the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Soil priming effects and involved microbial community along salt gradients
Haoli Zhang
Doudou Chang
Zhifeng Zhu
Chunmei Meng
Kaiyong Wang
Soil salinity mediates microorganisms and soil processes, like soil organic carbon (SOC) cycling. Yet, how soil salinity affects SOC mineralization via shaping bacterial community diversity and composition remains elusive. Therefore, soils were sampled along a salt gradient (salinity at 0.25 %, 0.58 %, 0.75 %, 1.00 %, and 2.64 %) and incubated for 90 d to investigate (i) SOC mineralization (i.e., soil priming effects induced by cottonseed meal, as substrate) and (ii) the responsible bacteria community by using high-throughput sequencing and natural abundance of 13C isotopes (to partition cottonseed-meal-derived CO2 and soil-derived CO2). We observed a negative priming effect during the first 28 d of incubation that turned to a positive priming effect after day 56. Negative priming at the early stage might be due to the preferential utilization of cottonseed meal. The followed positive priming decreased with the increase in salinity, which might be caused by the decreased α diversity of microbial communities in soil with high salinity. Specifically, soil pH and electrical conductivity (EC) along the salinity gradient were the dominant variables modulating the structure of the microbial community and consequently SOC priming (estimated by distance-based multivariate analysis and path analysis). By adopting two-way orthogonal projections to latent structures (O2PLS), priming effects were linked with specific microbial taxa; e.g., Proteobacteria (Luteimonas, Hoeflea, and Stenotrophomonas) were the core microbial genera that were attributed to the substrate-induced priming effects. Here, we highlight that the increase in salinity reduced the diversity of the microbial community and shifted dominant microorganisms (Actinobacteria and Proteobacteria: Luteimonas, Hoeflea, and Stenotrophomonas) that determined SOC priming effects, which provides a theoretical basis for understanding SOC dynamics and microbial drivers under the salinity gradient.
- Article
(2106 KB) - Full-text XML
-
Supplement
(732 KB) - BibTeX
- EndNote
Soil salinization is an increasing environmental problem caused by natural and human activities in arid and semi-arid areas (Wichern et al., 2006). Salinization is often a major threat to crop productivity in agricultural land. Soil microorganisms suffer from osmotic stress. Soil salinity often causes microbial death or dormancy. It was widely reported that the increased salinity decreases microbial biomass, enzymatic activity, and α diversity of microbial communities (Laura, 1974; Pathak and Rao, 1998; Rietz and Haynes, 2003). Soil salinity is reported to be the major determinant of the composition and activity of microbial communities (Kamble and Bååth, 2014; Ling et al., 2021, 2022). Although salinity is reported to be a vital factor in influencing microorganisms in arid and semi-arid areas, few studies have investigated C processes (e.g., priming effect) driven by microbial communities in saline soils (Sardinha et al., 2003).
Soil organic carbon (SOC) is the largest pool (1500 Pg C) in the terrestrial carbon (C) cycle and contains twice as much C as the atmosphere (Filley and Boutton, 2006; Wiesmeier et al., 2019; Jeewani et al., 2021). The input of substrate C can influence the output (i.e., CO2 release) through a phenomenon called priming effect, which was first discovered by Löhnis (1926). Substrate additions accelerate or decrease soil organic C mineralization, referred to as positive or negative priming effects (Kuzyakov et al., 2000). The intensity of the priming effect affects the turnover of SOC and thus storage pool (Sullivan and Hart, 2012). Soil priming effects are affected by many biotic and abiotic factors (Lavelle et al., 2006; Martin et al., 2019); investigating abiotic and biotic mechanisms underlying SOC priming enhances our strong understanding of SOC cycling.
