routes (Fig. 5). Metagenomics suggests that the fermenters of amino acids and other products of necromass degradation are the abundant Anaerolineaceae, Thermodesulfobrionia, SVA0485, and Bathyarchaeia. One of the major end products of fermentation is acetate, which can be used as a substrate for the acetoclastic methanogens. Both the acetoclastic methanogenesis and CO2 reduction to methane in these species are likely driven by syntrophy with Desulfuromondales spp., through DIET (Rotaru et al., 2014; Inaba et al., 2019; Wang et al., 2020). The syntrophy between Methanosarcinales with Desulfuromondales is likely in Lake Kinneret deep sediments, based on the fact that the vast majority of ORFs that are needed for the extracellular transfer of electrons were assigned to Desulfuromondales.
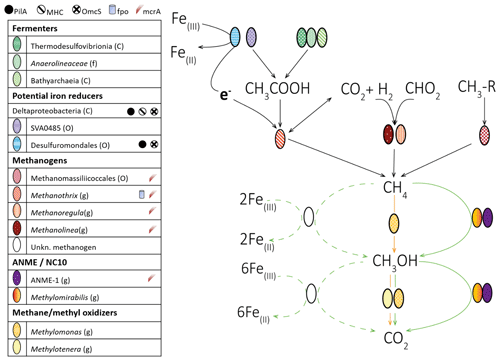
Figure 5The reaction model of possible methane and iron cycling routes in Lake Kinneret deep sediments (>20 cm sediment depth). Bacteria and archaea lineages are represented by ellipses. Functions associated with iron reduction (multiheme cytochromes (MHCs), PilA, OmcS, and the complex Fpo) or methanogenesis (McrA: methyl coenzyme M reductase) are represented by different symbols, and these symbols are shown next to the name of the lineages which possess them. Aerobic (orange), anaerobic (green), and unconfirmed anaerobic (dashed green) routes are displayed by arrows. These routes of methane oxidation may occur in parallel. We note that the CO2 can be produced by back-flux reactions in methanogenesis. The highest assigned taxonomic levels are shown: class (C), order (O), family (F), and genus (G).
Our geochemical isotope labeling experiments suggest that AOM is plausible in the deep methanogenic sediments. AOM may be mediated by ANME, such as ANME-1 that were scarce may be coupled to iron reduction and may involve methanogens which have not been considered as bona fide ANME (Fig. 5). However, we cannot rule out the role of methanogenesis back-flux reactions, which could have produced the isotopically labeled DIC. Both modes of methane oxidation, via the Fe-AOM by an unknown methanogen and the methanogenesis back flux, are justified from the thermodynamic and kinetic perspectives.
Our results, as well as the previous analyses of fatty acids (Bar-Or et al., 2017), suggest that the aerobic methane-oxidizing Methylomonas and its aerobic methylotrophic partner Methylotenera also have a role in methane oxidation in the anaerobic environment. However, the mechanism behind this process is unclear. One possibility is that a slow release of oxygen from particulate matter (Wang et al., 2018) could have fueled methane oxidation by Methylococcales, given that oxidation of methane in the absence of oxygen is unlikely. The alternative is that these lineages may be able to incorporate methanol derived from the incomplete process of reverse methanogenesis (Fig. 5), as was shown for ANME (Xin et al., 2004; Wegener et al., 2016). Another possible explanation for the methylated compound leakage is the reversibility of the enzymes involved in AOM (Thauer and Shima, 2008; Holler et al., 2011), which may lead to the formation of trace amounts of methylated substrates (Wegener et al., 2016). This option of methanol as an intermediate produced by the methanogenic archaea fits with the reported inhibition of methane oxidation upon the addition of BES (Bar-Or et al., 2017). This, as well as the functions of numerous other lineages that comprise the diverse consortia of Lake Kinneret deep iron-rich sediments, remains to be elucidated through further sequencing efforts, cultivation, and experimental studies.
The metagenome and short reads are available as NCBI BioProject accession number PRJNA637457.
The datasets mentioned in the text have been deposited at Figshare in an open-access respiratory and can by accessed at https://doi.org/10.6084/m9.figshare.c.5245157.v1 (Elul et al., 2020; separate links to each dataset can be found in the Supplement).
The supplement related to this article is available online at: https://doi.org/10.5194/bg-18-2091-2021-supplement.
OS and ZR designed the project. ME prepared the samples, extracted DNA, analyzed the data, designed and created the figures, and took the lead in writing the manuscript. IBO provided the samples and analyzed the 16S rRNA amplicons. MRB carried out the bioinformatics analyses and contributed considerably to the interpretation of the results and writing of the manuscript. All co-authors provided critical feedback and helped to shape and write the manuscript.
The authors declare that they have no conflict of interest.
We would like to express our gratitude to Efrat Eliani Russak, Damiana Diaz Reck, and Chen Hargil for their help in sampling and lab work. Special thanks go to Hanni Vigderovich and Noam Lotem for the helpful scientific discussions and advice. Many thanks to Ariel Kushmaro, Eitan Ben Dov, Marcus Elvert, and Jonathan Groop for their collaboration, assistance, and important insight on the research. Much appreciation goes to André Pellerin for his assistance with the thermodynamic calculations and helpful comments on previous versions of the manuscript. We also wish to thank Benny Sulimani and Oz Tzabari from the Yigal Allon Kinneret Limnological Laboratory for their onboard technical assistance. Many thanks to the reviewers for their thorough and constructive reviews.
This work was supported by the European Research Council, H2020 Research Infrastructures (MERIR (grant no. 818450)), and the Israel Science Foundation (grant no. 857-2016 to Orit Sivan and grant no. 913/19 to Maxim Rubin-Blum).
This paper was edited by Tina Treude and reviewed by four anonymous referees.
Adhikari, R. Y., Malvankar, N. S., Tuominen, M. T., and Lovley, D. R.: Conductivity of individual Geobacter pili, RSC Adv., 6, 8354–8357, https://doi.org/10.1039/c5ra28092c, 2016.
