the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Differentiation of cognate bacterial communities in thermokarst landscapes: implications for ecological consequences of permafrost degradation
Shudan Ye
Hongxuan Li
Xilei Huang
Luyao Chen
Thermokarst processes likely result in new habitats harboring novel bacterial communities in degraded permafrost soil (PB), thermokarst lake sediments (SB), and lake water (WB). Our study aimed to investigate the paired PB, SB, and WB across the Qinghai–Tibet Plateau (QTP) by assessing the spatial pattern of diversity as well as assembly mechanisms of these bacterial communities. Each habitat had distinct bacterial assemblages, with lower α diversity and higher β diversity in WB than in SB and PB. However, up to 41 % of the operational taxonomic units (OTUs) were shared by PB, SB, and WB, suggesting that many taxa originate from the same sources via dispersal. SB and WB had reciprocal dispersal effects, and both were correlated with PB. Dispersal limitation was the most dominant assembly process shaping PB and SB, while homogeneous selection was the most dominant for WB. Bacterial communities of the three habitats correlated differently with environmental variables, but latitude, mean annual precipitation, and pH were the common factors associated with their β diversity, while total phosphorus was the common factor associated with their assembly processes. Our results imply that thermokarst processes result in diverse habitats that have distinct bacterial communities that differ in diversity, assembly mechanisms, and environmental drivers.
- Article
(5355 KB) - Full-text XML
-
Supplement
(906 KB) - BibTeX
- EndNote
Permafrost is an important landscape in high-latitude and high-altitude regions, covering 15 % of the land area of the Northern Hemisphere (Obu, 2021) and 40 % of the Qinghai–Tibet Plateau (QTP) (Zou et al., 2017; Gao et al., 2021) and containing twice as much carbon as is currently present in the atmosphere (Schuur et al., 2009; Hugelius et al., 2014; Mishra et al., 2021). Permafrost is highly sensitive to climate warming (Wu et al., 2007; Jorgenson et al., 2010; Biskaborn et al., 2019), which is expected to reduce 50 %–90 % of permafrost cover by 2100 (Lawrence et al., 2012; Chadburn et al., 2017). As a result of ice-rich permafrost thaw, thermokarst lakes and ponds are formed (Kokelj and Jorgenson, 2013; Farquharson et al., 2016) and are extensively distributed across the Arctic and sub-Arctic regions (de Jong et al., 2018) as well as the QTP (Niu et al., 2011; Luo et al., 2020). The initial sediment and water in thermokarst lakes originate from the melting of permafrost, and they are continuously replenished through the collapse of permafrost and precipitation (West and Plug, 2008; de Jong et al., 2018). Thus, thermokarst lake sediments and water, as well as the surrounding degraded permafrost soil, represent three distinct habitats derived from the original permafrost during the process of thermokarst formation (Fig. 1). It is well known that thermokarst processes substantially influence regional hydrological, ecological, and biogeochemical processes (Chin et al., 2016; In'T Zandt et al., 2020; Manasypov et al., 2021) and initiate a strong positive climate feedback to global warming (Walter et al., 2006; Schuur et al., 2008; Schaefer et al., 2011; Anthony et al., 2018). However, the microbial differences and relationships among these distinct habitats in thermokarst landscapes are largely unknown.
Understanding microbes in thermokarst landscapes, and elsewhere, is important because microbial communities play pivotal roles in driving biogeochemical and ecological processes. To understand thermokarst microbial communities, we need to understand the assembly mechanisms structuring them, a central research topic in microbial ecology (Stegen et al., 2012; Nemergut et al., 2013; Zhou et al., 2014; Zhou and Ning, 2017). In the assembly of microbial communities, both deterministic and stochastic processes occur simultaneously but with contributions that can vary (Chase, 2010; Zhou et al., 2013; Vellend et al., 2014; Makoto et al., 2019). Typically, deterministic processes place a strong emphasis on niche-based mechanisms, including ecological selection, environmental filtering, and biotic interactions (Zhou and Ning, 2017). Conversely, stochastic processes involve neutral mechanisms like random birth and death, unforeseen disturbance, probability-based dispersal, and ecological drift (Chave, 2004; Chase, 2010; Zhou et al., 2014). In various ecosystems or habitats, the significance of deterministic and stochastic processes can differ greatly and be shaped by a multitude of environmental factors (Tripathi et al., 2018; Aguilar and Sommaruga, 2020; Jiao and Lu, 2020; She et al., 2021). During thermokarst formation, vast areas of permafrost have been transformed to thermokarst lakes, leading to major changes in physicochemical environments as well as in the biological communities of these regions. Thus, it is also expected that the microbial communities experience major changes in occupying degraded permafrost soil, thermokarst lake sediments, and lake water and, in doing so, display different assembly mechanisms (Fig. 1).
Better understanding of community assembly in these systems is important because thawing permafrost and thermokarst lakes are greenhouse gas emission hotspots (In'T Zandt et al., 2020; Mu et al., 2020; Elder et al., 2021). Close relationships between biogeochemical processes and microbial community assembly have generally been demonstrated (Bier et al., 2015; Graham et al., 2016; Le Moigne et al., 2020; Ren et al., 2022a). Assembly processes inevitably influence biogeochemical functions by shaping community diversity and composition (Graham et al., 2016; Leibold et al., 2017; Mori et al., 2018). For example, dispersal (a stochastic process) can suppress biogeochemical functioning by increasing the proportion of maladapted taxa (Strickland et al., 2009; Nemergut et al., 2013; Graham and Stegen, 2017), while selection (a deterministic process) may have a positive influence on biogeochemical function by facilitating locally adapted taxa (Graham et al., 2016). In particular, stochastic dispersal has been suggested to suppress the mineralization of organic carbon in soil and water (Le Moigne et al., 2020; Luan et al., 2020). Therefore, it is hypothesized that the relative influence of deterministic and stochastic processes on community assembly could impact the biogeochemical functions of microbial communities (Strickland et al., 2009; Nemergut et al., 2013; Pholchan et al., 2013; Graham and Stegen, 2017). Given the importance of understanding how microbial community variations affect the biogeochemical cycles in permafrost and thermokarst landscapes, it is necessary to have a deeper understanding of the assembly mechanisms shaping the microbial communities that form following permafrost degradation.
In this paper we evaluated these ideas on the Qinghai–Tibet Plateau (QTP), which is known as the “Third Pole” of the Earth and is therefore uniquely positioned as an indicator of global change (Yao et al., 2012). Pronounced environmental changes in response to climate warming on the QTP have been observed and documented, especially in the past half century (Piao et al., 2012; Zhang et al., 2018; Ren et al., 2019a). Major changes are predicted to continue on the QTP, and permafrost thawing is among the most prominent, but little is known about the microbial communities in these rapidly emerging ecosystems. To fill this gap, we investigated water and sediment in thermokarst lakes across the QTP as well as permafrost soil around the lakes (Fig. 1). Our aims were to (1) assess the spatial pattern of α and β diversity of bacterial communities and (2) evaluate the community assembly mechanisms and environmental responses of the bacterial communities in degraded permafrost soil, as well as in the sediment and water of thermokarst lakes.
2.1 Study area, field sampling, and chemical analysis
This work was conducted across the QTP in July 2021 (Fig. S1 in the Supplement). In total, 44 sites were investigated by collecting paired samples of lake water, lake sediment, and surrounding permafrost soil (Fig. 1a) (Ren et al., 2022a). The sampling strategy and chemical analysis methods were described in detail in our previous publications (Ren et al., 2022a, c). For water sampling of each lake, surface water samples were collected at a depth of 0.3 to 0.5 m in three acid-clean bottles. For microbial analysis, 200 mL of water from each bottle was filtered using a 0.2 µm polycarbonate membrane filter (Whatman, UK) for DNA extraction. For each lake, three filters were combined into one composite sample for DNA extraction. The remaining water samples were transported to the lab for other physicochemical measurements. For sediment sampling, the top 15 cm of sediment was collected from three points. Sediment samples for microbial analysis were collected in a 45 mL sterile centrifuge tube, and the remaining samples were air-dried for analyzing physicochemical properties. For permafrost sampling, five topsoil cores were collected along three 25 m transects with increasing distances to the lake shore, respectively. The soils from one transect were homogenized. Soil samples for microbial analysis were stored in 45 mL sterile centrifuge tubes, and the remaining soils were used for analyzing physicochemical properties. For each sampling site, pH, conductivity (Cond), organic carbon (dissolved organic carbon (DOC) in water and soil organic carbon (SOC) in sediment and soil), total nitrogen (TN), and total phosphorus (TP) were measured according to our previous publications (Ren et al., 2022a, c). Moreover, the QTP climate dataset (Zhou, 2018) was obtained from the National Tibetan Plateau Data Center (https://data.tpdc.ac.cn/en/, last access: 10 December 2022) and was utilized to extract the mean annual temperature (MAT) and mean annual precipitation (MAP) for each of the study sites.
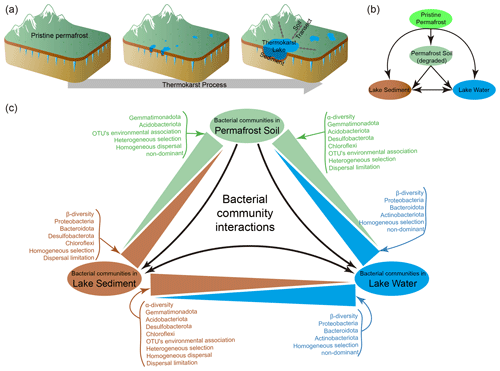
Figure 1(a) The process of thermokarst lake formation in ice-rich permafrost (modified from Ren et al, 2022a). (b) A schematic view of the relationships between permafrost soil, lake sediment, and lake water. (c) Summary of the differences between distinct habitats of the bacterial communities in permafrost soil, lake sediment, and lake water.