Soil priming effects are affected by soil animal fauna (Scheu and Parkinson, 1994) and the activities, diversity, and composition of microbial communities (Fontaine et al., 2011). The microbial decomposers are the major player in the decomposition process of added C sources. The addition of substrate, such as composts (Xun et al., 2016), animal sludges (Hartmann et al., 2015), sewage sludges (Su et al., 2017; Wagner and Raquel, 2011), and plant residues (Dai et al., 2017), generally increases soil microbial biomass C and stimulates the microbial activities, thus enhancing the loss of SOC (positive priming effects) (Fontaine et al., 2003; Bird et al., 2011; Li et al., 2018; Ali et al., 2019).
Concerning abiotic factors, the priming effect can be controlled by climate variables (Hagemann, 2008) and soil properties, like pH, electrical conductivity (EC), and total nitrogen (TN) (Blagodatskaya and Kuzyakov, 2008; Luo et al., 2017). To understand how environmental and edaphic factors affect the processes of SOC mineralization, it is important to estimate the terrestrial C pool (Lehmann and Kleber, 2015). Although many studies have tested the effects of soil pH, SOC content, and other edaphic variables on soil priming effects, few studies have investigated soil priming effects in saline soil (Asghar et al., 2012), especially those linked with soil microbial community structure and their functions in C decomposition (Soina et al., 2018).
Thus, we sampled the soils along natural salinity gradients (0.25 %, 0.58 %, 0.75 %, 1.00 %, and 2.64 % total water-soluble salt). Based on these soils, we conducted a 90 d indoor incubation study applying C3 substrate of cottonseed meal (δ13C = −23.47 ‰) to C4 soils with salt gradients (δ13C) between −14.21 ‰ and −16.01 ‰ to (1) investigate the mineralization rate of cottonseed meal and induced soil priming effects along salt gradients, (2) investigate the diversity of microbial communities in the soils with increased salinity, and (3) identify the bacteria-taxa-associated soil priming. We hypothesized that (i) soil microbial community diversity and composition will be different with the different soil variables, particularly pH and EC along salinity gradients, and (ii) soil C processes like priming effects will be regulated mainly by microbial communities and especially the core microbial species. To clarify the priming effects and involved microbial groups would help us better understanding C sequestration potential and underlying mechanisms in saline soils.
2.1 Soil sampling and cottonseed meal production
The soil type was gray desert soil, which was collected from farmlands (44.96∘ N, 82.90∘ E) in the town of Xiao Yinpan, city of Bole, prefecture of Bortala, northern Xinjiang Uygur Autonomous Region, northwest China. The farmland soil is naturally formed original saline-sodic soil with a continuous 30-year planting of maize (C4 crop) and maize straw returning to soil for 7–8 years. In September 2021, we determined the sampling area and used the five-point sampling method to collect non-rhizosphere soil. The soil samples were air-dried indoors, hand-picked to remove visible debris and animal and plant residues, sieved at field moisture (<2 mm), and subsequently adjusted to 40 % water-holding capacity (WHC). Five salinity gradients at 0.25 %, 0.58 %, 0.75 %, 1.00 %, and 2.64 % were determined through soil salinity measurements. Texture was determined by the pipette method without carbonate in all soil samples. They were then incubated at 25 ∘C for 7 d before starting the experiments to allow any early sampling and sieving effects to subside.
Cottonseed meal is a kind of reddish or yellow granular material obtained by pressing and leaching other cottonseed. The cottonseed meal was purchased from the market, dried at 105 ∘C for 24 h indoor, and then further pulverized by a ball mill and passed through a <2 mm sieve.
