the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Potential contributions of nitrifiers and denitrifiers to nitrous oxide sources and sinks in China's estuarine and coastal areas
Xiaofeng Dai
Mingming Chen
Xianhui Wan
Ehui Tan
Jialing Zeng
Nengwang Chen
Shuh-Ji Kao
Yao Zhang
Nitrous oxide (N2O) is an important ozone-depleting greenhouse gas produced and consumed by microbially mediated nitrification and denitrification pathways. Estuaries are intensive N2O emission regions in marine ecosystems. However, the potential contributions of nitrifiers and denitrifiers to N2O sources and sinks in China's estuarine and coastal areas are poorly understood. The abundance and transcription of six key microbial functional genes involved in nitrification and denitrification, as well as the clade II-type nosZ gene-bearing community composition of N2O reducers, were investigated in four estuaries spanning the Chinese coastline. The results showed that the ammonia-oxidizing archaeal amoA genes and transcripts were more dominant in the northern Bohai Sea (BS) and Yangtze River estuaries, which had low nitrogen concentrations, while the denitrifier nirS genes and transcripts were more dominant in the southern Jiulong River (JRE) and Pearl River estuaries, which had high levels of terrestrial nitrogen input. Notably, the nosZ clade II gene was more abundant than the clade I-type throughout the estuaries except for in the JRE and a few sites of the BS, while the opposite transcript distribution pattern was observed in these two estuaries. The gene and transcript distributions were significantly constrained by nitrogen and oxygen concentrations as well as by salinity, temperature, and pH. The nosZ clade II gene-bearing community composition along China's coastline had a high level of diversity and was distinctly different from that in the soil and in marine oxygen-minimum-zone waters. By comparing the gene distribution patterns across the estuaries with the distribution patterns of the N2O concentration and flux, we found that denitrification may principally control the N2O emissions pattern.
- Article
(8317 KB) - Full-text XML
-
Supplement
(1040 KB) - BibTeX
- EndNote
Nitrous oxide (N2O) is a kind of ozone-depleting substance and an important, long-lived greenhouse gas with a global warming potential 298 times that of carbon dioxide (CO2) (Solomon et al., 2007; Ravishankara et al., 2009; Rowley et al., 2013). Prokaryotic microorganisms play an important role in N2O production and consumption through the nitrification and denitrification pathways (Babbin et al., 2015; Domeignoz-Horta et al., 2015; Ji et al., 2018b; Meinhardt et al., 2018; Santoro et al., 2011; Silvennoinen et al., 2008). N2O is produced as a byproduct in the first step (NHNO) of nitrification, which is catalyzed by ammonia monooxygenase in ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) (Codispoti and Christensen, 1985). The ammonia monooxygenase subunit A gene (amoA) is frequently used as a functional gene marker for AOA and AOB analysis. N2O is also produced as a kind of intermediate product in the denitrification process, in which nitrite (NO) is reduced to nitric oxide (NO) and then further reduced to N2O. Usually, the nitrite reductase genes nirS and nirK are used to evaluate the N2O production potential through denitrification (Hallin et al., 2018; Shaw et al., 2006; Wrage et al., 2001). Some bacterial nitrifiers can also reduce NO to N2O through a nitrifier denitrification pathway. The last step of denitrification is the only known biological N2O consumption pathway, reducing N2O into nitrogen (N2) under the catalysis of nitrous oxide reductase encoded by the nosZ gene. This gene is divided into two clades according to the differences in the signal peptides of nitrous oxide reductase (Henry et al., 2006; Jones et al., 2013). The microorganisms possessing clade I-type nosZ genes are mainly affiliated with alpha-, beta-, and gamma-proteobacteria, and the clade I gene has a higher frequency of co-occurrence with nir and nor genes than the clade II gene. The nosZ clade II genes are present in a much larger range of archaeal and bacterial phyla (Jones et al., 2013), and intergenomic comparisons have revealed that more than half of the microorganisms possessing clade II genes lack nitrite reductase or nitric oxide reductase, do not produce N2O, and thus are expected to drive potential N2O sinks (Graf et al., 2014; Jones et al., 2008; Marchant et al., 2017; Sanford et al., 2012). The community composition of microorganisms with nosZ clade II genes is considered important for the N2O : N2 end-product ratio of denitrification, influencing the regional N2O source or sink characteristics (Domeignoz-Horta1 et al., 2015; Philippot, 2013). However, there are few studies on nosZ clade II gene diversity and community composition in Chinese estuarine and coastal areas.
Decades of research have revealed that the ocean, contributing one-third of the N2O emission fluxes to the atmosphere, is the second most important source of N2O emissions following arable soils (Nevison et al., 2003). Estuaries, as important bioreactors, are the most active N2O exchange areas in the ocean, accounting for 33 % of oceanic N2O emissions with only approximately 0.4 % of the area (Bange et al., 1996; Zhang et al., 2010). Denitrification is the major contributor to N2O production in terrestrial ecosystems and stream and river networks (Beaulieu et al., 2011; Marzadri et al., 2017). However, complete denitrification can consume N2O (Jones et al., 2014). A recent study reports a fourfold increase in global riverine N2O emissions influenced by human activities (Yao et al., 2020). Marine nitrification supported by ammonia-oxidizing archaea is largely responsible for oceanic N2O emissions, especially in the open ocean (Löscher et al., 2012; Santoro et al., 2011), while nitrate reduction is the dominant N2O source in oxygen-minimum zones (OMZs) (Frey et al., 2020; Ji et al., 2018a; Yamagishi et al., 2007). In estuaries, which are the transition zones between the land and sea, both nitrification and denitrification could be the dominant driving processes of active N2O exchange. For example, nitrification was credited as the dominant N2O production pathway in the Scheldt Estuary as well as in some other European estuaries (Barnes and Upstill-Goddard, 2011; Brase et al., 2017; De Wilde and De Bie, 2000; Li et al., 2015), while an inverse correlation between N2O concentrations and oxygen indicates that denitrifiers might be the dominant N2O contributor in the Potomac River estuary (McElroy et al., 1978). In addition, the incubation experiments with nitrogen-stable isotope tracers reveal active N2O production by denitrification in the Chesapeake Bay (Ji et al., 2018b). Another research project in the Chesapeake Bay reveals that physical processes such as wind events and vertical mixing affect the net balance between N2O production and consumption, resulting in a variable source and sink for N2O (Laperriere et al., 2019).
The four main estuaries along the Chinese coastline include the Bohai Sea (BS) in the north, the Yangtze River estuary (YRE) and the adjacent East China Sea (ECS) in the middle, as well as the Jiulong River estuary (JRE) and Pearl River estuary (PRE) in the south (Fig. 1). The BS is a semi-enclosed sea located in the north temperate zone of China. Influenced by frequent human activities and seasonal variability in inputs from the Yellow River, Liao River, Luan River, and Hai River, seasonal hypoxia is an important characteristic of the BS (Chen, 2009). The YRE and the adjacent ECS, which receive a large amount of nutrients from the largest river in Asia (Yangtze River: runoff 9.2 × 1011 m3 yr−1) (Zhang, 2002), also exhibit seasonal hypoxia off the estuary from July to September because of the enhanced primary productivity (Zhu et al., 2011). Both the JRE and PRE are located in densely populated and industrialized subtropical areas, with runoffs of 1.44 × 1010 and 3.26 × 1011 m3 yr−1, respectively (Cao et al., 2005; He et al., 2014). To clarify the potential contributions of nitrification and denitrification to sources and sinks of N2O in China's estuarine and coastal areas, the abundance and transcription activity of six key microbial functional genes involved in nitrification and denitrification (AOA and AOB amoA, nirS, nirK, nosZ clade I and II genes) were investigated in the four estuarine areas. In addition, the nosZ clade II gene diversity and N2O-reducing community composition were analyzed based on clone libraries in order to assess the local N2O sink potential.
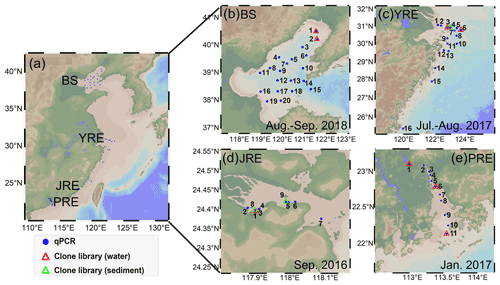
Figure 1(a) Sampling sites in the four estuaries along China's coastline; (b) Bohai Sea (BS); (c) Yangtze River estuary (YRE); (d) Jiulong River estuary (JRE); (e) Pearl River estuary (PRE). The sampling time for each region is shown in the subplots. The figure was produced by Ocean Data View 5.2.0 (http://odv.awi.de/, last access: 19 August 2020).
2.1 Sampling and biogeochemical parameter measurements
A total of 228 (130 for DNA and 98 for RNA) samples from 54 sites were collected (Fig. 1). A total of 116 samples (58 for DNA and 58 for RNA) were collected from 20 stations with two or three depth layers (3–63 m; Table S1) in the BS on the R/V Dongfanghong #2 from August to September 2018. A total of 74 (41 for DNA and 33 for RNA) samples were collected from 16 stations with one to four depth layers (3–55 m) in the YRE on the R/V Yanping II from July to August 2017. Water samples were collected using a rosette sampler fitted with Niskin bottles (SBE 911, Sea-Bird Co). A total of 16 surface samples (9 for DNA and 7 for RNA) from a water depth of ∼ 0.5 m were collected from the JRE on the R/V Ocean II during September 2016. A total of 22 samples for DNA were collected from 11 stations with two depth layers (0.5–18 m) in the PRE on the R/V Wanyu during January 2017. Water samples were collected using an organic glass hydrophore (1 L; Kedun Co., China). In addition, two and one surface sediment samples were acquired using a grab sampler from the JRE in December 2015 and from the YRE from July to August 2017, respectively. Water samples of 0.2–2.5 L were filtered through 0.22 µm pore-sized polycarbonate membranes (Millipore, USA) within 1 h at a < 0.03 MPa pressure for quantitative PCR (qPCR) analysis. Water samples were serially filtered through 10, 3, and 0.22 µm pore size polycarbonate membranes (Millipore, USA) for clone library analysis (Table S1). The membranes for RNA extraction were immediately fixed with 1.5 mL of RNAlater (Invitrogen, Life Technologies). All filters and sediment samples were quick-frozen in liquid nitrogen and then stored at −80 ∘C for laboratory analysis.
Temperature, salinity, and depth were measured using conductivity temperature depth (CTD) (SBE 911, Sea-Bird Co.) in the BS, YRE, and PRE. In the JRE, water temperature and salinity were continuously measured (every 3 s for 1 min) using a YSI6600D salinometer installed on an underway pumping system (Yan et al., 2019). Dissolved oxygen (DO) concentrations were measured using a WTW multiparameter portable meter (Multi 3430, Germany). Ammonia was analyzed on deck using the indophenol blue spectrophotometric method. Nitrate, nitrite, and silicate were measured using an AA3 Autoanalyzer (Bran + Luebbe Co., Germany) (Dai et al., 2008).