2.2 DNA extraction, PCR, and sequencing
The Magen HiPure Soil DNA Kit (Magen, China) was used to extract DNA from soil (0.5 g frozen soil), sediment (0.5 g frozen sediment), and water (membrane filters by filtering 600 mL lake water in total) samples according to the manufacturer's protocols. Extraction blanks were routinely performed in parallel. The prokaryotic 16S rRNA gene's V3–V4 hypervariable regions were amplified using universal primers 343F-TACGGRAGGCAGCAG and 798R-AGGGTATCTAATCCT (Nossa et al., 2010). PCRs were conducted in a 25 µL reaction mixture containing 2.5 µL of TransStart buffer, 2 µL of dNTPs, 1 µL of each primer, 0.5 µL of TransStart Taq DNA polymerase, and 20 ng template DNA. The PCR reactions were conducted on a thermal cycler (ABI GeneAmp® 9700, USA) using the following procedure: initial denaturation at 94 ∘C for 5 min, 24 cycles of denaturation at 94 ∘C for 30 s followed by annealing at 56 ∘C for 30 s and extension at 72 ∘C for 20 s, and a final extension at 72 ∘C for 5 min. To reduce amplification bias, three individual PCR amplifications were performed for each sample, and the triplicate PCR products were combined and purified. DNA libraries were verified on 2 % agarose gels and quantified using a Qubit 4 Fluorometer (Thermo Fisher Scientific, Waltham, USA). Next-generation sequencing of the amplicon products was conducted on an Illumina MiSeq Platform (Illumina, San Diego, CA, USA). Automated cluster generation and 250/300 paired-end sequencing with dual reads were performed following the manufacturer's instructions. The forward and reverse reads were joined and assigned to samples based on barcode and truncated by cutting off the barcode and primer sequence. Raw sequences were trimmed of ambiguous bases and low-quality sequences (quality score lower than 20). After trimming, the paired-end reads were joined and de-noised using QIIME1.9.1 (Caporaso et al., 2010). The sequences were subjected to the following de-noising criteria: sequences with ambiguous or homologous regions, as well as those below 200 bp in length, were excluded; sequences with at least 75 % of bases having a quality score above Q20 were retained; and chimeric sequences were identified and eliminated. All sequences from extraction blanks were removed. The effective sequences were grouped into operational taxonomic units (OTUs) using a 97 % sequence similarity threshold, and the taxonomic classifications were annotated against the SILVA 138 database (released on 2 November 2020) (Quast et al., 2013). The singletons were removed, and the sequences were rarefied to the lowest number of sequences per sample (24 251 sequences) to eliminate the bias from the sampling effort.
2.3 Analyses
Three α diversity indices, including the observed number of OTUs (OTU richness), Shannon diversity, and phylogenetic diversity (PD), were calculated using QIIME 1.9.1 (Caporaso et al., 2010). The “ses.mntd” function in the picante 1.8.2 package was used to calculate the standardized effect size measure of the mean nearest taxon distance (SES.MNTD) for assessing the phylogenetic clustering of bacterial communities (Kembel et al., 2010). β diversity was calculated as the Bray–Curtis distance based on the relative abundance of OTUs. In order to determine the habitat niche occupied by each taxon, we utilized the “spaa” package (Zhang, 2016) in R to calculate the Levins niche width (Levins, 1968). The formula of niche breadth is , where Bi represents the niche breadth of OTUi across the communities, n is the total number of communities, and pi is the proportion of OTUi in each community. Differences in α diversity and β diversity among bacterial communities in different habitats, including permafrost soil bacterial communities (PB), lake sediment bacterial communities (SB), and lake water bacterial communities (WB), were assessed using the Wilcoxon rank-sum test. The relationships between taxonomic and environmental variables were assessed using Spearman correlation, and the P values were corrected using the false discovery rate (FDR) method (Benjamini and Hochberg, 1995). Mantel tests were performed to examine the correlation between environmental variables and β diversity. A nonmetric multidimensional scaling (NMDS) analysis was conducted to examine the distribution of PB, SB, and WB using the “metaMDS” function in the vegan 2.5-7 package based on the Bray–Curtis distance using the relative abundance of OTUs (Oksanen et al., 2020). The distinctiveness of these communities was confirmed through a non-parametric statistical test (ANOSIM) using the “anosim” function in the vegan package. The habitat niche occupied by each species was estimated by calculating the Levins niche breadth (Levins, 1968) with the use of the spa 0.2.2 package (Zhang, 2016). Species with a broader niche breadth were distributed more evenly across a wider range of habitats, compared to those with a narrower niche breadth.
Structural equation modeling (SEM) was conducted to assess the relationships between location (including latitude, longitude, and elevation), climate (including mean annual temperature and mean annual precipitation), and physicochemical variables (including pH, conductivity, nutrient concentrations, and stoichiometric ratios) of each habitat (permafrost soil, lake sediment, and lake water), as well as their bacterial communities (PB, SB, and WB). In model building, the SEM incorporated the following prior knowledge: (a) location and climate factors potentially influence all the studied bacterial communities; (b) the physicochemical factor of each habitat potentially influences the corresponding bacterial communities; and (c) permafrost soil potentially influences thermokarst lake sediment and water, while lake sediment and water interact with each other. In the SEM, location, climate, and physicochemical environments were reduced in dimension by principal component analysis (PCA), respectively, using the “prcomp” function of the vegan package, and the first axis (PCA1) was used in SEM. For community structure, the first axis of NMDS was used. SEM was constructed using the lavaan package (Rosseel, 2012). The fit of SEM was assessed using standard indices, including chi-square (χ2), the goodness-of-fit index (GFI), the comparative fit index (CFI), the root mean square residual (RMR), and the root mean squared error of approximation (RMSEA) (Hu and Bentler, 1999; Barrett, 2007).
Phylogenetic trees of bacteria were constructed in the R package ggtree 3.2.1 (Yu et al., 2017) using the top 1000 abundant OTUs in PB, SB, and WB, respectively. For each phylogenetic tree, a heatmap was built in the inner ring to represent Spearman's correlation between OTUs and environmental variables. The middle ring was built to represent the frequency of the OTUs in our studied sites. The outer ring was built to represent the relative abundance of the OTUs.
A null model analysis was performed to investigate the processes shaping the assembly of bacterial communities in permafrost soil, lake sediment, and lake water using the R package picante 1.8.2 (Kembel et al., 2010). This analysis was based on the calculation of the β nearest taxon index (βNTI) to measure the extent of deterministic processes in shaping the phylogenetic composition of the communities, as well as a Bray–Curtis-based Raup–Crick matrix (RCBray) to assess the relative influences of stochastic processes (Stegen et al., 2013; Zhou and Ning, 2017). Because homogeneous selection results in communities that share greater phylogenetic similarity, the proportion of homogeneous selection was calculated as the fraction of pairwise comparisons with βNTI . On the other hand, heterogeneous selection, leading to communities with lesser phylogenetic similarity, was measured as the fraction of pairwise comparisons with βNTI . Because homogeneous dispersal results in communities exhibiting greater taxonomic resemblance, the extent of its impact was measured as the proportion of pairwise comparisons with NTI <2 and RC. Conversely, communities constrained by dispersal limitation display lesser taxonomic similarity, and the measure of dispersal limitation was derived from the fraction of pairwise comparisons with NTI <2 and RCBray>0.95. Finally, the fraction of the pairwise comparisons with NTI <2 and was identified as “undominated”. Mantel tests were conducted to test the relationships between environmental variables and βNTI.
All the statistical analyses were carried out in R 4.1.2 (R Core Team, 2020).
3.1 General distribution patterns of α diversity
After quality filtering, 3 201 132 high-quality sequences were obtained and clustered into 9361 OTUs, of which 3870 OTUs were core OTUs shared by bacterial communities in permafrost soil, lake sediment, and lake water (Fig. S2). Moreover, a large number of OTUs were shared by PB and SB (n=7053), of which 16.4 % were enriched in lake sediment and 19.3 % were enriched in permafrost soil (Fig. S2). However, a relatively small number of OTUs were shared by PB and WB (n=4007) and by SB and WB (n=4431), and only a very small proportion of OTUs were enriched in lake water (Fig. S2). Bacterial communities had a significantly lower α diversity in lake water than in lake sediment and permafrost soil (Fig. 2a). α diversity was not significantly different between PB and SB (Fig. 2a). Correlation analyses showed that the phylogenetic diversity of PB was positively correlated with SOC, TN, and ratios (Fig. 2b). For SB, α diversity indices were positively correlated with MAT, MAP, SOC, TN, C:P, and N:P, while they were negatively correlated with latitude and pH (Fig. 2b). For WB, α diversity indices were negatively correlated with pH, and Shannon diversity was negatively correlated with DOC, TN, and C:P (Fig. 2b).
PB and SB had a significantly greater phylogenetic diversity than WB (Figs. 2a and 3). The OTUs in PB had significantly higher frequencies than those of SB and WB (Fig. 3). The top 1000 abundant OTUs in PB were highly correlated with environmental variables, particularly with latitude, MAP, SOC, TN, TP, and ratios (Fig. 3a). The top 1000 abundant OTUs in SB were more commonly positively correlated with MAP, SOC, TN, and ratios but more commonly negatively correlated with latitude and pH (Fig. 3b). The top 1000 abundant OTUs in WB had relatively fewer significant relationships with environmental variables in general but were negatively correlated with latitude, conductivity, pH, DOC, TN, and ratios, while more positively correlated with MAP (Fig. 3c). In addition, WB had significantly higher SES.MNTD than PB and SB (Fig. 3d), suggesting higher phylogenetic clustering of bacterial taxa in WB.
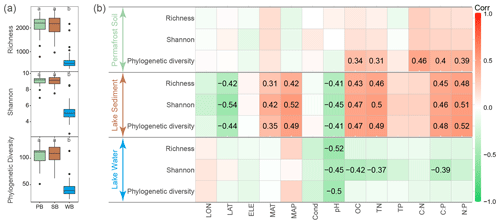
Figure 2(a) α diversity of bacterial communities in permafrost soil (PB), lake sediment (SB), and lake water (WB). The different lower-case letters represent significant differences assessed using the Wilcoxon rank-sum test. (b) Spearman correlations show the relationships between α diversity and environmental factors. The color represents the correlation coefficient, which is shown in detail when the result is statistically significant (p<0.05).
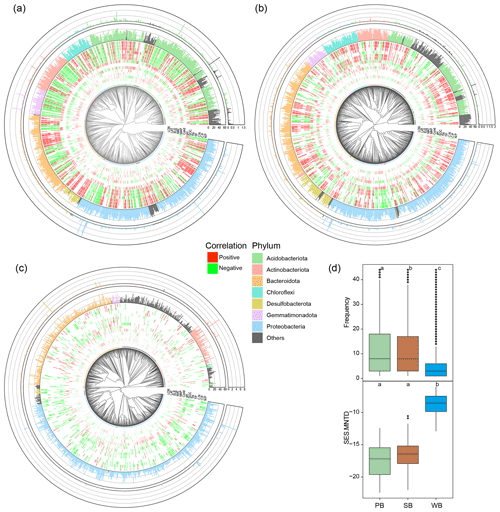
Figure 3Phylogenetic tree of the top 1000 OTUs in bacterial communities in (a) permafrost soil, (b) lake sediment, and (c) lake water. The tree tips are colored by major phylum. The inner ring of the heatmap represents Spearman's correlation between OTUs and environmental variables. Only significant (p<0.05) results are shown. The middle ring represents the frequency of the OTUs in our studied sites. The outer ring represents the relative abundance of the OTUs. (d) Boxplots showing differences of OTU's frequency and SES.MNTD values among bacterial communities in permafrost soil (PB), lake sediment (SB), and lake water (WB). The different lower-case letters represent significant differences assessed using the Wilcoxon rank-sum test.