2.2 Soil and substrate analyses
The EC and pH of soil and cottonseed meal were measured at a soil : water ratio of 1 : 5 (weight weight) (Bao, 2000). Air-dried soil (5 g, <2 mm) and 25 mL of deionized water were shaken together for 1 min and left to settle for 30 min, which was repeated once more before pH was determined with a pH electrode. Soil water-soluble salt was analyzed by being weighted at a soil : water ratio of 1 : 5 (weight weight). Air-dried soil (5 g, <1 mm) and 25 mL of deionized water were shaken together for 30 min and filtered to obtain a clear filtrate using a thermostat water bath to evaporate and weigh the sample (Bao, 2000). Soil total carbon (TC) and TN were collected to be tested, which were dried and ground through a 0.15 mm screen, and a certain amount of treated soil sample (air-dried, milled <150 µm) was wrapped in tin foil and placed in an element analyzer for determination by dry combustion (LECO CNS-2000, LECO Corporation, Michigan, USA). Soil microbial biomass C was determined by fumigation extraction (Vance et al., 1987; Wu et al., 1990). The K2SO4 extractable organic C was determined using an organic carbon autoanalyzer (Shimadzu, Analytical Sciences, Kyoto, Japan). Soil microbial biomass C (BC) was calculated from BC = 2.22 EC, where EC = ((organic C extracted from fumigated soil) minus (organic C extracted from non-fumigated soil)). The natural δ13C (‰) abundance in the soils (air-dried, milled <200 µm) was determined using an elemental analyzer isotope ratio mass spectrometer (Sercon Ltd, Crewe, UK). All measurements are given on an oven dry weight basis (105 ∘C, 24 h).
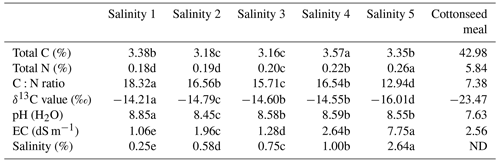
The δ13C (‰) abundance of the cottonseed meal (air-dried, milled <200 µm) was determined using an elemental analyzer isotope ratio mass spectrometer (Sercon Ltd, Crewe, UK). The main elemental composition of the substrate was determined using elemental analysis (vario EL cube, Hanau, Germany), with the samples combusted at 1200 ∘C. Natural δ13C (‰) abundance, the total carbon, total nitrogen contents, and C:N of the cottonseed meal are presented in Table 1.
2.3 Experiment design
After pre-incubation, five soils with different salinity gradients (salinity at 0.25 %, 0.58 %, 0.75 %, 1.00 %, and 2.64 %) were thoroughly mixed with cottonseed meal at 20 mg C g−1 soil (dry weight, d.w., basis) and incubated over 90 d following moisture adjustment to 40 % water-holding capacity (WHC) to investigate the substrate mineralization and priming effects. Each soil sample (40 g d.w. basis) was incubated in a 100 mL beaker inside a 1 L brown glass jar. Three jars with only water and NaOH were set as blanks. All the jars were sealed with a rubber bung and incubated in a randomized block design at 25 ∘C for the 90 d of incubation. The NaOH vials were changed after 1, 3, 5, 7, 14, 28, 56, and 90 d for the determination of evolved CO2 and 13C–CO2 (‰). Meanwhile, soil biomass C, NH, NO, pH, EC, TC, TN, and DNA extraction were measured at day 28.
2.4 Soil CO2–C and its isotopic composition
Soil C evolved as CO2–C in jars was measured by trapping CO2 in 1 M NaOH (20 mL) during soil incubation. After the NaOH (20 mL) trapped CO2 at different periods of soil incubation, 5 mL of 1 M NaOH of each sample was mixed with 10 mL deionized water and titrated with 0.05 M standardized HCl by the TIM840 autotitrator (Radiometer Analytical, Villeurbanne Cedex, France). Meanwhile, the δ13C (‰) of trapped CO2–C was precipitated with 8 mL of 1 M NaOH (20 mL) mixed with 8 mL of 1.5 M BaCl2 in vials (Aoyama et al., 2000). The BaCO3 precipitate was trapped on the glass fiber of the filter, rinsed with deionized water several times, dried overnight (80 ∘C), weighed (0.100–0.200 mg) into tin capsules, and analyzed for δ13C on an elemental analyzer isotope ratio mass spectrometer (Sercon Ltd, Crewe, UK).
2.5 DNA exaction and sequencing
The total soil DNA was extracted from 0.50 g of moist soil using a FastDNA Spin Kit (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer's protocol. The extracted DNA was dissolved in 50 µL of TE buffer, quantified using a spectrophotometer, and stored at −20 ∘C until sequencing.