2.2 Nucleic acid extraction, clone library, and phylogenetic analysis
DNA from water samples was extracted using the phenol-chloroform-isoamyl alcohol method (Massana et al., 1997) with minor modifications to maximize the DNA output. Briefly, tubes containing shredded filters, approximately 0.5 g of 0.1 mm glass beads, and 800 µL of STE lysis buffer (0.75 M sucrose, 50 mM Tris-HCl, 40 mM EDTA) were first agitated for 60 s on a FastPrep machine (MP Biomedicals, Solon, OH, USA) at 4.5 m s−1. Then, the mixture was processed with lysozyme (1 mg mL−1), proteinase K (0.5 mg mL−1), and sodium dodecyl sulfate (SDS) (1 %) sequentially. At last, the lysate was extracted twice with phenol-chloroform-isoamyl alcohol and once with chloroform-isoamyl alcohol. The DNA was precipitated with isopropyl alcohol and washed with 75 % ethyl alcohol before being dissolved in 50 µL sterile water. DNA from sediment samples was extracted using a FastDNA SPIN Kit for Soil (MP Biomedicals, USA). RNA from water samples was extracted using the RNeasy Mini kit according to the manual (Qiagen, USA). Clean RNA, which was verified by the amplification of the bacterial 16S rRNA gene with the primer set 342F/798R, was reverse-transcribed to cDNA by the SuperScript III first strand synthesis system (Invitrogen, Life Technologies) using random hexamers following the user manual. The quality of both the DNA and cDNA was checked by amplifying the full-length bacterial 16S rRNA gene before storage at −80 ∘C.
A total of 19 DNA samples (16 from water and 3 from sediment) from the four estuaries (Figs. 1 and 4) were used to construct clone libraries for the clade II-type nosZ gene. A PCR was run with the primer set nosZ-II-F (5′-CTIGGICCIYTKCAYAC-3′) and nosZ-II-R (5′-GCIGARCARAAITCBGTRC-3′) according to a previously reported reaction mixture and program (Jones et al., 2013) with the minor modification of using 10 µg of bovine serum albumin (BSA; Takara, Bio Inc.) instead of T4 gp32. PCR products were purified using an agarose gel DNA purification kit (Takara, Bio Inc.), ligated into the pMD19-T vector (Takara, Bio Inc.), and transformed into high-efficiency competent cells of Escherichia coli according to the manufacturer's instructions. A total of 40–127 positive nosZ clones were randomly selected from each library, reamplified using the vector primers M13-F and RV-M, and sequenced using the ABI 3730 automated DNA sequence analyzer (Applied Biosystems). Poor-quality sequences with termination codons were manually checked and removed, and chimeras were removed using UCHIME (Edgar et al., 2011). All sequences were clustered into operational taxonomic units (OTUs) based on a 3 % sequence divergence cutoff (Jones et al., 2014; Wittorf et al., 2020). The coverage (C) of each clone library was calculated by C = 100 % [1− ()] (Mullins et al., 1995), where n is the number of unique OTUs and N the total number of clones in a library. Alpha diversity indices (Shannon, Simpson, and Chao1) of the clade II-type nosZ gene were calculated using the Usearch package (Edgar et al., 2010). The representative sequences of OTUs were translated and analyzed with the BLASTp tool (e value < 10−5). The top 10 most similar sequences of each OTU were used as references. The deduplicated reference sequences and the representative sequences of OTUs were aligned using MAFFT (Katoh and Standley, 2013) and automatically trimmed using trimAl (Capella-Gutiérrez et al., 2009). A maximum likelihood (ML) phylogenetic tree was constructed using Fasttree (v2.7.1, default parameters) (Price et al., 2010) with 500 bootstrap replicates for node support determination. The taxonomy of the OTU was assigned according to the phylogenetic relationship.
2.3 Quantitative PCR of six functional genes
Archaeal amoA, bacterial amoA, nirS, nirK, nosZ clade I, and nosZ clade II genes were quantified by qPCR with DNA and cDNA as templates using a CFX96 (Bio-Rad Laboratories, Singapore). Given the relatively high ammonia concentration in the estuaries, the ammonia-oxidizing archaea (AOA) shallow cluster (Water Column Cluster A; Francis et al., 2005) was targeted with the primer set Arch-amoAFA and Arch-amoAR (Beman et al., 2008). Ammonia-oxidizing bacteria (AOB) are mostly affiliated with two groups: Betaproteobacteria (β-AOB) and Gammaproteobacteria (γ-AOB) (Lam et al., 2007). Since the latter was below the detection limit in previous studies of Chinese estuaries (Zheng et al., 2017; Hou et al., 2018), only β-AOB was targeted with the primer set amoA-1F and amoA-r New (Rotthauwe and Witzel, 1997; Hornek et al., 2006). Bacterial nirS and nirK genes were quantified with the primer sets nirS-1F and nirS-3R (Braker et al., 1998) and nirK876 and nirK1040 (Henry et al., 2004). Bacterial clade I-type nosZ genes were quantified with the primer set nosZ2F and nosZ2R (Henry et al., 2006).
For the clade II-type nosZ gene quantification, the previously published primer sets were found to have less than 80 % amplification efficiency (Jones et al., 2013, 2014; Chee-Sanford et al., 2020). Here, we designed a new primer set for use in our estuarine samples to quantify this gene. Representative nucleotide sequences of each OTU obtained from the clone libraries derived from the PRE samples (n=48) were translated into amino acid sequences and then aligned with the representative reference sequences (n=116; covering 87 genera) obtained from the Functional Gene Repository (http://fungene.cme.msu.edu/index.spr, last access: 27 April 2017, Fish et al., 2013) by Clustal W. Two highly conserved regions containing five and three amino acids in length were chosen to design new primer fragments. The new primer pairs and the previously published nosZ-II-F and nosZ-II-R primer sets (Jones et al., 2013) were all evaluated by Primer Premier 6.0, and eligible primer sets (GC content: 40 %–60 %; optimal melting temperatures: 52–58 ∘C; stable 5′ end and specific 3′ end with no clamp or complementary structure) were tested by qPCR. The best primer combination was nosZ-II-F and the newly designed reverse primer (nosZ-II-Rnew: KGCRTAGTGIGGYTCDCC) with a ∼ 325 bp target fragment length (Fig. S1). The qPCR system is shown in Table S2, and the optimized qPCR program was as follows: an initial 5 min denaturing step at 95∘C, followed by 35 cycles of 95∘C for 30 s, annealing at 53∘C for 60 s, 72∘C extension for 60 s, and a final extension at 72∘C for 10 min. The coverage of the primer sets was evaluated using the Search_pcr2 command of Usearch with the 116 reference sequences mentioned above and all clone sequences (n=1378) obtained from the clone libraries. A coverage of 93.5 % (≤2 mismatches) was obtained for the new primer set.
The presence of PCR inhibitors in DNA extracts was examined by qPCR with different dilutions of DNA (1-, 10-, and 100-fold dilutions). The samples with inhibitors were diluted 10 times to overcome the inhibitor effect according to our evaluation. Standard curves were constructed for the six genes using plasmid DNA from clone libraries generated from the PCR products. qPCRs were performed in triplicate and analyzed against a range of standards (101 to 108 copies µL−1). All specific primer sequences, reactions, and programs for qPCR/PCR used in this study are shown in Table S2. The amplification efficiencies ranged from 87 % to 109 % with R2>0.99 for each qPCR run. The specificity of qPCR products was verified by melting curves, agarose gel electrophoresis, and sequencing.
2.4 Statistical analysis
Redundancy analysis (RDA) based on qPCR or clone library data was used to analyze variations in the gene/transcription distribution and nosZ clade II community composition under environmental constraints using R (R Core Team, 2017). The qPCR or clone library-based relative abundances and environmental factors were normalized via Z transformation (Fayazbakhsh et al., 2009; Magalhães et al., 2008). The collinearity between environmental parameters was excluded (variance inflation factors >10; Palacin-Lizarbe et al., 2019). The null hypothesis that the community structure was independent of environmental parameters was tested using constrained ordination with a Monte Carlo permutation test (999 permutations). Since a normal distribution of the individual datasets was not always met, we used the nonparametric Wilcoxon rank-sum tests for comparing two variables in GraphPad Prism software (San Diego, CA, USA). The bivariate correlations were described by Spearman's (ρ value) or Pearson's (r value) correlation coefficients. False discovery rate-based multiple comparison procedures were applied to evaluate the significance of multiple hypotheses and to identify truly significant comparisons (false discovery rate-adjusted P value) (Pike, 2011).
3.1 Environmental characteristics of the four estuaries
Water temperature increased with decreasing latitude from the BS (16.1–26.4 ∘C) to the YRE (19.2–29.1 ∘C) and JRE (28.7–30.8 ∘C), where samples were all collected in summer. Samples were collected in winter in the southernmost PRE, where the water temperature was 19.7–20.5 ∘C (Fig. 2). Salinity exhibited consistently high values in all sites of the BS and YRE (26.4–34.6 ppt), except for two low values (14.34 and 21.66 ppt) observed in the river mouth. In the JRE and PRE, obvious salinity gradients were detected from 0.1 to 30.7. The DO concentration varied in the range of 4.25–8.46 mg L−1 in the BS, 1.25–8.71 mg L−1 in the YRE, 4.04–6.89 mg L−1 in the JRE, and 2.22–9.22 mg L−1 in the PRE. There was a distinct DO gradient from upstream to downstream in the PRE (Fig. 2). The dissolved inorganic nitrogen (DIN: ammonium, nitrite, and nitrate) concentrations were generally lower in the BS and YRE compared to those in the JRE and PRE. The ammonium concentration was in the range of 0.006–1.27 µM in the BS, below detection (BD) to 1.99 µM in the YRE, 7.01–36.78 µM in the JRE, and 1.71–417.38 µM in the PRE. The nitrite concentration was in the range of BD–5.65 µM in the BS and 0.004–2.5 µM in the YRE, 7.24–30.87 µM in the JRE, and 0.41–69.23 µM in the PRE. The nitrate concentration ranged from 0.067–13.97 µM in the BS, 0.23–65.09 µM in the YRE, 24.94–241.32 µM in the JRE, and 3.0–320.53 µM in the PRE. Clear DIN concentration gradients were observed from upstream to downstream in the JRE and PRE, particularly in the PRE.
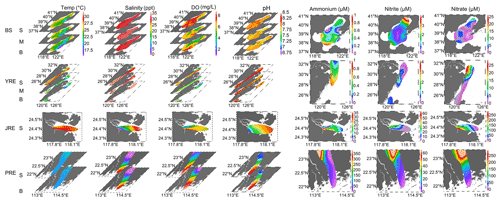
Figure 2Temperature (Temp), salinity, dissolved oxygen (DO), pH, ammonium, nitrite, and nitrate concentration distributions in the Bohai Sea (BS), Yangtze River estuary (YRE), Jiulong River estuary (JRE), and Pearl River estuary (PRE). Data from the surface layer (S), middle layer (M), and bottom layer (B) were shown for temperature, salinity, DO, and pH. Details about the depth of samples are listed in Table S1. Depth-integrated mean values were used for ammonium, nitrite, and nitrate concentration distributions.