3.2 Community composition and β diversity patterns
PB were dominated by Proteobacteria (30.4 %), Acidobacteriota (25.3 %), Bacteroidota (11.4 %), Actinobacteriota (6.8 %), Chloroflexi (5.2 %), and Gemmatimonadota (5.2 %) (Fig. 4a). SB were dominated by Proteobacteria (35.2 %), Bacteroidota (20.0 %), Acidobacteriota (11.3 %), Desulfobacterota (6.4 %), Chloroflexi (6.3 %), and Actinobacteriota (5.8 %) (Fig. 4a). WB were dominated by Proteobacteria (46.9 %), Bacteroidota (29.2 %), and Actinobacteriota (17.4 %) (Fig. 4a). While Proteobacteria were predominant in all three habitat types, these dominant phyla had significantly different relative abundances among these habitats. Proteobacteria and Bacteroidota had a significantly higher relative abundance in WB than in SB and PB (Fig. 4a). The relative abundance of Actinobacteriota was the highest in WB but was not significantly different between PB and SB (Fig. 4a). Gemmatimonadota and Acidobacteriota were more significantly enriched in PB than in SB and WB. Desulfobacterota and Chloroflexi were more significantly enriched in SB than in PB and WB (Fig. 4a).
These phyla responded differently to environmental variables (Fig. 4b). For example, Actinobacteriota and Gemmatimonadota in PB and Actinobacteriota and Desulfobacterota in SB were negatively correlated with nutrient concentrations and ratios, while Desulfobacterota in PB and Acidobacteriota in SB were positively correlated with nutrient concentrations and ratios (Fig. 4b). The pH frequently correlated with taxa in various taxonomic groups across all three habitats (Fig. 4b).
A nonmetric multidimensional scaling (NMDS) analysis along with non-parametric statistical tests showed that bacteria in different habitats formed distinct communities (Fig. 5a). The extent of difference was larger for WB vs. PB (β=0.98; RANOSIM=0.989, P<0.001) than the differences for WB vs. SB (β=0.96; RANOSIM=0.967, P<0.001). There was the least dissimilarity between PB and SB (β=0.81; RANOSIM=0.384, P<0.001). The fitted SEM model showed that PB had direct effects on SB and WB, and the latter two had reciprocal effects on each other (Fig. 5b). In addition, location, climate, and permafrost soil physicochemical environments had direct effects on PB. Climate had direct effects on SB, while lake water physicochemical environments had direct effects on WB (Fig. 5b).
WB had higher β diversity than SB and PB, suggesting that bacterial communities were more spatially heterogeneous in lake water than in lake sediment and permafrost soil (Fig. 5c). Taxa in PB had higher habitat niche breadths than taxa in SB and WB (Fig. 5d). We estimated the distance decay relationship of bacterial community similarity. Significant distance–decay relationships were observed for all communities, but the fitness values were relatively low (Fig. S3), indicating a weak decay of community similarity with geographic distance in the thermokarst landscape. We also explored the main environmental variables that influence the variations of the bacterial communities (Fig. 6). β diversities of PB, SB, and WB were all significantly correlated with latitude, MAP, and pH (Fig. 6). In addition, β diversity of PB was also significantly correlated with all the other environmental variables, except MAT and conductivity. β diversity of SB was also significantly correlated with conductivity and C:N (Fig. 6). β diversity of WB was also significantly correlated with elevation, MAT, conductivity, DOC, TN, and TP (Fig. 6). The results suggested that the compositional variation among PB, SB, and WB was differentially structured by spatial, climatic, and physicochemical variables.
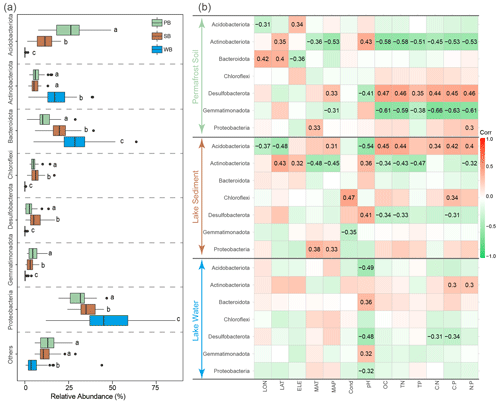
Figure 4(a) Relative abundances of major phyla in bacterial communities in permafrost soil (PB), lake sediment (SB), and lake water (WB). The different lower-case letters represent significant differences assessed using the Wilcoxon rank-sum test. (b) Spearman correlations show the relationships between the relative abundance of major phyla and environmental factors. The color represents the correlation coefficient, which shown in number when the result is significant (p<0.05).
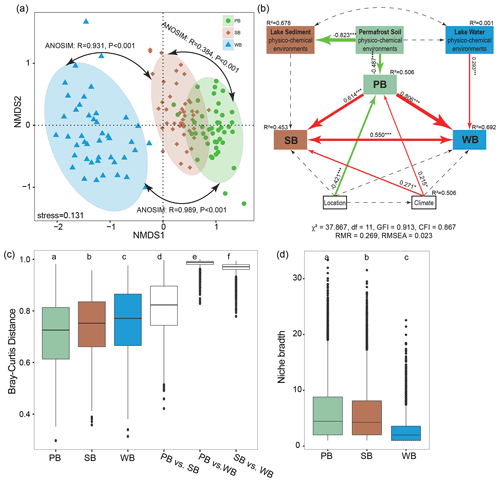
Figure 5(a) Nonmetric multidimensional scaling (NMDS) ordination showing the distribution of bacterial communities in permafrost soil (PB), lake sediment (SB), and lake water (WB). The differences between these communities are confirmed by the non-parametric statistical test (ANOSIM). (b) Structural equation modeling analysis depicting the relationships between location (including latitude, longitude, and elevation), climate (including mean annual temperature and mean annual precipitation), and physicochemical environments (pH, conductivity, nutrient concentrations, and stoichiometric ratios) of each habitat. The solid and dashed arrows represent the significant and nonsignificant relationships, respectively. The red and green arrows represent positive and negative relationships, respectively. Significant path coefficients are shown adjacent to the path, with *, **, and *** denoting the significant level of p<0.05, p<0.01, and p<0.001, respectively. (c) β diversities within and between PB, SB, and WB. (d) Habitat niche breadth of the bacterial communities.
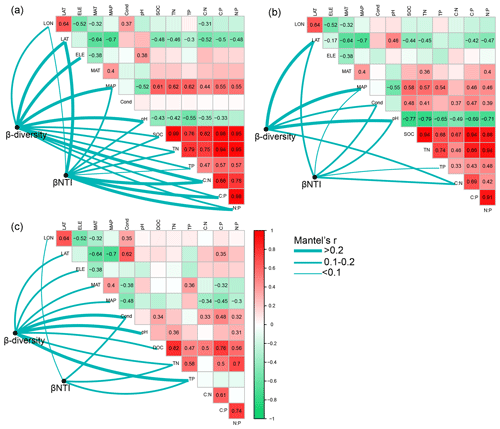
Figure 6Pairwise correlations between environmental variables as well as the Mantel tests between environmental variables and β diversity and β nearest taxon index (βNTI) for (a) bacterial communities in permafrost soil, (b) bacterial communities in lake sediment, and (c) bacterial communities in lake water. β diversity was calculated as Bray–Curtis distance. The lines denote significant relationships, while the line width represents Mantel's r statistic. Pairwise correlations between environmental variables are shown in a color gradient matrix. The color represents Pearson's correlation coefficient, which is shown in number when the result is significant (p<0.05). The abbreviations of the environmental variables are explained in the Methods section.
3.3 Assembly processes
To explore the mechanisms underlining the observed distribution patterns, a null-model-based framework was employed to quantify the deviation of phylogenetic turnover. PB had significantly higher βNTI than SB and WB (Fig. 7a). Deterministic processes contributed 51.3 %, 41.2 %, and 44.9 % to community variations for the bacterial communities in permafrost soil, lake sediment, and lake water, respectively (Fig. 7b). In particular, the results showed that homogeneous selection contributed a larger fraction to the assembly of the WB (44.8 %), followed by SB (35.2 %) and PB (29.7 %) (Fig. 7b). Heterogeneous selection influenced PB (21.6 %) more than SB (6.0 %) and WB (0.1 %). Dispersal limitation contributed a larger fraction to SB (57.2 %) than to PB (45 %) and WB (37.5 %).
The relationships between βNTI and major environmental variables were used to estimate changes in the relative influences of deterministic and stochastic assembly processes. Mantel tests showed that the assembly processes of bacterial communities in permafrost soil, lake sediment, and lake water had similarities and differences in the responses to environmental variables (Fig. 6). Particularly, the differences of TP were significantly associated with βNTI of PB, SB, and WB, implying that an increasing divergence of TP could contribute to a shift from homogeneous selection to heterogeneous selection in the assembly of bacterial communities in the QTP thermokarst landscape. Moreover, βNTI of PB was also significantly associated with other environmental variables, except elevation, MAT, and conductivity. βNTI of SB was also significantly associated with latitude, MAP, conductivity, pH, and C:N, while βNTI of WB was significantly associated with longitude, conductivity, and TN.
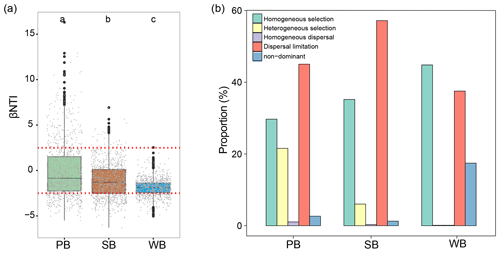
Figure 7(a) The values of βNTI with horizontal dashed red lines indicate upper- and lower-significance thresholds at βNTI and −2, respectively, in the three habitat types. (b) The contribution of deterministic (homogeneous and heterogeneous selection) and stochastic (dispersal limitations and homogenizing dispersal) processes to turnover in the assembly of bacterial communities in permafrost soil (PB), lake sediment (SB), and lake water (WB). “Non-dominant” indicates that the fraction was not dominated by any single process.
4.1 α diversity and community composition
Thermokarst lakes are known to have sediments that derive from the permafrost soil and are constantly replenished by the collapse of nearby permafrost (Payette et al., 2004; West and Plug, 2008; Veremeeva et al., 2021). This suggests that permafrost soil and lake sediments are likely to have high levels of similarity in bacterial diversity and community composition. Thus, there is no doubt that permafrost soil, lake sediments, and lake water should share a certain number of bacteria. Indeed, our study showed that 41 % of OTUs were shared among PB, SB, and WB, while 75 % of OTUs were shared between PB and SB. Additionally, our prior research has shown that there are close correlations between the abiotic features of the two environments (Ren et al., 2022c). However, despite these similarities and connections, we found substantial differences in the bacterial communities of permafrost soil and lake sediments. As proposed by the Baas-Becking hypothesis (Baas-Becking, 1934), environmental selection is partially responsible for variation in microbial communities, which are also shaped by other ecological processes such as diversification and dispersal limitation. In our study, α diversity and the dominant phyla found in PB and SB responded differently to various environmental variables. In addition, there were significant differences in composition and structure among PB, SB, and WB, while the dissimilarities between PB and SB were the lowest.