V3–V4 hypervariable regions of the bacterial 16S rRNA gene were amplified with primers 341F (5′-CCTAYGGRBGCASCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The PCR reactions were conducted with a thermocycler PCR system (GeneAmp 9700, ABI, USA) by using the following programs: 3 min of denaturation at 95 ∘C; followed by 27 cycles of 30 s at 95 ∘C, 30 s at 55 ∘C, and 45 s at 72 ∘C; and a final extension at 72 ∘C for 10 min with a thermocycler PCR system (GeneAmp 9700, ABI, USA). PCR amplicons pooled from the triplicate reactions were purified using a QIAquick PCR purification kit (Qiagen, Shenzhen, China) and quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). The PCR products were purified, mixed, and sent to Majorbio, Inc. (Shanghai, China) for sequencing based on the Illumina MiSeq platform.
2.6 Calculations
The mineralization of cottonseed meal was separated from SOC mineralization according to the change in stable isotopic composition (δ13C) with time. The standard equation for determining δ13C (‰) is derived with
where Rsample is the mass ratio of 13C to 12C of each sample, and RVPDB is the international PDB (Peedee Belemnite) limestone standard. The labeled 13C (%) of cottonseed meal was then estimated with
where CO2–13C (%) is the proportion of evolved CO2 from C3 (cottonseed meal) matter, δtreatment is the δ13C (‰) in treatments of soil with cottonseed meal, δC4 is the δ13C (‰) in the control soil, and δC3 is the δ13C (‰) from cottonseed meal. Thus, the CO2–C produced from cottonseed meal during the incubation was calculated with
CO2 from SOC was CO2–13C subtracted from total evolved CO2–C. The absolute soil priming effect (or primed soil CO2–C) with the addition of cottonseed meal was calculated from
where CO2–Ctreatment is the non-isotopically labeled CO2–C evolved from cottonseed-meal-amended soil, and CO2–Ccontrol is the non-isotopically labeled CO2–C evolved from soil without cottonseed meal.
2.7 Statistics
The data of 16S gene sequencing were processed using the Quantitative Insights Into Microbial Ecology (QIIME) 1.9.0 pipeline (Caporaso et al., 2010). In brief, reads with a length less than 200 bp and ambiguous bases were discarded. The sequences were then binned into operational taxonomic units (OTUs) by UCLUST (Edgar, 2010) based on 97 % pairwise identity. Chimeric OTUs identified by USEARCH (Edgar et al., 2011) in QIIME were removed. The most abundant sequence from each OTU was selected to represent that OTU. Taxonomy was assigned to 16S OTUs against a subset of the SILVA 104 database. The representative OTU sequences were aligned using PyNAST (Caporaso et al., 2012). We obtained between 64 425 and 89 989 clean_reads per sample for all experimental samples.
To avoid potential bias caused by sequencing depth, all sample datasets were rarefied for the bacteria α-diversity and β-diversity analyses. Faith's phylogenetic diversity was calculated to provide an integrated index of the phylogenetic breadth across taxonomic levels (Faith, 1992). To compare β diversity between samples, principal coordinate analyses based on the unweighted and weighted UniFrac (Lozupone et al., 2007) distances were calculated using the function “pcoa” in the R package “Ape”. Additionally, permutational multivariate analysis of variance (PERMANOVA) was carried out using the function “adonis” in the R package “vegan” to measure effect size and significance of β diversity. The variable influence projection (VIP) value was processed using the two-way orthogonal projections to latent structures (O2PLS) analysis by SIMCA P 14 (Version 14.1.0.2047) (Wang et al., 2016). The y matrix was defined as the dataset of environmental factors, and the x matrix was defined as the dataset of the microbial community on genus level.
Data were logarithmically transformed and analyzed by ANOVA. All analyses were performed using SPSS software (13th edition). Pearson's correlation analyses were performed to assess the linear correlation among soil physico-chemical properties and the microbial community. Multivariate analysis was operated to investigate the interaction of salinity treatments on bacteria community parameters.