3.2 Distribution of six key functional genes
The abundances of archaeal amoA, bacterial amoA, nirS, nirK, nosZ I, and nosZ II genes showed distinct distribution patterns among the four estuaries (Fig. 3a–h). We divided the six genes into two groups for analysis: one group included archaeal and bacterial amoA, nirS, and nirK genes indicating nitrification and denitrification related to N2O production (Fig. 3a–d), and the other included bacterial nosZ I and nosZ II genes indicating N2O consumption (Fig. 3e–h). In the gene group of N2O production-related processes, archaeal amoA was the most dominant in the BS (2.66 × 104–3.68 × 108 copies L−1) and YRE (4.86 × 103–9.47 × 107 copies L−1) (Wilcoxon test, P<0.01; Fig. 3a, b and Table S3), accounting for 3.96 %–96.2 % and 2.84 %–99.67 % of N2O production-related gene abundance, respectively. In contrast to the northern estuaries, archaeal amoA (5.28 × 105–4.40 × 106 copies L−1) and bacterial nirS (2.57 × 105–6.29 × 106 copies L−1) genes codominated the gene group of N2O production-related processes in the JRE (Fig. 3c), accounting for 2.43 %–72.93 % and 25.03 %–93.77 %, respectively. In the southernmost PRE, the nirS gene was the most abundant (3.48 × 104–1.66 × 109 copies L−1), especially upstream (P<0.05), accounting for 4.24 %–99.91 % (Fig. 3d). Generally, archaeal amoA was widespread in all samples, and its abundance decreased from north to south with differences of 1 to 2 orders of magnitude. A similar pattern was observed for bacterial amoA, with lower abundances than archaeal amoA (Table S3). The abundance of the nirS gene was highest in the PRE among the four estuaries, while the highest number of copies of the nirK gene was present in the BS (Table S3). Among the different water depths, only the bacterial amoA and nirS genes in the BS were observed to be more highly distributed in the middle and bottom layers than in the surface layer by one to three orders of magnitude (P<0.05).
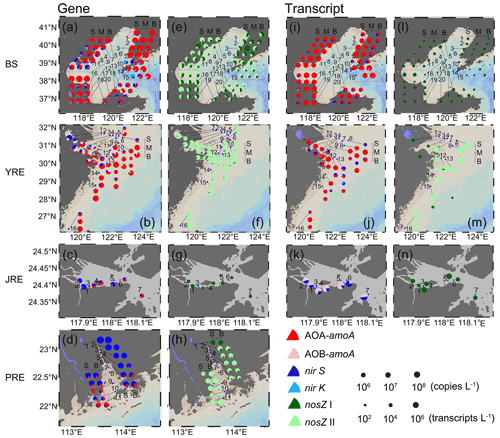
Figure 3Six key functional gene and transcript abundance distributions in the Bohai Sea (BS), Yangtze River estuary (YRE), Jiulong River estuary (JRE), and Pearl River estuary (PRE). S: surface layer; M: middle layer; B: bottom layer. (a–d) Gene related to N2O production; (e–h) gene related to N2O consumption; (i–k) transcript related to N2O production; (l–n) transcript related to N2O consumption.
In the N2O-consuming genes, the abundances of the clade II-type nosZ gene were 6.55 × 103–2.24 × 107 copies L−1 in the BS (Fig. 3e), 6.14 × 103–8.11 × 106 copies L−1 in the YRE (Fig. 3f), and BD–1.17 × 107 copies L−1 in the PRE (Fig. 3h), outnumbering the clade I-type (P<0.01) with no significant differences among the three estuaries. However, the clade II-type nosZ gene was below the detection limit in the JRE, and only the clade I-type was detected with a range of 7.15 × 103–2.32 × 105 copies L−1 (Fig. 3g and Table S3). The numbers of copies of the clade I-type nosZ gene were higher in the BS estuary than in the other three estuaries (P<0.01).
3.3 Transcription activity of six key functional genes
For the four genes of N2O production-related processes, a generally similar relative abundance distribution pattern was observed between transcripts and genes in the BS (Fig. 3i). Archaeal amoA gene transcripts (3.51 × 103–1.62 × 106 transcripts L−1) were significantly more abundant than other transcripts (P<0.01), accounting for 37.94 %–99.30 % of the total abundance of gene transcripts (Table S4). Slightly different from the gene distribution in which the number of copies of the bacterial amoA gene was relatively more abundant than that of the archaeal amoA gene in the river mouth of the YRE (Fig. 3b), the archaeal amoA gene transcript was abundant in the whole YRE, accounting for 9.1 %–100 % of the total abundance of gene transcripts, with a dominant abundance of nirS gene transcripts in a few samples (Fig. 3j). A different distribution pattern was also observed between transcripts and genes in the JRE (Fig. 3c, k). Bacterial amoA (7.06 × 105–8.22 × 107 transcripts L−1) rather than archaeal amoA transcripts (P<0.05) were codominant with nirS transcripts (5.96 × 105–2.31 × 107 transcripts L−1) (Fig. 3k). Notably, the total gene transcript abundance of N2O production-related processes was higher in the JRE (1.31 × 106–9.76 × 107 transcripts L−1) than in the BS and YRE (3.03 × 102–1.12 × 106 transcripts L−1) (P<0.01; Table S4). Bacterial amoA gene transcripts, consistent with the gene distribution, significantly increased with depth in the BS (P<0.05). No significant differences in transcript abundance were observed among different depths for the six functional genes in the YRE.
For the N2O-consuming genes, only the clade I-type nosZ gene transcript was determined (26.2–2.34 × 103 transcripts L−1), while the clade II-type nosZ gene transcript was below the detection limit in the BS (Fig. 3l; Table S4). However, the nosZ II gene transcripts (below detection–1.81 × 105 transcripts L−1) dominated most stations in the YRE, except for a dominant distribution of the nosZ I gene transcript in the river mouth (Fig. 3m). Similar to the gene distribution, in the JRE, only the nosZ I gene transcript was determined (1.23 × 103–5.37 × 104 transcripts L−1) (Fig. 3n). No RNA samples were obtained in the PRE.
3.4 Phylogenetic diversity of the clade II nosZ gene
Clone libraries of nosZ clade II were constructed for 19 samples from the four estuaries, resulting in a total of 1378 quality-controlled sequences that were clustered into 441 OTUs at a similarity level of 97 %. The coverage of each clone library ranged from 73.9 %–96.2 %. Higher gene diversity of nosZ clade II was observed in the water and sediment samples from the JRE and the sediment sample from the YRE than in the other samples (Fig. S2a). The rarefaction curves of the samples from JRE and the sediment sample from YRE did not reach a plateau (data not shown), suggesting that some of the diversity of nosZ clade II remained unsampled. Phylogenetic analysis of the representative sequences of all the OTUs indicated that the clade II nosZ gene sequences were grouped with Bacteroidetes, Proteobacteria, Actinobacteria, Chloroflexi, Chlorobi, Ignavibacteriae, Gemmatimonadetes, Cyanobacteria, and Acidobacteria, in which the OTUs affiliated with Bacteroidetes, Proteobacteria, Chloroflexi, and Actinobacteria were generally abundant among all samples (Fig. 4b). The OTUs belonging to Bacteroidetes were divided into two clusters according to the topological structure of the phylogenetic tree. One cluster contained the reference sequences mainly derived from marine habitats and the OTU sequences retrieved from the four estuaries, while the other cluster included the reference sequences mainly derived from terrestrial habitats and the OTU sequences retrieved only from the low-latitude subtropical estuaries JRE and PRE. The OTU sequences affiliated with Alpha-, Gamma-, Delta-, Epsilonproteobacteria, and Actinobacteria were retrieved from the four estuaries, and the reference sequences were mainly from marine habitats, while the OTUs related to Betaproteobacteria, Oligoflexia, Chlorobi, and Candidatus Melainabacteria were retrieved only from the subtropical estuaries (JRE and PRE), and the reference sequences were mainly from terrestrial habitats (Fig. 4a). Most known clusters of nosZ clade II can be found in our libraries, including a recently identified widespread clade II-type nosZ gene affiliated with the class Oligoflexia (Nakai et al., 2014).
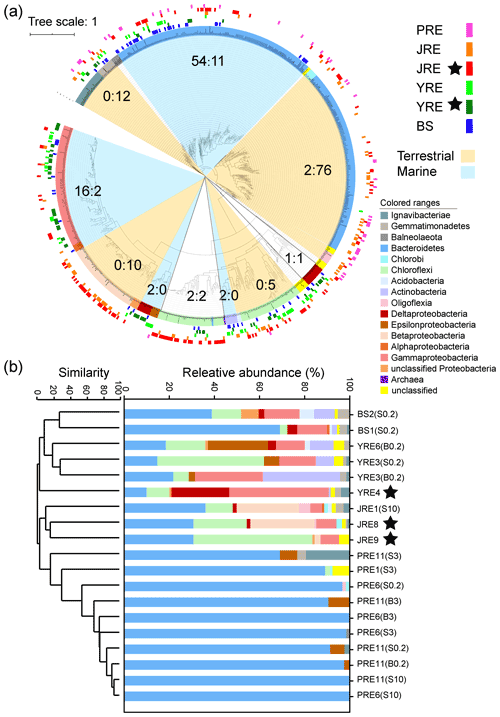
Figure 4(a) Maximum likelihood phylogenetic tree of amino acid sequences of the clade II-type nosZ. The colors of the inner circle indicate taxonomic affiliations based on reference sequences. The colors of the outer circles represent the sources of clone sequences. The phylogenetic tree was bootstrapped 500 times. The scale bar represents the number of amino acid substitutions per site. Numbers before and after the colons indicate the number of reference sequences from marine and terrestrial habitats, respectively. The figure was produced using the interactive tree of life (http://itol.embl.de/, last access: 20 September 2019; Letunic and Bork, 2016). (b) Relative abundances of community compositions of the clade II-type nosZ gene clone libraries in the four estuaries. The colors of the bars indicate taxonomic affiliations. The similarity was calculated from Bray–Curtis similarity. Black stars indicate sediment samples.
A community structure shift of nosZ clade II was observed among the four estuaries (Fig. 4b). Bacteroidetes was the most dominant group in the samples from the BS (39.0 %–68.5 %), followed by Proteobacteria (Gamma-, Delta-, and Alphaproteobacteria; 18.7 %–26.0 %). The sequences phylogenetically grouped into Proteobacteria (Gamma-, Delta-, and Epsilonproteobacteria; 23.0 %–70.6 %) dominated the clone libraries from the YRE, followed by Chloroflexi (6.9 %–47.3 %). The sequences from the JRE were also mainly affiliated with Proteobacteria (Beta-, Gamma-, Delta-, and Alphaproteobacteria and Oligoflexia; 11.8 %–40.5 %), Bacteroidetes (30.9 %–37.9 %), and Chloroflexi (12.1 %–50.9 %). In contrast to the three estuaries, the sequences affiliated with Bacteroidetes were absolutely dominant in the clone libraries of the PRE (>69.2 %). A nonmetric multidimensional scaling (NMDS) analysis indicated that nosZ clade II communities from the same estuary were clustered together at a >10 % Bray–Curtis similarity level, except for a separate cluster of the sediment community from the YRE (Fig. S2b). The nosZ clade II communities from the southern estuaries (JRE and PRE) and northern estuaries (YRE and BS) were clustered separately at a >3 % Bray–Curtis similarity level.