Adler, M.: Geochemical Control Mechanisms Regulating the Initiation and Limitation of Methanogenesis in Lacustrine and Estuarine Sediments: A Case Study of Lake Kinneret and the Yarqon River, PhD thesis, Ben-Gurion University of the Negev, Israel, 103 pp., 2015.
Adler, M., Eckert, W., and Sivan, O.: Quantifying rates of methanogenesis and methanotrophy in Lake Kinneret sediments (Israel) using pore-water profiles, Limnol. Oceanogr., 56, 1525–1535, https://doi.org/10.4319/lo.2011.56.4.1525, 2011.
Aepfler, R. F., Bühring, S. I., and Elvert, M.: Substrate characteristic bacterial fatty acid production based on amino acid assimilation and transformation in marine sediments, FEMS Microbiol. Ecol., 95, fiz131, https://doi.org/10.1093/femsec/fiz131, 2019.
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., von Heijne, G., and Nielsen, H.: SignalP 5.0 improves signal peptide predictions using deep neural networks, Nat. Biotechnol, 37, 420–423, https://doi.org/10.1038/s41587-019-0036-z, 2019.
Angle, J. C., Morin, T. H., Solden, L. M., Narrowe, A. B., Smith, G. J., Borton, M. A., Rey-Sanchez, C., Daly, R. A., Mirfenderesgi, G., Hoyt, D. W., Riley, W. J., Miller, C. S., Bohrer, G., and Wrighton, K. C.: Methanogenesis in oxygenated soils is a substantial fraction of wetland methane emissions, Nat. Commun., 8, 1567, https://doi.org/10.1038/s41467-017-01753-4, 2017.
Bai, Y. N., Wang, X. N., Wu, J., Lu, Y. Z., Fu, L., Zhang, F., Lau, T. C., and Zeng, R. J.: Humic substances as electron acceptors for anaerobic oxidation of methane driven by ANME-2d, Water Res., 164, 114935, https://doi.org/10.1016/j.watres.2019.114935, 2019.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., Lesin, V. M., Nikolenko, S. I., Pham, S., Prjibelski, A. D., Pyshkin, A. V., Sirotkin, A. V., Vyahhi, N., Tesler, G., Alekseyev, M. A., and Pevzner, P. A.: SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing, J. Comput. Biol., 19, 455–477, https://doi.org/10.1089/cmb.2012.0021, 2012.
Bar-Or, I., Ben-Dov, E., Kushmaro, A., Eckert, W., and Sivan, O.: Methane-related changes in prokaryotes along geochemical profiles in sediments of Lake Kinneret (Israel), Biogeosciences, 12, 2847–2860, https://doi.org/10.5194/bg-12-2847-2015, 2015.
Bar-Or, I., Elvert, M., Eckert, W., Kushmaro, A., Vigderovich, H., Zhu, Q., Ben-Dov, E., and Sivan, O.: Iron-coupled anaerobic oxidation of methane performed by a mixed bacterial-archaeal community based on poorly reactive minerals, Environ. Sci. Technol., 51, 12293–12301, https://doi.org/10.1021/acs.est.7b03126, 2017.
Bastviken, D., Tranvik, L. J., Downing, J. A., Crill, P. M., and Enrich-Prast, A.: Freshwater methane emissions offset the continental carbon sink, Science, 331, 50, https://doi.org/10.1126/science.1196808, 2011.
Beck, D. A. C., Kalyuzhnaya, M. G., Malfatti, S., Tringe, S. G., Glavina del Rio, T., Ivanova, N., Lidstrom, M. E., and Chistoserdova L.: A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between the Methylococcaceae and the Methylophilaceae, PeerJ, 1, e23, https://doi.org/10.7717/peerj.23, 2013.
Bond, D. R. and Lovley, D. R.: Reduction of Fe(III) oxide by methanogens in the presence and absence of extracellular quinones, Environ. Microbiol., 4, 115–124, https://doi.org/10.1046/j.1462-2920.2002.00279.x, 2002.
Boyd, J. A., Jungbluth, S. P., Leu, A. O., Evans, P. N., Woodcroft, B. J., Chadwick, G. L., Orphan, V. J., Amend, J. P., Rappé, M. S., and Tyson, G. W.: Divergent methyl-coenzyme M reductase genes in a deep-subseafloor Archaeoglobi, ISME J., 13, 1269–1279, https://doi.org/10.1038/s41396-018-0343-2, 2019.
Bräuer, S., Cadillo-Quiroz, H., Kyrpides, N., Woyke, T., Goodwin, L., Detter, C., Podell, S., Yavitt, J. B., and Zinder, S. H.: Genome of Methanoregula boonei 6A8 reveals adaptations to oligotrophic peatland environments, Microbiology+, 161, 1572–1581, https://doi.org/10.1099/mic.0.000117, 2015.
Bray, M. S., Wu, J., Padilla, C. C., Stewart, F. J., Fowle, D. A., Henny, C., Simister, R. L., Thompson, K. J., Crowe, S. A., and Glass, J. B.: Phylogenetic and structural diversity of aromatically dense pili from environmental metagenomes, Environ. Microbiol. Rep., 12, 49–57, https://doi.org/10.1111/1758-2229.12809, 2020.
Buckel, W. and Thauer, R. K.: Flavin-based electron bifurcation, ferredoxin, flavodoxin, and anaerobic respiration with protons (Ech) or NAD+ (Rnf) as electron acceptors: A historical review, Front. Microbiol., 9, 401, https://doi.org/10.3389/fmicb.2018.00401, 2018.
Cai, C., Leu, A. O., Xie, G.-J., Guo, J., Feng, Y., Zhao, J.-X., Tyson, G. W., Yuan, Z., and Hu, S.: A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction, ISME J., 12, 1929–1939, https://doi.org/10.1038/s41396-018-0109-x, 2018.