3.1 Soil physicochemical properties along salt gradients
The major soil physicochemical properties along salt gradients are presented (Table 1), and all of soil physicochemical properties have significant differences (p<0.05). The total soluble salinity content in the soils ranged from 0.25 % to 2.64 %, and soil salt gradients increased gradually from the Salinity 1 samples to the Salinity 5 samples. The pH and EC in soils ranged from 8.45 to 8.85 and from 1.06 to 7.75 dS m−1. Soil total C and N increased with salinity, ranging from 3.16 % to 3.57 % and from 0.18 % to 0.26 %. The δ13C values for soils are between −14.21 % and −16.01 ‰, which were relatively enriched compared to cottonseed meal (−23.47 ‰). This allowed the separation of soil-derived CO2 from total evolved CO2, according to the classic mixed modeling.
3.2 Total CO2 evolution
During the whole 90 d of incubation, the cumulative evolved CO2 had similar trends, in which the amount of CO2 increased with the incubation time (Fig. S1 in the Supplement). The cumulative evolved CO2 increased more rapidly with the addition of cottonseed meal before 14 d compared to non-amended soils. At 90 d of incubation, the cumulative evolved CO2 in the soil with the lowest salinity (Salinity 1) gave the lowest CO2 emission (597 µg C g−1) in the non-amended soils (Fig. S1, p<0.001).
3.3 Cottonseed-derived 13CO2 and soil priming effects
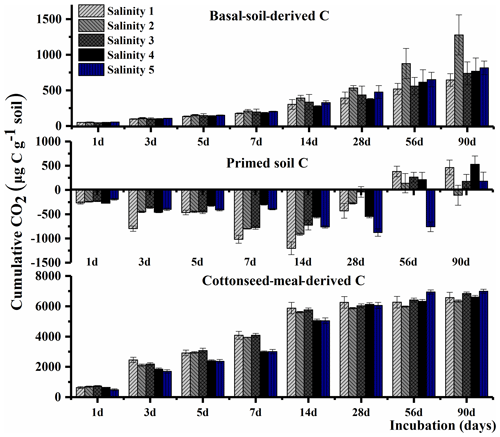
The total cumulative CO2–C was divided into three parts based on the δ13C value, including basal-soil-derived CO2, cottonseed-meal-derived CO2, and primed soil CO2 (Fig. 1). The cottonseed-meal-derived CO2 made a significant contribution to the total evolved CO2 during the early incubation period. The cottonseed-meal-derived CO2 was significantly higher in Salinity 1, Salinity 2, and Salinity 3 than in Salinity 4 and Salinity 5 before 28 d incubation. Meanwhile, the soil priming effects was negative in all amended soil treatments before 28 d incubation, and the direction of priming effects in most of the soil samples turned positive after 28 d. During the whole 90 d incubation, there was a negative correlation between cottonseed-meal-derived CO2 and primed soil CO2 (Fig. 2).
3.4 Bacterial diversity and community structure
The number of sequences ranged from 64 425 to 91 261 per sample (average valve of 80 602). About 27 990 OTUs in total were obtained under the five different treatments. Bacterial community diversity was measured by a series of OTU-based analyses of α diversity, including the chao1 estimator and observed_species in the QIIME pipeline (Fig. 3). The chao1 diversity estimator and observed_species were significantly different in treatments, being the highest in Salinity 1 and followed by Salinity 3, Salinity 2, Salinity 4, and Salinity 5 (p<0.01). In general, bacterial community diversity decreased with increasing salinity (Fig. 3).
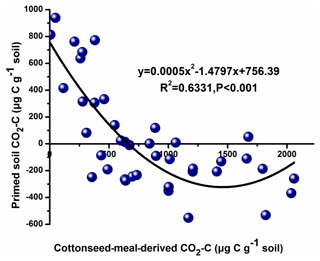
Figure 2Correlation between primed soil mineralization and cottonseed meal mineralization in the five saline soils during the 90 d incubation.