3.5 Correlations between six key functional genes and environmental factors
Variations in the gene–transcript distributions under environmental constraints were analyzed by RDA. The first two RDA axes explained 19.98 % and 5.36 % of the total variation in the gene–environment relationship (Fig. 5a). Salinity, DO, nitrite, and ammonium concentrations were significantly correlated with gene distribution (P<0.01). The main variation in N2O source or sink process-related genetic potentials was across a nirS vs. archaeal amoA abundance gradient. The nirS-rich samples corresponded to those from the southern estuaries (JRE and PRE) with higher ammonium and nitrite concentrations. In contrast, the samples with the highest abundance of archaeal amoA were located in sites with high salinity and low ammonium concentrations in the northern estuaries (BS and YRE). Notably, RDA of the gene transcripts and environmental variables clearly separated the transcripts from different estuaries along the axes, which explained 26.4 % and 8.27 % of the total variation (Fig. 5b). Variation in transcript distribution was significantly correlated with pH, temperature, nitrite, and nitrate concentration (P<0.01). The main variation of these transcripts was distributed across archaeal and bacterial amoA vs. nosZ clade II abundance gradients. The archaeal amoA transcript-rich samples corresponded to those from the BS and YRE sites with lower temperatures. The bacterial amoA gene was actively transcribed in the JRE and positively correlated with nitrite and nitrate concentrations. The nosZ clade II transcript-rich samples corresponded to those from the YRE sites with relatively higher pH and temperature. The nosZ clade I and nirS transcript distributions were also positively correlated with pH and temperature, respectively.
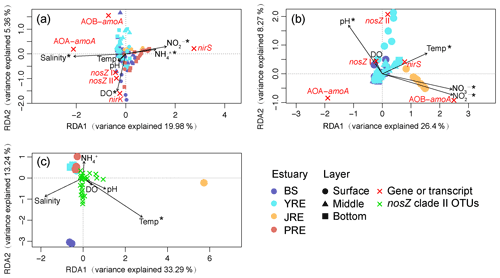
Figure 5Redundancy analysis of the relative abundances of ammonia-oxidizing archaeal amoA (AOA-amoA), bacterial amoA (AOB-amoA), nirS, nirK, and nosZ clade I and II (a) genes and (b) transcripts, as well as of (c) the community composition of the nosZ clade II clone libraries under biogeochemical constraints. Each circle, triangle, or square represents an individual sample from the surface, middle, or bottom layer, respectively. The fork-shaped symbol represents the functional gene, transcript, or nosZ clade II OTU. Vectors represent environmental variables. Asterisks indicate statistically significant variables. Temp, temperature; DO, dissolved oxygen.
RDA based on the clone library data of the clade II-type nosZ gene revealed that the nosZ II community composition was significantly affected by temperature (P<0.01; Fig. 5c). The first two RDA axes explained 33.29 % and 13.24 % of the total variation. The nosZ II gene community compositions in the BS may prefer environments with relatively high salinity and temperature. The community compositions in the JRE water may prefer environments with a high temperature (the sediment samples were not included in this analysis due to a lack of biogeochemical parameters). The nosZ clade II microbes in the PRE and YRE may prefer to distribute in environments with high ammonium concentrations.
4.1 Niche differentiation of functional genes controlled by environmental factors
There was a distinct large-scale spatial structure among the detected genes, as shown in Fig. 3. The different sampling seasons between the PRE (January) and the other three estuaries (June to September) may influence the spatial distribution of functional genes across the four estuaries. However, the niche differentiation of functional genes, spatially or temporally, is essentially controlled by environmental factors, such as temperature, salinity, oxygen and nutrient availabilities, and primary productivity. Comparing the relative contributions of these functional genes to the total number of gene copies across the study regions, there was a strong negative correlation between the relative abundances of the archaeal amoA gene and bacterial nirS gene (, P<0.01), and they showed contrasting patterns along salinity and DIN gradients (Fig. S3). Samples from the BS and YRE exhibited high salinity and low DIN concentrations. The high abundance of the archaeal amoA gene in these areas is consistent with previous findings of nitrifiers comprised predominantly of AOA in estuarine environments with higher salinity and lower ammonia concentrations, since archaeal nitrifiers exhibit a high ammonia affinity and salinity tolerance (Martens-Habbena et al., 2009; Abell et al., 2010; Bernhard et al., 2010; Zhang et al., 2014; Hou et al., 2018; Hink et al., 2018; Ma et al., 2019). In contrast, both the JRE and PRE are typical subtropical, eutrophic estuaries with high DIN inputs from surrounding environments (Cao et al., 2005; He et al., 2014; X. Yan et al., 2012). Denitrifying bacteria are more adaptable to environments with high organic carbon and nitrogen concentrations because they usually have high requirements for substrates (Braker et al., 2000; Smith et al., 2007; Mosier and Francis, 2010; Wang et al., 2014; Wei et al., 2015; Lee and Francis, 2017). The presence of nitrogen oxides was also shown to activate nirK and nirS gene expression under anoxic conditions (Riya et al., 2017). Thus, the nirS-containing group was more abundant upstream in the JRE and PRE. The significant correlations between DIN and the nirS gene (Fig. S3) and transcript (ρ=0.341, P<0.01; data not shown) are consistent with a previous conclusion that high anthropogenic N loading stimulates denitrification (Beaulieu et al., 2011; Cole and Caraco, 2001; Garnier et al., 2006; W. Yan et al., 2012).
Previous studies of N2O-consuming gene abundance have mainly focused on terrigenous ecosystems, e.g., in soil samples, the clade I- and II-type nosZ genes ranged from 104 to 108 and 104 to 107 copies g of dry soil−1, respectively (Jones et al., 2013, 2014). In marine ecosystems, only the oxygen-depleted waters and coastal sediments have been investigated, where the clade I-type was approximately 105 copies L−1 and both clades I and II ranged from 105 to 107 copies g of wet sediment−1, respectively (Wittorf et al., 2020; Sun et al., 2021). We detected that the number of copies of the nosZ gene ranged from 6.59 × 103 to 2.35 × 108 copies L−1, with an average of 4.94 × 106 copies L−1 in China's estuarine and coastal areas. There was a strong negative correlation between the relative abundance of the clade I- and II-type nosZ genes (, P<0.01), indicating that the two types were affiliated with different groups. The distribution of nosZ (clades I and II) gene transcripts was significantly positively correlated with pH (Fig. 5b), suggesting that acidification of the ocean may decrease N2O consumption potential. N2O production influenced by pH has been observed in N-cycling water engineering systems and terrestrial ecosystems (Mørkved et al., 2007; Blum et al., 2018). Therefore, some studies suggest that liming for acidic soils could mitigate N2O emissions (McMillan et al., 2016; Wang et al., 2017; Senbayram et al., 2019). The nosZ genes and transcripts showed significantly negative correlations with nitrate and/or nitrite (Fig. 5a and b), and similar correlations were also found in mountain lake habitats (Palacin-Lizarbe et al., 2019). It is possible that high abundances of the nosZ gene and transcript activity lead to high consumption of nitrate and nitrite. In addition, it was reported that the presence of nitrate can inhibit the reduction of N2O to N2 (Blackmer and Bremner, 1978). DO also showed an important influence on denitrifying genes, which is consistent with a previous conclusion that O2 concentrations can impact the expression and metabolism of denitrification genes through protein sensing of oxygen conditions (Qu et al., 2016; Riya et al., 2017). Notably, we found that the distribution and abundance of the nosZ gene and the nirS or nirK genes were distinctly different, indicating that these functional genes were affiliated with different denitrifiers. This may be because not all N2O-consuming bacteria contain all denitrification genes (Sanford et al., 2012).
4.2 Gene transcription expression controlled by environmental factors
The gene transcript abundance showed a certain regional distribution difference with gene abundance (Fig. 3), suggesting that environmental factors might have different influences on gene distribution and transcript activity. The bacterial amoA gene was transcribed actively in the JRE, although the archaeal amoA gene prevailed in gene abundance. Frequent water exchange may result in a large amount of the archaeal amoA gene from the ocean, but AOB are more active under high ammonium and low salinity conditions. AOB have been indicated to be the primary N2O producer, even in an AOA-dominated environment (Meinhardt et al., 2018). A meta-analysis also revealed that AOB respond more strongly than AOA to nitrogen addition (Carey et al., 2016). High abundances of bacterial amoA and nirS gene transcripts make the JRE a potentially more active area of N2O production compared to the northern estuarine and coastal areas, which may be attributed to the nitrogen input of the JRE's surrounding environment. In contrast, in the mouth of the YRE, although the bacterial amoA gene contributed a large proportion of the gene abundance, the archaeal amoA gene was transcribed more actively. Flushing water from the Yangtze River may transport a large amount of the bacterial amoA gene, but the archaeal amoA gene is more competitive in low ammonium and oxygen environments (Fig. 2) since the enzyme ammonia monooxygenase in AOA has a higher affinity for ammonia and a lower oxygen requirement than the AOB (Park et al., 2010; Molina et al., 2010; Martens-Habbena and Stahl, 2011). The nosZ clade I gene was transcribed more actively even though the nosZ clade II gene was more abundant (e.g., the case in the BS shown in Fig. 3e and l). The higher growth yields of clade II-type N2O-reducing bacteria than those of clade I-type (Yoon et al., 2016) may lead to a preponderance of the nosZ clade II gene. However, a microbial culture of clade I-type N2O-reducing bacteria has been reported to have the capability of continually synthesizing N2O reductase enzymes under oxic conditions to allow for a rapid transition into anoxic environments (Lycus et al., 2018). Such a strategy could result in the more abundant nosZ clade I transcripts observed in the estuaries.
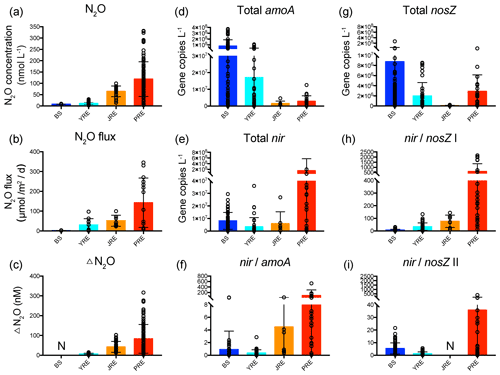
Figure 6The ranges of (a) N2O concentration; (b) N2O flux; (c) ΔN2O (data from Chen et al., 2008; Lin et al., 2016, 2020; Ma et al., 2019; Song et al., 2015; Wang et al., 2014, 2016; Wu et al., 2013; Xu et al., 2005; Zhan et al., 2011; Zhang et al., 2008, 2010); (d) total archaeal and bacterial amoA gene abundance; (e) total nirS and nirK gene abundance; (f) the ratio of total nir to amoA gene abundance; (g) total nosZ clade I and II gene abundance; (h) the ratio of total nir to nosZ clade I gene abundance; and (i) ratio of total nir to nosZ clade II gene abundance in the Bohai Sea (BS), Yangtze River estuary (YRE), Jiulong River estuary (JRE), and Pearl River estuary (PRE). Black circles represent the value of each sample. Bars represent the mean values. Error bars indicate standard deviation. N: no data or not determined.