Cao, Q., Liu, X., Li, N., Xie, Z., Li, Z., and Li, D.: Stable-isotopic analysis and high-throughput pyrosequencing reveal the coupling process and bacteria in microaerobic and hypoxic methane oxidation coupled to denitrification, Environ. Pollut., 250, 863–872, https://doi.org/10.1016/j.envpol.2019.04.111, 2019.
Carr, S. A., Schubotz, F., Dunbar, R. B., Mills, C. T., Dias, R., Summons, R. E., and Mandernack, K. W.: Acetoclastic Methanosaeta are dominant methanogens in organic-rich antarctic marine sediments, ISME J., 12, 330–342, https://doi.org/10.1038/ismej.2017.150, 2018.
Christensen, L. F. B., Hansen, L. M., Finster, K., Christiansen, G., Nielsen, P. H., Otzen, D. E., and Dueholm, M. S.: The sheaths of Methanospirillum are made of a new type of amyloid protein, Front. Microbiol., 9, 1–11, https://doi.org/10.3389/fmicb.2018.02729, 2018.
Dueholm, M. S., Larsen, P., Finster, K., Stenvang, M. R., Christiansen, G., Vad, B. S., Bøggild, A., Otzen, D. E., and Nielsen, P. H.: The tubular sheaths encasing Methanosaeta thermophila filaments are functional amyloids, J. Biol. Chem., 290, 20590–20600, https://doi.org/10.1074/jbc.M115.654780, 2015.
Eckert, W. and Conrad, R.: Sulfide and methane evolution in the hypolimnion of a subtropical lake: A three-year study, Biogeochemistry, 82, 67–76, https://doi.org/10.1007/s10533-006-9053-3, 2007.
Elul, M., Rubin-Blum, M., Ronen, Z., Bar-Or, I., Sivan, O., and Eckert, W.: Metagenomic insights into the metabolism of microbial communities that mediate iron and methane cycling in Lake Kinneret iron-rich methanic sediments, figshare, data set Collection, https://doi.org/10.6084/m9.figshare.c.5245157.v1, 2020.
Ettwig, K. F., Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M. M., Schreiber, F., Dutilh, B. E., Zedelius, J., de Beer, D., Gloerich, J., Wessels, H. J. C. T., van Alen, T., Luesken, F., Wu, M. L., van de Pas-Schoonen, K. T., Op den Camp, H. J. M., Janssen-Megens, E. M., Francoijs, K.-J., Stunnenberg, H., Weissenbach, J., Jetten, M. S. M., and Strous, M.: Nitrite-driven anaerobic methane oxidation by oxygenic bacteria, Nature, 464, 543–548, https://doi.org/10.1038/nature08883, 2010.
Ettwig, K. F., Zhu, B., Speth, D., Keltjens, J. T., Jetten, M. S. M., and Kartal, B.: Archaea catalyze iron-dependent anaerobic oxidation of methane, P. Natl. Acad. Sci. USA, 113, 12792–12796, https://doi.org/10.1073/pnas.1609534113, 2016.
Evans, P. N., Parks, D. H., Chadwick, G. L., Robbins, S. J., Orphan, V. J., Golding, S. D., and Tyson, G. W.: Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics, Science, 350, 434–438, https://doi.org/10.1126/science.aac7745, 2015.
Evans, P. N., Boyd, J. A., Leu, A. O., Woodcroft, B. J., Parks, D. H., Hugenholtz, P., and Tyson, G. W.: An evolving view of methane metabolism in the Archaea, Nat. Rev. Microbiol., 17, 219–232, https://doi.org/10.1038/s41579-018-0136-7, 2019.
Filman, D. J., Marino, S. F., Ward, J. E., Yang, L., Mester, Z., Bullitt, E., Lovley, D. R., and Strauss, M.: Cryo-EM reveals the structural basis of long-range electron transport in a cytochrome-based bacterial nanowire, Commun. Biol., 2, 19–24, https://doi.org/10.1038/s42003-019-0448-9, 2019.
Frank, Y. A., Kadnikov, V. V., Lukina, A. P., Banks, D., Beletsky, A. V., Mardanov, A. V., Sen'kina, E. I., Avakyan, M. R., Karnachuk, O. V., and Ravin, N. V.: Characterization and genome analysis of the first facultatively alkaliphilic Thermodesulfovibrio isolated from the deep terrestrial subsurface, Front. Microbiol., 7, 1–11, https://doi.org/10.3389/fmicb.2016.02000, 2016.
Froelich, P. N., Klinkhammer, G. P., Bender, M. L., Luedtke, N. A., Heath, G. R., Cullen, D., Dauphin, P., Hammond, D., Hartman, B., and Maynard, V.: Early oxidation of organic matter in pelagic sediments of the eastern equatorial Atlantic: suboxic diagenesis, Geochim. Cosmochim. Ac., 43, 1075–1090, https://doi.org/10.1016/0016-7037(79)90095-4, 1979.
Fu, L., Li, S.-W., Ding, Z.-W., Ding, J., Lu, Y.-Z., and Zeng, R. J.: Iron reduction in the DAMO/Shewanella oneidensis MR-1 coculture system and the fate of Fe(II), Water Res., 88, 808–815, https://doi.org/10.1016/J.WATRES.2015.11.011, 2016.
Glöckner, F. O., Yilmaz, P., Quast, C., Gerken, J., Beccati, A., Ciuprina, A., Bruns, G., Yarza, P., Peplies, J., Westram, R., and Ludwig, W.: 25 years of serving the community with ribosomal RNA gene reference databases and tools, J. Biotechnol., 261, 169–176, https://doi.org/10.1016/j.jbiotec.2017.06.1198, 2017.
Gralnick, J. A. and Newman. D. K.: Extracellular respiration, Mol. Microbiol., 65, 1–11, https://doi.org/10.1111/j.1365-2958.2007.05778.x, 2007.