Bacterial communities in lake water had significantly lower α diversity as well as distinct community composition and structure in comparison to bacterial communities in permafrost soil and lake sediment. However, PB and SB had direct influence on WB. For thermokarst lakes, the water first originates from the thawing of the ice-rich permafrost, and the lake is then fed by precipitation-derived and permafrost-derived water (Y. Yang et al., 2016; Narancic et al., 2017; Wan et al., 2019). Microorganisms present in lake water have a diverse range of sources, including terrestrial inputs and other sources such as bacteria distributed within the atmosphere, associated with plants and animals, and carried by migratory birds and animals (Ruiz-Gonzalez et al., 2015). Thus, there was a relatively small proportion of OTUs shared between permafrost soil and lake water, as well as between lake sediment and water, and only a few shared OTUs were enriched in lake water. It is a well-established fact that different habitats often support distinct microbial communities (Fierer et al., 2012; Hugerth et al., 2015; Louca et al., 2016). The contrast in bacterial community composition between lake sediments and water has been extensively documented (Briee et al., 2007; Gough and Stahl, 2011; J. Yang et al., 2016; Ren et al., 2017). In addition, sediments generally harbor higher species-level diversity of bacteria compared to lake water (Lozupone and Knight, 2007; Ren et al., 2019b). For example, in a permafrost thaw pond of the Andes, it was also found that water samples had lower α diversity than lake sediment and permafrost samples (Aszalós et al., 2020). Permafrost soil and lake sediment may provide more habitat heterogeneity for bacterial taxa than the water column, supported by our observation that the bacterial taxa had higher niche breadth in permafrost soil and lake sediment than in lake water. Moreover, in hydrologically connected terrestrial–aquatic ecosystems, bacterial communities can present distinct but directional spatial structure driven by terrestrial recruited taxa (Ruiz-Gonzalez et al., 2015). Thus, these community similarities between distinct bacterial habitats might be the result of a common bacterial source (original permafrost), and the differences are likely caused by subsequent environmental selection, colonization from multiple other bacterial sources, and distinct assembly mechanisms.
Despite connections driven by dispersal, distinct thermokarst habitats had different bacterial community compositions, as seen in previous work (Ottoni et al., 2022). All the dominant phyla were significantly different in relative abundance among PB, SB, and WB. In this study, Proteobacteria, Bacteroidota, Actinobacteriota, Gemmatimonadota, Acidobacteriota, Desulfobacterota, and Chloroflexi dominated bacterial communities in permafrost soil and/or thermokarst lakes despite high variabilities. Similar dominance of these taxa has also been found in permafrost and thermokarst landscapes in other areas (Aszalós et al., 2020; Belov et al., 2020; Wu et al., 2022). The most commonly reported bacterial groups in permafrost environments include members of Proteobacteria, Acidobacteria, Actinobacteria, Bacteroidetes, Firmicutes, and Chloroflexi (Steven et al., 2009; Altshuler et al., 2017; Ottoni et al., 2022), as observed in our samples.
4.2 β diversity and assembly processes
In our studied regions across the QTP, PB, SB, and WB all had high β diversity (average values >0.7), with WB showing the highest, suggesting that bacterial communities shifted substantially across the large spatial scale of our sampling. Moreover, β diversities of PB, SB, and WB were significantly correlated with each other, further suggesting that the bacteria in different habitats had a considerable proportion of members from the same source, the original pristine permafrost soil. The significantly lower mean SES.MNTD for PB indicates that bacterial communities in permafrost soil were more closely phylogenetically clustered and suffered stronger environmental filtering than those in lake sediment and water (Langenheder et al., 2017), consistent with the observation that PB had lower β diversity than SB and WB. SES.MNTD is sensitive to changes in lineage close to the phylogenetic tips (Kembel et al., 2010). The higher SES.MNTD observed for SB and WB suggests the possibility that the bacteria in lake sediment and water exhibit a substantial divergence in the co-occurring species, and thermokarst lakes have experienced colonization by bacterial species originating from distinct clades or lineages from external sources following permafrost thaw (Webb et al., 2002; Stegen et al., 2013).
The structure of bacterial communities can vary across spatiotemporal scales and different habitats (Ren et al., 2017; Aguilar and Sommaruga, 2020; Pearman et al., 2020). A key objective in the field of microbial ecology is to determine the relative influence of stochastic and deterministic processes in shaping the assembly of communities (Stegen et al., 2013; Zhou and Ning, 2017). In this study, bacterial communities in lake water displayed a higher influence of homogeneous selection but a lower influence of heterogeneous selection compared to those in lake sediments and permafrost soil. Long-term changes in thermokarst lakes result in homogenized habitats and, consequently, strong homogenous selection in bacterial communities (Ning et al., 2019). In contrast, permafrost soil is a highly heterogeneous environment across spatial scales (Etzelmüller, 2013; Nitzbon et al., 2021), creating a wide range of habitats which can impose strong heterogeneous selection pressures on bacterial communities. Furthermore, permafrost soil is characterized by limited nutrient availability due to the frozen state of organic matter (Beermann et al., 2017; Zhang et al., 2023), while lake water offers a more diverse and abundant array of dissolved organic compounds and nutrients. As a result, bacterial communities in permafrost soil might be more sensitive to variations in resource availability, rendering them more strongly influenced by heterogeneous selection. In addition, dispersal limitation contributed a larger fraction to SB (57.2 %) than to PB (45 %) and WB (37.5 %). The dispersal of microorganisms is often considered as a passive process that results in community variation and turnover coupled with the function of environmental filtering (Cline and Zak, 2014; Stegen et al., 2015; Custer et al., 2022). The high dispersal limitation of microbial communities in thermokarst lakes may be attributed to several factors. These include the isolated nature of thermokarst lakes, which are endorheic and therefore have limited connectivity, as well as the strong restriction on microbial dispersal and the presence of strong environmental filtering processes. Additionally, the prolonged frozen phase of thermokarst lakes and permafrost soil restricts the movement of microorganisms (Vargas Medrano, 2019; Vigneron et al., 2019). Furthermore, geographical barriers, exemplified by prominent mountain ranges like the Tanggula Mountains, Kunlun Mountains, Nyenchen Tanglha Mountains, and Bayan Har Mountains, serve as impediments to the dispersal of both macro- and microorganisms (Wan et al., 2016; Yu et al., 2019; Ren et al., 2022b), particularly in lake sediment, where bacterial communities are more isolated over distances and will not disperse as far as those in lake water and permafrost soil, resulting in a stronger influence of dispersal limitation (Martiny et al., 2006; Xiong et al., 2012). Although the “everything is everywhere” hypothesis suggests that many microorganisms have a cosmopolitan distribution, their slow mobility allows for the development of regional phylogenetic differences and the emergence of specialized, endemic taxa in isolated habitats, resulting in a low likelihood of microorganisms dispersing to suitable distant sites (Telford et al., 2006). Therefore, dispersal processes in this thermokarst landscape may be restricted by the lack of hydrological connection, limited movement of water, short duration since thawing, and strong environmental filtering, contributing to the observed high dispersal limitation in the studied permafrost soil and thermokarst lakes. This inference is supported by many previous studies showing that dispersal limitation plays a major role in structuring microbial communities in lakes (Telford et al., 2006). Strong dispersal limitation for bacterial communities in permafrost has also been documented across an Alaskan boreal forest landscape (Bottos et al., 2018).
4.3 Environmental influences
Understanding how environmental factors shape bacterial communities is a crucial aspect in the field of microbial ecology (Fierer and Jackson, 2006; Pla-Rabes et al., 2011). In our study, bacterial communities were differentially correlated with various measured environmental variables. In our data, pH was consistently identified as a strong correlate of microbial community structure and diversity, as is often observed in terrestrial and aquatic ecosystems worldwide (Fierer and Jackson, 2006; Xiong et al., 2012). While such correlations between pH and bacterial communities have been widely found, the regulation mechanisms still remain unknown (Malard and Pearce, 2018). Moreover, the influences of pH are often species- and location-specific (Malard and Pearce, 2018; Egelberg et al., 2021). In this study, pH had significant negative relationships with α diversity of bacterial communities in lake sediment and water and had negative or positive correlations with some phyla. Moreover, differences in pH might drive the community variation observed between PB, SB, and WB and shift the community assembly processes for PB and SB. Moreover, with permafrost degrading and thermokarst developing, nutrient status will be strongly altered in permafrost areas. Organic carbon and nutrient stocks in permafrost are decreasing (Turetsky et al., 2020; Wu et al., 2021), and thermokarst lakes are developing, leading to dynamic environmental change (Luo et al., 2015; Vucic et al., 2020). These environmental disruptions likely impose strong influences on bacterial communities in thermokarst landscapes. Our study showed that nutrient (C, N, and P) concentrations and stoichiometric ratios were strongly related to α diversity (particularly for SB) and community variation and assembly (particularly for PB). High organic matter content, for instance, has been shown to support diverse and complex microbial communities (Garrido-Benavent et al., 2020; Ren and Gao, 2022). The role of nutrient availability in shaping bacterial communities has also been well established (Torsvik et al., 2002; Lee et al., 2017; Zhou et al., 2020). For example, Actinobacteria and Gemmatimonadota have a negative, while Gemmatimonadota has a positive, relationship with organic carbon and nutrients in permafrost (Romanowicz and Kling, 2022; Fu et al., 2023), in line with our results. The fact that different bacterial phyla exhibited varied responses to changes in organic carbon and nutrients further emphasizes the intricate interplay between microorganisms and their environment. Due to their ecological strategies, metabolic features, and environmental preferences, bacteria in permafrost respond differentially to nutrient status and other stressors, driving adaptive changes in community composition and function (Mackelprang et al., 2017). In addition, compared to permafrost soil and lake water, lake sediment can exhibit more stable physicochemical conditions. However, permafrost soil and lake water experience more dynamic and extreme environmental changes, which drive the bacterial communities. The results of SEM are also in line with bacterial community assembly, showing that deterministic processes had stronger influences on PB and WB than on SB. In addition, thermokarst lakes have sediment directly formed from permafrost soil, and thus permafrost soil environments and bacterial communities had strong associations with the corresponding components of lake sediment.
In addition to physicochemical environments, location and climate were also suggested as influences of bacterial communities in distinct habitats. On the QTP in particular, air temperature and precipitation are increasing in most regions (Xu et al., 2008; Lu et al., 2018). Warming and altered precipitation regimes under climate change have been demonstrated to affect α diversity and the composition of stream microbial communities at continental scales (Picazo et al., 2020). Our study indicates that location (particularly latitude) and climate (particularly MAP) factors are important in shifting bacterial communities in thermokarst landscapes. Particularly for bacterial communities in permafrost soil, location and climate have been evidenced as strong factors in shaping microbial communities (Taş et al., 2018; Barbato et al., 2022). Understanding the large-scale pattern of bacterial communities is increasingly important for offering insights into the impacts of climate change (Picazo et al., 2020; Ren et al., 2021). As the global climate changes, the QTP is getting warmer and more humid (Xu et al., 2008; Lu et al., 2018). Therefore, significant alterations to the physical, chemical, and biological properties of thermokarst lakes on the QTP can be expected in the coming decades. Based on the “space-for-time” substitution, our study serves as a foundation for predicting the potential impact of climate change on bacterial communities in thermokarst landscapes.