4.3 N2O emissions potential implied by functional gene distribution
The community structure of nitrifiers and denitrifiers is thought to have an important influence on N2O emissions. For example, the abundance and expression of the archaeal amoA gene showed comparable patterns with N2O production in the OMZ of the eastern tropical North Atlantic (Löscher et al., 2012). Reduction of the abundance of bacterial amoA genes in hyperthermophilic composting was proven to decrease N2O emissions (Cui et al., 2019). The expression of the nirK gene induced by the addition of nitrate caused an increase in N2O production in an anoxic soil slurry experiment (Riya et al., 2017). Transcription of clade I-type nosZ mRNA in the lower N2O emission system was one order of magnitude higher than that in the higher N2O emission system in wastewater treatment plants (Song et al., 2014). To assess how community structure controls the regional N2O source or sink potential across China's estuaries, we collected the data on N2O concentration, N2O flux, and ΔN2O in the four estuaries from the literature, covering January to November from 2002 to 2015 (Table S5; Chen et al., 2008; Lin et al., 2016, 2020; Ma et al., 2019; Song et al., 2015; Wang et al., 2014, 2016; Wu et al., 2013; Xu et al., 2005; Zhan et al., 2011; Zhang et al., 2008, 2010) and analyzed their relationships with the six functional gene distributions. The N2O concentration, N2O flux, and ΔN2O all showed an increasing distribution pattern from the northern, high-latitude to the southern, low-latitude estuaries (Fig. 6a–c), with hot spots in the north and center of the BS, nearshore of the YRE, and upstream of the JRE and PRE. Notably, total amoA gene abundances displayed a contrary pattern, while total nir gene abundances and the ratio of total nir to amoA gene abundances (nir amoA) had generally consistent patterns with the N2O concentration, N2O flux, and ΔN2O across the four estuaries (Fig. 6d–f). A significant correlation was even observed between the N2O flux and the nir amoA ratio based on the four averages of the four estuaries (r=0.95, n=4, P<0.05). Therefore, the nir amoA ratio can indicate the N2O emission potential in China's estuaries, which is consistent with previous findings that the N2O production yield of denitrification is higher than that of nitrification in the lab and in situ experiments (Frey et al., 2019; Kester et al., 1997; Löscher et al., 2012; Stieglmeier et al., 2014).
Notably, the total nosZ gene abundance of N2O-reducing denitrifiers seemed to have a contrasting distribution pattern with the N2O concentration, N2O flux, and ΔN2O across the four estuaries, with higher abundances in the high-latitude BS and lower abundances in the low-latitude JRE (Fig. 6g). The total nosZ gene abundances were one to two orders of magnitude lower than the total nir gene abundances in the JRE and PRE, where the N2O concentration and flux were higher than those in the BS and YRE. This indicated a distinctly higher denitrification-derived N2O emission potential in the JRE and PRE. The ratio of total nir to nosZ clade I gene abundances (nir nosZ I) had a highly similar pattern with the N2O concentration, N2O flux, and ΔN2O across the four estuaries in general (Fig. 6h), and significant correlations were also observed between the N2O flux and nir nosZ I (r=0.97, n=4, P<0.05). Therefore, the nir nosZ I ratio could be a better indicator of N2O emission potential in China's estuaries. Abundances and activities of the N2O-producing (nirS or nirK–bearing) community relative to the N2O-reducing (nosZ-bearing) community have also been used to assess the N2O emission potential of soils (Thompson, 2016; Zhao et al., 2018). Similarly, the functional gene transcript distribution indicated that the nir nosZ I and nir amoA gene transcript abundance ratios also had consistent patterns with the N2O concentration, N2O flux, and ΔN2O across the four estuaries in general (Fig. S4). The high load of DIN in estuaries could be responsible for the high denitrification-derived N2O emission potential. Both the nir nosZ and nir amoA ratios were positively correlated with the NH, NO, and NO concentrations (Spearman's ρ=0.32–0.68, n=114–122, P<0.01 for each) and negatively correlated with salinity (Spearman's to −0.66, n=114–122, P<0.01 for each). Previous studies in the YRE have proven that nitrogen input accelerates N2O production in estuaries (W. Yan et al., 2012; Zhang et al., 2010). Therefore, sufficient supplies of substrates may support high rates of denitrification and thus high N2O emissions.
4.4 Influence of N2O emissions by N2O reducer composition
The community structure and diversity of the clade II nosZ gene retrieved from China's estuaries are different from those previously reported in soil and marine OMZ water (Jones et al., 2013, 2014; Sun, 2021). The dominant nosZ clade II-bearing groups are affiliated with Bacteroidetes, Chloroflexi, Gamma-, and Betaproteobacteria in our four estuarine and coastal areas. However, the most abundant nosZ clade II groups found in the OMZs of the eastern tropical South and North Pacific and the Arabian Sea are affiliated with Anaeromyxobacter (Deltaproteobacteria) (Sun et al., 2017; Sun et al., 2021) and those in the coastal OMZ waters of the Golfo Dulce, Costa Rica, are affiliated with Gammaproteobacteria, Marinimicrobia, Bacteroidetes, and SAR324 (Bertagnolli et al., 2020). The nosZ clade II organisms from terrestrial systems show distinctly higher diversity (Sanford et al., 2012; Jones et al., 2014; Hallin et al., 2018; Zhao et al., 2018; Kato et al., 2018). The phylogenetically distinct predominant N2O reducers can influence N2O emissions directly or indirectly (Song et al., 2014). According to genomic information, nosZ clade II carriers affiliated with Deltaproteobacteria and Chlorobi have neither the nirK nor nirS gene, and less than half of nosZ clade II organisms affiliated with Bacteroidetes, Chloroflexi, Gamma-, and Epsilonproteobacteria harbor the nirK or nirS gene, while all of the nosZ clade II microbes affiliated with Alpha- and Betaproteobacteria also have the nirS gene (Hallin et al., 2018). Therefore, the distinct nosZ clade II community structure among the four estuaries may contribute to their different N2O emissions potential. For example, distinctly high diversity of the nosZ clade II gene was retrieved from the JRE water and sediment samples as well as the YRE sediment sample compared to the other estuaries. The high diversity of the nosZ clade II gene may be caused by the high temperature (e.g., in the low-latitude JRE) and sufficient nutrients at those sites. Previous studies have also indicated that the biodiversity of denitrifying bacteria increases in high-temperature seasons (Castellano-Hinojosa et al., 2017) and that nitrogen availability has a positive effect on denitrifying bacteria in boreal lakes (Rissanen et al., 2011). In addition, the habitat type may also affect the abundance and diversity of N2O-reducing communities, e.g., silty mud and sandy sediments have higher genetic potentials for N2O reduction than cyanobacterial mat and Ruppia maritima meadow sediments (Wittorf et al., 2020).
This study revealed the distinct distribution patterns of six key microbial functional genes and transcripts related to N2O production and consumption pathways in the BS, the YRE, the adjacent ECS, the JRE, and the PRE. The archaeal amoA genes and transcripts were more abundant in the northern BS, YRE, and the adjacent ECS, while the denitrifier nirS genes and transcripts were more abundant in the southern JRE and PRE. The nosZ clade II gene was more abundant than the clade I-type throughout the estuaries except for in the JRE and a few sites of the BS, while the opposite transcript distribution pattern was observed in these two estuaries. Water mass parameters (temperature and salinity), substrates (ammonia/ammonium, nitrite, and nitrate), and influencing parameters of substrate availability (DO and pH) regulated the gene, transcript, and community composition distribution patterns. The community structure of the clade II-type nosZ gene retrieved from China's estuaries was distinctly different from those of the soil and marine OMZ. Furthermore, combined with the N2O concentration, flux, and ΔN2O data collected from previous studies, our analysis found that, although both the clade I- and II-type nosZ genes of N2O reducers were widely distributed in these estuaries, N2O production by the denitrification pathway may be more important in determining the N2O emissions patterns across the estuaries. Nitrogen loads may influence the N2O source and sink processes by regulating the distribution of the related functional microbial groups.
All quality-controlled sequences were submitted to GenBank with accession numbers OM567739–OM568649. All other data can be accessed in the form of Excel spreadsheets via the corresponding author.
The supplement related to this article is available online at: https://doi.org/10.5194/bg-19-3757-2022-supplement.
YZ conceived and designed the study. XD, XW, MC, ET, and NC performed the experiments and auxiliary data collection. XD analyzed the data. XD and YZ wrote the paper. All authors contributed to the interpretation of the results and critical revision.
The contact author has declared that none of the authors has any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
We thank Zuhui Zuo, Yufang Li, and Minyuan Liu for their assistance in sampling and DNA/RNA extraction, as well as Jiaming Shen for his valuable comments and suggestions in the preparation of the manuscript. Thanks are also given to CEES Open Cruise for the Jiulong River estuary – Xiamen Bay and Shuiying Huang and Jiezhong Wu for their organizational help.
This research has been supported by the National Natural Science Foundation of China (grant nos. 42125603, 92051114, 91751207, 41721005, and 42188102).
This paper was edited by Denise Akob and reviewed by two anonymous referees.
Abell, G. C. J., Revill, A. T., Smith, C., Bissett, A. P., Volkman, J. K., and Robert, S. S.: Archaeal ammonia oxidizers and nirS-type denitrifiers dominate sediment nitrifying and denitrifying populations in a subtropical macrotidal estuary, ISME J., 4, 286–300, https://doi.org/10.1038/ismej.2009.105, 2010.
Babbin, A. R., Bianchi, D., Jayakumar, A., and Ward, B. B.: Rapid nitrous oxide cycling in the suboxic ocean, Science, 348, 1127–1129, https://doi.org/10.1126/science.aaa8380, 2015.
Bange, H. W., Rapsomanik, S., and Andreae, M. O.: Nitrous oxide in coastal waters, Global Biogeochem. Cy., 10, 197–207, https://doi.org/10.1029/95GB03834, 1996.
Barnes, J. and Upstill-Goddard, R. C.: N2O seasonal distributions and air-sea exchange in UK estuaries: Implications for the tropospheric N2O source from European coastal waters, J. Geophys. Res.-Biogeo., 116, G01006, https://doi.org/10.1029/2009JG001156, 2011.