Gruber-Vodicka, H. R., Seah, B. K., and Pruesse, E.: phyloFlash – Rapid SSU rRNA profiling and targeted assembly from metagenomes, bioRxiv, 521922, https://doi.org/10.1101/521922, 2019.
Haroon, M. F., Hu, S., Shi, Y., Imelfort, M., Keller, J., Hugenholtz, P., Yuan, Z., and Tyson, G. W.: Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage, Nature, 500, 567–570, https://doi.org/10.1038/nature12375, 2013.
He, Q., Yu, L., Li, J., He, D., Cai, X., and Zhou, S.: Electron shuttles enhance anaerobic oxidation of methane coupled to iron(III) reduction, Sci. Total Environ., 688, 664–672, https://doi.org/10.1016/J.SCITOTENV.2019.06.299, 2019.
He, Y., Li, M., Perumal, V., Feng, X., Fang, J., Xie, J., Sievert, S. M., and Wang, F.: Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments, Nat. Microbiol., 1, 16035, https://doi.org/10.1038/nmicrobiol.2016.35, 2016.
He, Z., Zhang, Q., Feng, Y., Luo, H., Pan, X., and Gadd, G. M.: Microbiological and environmental significance of metal-dependent anaerobic oxidation of methane, Sci. Total Environ., 610–611, 759–768, https://doi.org/10.1016/j.scitotenv.2017.08.140, 2018.
Holler, T., Wegener, G., Niemann, H., Deusner, C., Ferdelman, T. G., Boetius, A., Brunner, B., and Widdel, F.: Carbon and sulfur back flux during anaerobic microbial oxidation of methane and coupled sulfate reduction, P. Natl. Acad. Sci. USA, 108, E1484–E1490, https://doi.org/10.1073/pnas.1106032108, 2011.
Holmes, D. E., Rotaru, A. E., Ueki, T., Shrestha, P. M., Ferry, J. G., and Lovley, D. R.: Electron and proton flux for carbon dioxide reduction in methanosarcina barkeri during direct interspecies electron transfer, Front. Microbiol., 9, 1–11, https://doi.org/10.3389/fmicb.2018.03109, 2018.
Holmes, D. E., Ueki, T., Tang, H., Zhou, J., Smith, J. A., Chaput, G., and Lovley, D. R.: A membrane-bound cytochrome enables Methanosarcina acetivorans to conserve energy from extracellular electron transfer, MBio, 10, 1–12, https://doi.org/10.1128/mBio.00789-19, 2019.
Imachi, H. and Sakai, S.: Methanoregulaceae, in: Bergey's manual of systematics of Archaea and Bacteria, edited by: Bergey, D. H., Harrison, F. C., Breed, R. S., Hammer, B. W., Huntoon, F. M., Wiley, New York, NY, 1–4, https://doi.org/10.1002/9781118960608.fbm00271, 2016.
Inaba, R., Nagoya, M., Kouzuma, A., and Watanabe K.: Metatranscriptomic evidence for magnetite nanoparticle-stimulated acetoclastic methanogenesis under continuous agitation, Appl. Environ. Microb., 85, e01733-19, https://doi.org/10.1128/AEM.01733-19, 2019.
Kadnikov, V. V., Savvichev, A. S., Mardanov, A. V., Beletsky, A. V., Merkel, A. Y., Ravin, N. V., and Pimenov, N. V.: Microbial communities involved in the methane cycle in the near-bottom water layer and sediments of the meromictic subarctic Lake Svetloe, Antonie van Leeuwenhoek, 112, 1801–1814, https://doi.org/10.1007/s10482-019-01308-1, 2019.
Kang, D. D., Froula, J., Egan, R., and Wang, Z.: MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities, PeerJ, 3, e1165, https://doi.org/10.7717/peerj.1165, 2015.
Lang, K., Schuldes, J., Klingl, A., Poehlein, A., Daniel, R., and Brune, A.: New mode of energy metabolism in the seventh order of methanogens as revealed by comparative genome analysis of “Candidatus Methanoplasma termitum.”, Appl. Environ. Microb., 81, 1338–1352, https://doi.org/10.1128/AEM.03389-14, 2015.
Lazar, C. S., Biddle, J. F., Meador, T. B., Blair, N., Hinrichs, K. U., and Teske, A. P.: Environmental controls on intragroup diversity of the uncultured benthic archaea of the miscellaneous Crenarchaeotal group lineage naturally enriched in anoxic sediments of the White Oak River estuary (North Carolina, USA), Environ. Microbiol., 17, 2228–2238, https://doi.org/10.1111/1462-2920.12659, 2015.
Lazar, C. S., Baker, B. J., Seitz, K. W., and Teske, A. P.: Genomic reconstruction of multiple lineages of uncultured benthic archaea suggests distinct biogeochemical roles and ecological niches, ISME J., 11, 1118–1129, https://doi.org/10.1038/ismej.2016.189, 2017.
Leang, C., Coppi, M. V., and Lovley, D. R.: OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens, J. Bacteriol., 185, 2096–2103, https://doi.org/10.1128/JB.185.7.2096-2103.2003, 2003.
Leu, A. O., Cai, C., McIlroy, S. J., Southam, G., Orphan, V. J., Yuan, Z., Hu, S., and Tyson, G. W.: Anaerobic methane oxidation coupled to manganese reduction by members of the Methanoperedenaceae, ISME J., 1–12, https://doi.org/10.1038/s41396-020-0590-x, 2020.
Liang, B., Wang, L.-Y., Mbadinga, S. M., Liu, J.-F., Yang, S.-Z., Gu, J.-D., and Mu, B.-Z.: Anaerolineaceae and Methanosaeta turned to be the dominant microorganisms in alkanes-dependent methanogenic culture after long-term of incubation, AMB Express, 5, 37, https://doi.org/10.1186/s13568-015-0117-4, 2015.
Liu, C., Sun, D., Zhao, Z., Dang, Y., and Holmes, D. E.: Methanothrix enhances biogas upgrading in microbial electrolysis cell via direct electron transfer, Bioresour. Technol., 291, 121877, https://doi.org/10.1016/j.biortech.2019.121877, 2019a.