In this study, we investigated bacterial communities in paired water and sediment samples in thermokarst lakes as well as permafrost soil around the lakes across the QTP. Results showed that each habitat had distinct bacterial assemblages, with lower α diversity in lake water and higher β diversity in lake sediment and permafrost soil. There was considerable overlap in OTUs across habitats. Bacterial communities in permafrost soil and lake sediment were influenced by dispersal limitation, while those in lake water were driven by homogeneous selection. Environmental variables, including latitude, mean annual precipitation, and pH, affected bacterial community variations in all habitats. The study highlights the unique bacterial communities and ecological impacts of permafrost degradation in diverse habitats created by thermokarst processes.
The raw sequences were uploaded to the China National Center for Bioinformation (PRJCA009850, CRA007082, https://ngdc.cncb.ac.cn/search/?dbId=gsa&q=CRA007082&page=1, National Genomics Data Center, 2022).
The supplement related to this article is available online at: https://doi.org/10.5194/bg-20-4241-2023-supplement.
ZR designed the study, did the analyses, prepared the paper, and performed the fieldwork and laboratory work. All the authors prepared the paper.
The contact author has declared that none of the authors has any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
We are grateful to Yongming Deng and Xuan Jia for their assistance in the fieldwork and laboratory work.
This study was supported by the National Natural Science Foundation of China (grant no. 42301132) and the open funding of the State Key Laboratory of Lake Science and Environment (grant no. 2022SKL010).
This paper was edited by Lucia Fuchslueger and reviewed by Dajana Radujkovic and one anonymous referee.
Aguilar, P. and Sommaruga, R.: The balance between deterministic and stochastic processes in structuring lake bacterioplankton community over time, Mol. Ecol., 29, 3117–3130, https://doi.org/10.1111/mec.15538, 2020.
Altshuler, I., Goordial, J., and Whyte, L. G.: Microbial life in permafrost, in: Psychrophiles: from biodiversity to biotechnology, edited by: Margesin, R., Springer, Cham, https://doi.org/10.1007/978-3-319-57057-0_8, 153–179, 2017.
Anthony, K. M., von Deimling, T. S., Nitze, I., Frolking, S., Emond, A., Daanen, R., Anthony, P., Lindgren, P., Jones, B., and Grosse, G.: 21st-century modeled permafrost carbon emissions accelerated by abrupt thaw beneath lakes, Nat. Commun., 9, 3262, https://doi.org/10.1038/s41467-018-05738-9, 2018.
Aszalós, J. M., Szabó, A., Megyes, M., Anda, D., Nagy, B., and Borsodi, A. K.: Bacterial Diversity of a High-Altitude Permafrost Thaw Pond Located on Ojos del Salado (Dry Andes, Altiplano-Atacama Region), Astrobiology, 20, 754–765, https://doi.org/10.1089/ast.2018.2012, 2020.
Baas-Becking, L. G. M. (Ed.): Geobiologie of Inleiding Tot de Milieukunde, Van Stockkum & Zoon, The Hague, 1934.
Barbato, R. A., Jones, R. M., Douglas, T. A., Doherty, S. J., Messan, K., Foley, K. L., Perkins, E. J., Thurston, A. K., and Garcia-Reyero, N.: Not all permafrost microbiomes are created equal: Influence of permafrost thaw on the soil microbiome in a laboratory incubation study, Soil Biol. Biochem., 167, 108605, https://doi.org/10.1016/j.soilbio.2022.108605, 2022.
Barrett, P.: Structural equation modelling: Adjudging model fit, Pers. Individ. Differ., 42, 815–824, https://doi.org/10.1016/j.paid.2006.09.018, 2007.
Beermann, F., Langer, M., Wetterich, S., Strauss, J., Boike, J., Fiencke, C., Schirrmeister, L., Pfeiffer, E. M., and Kutzbach, L.: Permafrost thaw and liberation of inorganic nitrogen in Eastern Siberia, Permafrost Periglac. Process., 28, 605–618, 2017.
Belov, A. A., Cheptsov, V. S., Manucharova, N. A., and Ezhelev, Z. S.: Bacterial Communities of Novaya Zemlya Archipelago Ice and Permafrost, Geosciences, 10, 67, https://doi.org/10.3390/geosciences10020067, 2020.
Benjamini, Y. and Hochberg, Y.: Controlling the false discovery rate: a practical and powerful approach to multiple testing, J. Roy. Stat. Soc. B, 57, 289–300, https://doi.org/10.2307/2346101, 1995.
Bier, R. L., Bernhardt, E. S., Boot, C. M., Graham, E. B., Hall, E. K., Lennon, J. T., Nemergut, D. R., Osborne, B. B., Ruiz-Gonzalez, C., Schimel, J. P., Waldrop, M. P., and Wallenstein, M. D.: Linking microbial community structure and microbial processes: an empirical and conceptual overview, FEMS Microbiol. Ecol., 91, fiv113, https://doi.org/10.1093/femsec/fiv113, 2015.
Biskaborn, B. K., Smith, S. L., Noetzli, J., Matthes, H., Vieira, G., Streletskiy, D. A., Schoeneich, P., Romanovsky, V. E., Lewkowicz, A. G., Abramov, A., Allard, M., Boike, J., Cable, W. L., Christiansen, H. H., Delaloye, R., Diekmann, B., Drozdov, D., Etzelmuller, B., Grosse, G., Guglielmin, M., Ingeman-Nielsen, T., Isaksen, K., Ishikawa, M., Johansson, M., Johannsson, H., Joo, A., Kaverin, D., Kholodov, A., Konstantinov, P., Kroger, T., Lambiel, C., Lanckman, J. P., Luo, D., Malkova, G., Meiklejohn, I., Moskalenko, N., Oliva, M., Phillips, M., Ramos, M., Sannel, A., Sergeev, D., Seybold, C., Skryabin, P., Vasiliev, A., Wu, Q., Yoshikawa, K., Zheleznyak, M., and Lantuit, H.: Permafrost is warming at a global scale, Nat. Commun., 10, 264, https://doi.org/10.1038/s41467-018-08240-4, 2019.
Bottos, E. M., Kennedy, D. W., Romero, E. B., Fansler, S. J., Brown, J. M., Bramer, L. M., Chu, R. K., Tfaily, M. M., Jansson, J. K., and Stegen, J. C.: Dispersal limitation and thermodynamic constraints govern spatial structure of permafrost microbial communities, FEMS Microbiol. Ecol., 94, fiy110, https://doi.org/10.1093/femsec/fiy110, 2018.
Briee, C., Moreira, D., and Lopez-Garcia, P.: Archaeal and bacterial community composition of sediment and plankton from a suboxic freshwater pond, Res. Microbiol., 158, 213–227, https://doi.org/10.1016/j.resmic.2006.12.012, 2007.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., Fierer, N., Pena, A. G., Goodrich, J. K., Gordon, J. I., Huttley, G. A., Kelley, S. T., Knights, D., Koenig, J. E., Ley, R. E., Lozupone, C. A., Mcdonald, D., Muegge, B. D., Pirrung, M., Reeder, J., Sevinsky, J. R., Tumbaugh, P. J., Walters, W. A., Widmann, J., Yatsunenko, T., Zaneveld, J., and Knight, R.: QIIME allows analysis of high-throughput community sequencing data, Nat. Methods, 7, 335–336, https://doi.org/10.1038/nmeth.f.303, 2010.
Chadburn, S. E., Burke, E. J., Cox, P. M., Friedlingstein, P., and Hugelius, G.: An observation-based constraint on permafrost loss as a function of global warming, Nat. Clim. Chang., 7, 340–344, https://doi.org/10.1038/NCLIMATE3262, 2017.
Chase, J. M.: Stochastic community assembly causes higher biodiversity in more productive environments, Science, 328, 1388–1391, https://doi.org/10.1126/science.1187820, 2010.
Chave, J.: Neutral theory and community ecology, Ecol. Lett., 7, 241–253, 2004.
Chin, K. S., Lento, J., Culp, J. M., Lacelle, D., and Kokelj, S. V.: Permafrost thaw and intense thermokarst activity decreases abundance of stream benthic macroinvertebrates, Glob. Change Biol., 22, 2715–2728, https://doi.org/10.1111/gcb.13225, 2016.
Cline, L. C. and Zak, D. R.: Dispersal limitation structures fungal community assembly in a long-term glacial chronosequence, Environ. Microbiol., 16, 1538–1548, https://doi.org/10.1111/1462-2920.12281, 2014.
Custer, G. F., Bresciani, L., and Dini-Andreote, F.: Ecological and Evolutionary Implications of Microbial Dispersal, Front. Microbiol., 13, 855859, https://doi.org/10.3389/fmicb.2022.855859, 2022.
de Jong, A., In, T. Z. M., Meisel, O. H., Jetten, M., Dean, J. F., Rasigraf, O., and Welte, C. U.: Increases in temperature and nutrient availability positively affect methane-cycling microorganisms in Arctic thermokarst lake sediments, Environ. Microbiol., 20, 4314–4327, https://doi.org/10.1111/1462-2920.14345, 2018.
Egelberg, J., Pena, N., Rivera, R., and Andruk, C.: Assessing the geographic specificity of pH prediction by classification and regression trees, PLoS One, 16, e0255119, https://doi.org/10.1371/journal.pone.0255119, 2021.
Elder, C. D., Thompson, D. R., Thorpe, A. K., Chandanpurkar, H. A., Hanke, P. J., Hasson, N., James, S. R., Minsley, B. J., Pastick, N. J., Olefeldt, D., Walter Anthony, K. M., and Miller, C. E.: Characterizing Methane Emission Hotspots From Thawing Permafrost, Glob. Biogeochem. Cycle, 35, e2020GB006922, https://doi.org/10.1029/2020GB006922, 2021.
Etzelmüller, B.: Recent advances in mountain permafrost research, Permafrost Periglac. Process., 24, 99–107, 2013.
Farquharson, L. M., Mann, D. H., Grosse, G., Jones, B. M., and Romanovsky, V. E.: Spatial distribution of thermokarst terrain in Arctic Alaska, Geomorphology, 273, 116–133, https://doi.org/10.1016/j.geomorph.2016.08.007, 2016.
Fierer, N. and Jackson, R. B.: The diversity and biogeography of soil bacterial communities, P. Natl. Acad. Sci. USA, 103, 626–631, https://doi.org/10.1073/pnas.0507535103, 2006.
Fierer, N., Leff, J. W., Adams, B. J., Nielsen, U. N., Bates, S. T., Lauber, C. L., Owens, S., Gilbert, J. A., Wall, D. H., and Caporaso, J. G.: Cross-biome metagenomic analyses of soil microbial communities and their functional attributes, P. Natl. Acad. Sci. USA, 109, 21390–21395, https://doi.org/10.1073/pnas.1215210110, 2012.