Beaulieu, J. J., Tank, J. L., Hamilton, S. K., Wollheim, W. M., Hall, R. O., Mulholland, P. J., Peterson, B. J., Ashkenas, L. R., Cooper, L. W., Dahm, C. N., Dodds, W. K., Grimm, N. B., Johnson, S. L., McDowell, W. H., Poole, G. C., Maurice Valett, H., Arango, C. P., Bernot, M. J., Burgin, A. J., Crenshaw, C. L., Helton, A. M., Johnson, L. T., O'Brien, J. M., Potter, J. D., Sheibley, R. W., Sobota, D. J., and Thomas, S. M.: Nitrous oxide emission from denitrification in stream and river networks, P. Natl. Acad. Sci. USA, 108, 214–219, https://doi.org/10.1073/pnas.1011464108, 2011.
Beman, J. M., Popp, B. N., and Francis, C.: Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California, ISME J., 2, 429–441, https://doi.org/10.1038/ismej.2008.33, 2008.
Bernhard, A. E., Landry, Z. C., Blevins, A., De La Torre, J. R., Giblin, A. E., and Stahl, D. A.: Abundance of ammonia-oxidizing archaea and bacteria along an estuarine salinity gradient in relation to potential nitrification rates, Appl. Environ. Microbiol., 76, 1285–1289, https://doi.org/10.1128/AEM.02018-09, 2010.
Bertagnolli, A. D., Konstantinidis, K. T., and Stewart, F. J.: Non-denitrifier nitrous oxide reductases dominate marine biomes, Environ. Microbiol. Rep., 12, 681–692, https://doi.org/10.1111/1758-2229.12879, 2020.
Blackmer, A. M. and Bremner, J. M.: Inhibitory effect of nitrate on reduction of N2O to N2 by soil microorganisms, Soil Biol Biochem., 10, 187–191, https://doi.org/10.1016/0038-0717(78)90095-0, 1978.
Blum, J. M., Su, Q., Ma, Y., Valverde-Pérez, B., Domingo-Félez, C., Jensen, M. M., and Smets, B. F.: The pH dependency of N-converting enzymatic processes, pathways and microbes: effect on net N2O production, Environ. Microbiol., 20, 1623–1640, https://doi.org/10.1111/1462-2920.14063, 2018.
Braker, G., Zhou, J., Wu, L., Devol, A. H., and Tiedje, J. M.: Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific northwest marine sediment communities, Appl. Environ. Microbiol., 66, 2096–2104, 2000.
Brase, L., Bange, H. W., Lendt, R., Sanders, T., and Dähnke, K.: High resolution measurements of nitrous oxide (N2O) in the Elbe estuary, Front. Mar. Sci., 4, 162, https://doi.org/10.3389/fmars.2017.00162, 2017.
Cao, W., Hong, H., and Yue, S.: Modelling agricultural nitrogen contributions to the Jiulong River estuary and coastal water, Glob. Planet. Change, 47, 111–121, https://doi.org/10.1016/j.gloplacha.2004.10.006, 2005.
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T.: trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses, Bioinformatics, 25, 1972–1973, https://doi.org/10.1093/bioinformatics/btp348, 2009.
Carey, C. J., Dove, N. C., Beman, J. M., Hart, S. C., and Aronson, E. L.: Meta-analysis reveals ammonia-oxidizing bacteria respond more strongly to nitrogen addition than ammonia-oxidizing archaea, Soil Biol. Biochem., 99, 158–166, https://doi.org/10.1016/j.soilbio.2016.05.014, 2016.
Castellano-Hinojosa, A., Correa-Galeote, D., Carrillo, P., Bedmar, E. J., and Medina-Sánchez, J. M.: Denitrification and biodiversity of denitrifiers in a High-Mountain Mediterranean Lake, Front. Microbiol., 8, 1911, https://doi.org/10.3389/fmicb.2017.01911, 2017.
Chee-Sanford, J. C., Connor, L., Krichels, A., Yang, W. H., and Sanford, R. A.: Hierarchical detection of diverse Clade II (atypical) nosZ genes using new primer sets for classical- and multiplex PCR array applications, J. Microbiol. Methods., 172, 105908, https://doi.org/10.1016/j.mimet.2020.105908, 2020.
Chen, C. A., Wang, S., Lu, X., Zhang, S., Lui, H., Tseng, H., Wang, B., and Huang, H.: Hydrogeochemistry and greenhouse gases of the Pearl River, its estuary and beyond, Quaternary Int., 186, 79–90, https://doi.org/10.1016/j.quaint.2007.08.024, 2008.
Chen, C. T. A.: Chemical and physical fronts in the Bohai, Yellow and East China seas, J. Mar. Syst., 78, 394–410, https://doi.org/10.1016/j.jmarsys.2008.11.016, 2009.
Codispoti, L. A. and Christensen, J. P.: Nitrification, denitrification and nitrous oxide cycling in the eastern tropical South Pacific ocean, Mar. Chem., 16, 277–300, https://doi.org/10.1016/0304-4203(85)90051-9, 1985.
Cole, J. J. and Caraco, N. F.: Emissions of nitrous oxide (N2O) from a tidal, freshwater river, the Hudson River, New York, Environ. Sci. Technol., 35, 991–996, https://doi.org/10.1021/es0015848, 2001.
Cui, P., Chen, Z., Zhao, Q., Yu, Z., Yi, Z., Liao, H., and Zhou, S.: Hyperthermophilic composting significantly decreases N2O emissions by regulating N2O-related functional genes, Bioresour. Technol., 272, 433–441, https://doi.org/10.1016/j.biortech.2018.10.044, 2019.
Dai, M., Wang, L., Guo, X., Zhai, W., Li, Q., He, B., and Kao, S. J.: Nitrification and inorganic nitrogen distribution in a large perturbed river/estuarine system: The Pearl River Estuary, China, Biogeosciences, 5, 1227–1244, https://doi.org/10.5194/bg-5-1227-2008, 2008.
De Wilde, H. P. J. and De Bie, M. J. M.: Nitrous oxide in the Schelde estuary: Production by nitrification and emission to the atmosphere, Mar. Chem., 69, 203–216, https://doi.org/10.1016/S0304-4203(99)00106-1, 2000.
Domeignoz-Horta1, L. A., AyméSpor, D. B., Breuil, M.-C., Florian Bizouard1, J. L., and L. P.: The diversity of the N2O reducers matters for the N2O : N2 denitrification end-product ratio across an annual and a perennial cropping system, Front. Microbiol., 6, 971, https://doi.org/10.3389/fmicb.2015.00971, 2015.
Edgar, R. C.: Search and clustering orders of magnitude faster than BLAST, Bioinformatics, 26, 2460–2461, 2010.
Fayazbakhsh, K., Abedian, A., Manshadi, B. D., and Khabbaz, R. S.: Introducing a novel method for materials selection in mechanical design using Z-transformation in statistics for normalization of material properties, Mater. Des., 30, 4396–4404, https://doi.org/10.1016/j.matdes.2009.04.004, 2009.
Fish, J. A., Chai, B., Wang, Q., Sun, Y., Brown, C. T., Tiedje, J. M., and Cole, J. R.: FunGene: the functional gene pipeline and repository [data set], Front. Microbiol., 4, 291, https://doi.org/10.3389/fmicb.2013.00291, 2013.
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E., and Oakley, B. B.: Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean, P. Natl. Acad. Sci. USA, 102, 14683–14688, https://doi.org/10.1073/pnas.0506625102, 2005.
Frey, C., Bange, H. W., Achterberg, E. P., Jayakumar, A., Löscher, C. R., Arévalo-Martínez, D. L., León-Palmero, E., Sun, M., Sun, X., Xie, R. C., Oleynik, S., and Ward, B. B.: Regulation of nitrous oxide production in low-oxygen waters off the coast of Peru, Biogeosciences, 17, 2263–2287, https://doi.org/10.5194/bg-17-2263-2020, 2020.
Garnier, J., Cébron, A., Tallec, G., Billen, G., Sebilo, M., and Martinez, A.: Nitrogen behaviour and nitrous oxide emission in the tidal Seine River estuary (France) as influenced by human activities in the upstream watershed, Biogeochemistry, 77, 305–326, https://doi.org/10.1007/s10533-005-0544-4, 2006.
Graf, D. R., Jones, C. M., and Hallin, S.: Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions, PLoS ONE, 9, e114118, https://doi.org/10.1371/journal.pone.0114118, 2014.
Hallin, S., Philippot, L., Löf, F. E., Sanford, R. A., and Jones, C. M.: Genomics and ecology of novel N2O-Reducing microorganisms, Trends Microbiol., 26, 43–55, https://doi.org/10.1016/j.tim.2017.07.003, 2018.
He, B., Dai, M., Zhai, W., Guo, X., and Wang, L.: Hypoxia in the upper reaches of the Pearl River Estuary and its maintenance mechanisms: A synthesis based on multiple year observations during 2000–2008, Mar. Chem., 167, 13–24, https://doi.org/10.1016/j.marchem.2014.07.003, 2014.
Henry, S., Bru, D., Stres, B., Hallet, S., and Philippot, L.: Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils, Appl. Environ. Microbiol., 72, 5181–5189, https://doi.org/10.1128/AEM.00231-06, 2006.
Hink, L., Gubry-Rangin, C., Nicol, G. W., and Prosser, J. I.: The consequences of niche and physiological differentiation of archaeal and bacterial ammonia oxidisers for nitrous oxide emissions, ISME J., 12, 1084–1093, 2018.
Hornek, R., Pommerening-Roser, A., Koops, H. P., Farnleitner, A. H., Kreuzinger, N., Kirschner, A., and Mach, R.L.: Primers containing universal bases reduce multiple amoA gene specific DGGE band patterns when analysing the diversity of beta-ammonia oxidizers in the environment, J. Microbiol. Methods., 66, 147–155, 2006.
Hou, L., Xie, X., Wan, X., Kao, S. J., Jiao, N., and Zhang, Y.: Niche differentiation of ammonia and nitrite oxidizers along a salinity gradient from the Pearl River estuary to the South China Sea, Biogeosciences, 15, 5169–5187, https://doi.org/10.5194/bg-15-5169-2018, 2018.
Ji, Q., Buitenhuis, E., Suntharalingam, P., Sarmiento, J. L., and Ward, B. B.: Global nitrous oxide production determined by oxygen sensitivity of nitrification and denitrification, Global Biogeochem. Cy., 32, 1790–1802, https://doi.org/10.1029/2018GB005887, 2018a.
Ji, Q., Frey, C., Sun, X., Jackson, M., Lee, Y., Jayakumar, A., Jeffrey, C., and Ward, B. B.: Nitrogen and oxygen availabilities control water column nitrous oxide production during seasonal anoxia in the Chesapeake Bay, Biogeosciences, 15, 6127–6138, https://doi.org/10.5194/bg-15-6127-2018, 2018b.
Jones, C. M., Stres, B., Rosenquist, M., and Hallin, S.: Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification, Mol. Biol. Evol. 25, 1955–1966, https://doi.org/10.1093/molbev/msn146, 2008.