Liu, X., Ye, Y., Xiao, K., Rensing, C., and Zhou, S.: Molecular evidence for the adaptive evolution of Geobacter sulfurreducens to perform dissimilatory iron reduction in natural environments, Mol. Microbiol., 289, mmi.14443, https://doi.org/10.1111/mmi.14443, 2019b.
Lovley, D. R.: Live wires: direct extracellular electron exchange for bioenergy and the bioremediation of energy-related contamination, Energy Environ. Sci., 4, 4896–4906, https://doi.org/10.1039/c1ee02229f, 2011.
Lovley, D. R. and Walker, D. J. F.: Geobacter protein nanowires, Front. Microbiol., 10, 2078, https://doi.org/10.3389/fmicb.2019.02078, 2019.
Martinez-Cruz, K., Sepulveda-Jauregui, A., Casper, P., Anthony, K. W., Smemo, K. A., and Thalasso, F.: Ubiquitous and significant anaerobic oxidation of methane in freshwater lake sediments, Water Res., 144, 332–340, https://doi.org/10.1016/j.watres.2018.07.053, 2018.
Maus, I., Rumming, M., Bergmann, I., Heeg, K., Pohl, M., Nettmann, E., Jaenicke, S., Blom, J., Pühler, A., Schlüter, A., Sczyrba, A., and Klocke, M.: Characterization of Bathyarchaeota genomes assembled from metagenomes of biofilms residing in mesophilic and thermophilic biogas reactors, Biotechnol. Biofuels, 11, 167, https://doi.org/10.1186/s13068-018-1162-4, 2018.
McGlynn, S. E., Chadwick, G. L., Kempes, C. P., and Orphan, V. J.: Single cell activity reveals direct electron transfer in methanotrophic consortia, Nature, 526, 531–535, https://doi.org/10.1038/nature15512, 2015.
Menzel, P., Ng, K. L., and Krogh, A.: Fast and sensitive taxonomic classification for metagenomics with Kaiju, Nat. Commun. 7, 1–9, https://doi.org/10.1038/ncomms11257, 2016.
Moller, S., Croning, M. D. R., and Apweiler, R..: Evaluation of methods for the prediction of membrane spanning regions, Bioinformatics, 18, 218–218, https://doi.org/10.1093/bioinformatics/18.1.218, 2002.
Moran, J. J., House, C. H., Freeman, K. H., and Ferry, J. G.: Trace methane oxidation studied in several Euryarchaeota under diverse conditions, Archaea, 303–309, 2005.
Narihiro, T., Nobu, M. K., Tamaki, H., Kamagata, Y., Sekiguchi, Y., and Liu, W. T.: Comparative genomics of syntrophic branched-chain fatty acid degrading bacteria, Microbes Environ., 31, 288–292, https://doi.org/10.1264/jsme2.ME16057, 2016.
Newman, D. K. and Kolter, R.: A role for excreted quinones in extracellular electron transfer, Nature, 405, 94–97, https://doi.org/10.1038/35011098, 2000.
Nobu, M. K., Narihiro, T., Kuroda, K., Mei, R., and Liu, W.-T.: Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen, ISME J., 10, 2478–2487, https://doi.org/10.1038/ismej.2016.33, 2016.
Norði, K. À., Thamdrup, B., and Schubert, C. J.: Anaerobic oxidation of methane in an iron-rich Danish freshwater lake sediment, Limnol. Oceanogr, 58, 546–554, https://doi.org/10.4319/lo.2013.58.2.0546, 2013.
Nurk, S., Bankevich, A., and Antipov, D.: Assembling genomes and mini-metagenomes from highly chimeric reads, Res. Comput. Mol. Biol., 7821, 158–170, https://doi.org/10.1007/978-3-642-37195-0, 2013.
O'Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., Rajput, B., Robbertse, B., Smith-White, B., Ako-Adjei, D., Astashyn, A., Badretdin, A., Bao, Y., Blinkova, O., Brover, V., Chetvernin, V., Choi, J., Cox, E., Ermolaeva, O., Farrell, C. F., Goldfarb, T., Gupta, T., Haft, D., Hatcher, E., Hlavina, W., Joardar, V. S., Kodali, V. K., Li, W., Maglott, D., Masterson, P., McGarvey, K. M., Murphy, M. R., O'Neill, K., Pujar, S., Rangwala, S. H., Rausch, D., Riddick, L. D., Schoch, C., Shkeda, A., Storz, S. S., Sun, H., Thibaud-Nissen, F., Tolstoy, I., Tully, R. E., Vatsan, A. R., Wallin, C., Webb, D., Wu, W., Landrum, M. J., Kimchi, A., Tatusova, T., DiCuccio, M., Kitts, P., Murphy, T. D., and Pruitt, K. D.: Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation, Nucleic Acids Res., 44, D733–D745, https://doi.org/10.1093/nar/gkv1189, 2016.
Paquete, C. M., Fonseca, B. M., Cruz, D. R., Pereira, T. M., Pacheco, I., Soares, C. M., and Louro, R. O.: Exploring the molecular mechanisms of electron shuttling across the microbe/metal space, Front. Microbiol., 5, 1–12, https://doi.org/10.3389/fmicb.2014.00318, 2014.
Park, J., Park, S., and Kim, M.: Anaerobic degradation of amino acids generated from the hydrolysis of sewage sludge, Environ. Technol., 35, 1133–1139, https://doi.org/10.1080/09593330.2013.863951, 2014.
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P.-A., and Hugenholtz, P.: A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life, Nat. Biotechnol., 36, 996–1004, 2018.
Patel, G. B., Sprott, G. D., Humphrey, R. W., and Beveridge, T. J.: Comparative analyses of the sheath structures of Methanothrix concilii GP6 and Methanospirillum hungatei strains GP1 and JF1, Can. J. Microbiol., 32, 623–631, https://doi.org/10.1139/m86-117, 1986.