Fu, L., Xie, R., Ma, D., Zhang, M., and Liu, L.: Variations in soil microbial community structure and extracellular enzymatic activities along a forest–wetland ecotone in high-latitude permafrost regions, Ecol. Evol., 13, e10205, https://doi.org/10.1002/ece3.10205, 2023.
Gao, H., Wang, J., Yang, Y., Pan, X., Ding, Y., and Duan, Z.: Permafrost Hydrology of the Qinghai-Tibet Plateau: A Review of Processes and Modeling, Front. Earth Sci., 8, e576838, https://doi.org/10.3389/feart.2020.576838, 2021.
Garrido-Benavent, I., Pérez-Ortega, S., Durán, J., Ascaso, C., Pointing, S. B., Rodríguez-Cielos, R., Navarro, F., and de Los Ríos, A.: Differential colonization and succession of microbial communities in rock and soil substrates on a maritime antarctic glacier forefield, Front. Microbiol., 11, e126, https://doi.org/10.3389/fmicb.2020.00126, 2020.
Gough, H. L. and Stahl, D. A.: Microbial community structures in anoxic freshwater lake sediment along a metal contamination gradient, The ISME J., 5, 543–558, https://doi.org/10.1038/ismej.2010.132, 2011.
Graham, E. and Stegen, J.: Dispersal-based microbial community assembly decreases biogeochemical function, Processes, 5, 65, https://doi.org/10.3390/pr5040065, 2017.
Graham, E. B., Crump, A. R., Resch, C. T., Fansler, S., Arntzen, E., Kennedy, D. W., Fredrickson, J. K., and Stegen, J. C.: Coupling spatiotemporal community assembly processes to changes in microbial metabolism, Front. Microbiol., 7, 1949, https://doi.org/10.3389/fmicb.2016.01949, 2016.
Hu, L. and Bentler, P. M.: Cutoff criteria for fit indexes in covariance structure analysis: Conventional criteria versus new alternatives, Struct. Eq. Model., 6, 1–55, https://doi.org/10.1080/10705519909540118, 1999.
Hugelius, G., Strauss, J., Zubrzycki, S., Harden, J. W., Schuur, E. A. G., Ping, C.-L., Schirrmeister, L., Grosse, G., Michaelson, G. J., Koven, C. D., O'Donnell, J. A., Elberling, B., Mishra, U., Camill, P., Yu, Z., Palmtag, J., and Kuhry, P.: Estimated stocks of circumpolar permafrost carbon with quantified uncertainty ranges and identified data gaps, Biogeosciences, 11, 6573–6593, https://doi.org/10.5194/bg-11-6573-2014, 2014.
Hugerth, L. W., Larsson, J., Alneberg, J., Lindh, M. V., Legrand, C., Pinhassi, J., and Andersson, A. F.: Metagenome-assembled genomes uncover a global brackish microbiome, Genome Biol., 16, 279, https://doi.org/10.1186/s13059-015-0834-7, 2015.
In'T Zandt, M. H., Liebner, S., and Welte, C. U.: Roles of thermokarst lakes in a warming world, Trends Microbiol., 28, 769–779, https://doi.org/10.1016/j.tim.2020.04.002, 2020.
Jiao, S. and Lu, Y.: Abundant fungi adapt to broader environmental gradients than rare fungi in agricultural fields, Glob. Change Biol., 26, 4506–4520, https://doi.org/10.1111/gcb.15130, 2020.
Jorgenson, M. T., Romanovsky, V., Harden, J., Shur, Y., O'Donnell, J., Schuur, E. A. G., Kanevskiy, M., and Marchenko, S.: Resilience and vulnerability of permafrost to climate change, Can. J. For. Res., 40, 1219–1236, https://doi.org/10.1139/X10-060, 2010.
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., Blomberg, S. P., and Webb, C. O.: Picante: R tools for integrating phylogenies and ecology, Bioinformatics, 26, 1463–1464, https://doi.org/10.1093/bioinformatics/btq166, 2010.
Kokelj, S. V. and Jorgenson, M. T.: Advances in thermokarst research, Permafrost Periglac. Process., 24, 108–119, https://doi.org/10.1002/ppp.1779, 2013.
Langenheder, S., Wang, J., Karjalainen, S. M., Laamanen, T. M., Tolonen, K. T., Vilmi, A., and Heino, J.: Bacterial metacommunity organization in a highly connected aquatic system, FEMS Microbiol. Ecol., 93, fiw225, https://doi.org/10.1093/femsec/fiw225, 2017.
Lawrence, D. M., Slater, A. G., and Swenson, S. C.: Simulation of Present-Day and Future Permafrost and Seasonally Frozen Ground Conditions in CCSM4, J. Clim., 25, 2207–2225, https://doi.org/10.1175/JCLI-D-11-00334.1, 2012.
Lee, Z. M. P., Poret-Peterson, A. T., Siefert, J. L., Kaul, D., Moustafa, A., Allen, A. E., Dupont, C. L., Eguiarte, L. E., Souza, V., and Elser, J. J.: Nutrient stoichiometry shapes microbial community structure in an evaporitic shallow pond, Front. Microbiol., 8, 949, https://doi.org/10.3389/fmicb.2017.00949, 2017.
Leibold, M. A., Chase, J. M., and Ernest, S. K.: Community assembly and the functioning of ecosystems: how metacommunity processes alter ecosystems attributes, Ecology, 98, 909–919, https://doi.org/10.1002/ecy.1697, 2017.
Le Moigne, A., Bartosiewicz, M., Schaepman Strub, G., Abiven, S., and Pernthaler, J.: The biogeochemical variability of Arctic thermokarst ponds is reflected by stochastic and niche-driven microbial community assembly processes, Environ. Microbiol., 22, 4847–4862, https://doi.org/10.1111/1462-2920.15260, 2020.
Levins, R.: Evolution in changing environments: some theoretical explorations (No. 2), Princeton University Press, Princeton, USA, 1968.
Louca, S., Parfrey, L. W., and Doebeli, M.: Decoupling function and taxonomy in the global ocean microbiome, Science, 353, 1272–1277, https://doi.org/10.1126/science.aaf4507, 2016.
Lozupone, C. A. and Knight, R.: Global patterns in bacterial diversity, P. Natl. Acad. Sci. USA, 104, 11436–11440, https://doi.org/10.1073/pnas.0611525104, 2007.
Lu, W., Wang, W., Shao, Q., Yu, Z., Hao, Z., Xing, W., Yong, B., and Li, J.: Hydrological projections of future climate change over the source region of Yellow River and Yangtze River in the Tibetan Plateau; a comprehensive assessment by coupling RegCM4 and VIC model, Hydrol. Process., 32, 2096–2117, https://doi.org/10.1002/hyp.13145, 2018.
Luo, D., Jin, H., Du, H., Li, C., Ma, Q., Duan, S., and Li, G.: Variation of alpine lakes from 1986 to 2019 in the Headwater Area of the Yellow River, Tibetan Plateau using Google Earth Engine, Adv. Clim. Chang. Res., 11, 11–21, https://doi.org/10.1016/j.accre.2020.05.007, 2020.
Luo, J., Niu, F., Lin, Z., Liu, M., and Yin, G.: Thermokarst lake changes between 1969 and 2010 in the Beilu River Basin, Qinghai-Tibet Plateau, China, Sci. Bull., 60, 556–564, https://doi.org/10.1007/s11434-015-0730-2, 2015.
Luan, L., Liang, C., Chen, L., Wang, H., Xu, Q., Jiang, Y., and Sun, B.: Coupling Bacterial Community Assembly to Microbial Metabolism across Soil Profiles, mSystems, 5, e00298-20, https://doi.org/10.1128/mSystems.00298-20, 2020.
Mackelprang, R., Burkert, A., Haw, M., Mahendrarajah, T., Conaway, C. H., Douglas, T. A., and Waldrop, M. P.: Microbial survival strategies in ancient permafrost: insights from metagenomics, The ISME J., 11, 2305–2318, https://doi.org/10.1038/ismej.2017.93, 2017.
Makoto, K., Wilson, S. D., Turner, B., and Turner, B.: When and where does dispersal limitation matter in primary succession? J. Ecol., 107, 559–565, https://doi.org/10.1111/1365-2745.12988, 2019.
Malard, L. A. and Pearce, D. A.: Microbial diversity and biogeography in Arctic soils, Environ. Microbiol. Rep., 10, 611–625, https://doi.org/10.1111/1758-2229.12680, 2018.
Manasypov, R. M., Pokrovsky, O. S., Shirokova, L. S., Auda, Y., Zinner, N. S., Vorobyev, S. N., and Kirpotin, S. N.: Biogeochemistry of macrophytes, sediments and porewaters in thermokarst lakes of permafrost peatlands, western Siberia, Sci. Total Environ., 763, 144201, https://doi.org/10.1016/j.scitotenv.2020.144201, 2021.
Martiny, J., Bohannan, B., Brown, J. H., Colwell, R. K., Fuhrman, J. A., Green, J. L., Horner-Devine, M. C., Kane, M., Krumins, J. A., Kuske, C. R., Morin, P. J., Naeem, S., Ovreas, L., Reysenbach, A. L., Smith, V. H., and Staley, J. T.: Microbial biogeography: putting microorganisms on the map, Nat. Rev. Microbiol., 4, 102–112, https://doi.org/10.1038/nrmicro1341, 2006.
Mishra, U., Hugelius, G., Shelef, E., Yang, Y., Strauss, J., Lupachev, A., Harden, J. W., Jastrow, J. D., Ping, C. L., Riley, W. J., Schuur, E., Matamala, R., Siewert, M., Nave, L. E., Koven, C. D., Fuchs, M., Palmtag, J., Kuhry, P., Treat, C. C., Zubrzycki, S., Hoffman, F. M., Elberling, B., Camill, P., Veremeeva, A., and Orr, A.: Spatial heterogeneity and environmental predictors of permafrost region soil organic carbon stocks, Sci. Adv., 7, 5236, https://doi.org/10.1126/sciadv.aaz5236, 2021.
Mori, A. S., Isbell, F., and Seidl, R.: β-Diversity, Community Assembly, and Ecosystem Functioning, Trends Ecol. Evol., 33, 549–564, https://doi.org/10.1016/j.tree.2018.04.012, 2018.
Mu, C., Abbott, B. W., Norris, A. J., Mu, M., Fan, C., Chen, X., Jia, L., Yang, R., Zhang, T., Wang, K., Peng, X., Wu, Q., Guggenberger, G., and Wu, X.: The status and stability of permafrost carbon on the Tibetan Plateau, Earth-Sci. Rev., 211, 103433, https://doi.org/10.1016/j.earscirev.2020.103433, 2020.
Narancic, B., Wolfe, B. B., Pienitz, R., Meyer, H., and Lamhonwah, D.: Landscape-gradient assessment of thermokarst lake hydrology using water isotope tracers, J. Hydrol., 545, 327–338, 2017.