Jones, C. M., Graf, D. R. H., Bru, D., Philippot, L., and Hallin, S.: The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink, ISME J. 7, 417–26, https://doi.org/10.1038/ismej.2012.125, 2013.
Jones, C. M., Spor, A., Brennan, F. P., Breuil, M., Bru, D., Lemanceau, P., Griffiths, B., Hallin, S., and Philippot, L.: Recently identified microbial guild mediates soil N2O sink capacity, Nat. Clim. Change, 4, 801–805, https://doi.org/10.1038/nclimate2301, 2014.
Kato, S., Shibuya, T., Takaki, Y., Hirai, M., Nunoura, T., and Suzuki, K.: Genome-enabled metabolic reconstruction of dominant chemosynthetic colonizers in deep-sea massive sulfide deposits, Environ. Microbiol., 20, 862–877, https://doi.org/10.1111/1462-2920.14032, 2018.
Katoh, K. and Standley, D. M.: MAFFT multiple sequence alignment software version 7: Improvements in performance and usability, Mol. Biol. Evol., 30, 772–780, https://doi.org/10.1093/molbev/mst010, 2013.
Kester, R. A., De Boer, W., and Laanbroek, H. J.: Production of NO and N2O by pure cultures of nitrifying and denitrifying bacteria during changes in aeration, Appl. Environ. Microbiol., 63, 3872–3877, 1997.
Lam, P., Jensen, M. M., Lavik, G., McGinnis, D. F., Müller, B., Schubert, C. J., Amann, R., Thamdrup, B., and Kuypers, M. M. M.: Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea, P. Natl. Acad. Sci. USA, 104, 7104–7109, https://doi.org/10.1073/pnas.0611081104, 2007.
Laperriere, S. M., Nidzieko, N. J., Fox, R. J., Fisher, A. W., and Santoro, A. E.: Observations of variable ammonia oxidation and nitrous oxide flux in a eutrophic estuary, Estuar. Coast., 42, 33–44, https://doi.org/10.1007/s12237-018-0441-4, 2019.
Lee, J. A. and Francis, C. A.: Spatiotemporal characterization of San Francisco Bay denitrifying communities: a comparison of nirK and nirS diversity and abundance, Microb. Ecol., 73, 271–284, https://doi.org/10.1007/s00248-016-0865-y, 2017.
Li, J., Nedwell, D. B., Beddow, J., Dumbrell, A. J., McKew, B. A., Thorpe, E. L., and Whitby, C.: amoA gene abundances and nitrification potential rates suggest that benthic ammonia-oxidizing bacteria and not archaea dominate N cycling in the Colne estuary, United Kingdom, Appl. Environ. Microbiol., 81, 159–165, https://doi.org/10.1128/AEM.02654-14, 2015.
Lin, H., Dai, M., Kao, S. J., Wang, L., Roberts, E., Yang, J., Huang, T., and He, B.: Spatiotemporal variability of nitrous oxide in a large eutrophic estuarine system: The Pearl River Estuary, China, Mar. Chem., 182, 14–24, 2016.
Lin, J., Chen, N., Wang, F., Huang, Z., Zhang, X., and Liu, L.: Urbanization increased river nitrogen export to western Taiwan Strait despite increased retention by nitrification and denitrification, Ecol. Indic., 109, 105756, https://doi.org/10.1016/j.ecolind.2019.105756, 2020.
Löscher, C. R., Kock, A., Könneke, M., Laroche, J., Bange, H. W., and Schmitz, R. A.: Production of oceanic nitrous oxide by ammonia-oxidizing archaea, Biogeosciences, 9, 2419–2429, https://doi.org/10.5194/bg-9-2419-2012, 2012.
Lycus, P., Soriano-Laguna, M. J., Kjos, M., Richardson, D. J., Gates, A. J., Milligan, D. A., Frostegårda, Å., Bergausta, L., and Bakken, L. R.: A bet-hedging strategy for denitrifying bacteria curtails their release of N2O, P. Natl. Acad. Sci. USA, 115, 11820–11825, https://doi.org/10.1073/pnas.1805000115, 2018.
Ma, L., Lin, H., Xie, X., Dai, M., and Zhang, Y.: Major role of ammonia-oxidizing bacteria in N2O production in the Pearl River estuary, Biogeosciences, 16, 4765–4781, https://doi.org/10.5194/bg-16-4765-2019, 2019.
Magalhães, C., Bano, N., Wiebe, W. J., Bordalo, A. A., and Hollibaugh, J. T.: Dynamics of nitrous oxide reductase genes (nosZ) in intertidal rocky biofilms and sediments of the Douro River Estuary (Portugal), and their relation to N-biogeochemistry, Microb. Ecol., 55, 259–269, 2008.
Marchant, H. K., Ahmerkamp, S., Lavik, G., Tegetmeyer, H. E., Graf, J., Klatt, J. M., Holtappels, M., Walpersdorf, E., and Kuypers, M. M. M.: Denitrifying community in coastal sediments performs aerobic and anaerobic respiration simultaneously, ISME J. 11, 1799–1812, https://doi.org/10.1038/ismej.2017.51, 2017.
Martens-Habbena, W. and Stahl, D. A.: Nitrogen metabolism and kinetics of ammonia-oxidizing archaea, Methods Enzymol., 496, 465–487, https://doi.org/10.1016/B978-0-12-386489-5.00019-1, 2011.
Marzadri, A., Dee, M. M., Tonina, D., Bellin, A., and Tank, J. L.: Role of surface and subsurface processes in scaling N2O emissions along riverine networks, P. Natl. Acad. Sci. USA, 114, 4330–4335, https://doi.org/10.1073/pnas.1617454114, 2017.
Massana, R., Murray, A. E., Preston, C. M., and DeLong, E. F.: Vertical distribution and phylogenetic characterization of marine planktonic archaea in the Santa Barbara Channel, Appl. Environ. Microbiol., 63, 50–56, https://doi.org/10.1128/aem.63.1.50-56.1997, 1997.
McElroy, M. B., Elkins, J. W., Wofsy, S. C., Kolb, C. E., Durán, A. P., and Kaplan, W. A.: Production and release of N2O from the Potomac Estuary, Limnol. Oceanogr., 23, 1168–1182, https://doi.org/10.4319/lo.1978.23.6.1168, 1978.
Meinhardt, K. A., Stopnisek, N., Pannu, M. W., Strand, S. E., Fransen, S. C., Casciotti, K. L., and Stahl, D. A.: Ammonia-oxidizing bacteria are the primary N2O producers in an ammonia-oxidizing archaea dominated alkaline agricultural soil, Environ. Microbiol., 20, 2195–2206, https://doi.org/10.1111/1462-2920.14246, 2018.
Molina, V., Belmar, L., and Ulloa, O.: High diversity of ammonia-oxidizing archaea in permanent and seasonal oxygen-deficient waters of the Eastern South Pacific, Environ. Microbiol., 12, 2450–2465, https://doi.org/10.1111/1462-2920.14246, 2010.
Mosier, A. C. and Francis, C. A.: Denitrifier abundance and activity across the San Francisco Bay estuary, Environ. Microbiol Rep., 2, 667–676, https://doi.org/10.1111/j.1758-2229.2010.00156.x, 2010.
Mullins, T. D., Britschgi, T. B., Krest, R. L., and Giovannoni, S. J.: Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities, Limnol. Oceanogr., 40, 148–158, 1995.
Nakai, R., Nishijima, M., Tazato, N., Handa, Y., Karray, F., Sayadi, S., Isoda, H., and Naganuma, T.: Oligoflexus tunisiensis gen. nov., sp. nov., a Gram-negative, aerobic, filamentous bacterium of a novel proteobacterial lineage, and description of Oligoflexaceae fam. nov., Oligoflexales ord. nov. and Oligoflexia classis nov, Int. J. Syst. Evol. Microbiol, 64, 3353–3359, https://doi.org/10.1099/ijs.0.060798-0, 2014.
Nevison, C., Butler, J. H., and Elkins, J. W.: Global distribution of N2O and the ΔN2O-AOU yield in the subsurface ocean, Global Biogeochem. Cy., 17, 1–18, https://doi.org/10.1029/2003GB002068, 2003.
Palacin-Lizarbe, C., Camarero, L., Hallin, S., Jones, C., Caliz, J., Casamayor, E. O., and Catalan, J.: The DNRA-denitrification dichotomy differentiates nitrogen transformation pathways in mountain lake benthic habitats, Front. Microbiol., 10, 1229, https://doi.org/10.3389/FMICB.2019.01229, 2019.
Park, B. J., Park, S. J., Yoon, D. N., Schouten, S., Damsté, J. S. S., and Rhee, S. K.: Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria, Appl. Environ. Microb., 76, 7575–87, 2010.
Philippot, L.: Loss in microbial diversity affects nitrogen cycling in soil, ISME J., 11, 1609–1619, 2013.
Pike, N.: Using false discovery rates for multiple comparisons in ecology and evolution, Methods Ecol. Evol., 2, 278–282, 2011.
Price, M. N., Dehal, P. S., and Arkin, A. P.: FastTree 2 – Approximately maximum-likelihood trees for large alignments, PLoS One 5, 9490, https://doi.org/10.1371/journal.pone.0009490, 2010.
Xu, J., Wang, Y., Wang, Q., and Yin, J.: Nitrous oxide concentration and nitrification and denitrification in Zhujiang River Estuary, China. J. Environ. Sci., 18, 122–130, 2005.
Qu, Z., Bakken, L. R., Molstad, L., Frostegård, Å., and Bergaust, L. L.: Transcriptional and metabolic regulation of denitrification in Paracoccus denitrificans allows low but significant activity of nitrous oxide reductase under oxic conditions, Environ. Microbiol., 18, 2951–2963, https://doi.org/10.1111/1462-2920.13128, 2016.
R Core Team: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, https://www.R-project.org/ (last access: 5 July 2019), Vienna, Austria, 2017.
Ravishankara, A. R., Daniel, J. S., and Portmann, R. W.: Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century, Science, 326, 123–125, https://doi.org/10.1126/science.1176985, 2009.
Rissanen, A. J., Tiirola, M., and Ojala, A.: Spatial and temporal variation in denitrification and in the denitrifier community in a boreal lake, Aquat. Microbiol. Ecol., 64, 27–40, https://doi.org/10.3354/ame01506, 2011.
Riya, S., Takeuchi, Y., Zhou, S., Terada, A., and Hosomi, M.: Nitrous oxide production and mRNA expression analysis of nitrifying and denitrifying bacterial genes under floodwater disappearance and fertilizer application, Environ. Sci. Pollut. Res., 24, 15852–15859, https://doi.org/10.1007/s11356-017-9231-y, 2017.
Rotthauwe, J. H., Witzel, K. P., and Liesack, W.: The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations, Appl. Environ. Microb., 63, 4704-4712, 1997.
Rowley, G., Sullivan, M. J., Appia-Ayme, C., Gates, A. J., and Richardson, D. J.: Copper control of bacterial nitrous oxide emission and its impact on vitamin B12-dependent metabolism, P. Natl. Acad. Sci. USA, 110, 19926–19931, https://doi.org/10.1073/pnas.1314529110, 2013.