Pereira, I. A. C., Ramos, A. R., Grein, F., Marques, M. C., da Silva, S. M., and Venceslau, S. S.: A comparative genomic analysis of energy metabolism in sulfate reducing Bacteria and Archaea, Front. Microbiol., 2, 1–22, https://doi.org/10.3389/fmicb.2011.00069 2011.
Prestat, E., David, M. M., Hultman, J., Taş, N., Lamendella, R., Dvornik, J., Mackelprang, R., Myrold, D. D., Jumpponen, A., Tringe, S. G., Holman, E., Mavromatis, K., and Jansson, J. K: FOAM (Functional Ontology Assignments for Metagenomes): a Hidden Markov Model (HMM) database with environmental focus, Nucleic Acids Res., 42, e145–e145, https://doi.org/10.1093/nar/gku702, 2014.
Raghoebarsing, A. A., Pol, A., Van De Pas-Schoonen, K. T., Smolders, A. J. P., Ettwig, K. F., Rijpstra, W. I. C., Schouten, S., Sinninghe Damsté, J. S., Op den Camp, H. J. M., Jetten, M. S. M., and Strous, M.: A microbial consortium couples anaerobic methane oxidation to denitrification, Nature, 440, 918–921, https://doi.org/10.1038/nature04617, 2006.
Ramos, A. R., Grein, F., Oliveira, G. P., Venceslau, S. S., Keller, K. L., Wall, J. D., and Pereira, I. A. C.: The FlxABCD-HdrABC proteins correspond to a novel NADH dehydrogenase/heterodisulfide reductase widespread in anaerobic bacteria and involved in ethanol metabolism in Desulfovibrio vulgaris Hildenborough, Environ. Microbiol., 17, 2288–2305, https://doi.org/10.1111/1462-2920.12689, 2015.
Reguera, G., McCarthy, K. D., Mehta, T., Nicoll, J. S., Tuominen, M. T., and Lovley, D. R.: Extracellular electron transfer via microbial nanowires, Nature, 435, 1098–1101, https://doi.org/10.1038/nature03661, 2005.
Rotaru, A. E., Shrestha, P. M., Liu, F., Shrestha, M., Shrestha, D., Embree, M., Zengler, K., Wardman, C., Nevin, K. P., and Lovley, D. R.: A new model for electron flow during anaerobic digestion: Direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane, Energy Environ. Sci. 7, 408–415, https://doi.org/10.1039/c3ee42189a, 2014.
Saunois, M., Stavert, A. R., Poulter, B., Bousquet, P., Canadell, J. G., Jackson, R. B., Raymond, P. A., Dlugokencky, E. J., Houweling, S., Patra, P. K., Ciais, P., Arora, V. K., Bastviken, D., Bergamaschi, P., Blake, D. R., Brailsford, G., Bruhwiler, L., Carlson, K. M., Carrol, M., Castaldi, S., Chandra, N., Crevoisier, C., Crill, P. M., Covey, K., Curry, C. L., Etiope, G., Frankenberg, C., Gedney, N., Hegglin, M. I., Höglund-Isaksson, L., Hugelius, G., Ishizawa, M., Ito, A., Janssens-Maenhout, G., Jensen, K. M., Joos, F., Kleinen, T., Krummel, P. B., Langenfelds, R. L., Laruelle, G. G., Liu, L., Machida, T., Maksyutov, S., McDonald, K. C., McNorton, J., Miller, P. A., Melton, J. R., Morino, I., Müller, J., Murguia-Flores, F., Naik, V., Niwa, Y., Noce, S., O'Doherty, S., Parker, R. J., Peng, C., Peng, S., Peters, G. P., Prigent, C., Prinn, R., Ramonet, M., Regnier, P., Riley, W. J., Rosentreter, J. A., Segers, A., Simpson, I. J., Shi, H., Smith, S. J., Steele, L. P., Thornton, B. F., Tian, H., Tohjima, Y., Tubiello, F. N., Tsuruta, A., Viovy, N., Voulgarakis, A., Weber, T. S., van Weele, M., van der Werf, G. R., Weiss, R. F., Worthy, D., Wunch, D., Yin, Y., Yoshida, Y., Zhang, W., Zhang, Z., Zhao, Y., Zheng, B., Zhu, Q., Zhu, Q., and Zhuang, Q.: The Global Methane Budget 2000–2017, Earth Syst. Sci. Data, 12, 1561–1623, https://doi.org/10.5194/essd-12-1561-2020, 2020.
Scheller, S., Yu, H., Chadwick, G. L., McGlynn, S. E., and Orphan, V. J.: Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction, Science, 351, 703–707, https://doi.org/10.1126/SCIENCE.AAD7154, 2016.
Schwarz, J. I. K., Eckert, W., and Conrad, R.: Community structure of Archaea and Bacteria in a profundal lake sediment Lake Kinneret (Israel), Syst. Appl. Microbiol., 30, 239–254, https://doi.org/10.1016/J.SYAPM.2006.05.004, 2007.
Scully, S. M. and Orlygsson, J.: Branched-chain amino acid catabolism of Thermoanaerobacter pseudoethanolicus reveals potential route to branched-chain alcohol formation, Extremophiles, 24, 121–133, https://doi.org/10.1007/s00792-019-01140-5, 2019.
Seah, B. K. B. and Gruber-Vodicka, H. R.: gbtools: Interactive visualization of metagenome bins in R. Front, Microbiology+, 6, 1451, https://doi.org/10.3389/fmicb.2015.01451, 2015.
Segarra, K. E. A., Schubotz, F., Samarkin, V., Yoshinaga, M. Y., Hinrichs, K. U., and Joye, S. B.: High rates of anaerobic methane oxidation in freshwater wetlands reduce potential atmospheric methane emissions, Nat. Commun., 6, 1–8, https://doi.org/10.1038/ncomms8477, 2015.