National Genomics Data Center: CRA007082, National Genomics Data Center [data set], https://ngdc.cncb.ac.cn/search/?dbId=gsa&q=CRA007082&page=1, last access: 2 June 2022.
Nemergut, D. R., Schmidt, S. K., Fukami, T., O'Neill, S. P., Bilinski, T. M., Stanish, L. F., Knelman, J. E., Darcy, J. L., Lynch, R. C., Wickey, P., and Ferrenberg, S.: Patterns and Processes of Microbial Community Assembly, Microbiol. Mol. Biol. Rev., 77, 342–356, https://doi.org/10.1128/MMBR.00051-12, 2013.
Ning, D., Deng, Y., Tiedje, J. M., and Zhou, J.: A general framework for quantitatively assessing ecological stochasticity, P. Natl. Acad. Sci. USA, 116, 16892–16898, https://doi.org/10.1073/pnas.1904623116, 2019.
Nitzbon, J., Langer, M., Martin, L. C. P., Westermann, S., Schneider von Deimling, T., and Boike, J.: Effects of multi-scale heterogeneity on the simulated evolution of ice-rich permafrost lowlands under a warming climate, The Cryosphere, 15, 1399–1422, https://doi.org/10.5194/tc-15-1399-2021, 2021.
Niu, F., Lin, Z., Liu, H., and Lu, J.: Characteristics of thermokarst lakes and their influence on permafrost in Qinghai–Tibet Plateau, Geomorphology, 132, 222–233, https://doi.org/10.1016/j.geomorph.2011.05.011, 2011.
Nossa, C. W., Oberdorf, W. E., Yang, L., Aas, J. A., Paster, B. J., Desantis, T. Z., Brodie, E. L., Malamud, D., Poles, M. A. and Pei, Z.: Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome, World J. Gastroenterol., 16, 4135–4144, https://doi.org/10.3748/wjg.v16.i33.4135, 2010.
Obu, J.: How much of the Earth's surface is underlain by permafrost?, J. Geophys. Res.-Earth Surf., 126, 2021JF006123, https://doi.org/10.1029/2021JF006123, 2021.
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., Mcglinn, D., Minchin, P. R., O'Hara, R. B., Simpson, G. L., Solymos, P., Stevens, M. H. H., Szoecs, E., and Wagner, H.: vegan: Community Ecology Package, R package version 2.5-7, https://CRAN.R-project.org/package=vegan (last access: 28 November 2021), 2020.
Ottoni, J. R., de Oliveira, V. M., and Passarini, M. R. Z.: Microbes in thawing permafrost: contributions to climate change, in: Microbiome Under Changing Climate, Woodhead Publishing, 1–28, https://doi.org/10.1016/B978-0-323-90571-8.00001-8, 2022.
Payette, S., Delwaide, A., Caccianiga, M., and Beauchemin, M.: Accelerated thawing of subarctic peatland permafrost over the last 50 years, Geophys. Res. Lett., 31, L18208, https://doi.org/10.1029/2004GL020358, 2004.
Pearman, J. K., Biessy, L., Thomson-Laing, G., Waters, S., Vandergoes, M. J., Howarth, J. D., Rees, A., Moy, C., Pochon, X., and Wood, S. A.: Local factors drive bacterial and microeukaryotic community composition in lake surface sediment collected across an altitudinal gradient, FEMS Microbiol. Ecol., 96, fiaa070, https://doi.org/10.1093/femsec/fiaa070, 2020.
Pholchan, M. K., Baptista, J. D. C., Davenport, R. J., Sloan, W. T., and Curtis, T. P.: Microbial community assembly, theory and rare functions, Front. Microbiol., 4, e68, https://doi.org/10.3389/fmicb.2013.00068, 2013.
Piao, S., Tan, K., Nan, H., Ciais, P., Fang, J., Wang, T., Vuichard, N., and Zhu, B.: Impacts of climate and CO2 changes on the vegetation growth and carbon balance of Qinghai–Tibetan grasslands over the past five decades, Glob. Planet. Change, 98–99, 73–80, https://doi.org/10.1016/j.gloplacha.2012.08.009, 2012.
Picazo, F., Vilmi, A., Aalto, J., Soininen, J., Casamayor, E. O., Liu, Y., Wu, Q., Ren, L., Zhou, J., Shen, J., and Wang, J.: Climate mediates continental scale patterns of stream microbial functional diversity, Microbiome, 8, e92, https://doi.org/10.1186/s40168-020-00873-2, 2020.
Pla-Rabes, S., Flower, R. J., Shilland, E. M., and Kreiser, A. M.: Assessing microbial diversity using recent lake sediments and estimations of spatio-temporal diversity, J. Biogeogr., 38, 2033–2040, https://doi.org/10.1111/j.1365-2699.2011.02530.x, 2011.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glockner, F. O.: The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic. Acids. Res., 41, 590–596, https://doi.org/10.1093/nar/gks1219, 2013.
R Core Team: R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org (last access: 28 November 2021), 2020.
Ren, Z. and Gao, H. K.: Abundant and rare soil fungi exhibit distinct succession patterns in the forefield of Dongkemadi glacier on the central Qinghai-Tibet Plateau, Sci. Total Environ., 828, e154563, https://doi.org/10.1016/j.scitotenv.2022.154563, 2022.
Ren, Z., Wang, F., Qu, X., Elser, J. J., Liu, Y., and Chu, L.: Taxonomic and functional differences between microbial communities in Qinghai Lake and its input streams, Front. Microbiol., 8, e2319, https://doi.org/10.3389/fmicb.2017.02319, 2017.
Ren, Z., Niu, D., Ma, P., Wang, Y., Fu, H., and Elser, J. J.: Cascading influences of grassland degradation on nutrient limitation in a high mountain lake and its inflow streams, Ecology, 100, e02755, https://doi.org/10.1002/ecy.2755, 2019a.
Ren, Z., Qu, X. D., Peng, W. Q., Yu, Y., and Zhang, M.: Nutrients drive the structures of bacterial communities in sediments and surface waters in the river-lake system of Poyang Lake, Water, 11, e930, https://doi.org/10.3390/w11050930, 2019b.
Ren, Z., Wang, Z. M., Wang, Y., Ma, P. P., Niu, D. C., Fu, H., and Elser, J. J.: Soil bacterial communities vary with grassland degradation in the Qinghai Lake watershed, Plant Soil, 460, 541–557, https://doi.org/10.1007/s11104-020-04823-7, 2021.
Ren, Z., Cao, S. K., Chen, T., Zhang, C., and Yu, J. L.: Bacterial functional redundancy and carbon metabolism potentials in soil, sediment, and water of thermokarst landscapes across the Qinghai-Tibet Plateau: implications for the fate of permafrost carbon, Sci. Total Environ., 852, e158340, https://doi.org/10.1016/j.scitotenv.2022.158340, 2022a.
Ren, Z., Jia, X., Zhang, Y. T., Ma, K., Zhang, C., and Li, X.: Biogeography and environmental drivers of zooplankton communities in permafrost-affected lakes on the Qinghai-Tibet Plateau, Glob. Ecol. Conserv., 38, e02191, https://doi.org/10.1016/j.gecco.2022.e02191, 2022b.
Ren, Z., Li, X., Zhang, C., Wang, Q., Fang, L., Cao, S. K., and Yu, J. L.: From permafrost soil to thermokarst lake sediment: A view from stoichiometry, Front. Environ. Sci., 10, e986879, https://doi.org/10.3389/fenvs.2022.986879, 2022c.
Romanowicz, K. J. and Kling, G. W.: Summer thaw duration is a strong predictor of the soil microbiome and its response to permafrost thaw in arctic tundra, Environ. Microbiol., 24, 6220–6237, https://doi.org/10.1111/1462-2920.16218, 2022.
Rosseel, Y.: lavaan: An R Package for Structural Equation Modeling, J. Stat. Softw., 48, 1–36, https://doi.org/10.18637/jss.v048.i02, 2012.
Ruiz-Gonzalez, C., Pablo Nino-Garcia, J., and Del Giorgio, P. A.: Terrestrial origin of bacterial communities in complex boreal freshwater networks, Ecol. Lett., 18, 1198–1206, https://doi.org/10.1111/ele.12499, 2015.
Schaefer, K., Zhang, T., Bruhwiler, L., and Barrett, A. P.: Amount and timing of permafrost carbon release in response to climate warming, Tellus B, 63, 168–180, 2011.
Schuur, E. A., Vogel, J. G., Crummer, K. G., Lee, H., Sickman, J. O., and Osterkamp, T. E.: The effect of permafrost thaw on old carbon release and net carbon exchange from tundra, Nature, 459, 556–569, https://doi.org/10.1038/nature08031, 2009.
Schuur, E. A. G., Bockheim, J., Canadell, J. G., Euskirchen, E., Field, C. B., Goryachkin, S. V., Hagemann, S., Kuhry, P., Lafleur, P. M., Lee, H., Mazhitova, G., Nelson, F. E., Rinke, A., Romanovsky, V. E., Shiklomanov, N., Tarnocai, C., Venevsky, S., Vogel, J. G., and Zimov, S. A.: Vulnerability of permafrost carbon to climate change: Implications for the global carbon cycle, Bioscience, 58, 701–714, https://doi.org/10.1641/B580807, 2008.
She, Z., Pan, X., Wang, J., Shao, R., Wang, G., Wang, S., and Yue, Z.: Vertical environmental gradient drives prokaryotic microbial community assembly and species coexistence in a stratified acid mine drainage lake, Water Res., 206, 117739, https://doi.org/10.1016/j.watres.2021.117739, 2021.
Stegen, J. C., Lin, X., Konopka, A. E., and Fredrickson, J. K.: Stochastic and deterministic assembly processes in subsurface microbial communities, ISME J., 6, 1653–1664, https://doi.org/10.1038/ismej.2012.22, 2012.
Stegen, J. C., Lin, X., Fredrickson, J. K., Chen, X., Kennedy, D. W., Murray, C. J., Rockhold, M. L., and Konopka, A.: Quantifying community assembly processes and identifying features that impose them, The ISME J., 7, 2069–2079, https://doi.org/10.1038/ismej.2013.93, 2013.
Stegen, J. C., Lin, X., Fredrickson, J. K., and Konopka, A. E.: Estimating and mapping ecological processes influencing microbial community assembly, Front. Microbiol., 6, e370, https://doi.org/10.3389/fmicb.2015.00370, 2015.
Steven, B., Niederberger, T. D., and Whyte, L. G.: Bacterial and archaeal diversity in permafrost, in: Permafrost soils, edited by: Margesin, R., Springer, Berlin, Heidelberg, 59–72, https://doi.org/10.1007/978-3-540-69371-0_5, 2009.
Strickland, M. S., Lauber, C., Fierer, N., and Bradford, M. A.: Testing the Functional Significance of Microbial Community Composition, Ecology, 90, 441–451, https://doi.org/10.1890/08-0296.1, 2009.