Sanford, R. A., Wagner, D. D., Wu, Q. Z., Chee-Sanford, J. C., Thomas, S. H., Cruz-Garcia, C., Rodriguez, G., Massol-Deya, A., Krishnani, K. K., Ritalahti, K. M., Nissen, S., Konstantinidis, K. T., and Loffler, F. E.: Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils, P. Natl. Acad. Sci. USA, 109, 19709–19714, https://doi.org/10.1073/Pnas.1211238109, 2012.
Santoro, A. E., Buchwald, C., McIlvin, M. R., and Casciotti, K. L.: Isotopic Signature of N2O Produced by Marine Ammonia-Oxidizing Archaea, Science, 333, 1282–1285, https://doi.org/10.1126/science.1208239, 2011.
Senbayram, M., Budai, A., Bol, R., Chadwick, D., Marton, L., Gündogan, R., and Wu, D.: Soil NO level and O2 availability are key factors in controlling N2O reduction to N2 following long-term liming of an acidic sandy soil, Soil Biol. Biochem., 132, 165–173, https://doi.org/10.1016/j.soilbio.2019.02.009, 2019.
Shaw, L. J., Nicol, G. W., Smith, Z., Fear, J., Prosser, J. I., and Baggs, E. M.: Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway, Environ. Microbiol., 8, 214–222, https://doi.org/10.1111/j.1462-2920.2005.00882.x, 2006.
Silvennoinen, H., Liikanen, A., Torssonen, J., Florian Stange, C., and Martikainen, P. J.: Denitrification and nitrous oxide effluxes in boreal, eutrophic river sediments under increasing nitrate load: A laboratory microcosm study, Biogeochemistry, 91, 105–116, https://doi.org/10.1007/s10533-008-9262-z, 2008.
Smith, C. J., Nedwell, D. B., Dong, L. F., and Osborn, A. M.: Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments, Appl. Environ. Microbiol., 73, 3612–3622, https://doi.org/10.1128/AEM.02894-06, 2007.
Solomon, S. D., Qin, D., Manning, M., Chen, Z., Marquis, M., Averyt, K., Tignor, M., and Miller, H. L.: Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change (IPCC), Cambridge Univ. Press, Cambridge, 996 pp., UK, 2007.
Song, D., Zhang, G., Li, P., and Liu, S.: Distribution and fluxes of nitrous oxide in the Bohai Sea in summer, Adv. Mar. Sci., 2, 13–21, https://doi.org/10.12677/ams.2015.22003, 2015.
Song, K., Suenaga, T., Hamamoto, A., Satou, K., Riya, S., Hosomi, M., and Terada, A.: Abundance, transcription levels and phylogeny of bacteria capable of nitrous oxide reduction in a municipal wastewater treatment plant, J. Biosci. Bioeng., 118, 289–297, https://doi.org/10.1016/j.jbiosc.2014.02.028, 2014.
Stieglmeier, M., Mooshammer, M., Kitzler, B., Wanek, W., Zechmeister-Boltenstern, S., Richter, A., and Schleper, C.: Aerobic nitrous oxide production through N-nitrosating hybrid formation in ammonia-oxidizing archaea, ISME J., 8, 1135–1146, https://doi.org/10.1038/ismej.2013.220, 2014.
Sun, X., Jayakumar, A., and Ward, B. B.: Community composition of nitrous oxide consuming bacteria in the oxygen minimum zone of the Eastern Tropical South Pacific, Front. Microbiol., 8, 1183, https://doi.org/10.3389/fmicb.2017.01183, 2017.
Sun, X., Jayakumar, A., Tracey, J. C., Wallace, E., Kelly, C. L., Casciotti, K. L., and Ward, B. B.: Microbial N2O consumption in and above marine N2O production hotspots, ISME J., 15, 1434–1444, https://doi.org/10.1038/s41396-020-00861-2, 2021.
Thompson, K. A.: Abundance, activity and community structure of nitrifier and denitrifier communities in Agro-Ecosystems, PhD thesis, Land Resource Science, University of Guelph, Guelph, Ontario, Canada, 184 pp., 2016.
Wang, L., Zheng, B., Nan, B., and Hu, P.: Diversity of bacterial community and detection of nirS- and nirK-encoding denitrifying bacteria in sandy intertidal sediments along Laizhou Bay of Bohai Sea, China, Mar. Pollut. Bull., 88, 215–223, https://doi.org/10.1016/j.marpolbul.2014.09.002, 2014.
Wang, L., Zhang, G., Zhu, Z., Li, J., Liu, S., Ye, W., and Han, Y.: Distribution and sea-to-air flux of nitrous oxide in the East China Sea during the summer of 2013, Cont. Shelf Res., 123, 99–110, https://doi.org/10.1016/j.csr.2016.05.001, 2016.
Wang, Y., Guo, J., Vogt, R.D., Mulder, J., Wang, J., and Zhang, X.: Soil pH as the chief modifier for regional nitrous oxide emissions: new evidence and implications for global estimates and mitigation, Glob. Change Biol., 24, e617–e626, 2017.
Wei, W., Isobe, K., Nishizawa, T., Zhu, L., Shiratori, Y., Ohte, N., Koba, K., Otsuka, S., and Senoo, K.: Higher diversity and abundance of denitrifying microorganisms in environments than considered previously, ISME J., 9, 1954–1965, https://doi.org/10.1038/ismej.2015.9, 2015.
Wittorf, L., Roger, F., Alsterberg, C., Gamfeldt, L., Hulth, S., Sundback, K., Jones, C. M., and Hallin, S.: Habitat diversity and type govern potential nitrogen loss by denitrification in coastal sediments and differences in ecosystem-level diversities of disparate N2O reducing communities, FEMS Microbiol. Ecol. 96, 1–9, https://doi.org/10.1093/femsec/fiaa091, 2020.
Wrage, N., Velthof, G. L., Van Beusichem, M. L., and Oenema, O.: Role of nitrifier denitrification in the production of nitrous oxide, Soil Biol. Biochem. 33, 1723–1732, https://doi.org/10.1016/S0038-0717(01)00096-7, 2001.
Wu, J., Chen, N., Hong, H., Lu, T., Wang, L., and Chen, Z.: Direct measurement of dissolved N2 and denitrification along a subtropical river-estuary gradient, China. Mar. Pollut. Bull., 66, 125–134, https://doi.org/10.1016/j.marpolbul.2012.10.020, 2013.
Yamagishi, H., Westley, M. B., Popp, B. N., Toyoda, S., Yoshida, N., Watanabe, S., Koba, K., and Yamanaka, Y.: Role of nitrification and denitrification on the nitrous oxide cycle in the eastern tropical North Pacific and Gulf of California, J. Geophys. Res., 112, 1–15, https://doi.org/10.1029/2006JG000227, 2007.
Yan, W., Yang, L., Wang, F., Wang, J., and Ma, P.: Riverine N2O concentrations, exports to estuary and emissions to atmosphere from the Changjiang River in response to increasing nitrogen loads, Global Biogeochem. Cy., 26, GB4006, https://doi.org/10.1029/2010GB003984, 2012.
Yan, X., Wan, X. S., Liu, L., Xu, M. N., Tan, E., Zheng, Z., Zou, W., Tian, L., Li, D. W., Trull, T. W., and Kao, S. J.: Biogeochemical dynamics in a eutrophic tidal estuary revealed by isotopic compositions of multiple nitrogen species, J. Geophys. Res.-Biogeo., 124, 1849–1864, https://doi.org/10.1029/2018JG004959, 2019.
Yan, X., Zhai, W., Hong, H., Li, Y., and Guo, W.: Distribution, fluxes and decadal changes of nutrients in the Jiulong River Estuary, Southwest Taiwan Strait, Chinese Sci. Bull. 57, 2307–2318, https://doi.org/10.1007/s11434-012-5084-4, 2012.
Yao, Y., Tian, H., Shi, H., Pan, S., Xu, R., Pan, N., and Canadell, J. G.: Increased global nitrous oxide emissions from streams and rivers in the Anthropocene, Nat. Clim. Change, 10, 138–142, https://doi.org/10.1038/s41558-019-0665-8, 2020.
Yoon, S., Nissen, S., Park, D., Sanford, R. A., and Löffler, F. E.: Nitrous oxide reduction kinetics distinguish bacteria harboring clade I versus clade II NosZ, Appl. Environ. Microb., 82, 3793–3800, 2016.
Zhan, L., Chen, L., Zhang, J., and Zheng, A.: Distribution of N2O in the Jiulongjiang River Estuary and estimation of its air-sea flux during winter, Journal of Oceanography in Taiwan Strait, 30, 189–195, https://doi.org/10.3969/J.ISSN.1000-8160.2011.02.006, 2011. (in Chinese).
Zhang, G., Zhang, J., Ren, J., Li, J., and Liu, S.: Distributions and sea-to-air fluxes of methane and nitrous oxide in the North East China Sea in summer, Mar. Chem., 110, 42–55, https://doi.org/10.1016/j.marchem.2008.02.005, 2008.
Zhang, G., Zhang, J., Liu, S., Ren, J., and Zhao, Y.: Nitrous oxide in the Changjiang (Yangtze River) Estuary and its adjacent marine area: Riverine input, sediment release and atmospheric fluxes, Biogeosciences, 7, 3505–3516, https://doi.org/10.5194/bg-7-3505-2010, 2010.
Zhang, J.: Biogeochemistry of Chinese estuarine and coastal waters: nutrients, trace metals and biomarkers, J. Mater. Cycl. Waste Manag., 3, 65–76, https://doi.org/10.1007/s10113-001-0039-3, 2002.
Zhang, Y., Xie, X., Jiao, N., Hsiao, S. S. Y., and Kao, S. J.: Diversity and distribution of amoA-type nitrifying and nirS-type denitrifying microbial communities in the Yangtze River estuary, Biogeosciences, 11, 2131–2145, https://doi.org/10.5194/bg-11-2131-2014, 2014.
Zhao, S., Wang, Q., Zhou, J., Yuan, D., and Zhu, G.: Linking abundance and community of microbial N2O-producers and N2O-reducers with enzymatic N2O production potential in a riparian zone, Sci. Total Environ., 642, 1090–1099, https://doi.org/10.1016/J.SCITOTENV.2018.06.110, 2018.
Zheng, Z. Z., Wan, X., Xu, M. N., Hsiao, S. S. Y., Zhang, Y., Zheng, L. W., Wu, Y., Zou, W., and Kao, S. J.: Effects of temperature and particles on nitrification in a eutrophic coastal bay in southern China, J. Geophys. Res.-Biogeo., 122, 2325–2337, https://doi.org/10.1002/2017JG003871, 2017.
Zhu, Z. Y., Zhang, J., Wu, Y., Zhang, Y. Y., Lin, J., and Liu, S. M.: Hypoxia off the Changjiang (Yangtze River) Estuary: Oxygen depletion and organic matter decomposition, Mar. Chem., 125, 108–116, https://doi.org/10.1016/j.marchem.2011.03.005, 2011.