Sekiguchi, Y., Muramatsu, M., Imachi, H., Narihiro, T., Ohashi, A., Harada, H., Hanada, S., and Kamagata, Y.: Thermodesulfovibrio aggregans sp. nov. and Thermodesulfovibrio thiophilus sp. nov., anaerobic,thermophilic, sulfate-reducing bacteria isolated from thermophilic methanogenic sludge, and emended description of the genus Thermodesulfovibrio, Int. J. Syst. Evol. Microbiol., 58, 2541–2548, https://doi.org/10.1099/ijs.0.2008/000893-0, 2008.
Shi, L., Dong, H., Reguera, G., Beyenal, H., Lu, A., Liu, J., Yu, H.-Q., and Fredrickson, J. K.: Extracellular electron transfer mechanisms between microorganisms and minerals, Nat. Rev. Microbiol., 14, 651–662, https://doi.org/10.1038/nrmicro.2016.93, 2016.
Shi, L., Squier,T. C., Zachara, J. M., and Fredrickson, J. K.: Respiration of metal (hydr)oxides by Shewanella and Geobacter: A key role for multihaem c-type cytochromes, Mol. Microbiol., 65, 12–20, https://doi.org/10.1111/j.1365-2958.2007.05783.x, 2007.
Sieber, C. M. K., Probst, A. J., Sharrar, A., Thomas, B. C., Hess, M., Tringe, S. G., and Banfield, J. F.: Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy, Nat. Microbiol., 3, 836–843, https://doi.org/10.1038/s41564-018-0171-1, 2018.
Sieber, J. R., McInerney, M. J., and Gunsalus, R. P.: Genomic insights into syntrophy: The paradigm for anaerobic metabolic cooperation, Annu. Rev. Microbiol., 66, 429–452, https://doi.org/10.1146/annurev-micro-090110-102844, 2012.
Sivan, O., Adler, M., Pearson, A., Gelman, F., Bar-Or, I., John, S. G., and Eckert, W.: Geochemical evidence for iron-mediated anaerobic oxidation of methane, Limnol. Oceanogr., 56, 1536–1544, https://doi.org/10.4319/lo.2011.56.4.1536, 2011.
Sivan, O., Shusta, S. S., and Valentine, D. L.: Methanogens rapidly transition from methane production to iron reduction, Geobiology, 14, 190–203, https://doi.org/10.1111/gbi.12172, 2016.
Smith, K. S. and Ingram-Smith, C.: Methanosaeta, the forgotten methanogen? Trends Microbiol., 15, 150–155, https://doi.org/10.1016/j.tim.2007.02.002, 2007.
Soo, V. W. C., McAnulty, M. J., Tripathi, A., Zhu, F., Zhang, L., Hatzakis, E., Smith, P. B., Agrawal, S., Nazem-Bokaee, H., Gopalakrishnan, S., Salis, H. M., Ferry, J. G., Maranas, C. D., Patterson, A. D., and Wood, T. K.: Reversing methanogenesis to capture methane for liquid biofuel precursors, Microb. Cell Fact., 15, 11, https://doi.org/10.1186/s12934-015-0397-z, 2016.
Tamames, J. and Puente-Sánchez, F.: SqueezeMeta, a highly portable, fully automatic metagenomic analysis pipeline, Front. Microbiol., 10, https://doi.org/10.3389/fmicb.2018.03349, 2019.
Tan, S., Liu, J., Fang, Y., Hedlund, B. P., Lian, Z.-H., Huang, L.-Y., Li, J.-T., Huang, L.-N., Li, W.-J., Jiang, H.-C., Dong, H.-L., and Shu, W.-S.: Insights into ecological role of a new deltaproteobacterial order Candidatus Acidulodesulfobacterales by metagenomics and metatranscriptomics, ISME J., 13, 2044–2057, https://doi.org/10.1038/s41396-019-0415-y, 2019.
Thauer, R. K. and Shima, S.: Methane as fuel for anaerobic microorganisms, Ann. N.Y. Acad. Sci., 1125, 158–170, https://doi.org/10.1196/annals.1419.000, 2008.
Valenzuela, E. I., Prieto-Davó, A., López-Lozano, N. E., Hernández-Eligio, A., Vega-Alvarado, L., Juárez, K., Sarahí García-González, A., López, M. G., and Cervantes, F. J.: Anaerobic Methane Oxidation Driven by Microbial Reduction of Natural Organic Matter in a Tropical Wetland, Appl. Environ. Microbiol., 83, e00645-17, https://doi.org/10.1128/AEM.00645-17, 2017.
Valenzuela, E. I., Avendaño, K. A., Balagurusamy, N., Arriaga, S., Nieto-Delgado, C., Thalasso, F., and Cervantes, F. J.: Electron shuttling mediated by humic substances fuels anaerobic methane oxidation and carbon burial in wetland sediments, Sci. Total Environ., 650, 2674–2684, https://doi.org/10.1016/J.SCITOTENV.2018.09.388, 2019.
Vigderovich, H., Liang, L., Herut, B., Wang, F., Wurgaft, E., Rubin-Blum, M., and Sivan, O.: Evidence for microbial iron reduction in the methanic sediments of the oligotrophic southeastern Mediterranean continental shelf, Biogeosciences, 16, 3165–3181, https://doi.org/10.5194/bg-16-3165-2019, 2019.
Vuillemin, A., Horn, F., Friese, A., Winkel, M., Alawi, M., Wagner, D., Henny, C., Orsi, W. D., Crowe, S. A., and Kallmeyer, J.: Metabolic potential of microbial communities from ferruginous sediments, Environ. Microbiol., 20, 4297–4313, https://doi.org/10.1111/1462-2920.14343, 2018.
Wagner, T., Koch, J., Ermler U., and Shima, S.: Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction, Science, 357, 699–703, https://doi.org/10.1126/science.1252826, 2017.