Taş, N., Prestat, E., Wang, S., Wu, Y., Ulrich, C., Kneafsey, T., Tringe, S. G., Torn, M. S., Hubbard, S. S., and Jansson, J. K.: Landscape topography structures the soil microbiome in arctic polygonal tundra, Nat. Commun., 9, 777, https://doi.org/10.1038/s41467-018-03089-z, 2018.
Telford, R. J., Vandvik, V., and Birks, H. J. B.: Dispersal Limitations Matter for Microbial Morphospecies, Science, 312, 1015–1015, https://doi.org/10.1126/science.1125669, 2006.
Torsvik, V., Ovreas, L., and Thingstad, T. F.: Prokaryotic diversity – Magnitude, dynamics, and controlling factors, Science, 296, 1064–1066, https://doi.org/10.1126/science.1071698, 2002.
Tripathi, B. M., Stegen, J. C., Kim, M., Dong, K., Adams, J. M., and Lee, Y. K.: Soil pH mediates the balance between stochastic and deterministic assembly of bacteria, The ISME J., 12, 1072–1083, https://doi.org/10.1038/s41396-018-0082-4, 2018.
Turetsky, M. R., Abbott, B. W., Jones, M. C., Anthony, K. W., Olefeldt, D., Schuur, E. A. G., Grosse, G., Kuhry, P., Hugelius, G., Koven, C., Lawrence, D. M., Gibson, C., Sannel, A. B. K., and Mcguire, A. D.: Carbon release through abrupt permafrost thaw, Nat. Geosci., 13, 138–143, https://doi.org/10.1038/s41561-019-0526-0, 2020.
Vargas Medrano, M.: Recent Changes in the Zooplankton Communities of Arctic Tundra Ponds in Response to Warmer Temperatures and Nutrient Enrichment, Dissertation, University of Texas at El Paso, 2019.
Vellend, M., Srivastava, D. S., Anderson, K. M., Brown, C. D., Jankowski, J. E., Kleynhans, E. J., Kraft, N. J. B., Letaw, A. D., Macdonald, A. A. M., Maclean, J. E., Myers-Smith, I. H., Norris, A. R., and Xue, X.: Assessing the relative importance of neutral stochasticity in ecological communities, Oikos, 123, 1420–1430, https://doi.org/10.1111/oik.01493, 2014.
Veremeeva, A., Nitze, I., Günther, F., Grosse, G., and Rivkina, E.: Geomorphological and climatic drivers of thermokarst lake area increase trend (1999-2018) in the Kolyma Lowland Yedoma Region, North-Eastern Siberia, Remote Sens., 13, 178, https://doi.org/10.3390/rs13020178, 2021.
Vigneron, A., Lovejoy, C., Cruaud, P., Kalenitchenko, D., Culley, A., and Vincent, W. F.: Contrasting Winter Versus Summer Microbial Communities and Metabolic Functions in a Permafrost Thaw Lake, Front. Microbiol., 10, 1656, https://doi.org/10.3389/fmicb.2019.01656, 2019.
Vucic, J. M., Gray, D. K., Cohen, R. S., Syed, M., Murdoch, A. D., and Sharma, S.: Changes in water quality related to permafrost thaw may significantly impact zooplankton in small Arctic lakes, Ecol. Appl., 30, e02186, https://doi.org/10.1002/eap.2186, 2020.
Walter, K. M., Zimov, S. A., Chanton, J. P., Verbyla, D., and Chapin, F. S.: Methane bubbling from Siberian thaw lakes as a positive feedback to climate warming, Nature, 443, 71–75, https://doi.org/10.1038/nature05040, 2006.
Wan, C., Gibson, J. J., Shen, S., Yi, Y., Yi, P., and Yu, Z.: Using stable isotopes paired with tritium analysis to assess thermokarst lake water balances in the Source Area of the Yellow River, northeastern Qinghai-Tibet Plateau, China, Sci. Total Environ., 689, 1276–1292, https://doi.org/10.1016/j.scitotenv.2019.06.427, 2019.
Wan, D. S., Feng, J. J., Jiang, D. C., Mao, K. S., Duan, Y. W., Miehe, G., and Opgenoorth, L.: The Quaternary evolutionary history, potential distribution dynamics, and conservation implications for a Qinghai-Tibet Plateau endemic herbaceous perennial, Anisodus tanguticus (Solanaceae), Ecol. Evol., 6, 1977–1995, https://doi.org/10.1002/ece3.2019, 2016.
Webb, C. O., Ackerly, D. D., Mcpeek, M. A., and Donoghue, M. J.: Phylogenies and community ecology, Annu. Rev. Ecol. Syst., 33, 475–505, 2002.
West, J. J. and Plug, L. J.: Time-dependent morphology of thaw lakes and taliks in deep and shallow ground ice, J. Geophys. Res., 113, 1009, https://doi.org/10.1029/2006JF000696, 2008.
Wu, L., Yang, F., Feng, J., Tao, X., Qi, Q., Wang, C., Schuur, E. A. G., Bracho, R., Huang, Y., Cole, J. R., Tiedje, J. M., and Zhou, J.: Permafrost thaw with warming reduces microbial metabolic capacities in subsurface soils, Mol. Ecol., 31, 1403–1415, https://doi.org/10.1111/mec.16319, 2022.
Wu, M., Chen, S., Chen, J., Xue, K., Chen, S., Wang, X., Chen, T., Kang, S., Rui, J., Thies, J. E., Bardgett, R. D., and Wang, Y.: Reduced microbial stability in the active layer is associated with carbon loss under alpine permafrost degradation, P. Natl. Acad. Sci. USA, 118, e2025321118, https://doi.org/10.1073/pnas.2025321118, 2021.
Wu, Q., Dong, X., Liu, Y., Jin, H., and Zhang, T.: Responses of permafrost on the Qinghai-Tibet Plateau, China, to climate change and engineering construction, Arct. Antarct. Alp. Res., 39, 682–687, https://doi.org/10.1657/1523-0430(07-508)[WU]2.0.CO;2, 2007.
Xiong, J., Liu, Y., Lin, X., Zhang, H., Zeng, J., Hou, J., Yang, Y., Yao, T., Knight, R., and Chu, H.: Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau, Environ. Microbiol., 14, 2457–2466, https://doi.org/10.1111/j.1462-2920.2012.02799.x, 2012.
Xu, Z. X., Gong, T. L., and Li, J. Y.: Decadal trend of climate in the Tibetan Plateau-regional temperature and precipitation, Hydrol. Process., 22, 3056–3065, https://doi.org/10.1002/hyp.6892, 2008.
Yang, J., Ma, L., Jiang, H., Wu, G., and Dong, H.: Salinity shapes microbial diversity and community structure in surface sediments of the Qinghai-Tibetan Lakes, Sci. Rep., 6, e25078, https://doi.org/10.1038/srep25078, 2016.
Yang, Y., Wu, Q., Yun, H., Jin, H., and Zhang, Z.: Evaluation of the hydrological contributions of permafrost to the thermokarst lakes on the Qinghai-Tibet Plateau using stable isotopes, Glob. Planet. Change, 140, 1–8, https://doi.org/10.1016/j.gloplacha.2016.03.006, 2016.
Yao, T., Thompson, L. G., Mosbrugger, V., Zhang, F., Ma, Y., Luo, T., Xu, B., Yang, X., Joswiak, D. R., Wang, W., Joswiak, M. E., Devkota, L. P., Tayal, S., Jilani, R., and Fayziev, R.: Third Pole Environment (TPE), Environ. Dev., 3, 52–64, https://doi.org/10.1016/j.envdev.2012.04.002, 2012.
Yu, G., Smith, D. K., Zhu, H., Guan, Y., and Lam, T. T. Y.: ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data, Methods Ecol. Evol., 8, 28–36, 2017.
Yu, H., Favre, A., Sui, X., Chen, Z., Qi, W., Xie, G., Kleunen, M., and van Kleunen, M.: Mapping the genetic patterns of plants in the region of the Qinghai–Tibet Plateau: Implications for conservation strategies, Diversity & distributions, 25, 310–324, https://doi.org/10.1111/ddi.12847, 2019.
Zhang, D., Wang, L., Qin, S., Kou, D., Wang, S., Zheng, Z., Peñuelas, J., and Yang, Y.: Microbial nitrogen and phosphorus co-limitation across permafrost region, Glob. Change Biol., 29, 3910–3923, https://doi.org/10.1111/gcb.16743, 2023.
Zhang, J. L.: Species association analysis, Version 0.2.2, https://CRAN.R-project.org/package=spaa (last access: 28 November 2021), 2016.
Zhang, Z., Chang, J., Xu, C., Zhou, Y., Wu, Y., Chen, X., Jiang, S., and Duan, Z.: The response of lake area and vegetation cover variations to climate change over the Qinghai-Tibetan Plateau during the past 30 years, Sci. Total Environ., 635, 443–451, https://doi.org/10.1016/j.scitotenv.2018.04.113, 2018.
Zhou, C. P.: The spatial dataset of climate on the Tibetan Plateau (1961–2020). National Tibetan Plateau Data Center [data set], https://doi.org/10.11888/AtmosphericPhysics.tpe.49.file, 2018.
Zhou, J. and Ning, D.: Stochastic Community Assembly: Does It Matter in Microbial Ecology?, Microbiol. Mol. Biol. Rev., 81, e00002-17, https://doi.org/10.1128/MMBR.00002-17, 2017.
Zhou, J., Deng, Y., Zhang, P., Xue, K., Liang, Y., Van Nostrand, J. D., Yang, Y., He, Z., Wu, L., Stahl, D. A., Hazen, T. C., Tiedje, J. M., and Arkin, A. P.: Stochasticity, succession, and environmental perturbations in a fluidic ecosystem, P. Natl. Acad. Sci. USA, 111, E836–E845, https://doi.org/10.1073/pnas.1324044111, 2014.
Zhou, J., Liu, W., Deng, Y., Jiang, Y., Xue, K., He, Z., Van Nostrand, J. D., Wu, L., Yang, Y., Wang, A., and Handelsman, J.: Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community, mBio, 4, e00584-12, https://doi.org/10.1128/mBio.00584-12, 2013.
Zhou, L., Zhou, Y., Yao, X., Cai, J., Liu, X., Tang, X., Zhang, Y., Jang, K., and Jeppesen, E.: Decreasing diversity of rare bacterial subcommunities relates to dissolved organic matter along permafrost thawing gradients, Environ. Int., 134, 105330, https://doi.org/10.1016/j.envint.2019.105330, 2020.
Zou, D., Zhao, L., Sheng, Y., Chen, J., Hu, G., Wu, T., Wu, J., Xie, C., Wu, X., Pang, Q., Wang, W., Du, E., Li, W., Liu, G., Li, J., Qin, Y., Qiao, Y., Wang, Z., Shi, J., and Cheng, G.: A new map of permafrost distribution on the Tibetan Plateau, The Cryosphere, 11, 2527–2542, https://doi.org/10.5194/tc-11-2527-2017, 2017.