Walker, D. J. F., Nevin, K. P., Holmes, D. E., Rotaru, A. E., Ward, J. E., Woodard, T. L., Zhu, J., Ueki, T., Nonnenmann, S. S., McInerney, M. J., and Lovley, D. R.: Syntrophus conductive pili demonstrate that common hydrogen-donating syntrophs can have a direct electron transfer option, ISME J., 1–10, https://doi.org/10.1038/s41396-019-0575-9, 2020.
Wang, F., Gu, Y., O'Brien, J. P., Yi, S. M., Ebru Yalcin, S., Srikanth, V., Shen, C., Vu, D., Ing, N. L., Hochbaum, A. I., Egelman, E. H., and Malvankar, N. S.: Structure of microbial nanowires reveals stacked hemes that transport electrons over micrometers, Cell, 177, 361–369, https://doi.org/10.1016/j.cell.2019.03.029, 2019.
Wang, H., Byrne, J. M., Liu, P., Liu, J., Dong, X., and Lu, Y.: Redox cycling of Fe(II) and Fe(III) in magnetite accelerates aceticlastic methanogenesis by Methanosarcina mazei, Environ. Microbiol. Rep., 12, 97–109, https://doi.org/10.1111/1758-2229.12819, 2020.
Wang, L., Miao, X., Ali, J., Lyu, T., and Pan, G.: Quantification of oxygen nanobubbles in particulate matters and potential applications in remediation of anaerobic environment, ACS Omega, 3, 10624–10630, https://doi.org/10.1021/acsomega.8b00784, 2018.
Wang, Y. and Newman, D. K.: Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen, Environ. Sci. Technol., 42, 2380–2386, https://doi.org/10.1021/es702290a, 2008.
Wegener, G., Krukenberg, V., Riedel, D., Tegetmeyer, H. E., and Boetius, A.: Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria, Nature, 526, 587–590, https://doi.org/10.1038/nature15733, 2015.
Wegener, G., Krukenberg, V., Ruff, S. E., Kellermann, M. Y., and Knittel, K.: Metabolic capabilities of microorganisms involved in and associated with the anaerobic oxidation of methane, Front. Microbiol., 7, 46, https://doi.org/10.3389/fmicb.2016.00046, 2016.
Welte, C. and Deppenmeier, U.: Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens, Biochim. Biophys. Acta, 1837, 1130–1147, https://doi.org/10.1016/j.bbabio.2013.12.002, 2014.
Wu, Y. W., Simmons, B. A., and Singer, S. W.: MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets, Bioinformatics, 32, 605–607, https://doi.org/10.1093/bioinformatics/btv638, 2015.
Wuebbles, D. J. and Hayhoe, K.: Atmospheric methane and global change. Earth-Sci. Rev., 57, 177–210, https://doi.org/10.1016/S0012-8252(01)00062-9, 2002.
Xin, J. Y., Cui, J. R., Niu, J. Z., Hua, S. F., Xia, C. G., Ben Li, S., and Zhu, L. M.: Production of methanol from methane by methanotrophic bacteria, Biocatal. Biotransformation, 22, 225–229, https://doi.org/10.1080/10242420412331283305, 2004.
Yan, Z. and Ferry, J. G.: Electron bifurcation and confurcation in methanogenesis and reverse methanogenesis, Front. Microbiol., 9, 1322, https://doi.org/10.3389/fmicb.2018.01322, 2018.
Yan, Z., Joshi, P., Gorski, C. A., and Ferry, J. G.: A biochemical framework for anaerobic oxidation of methane driven by Fe(III)-dependent respiration, Nat. Commun., 9, 1642, https://doi.org/10.1038/s41467-018-04097-9, 2018.
Yee, M. O., and Rotaru, A. E.: Extracellular electron uptake in Methanosarcinales is independent of multiheme c-type cytochromes, Sci. Rep.-UK, 10, 372, https://doi.org/10.1038/s41598-019-57206-z, 2020.
Yu, T., Wu, W., Liang, W., Lever, M. A., Hinrichs, K. U., and Wang, F.: Growth of sedimentary Bathyarchaeota on lignin as an energy source, P. Natl. Acad. Sci. USA, 115, 6022–6027, https://doi.org/10.1073/pnas.1718854115, 2018.
Zehnder, A. J. B. and Brock, T. D.: Methane formation and methane oxidation by methanogenic Bacteria, J. Bacteriol., 137, 420–432, 1979.
Zhang, L., Zhao, T., Shen, T., and Gao G.: Seasonal and spatial variation in the sediment bacterial community and diversity of Lake Bosten, China, J. Basic Microbiol., 59, 224–233, https://doi.org/10.1002/jobm.201800452, 2019.
Zhao, Z., Wang, J., Li, Y., Zhu, T., Yu, Q., Wang, T., Liang, S., and Zhang, Y.: Why do DIETers like drinking: Metagenomic analysis for methane and energy metabolism during anaerobic digestion with ethanol, Water Res., 171, 115425, https://doi.org/10.1016/j.watres.2019.115425, 2020.
Zhou, Z., Pan, J., Wang, F., Gu, J.-D., and Li, M.: Bathyarchaeota: globally distributed metabolic generalists in anoxic environments, FEMS Microbiol. Rev., 023, 639–655, https://doi.org/10.1093/femsre/fuy023, 2018.
Zhu, Y., Purdy, K. J., Eyice, Ö., Shen, L., Harpenslager, S. F., Yvon-Durocher, G., Dumbrell, A. J., and Trimmer, M.: Disproportionate increase in freshwater methane emissions induced by experimental warming, Nat. Clim. Change, 10, 685–690, https://doi.org/10.1038/s41558-020-0824-y, 2020.
Ziels, R. M., Nobu, M. K., and Sousa, D. Z.: Elucidating syntrophic butyrate-degrading populations in anaerobic digesters using stable-isotope-informed genome-resolved metagenomics, mSystems, 4, 1–16, https://doi.org/10.1128/msystems.00159-19, 2019.