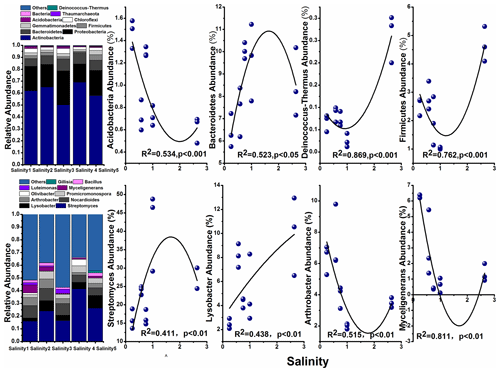
The most abundant phylum in the soils and their correlation with salinity are shown in Fig. 4. Among them, Actinobacteria was the dominant taxon in all soils, with the abundance ranging from 50.07 % (Salinity 3) to 68.99 % (Salinity 4). The relative abundance of Bacteroidetes, Firmicutes, and Deinococcus-Thermus increased with the salinity, while Acidobacteria decreased with salinity degree.
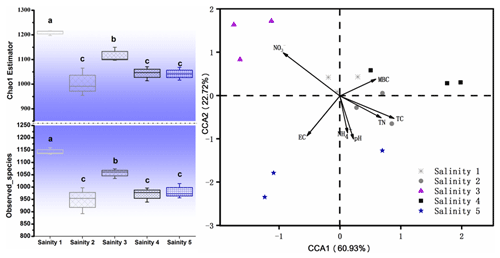
Figure 3Microbial community α diversity (chao1), observed_species, and β diversity. Within each panel, boxplot data refer to maximum date (top line), 99 % (the second line), mean (the third line), 1 % (the fourth line), and minimum date (bottom line) of the different treatments with statistical significance (p<0.05).
Based on OTUs of five salt-gradient treatments, the principal component analysis (PCA) showed that treatments from Salinity 2 and Salinity 4 clustered together. Meanwhile, soil samples of Salinity 1, Salinity 3, and Salinity 5 were distributed in the first, fourth, and third quadrants, which indicated that these treatments had large environmental heterogeneity (Fig. S4).
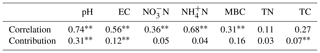
In order to visualize the relationship between environmental factors and microbial community, canonical correspondence analysis (CCA) was conducted, showing that NO–N, EC, and TC had a more obvious impact than other factors for the microbial community (Fig. 3). Soil EC was positively correlated with pH and NH–N and negatively correlated with TN, TC, and microbial biomass carbon (MBC). The Mantel test and distance-based multivariate analysis showed that the contribution rate of different environmental factors accounts for 78 % of the variability in microbial communities (Table 2). The value of pH (31 %) and EC (12 %) had a strong influence on the microbial community.
3.5 Relation between soil microbial community and C dynamics
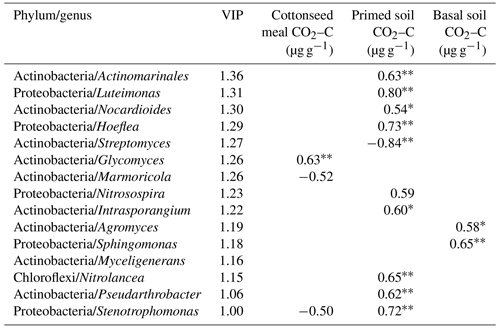
Based on the O2PLS analysis, the variable influence projection (VIP) values of bacterial genera of more than 1.00 % show their contributions to C decomposition of cottonseed-meal-derived C, basal-soil-derived C, and primed soil C (Table 3). There were many microbial taxa positively correlating to primed soil CO2; for instance, genera of Actinomarinales, Luteimonas, Nocardioides, Hoeflea, Intrasporangium, Nitrolancea, Pseudarthrobacter, and Stenotrophomonas had a positive correlation with primed CO2. The result showed that soil pH and EC made a negative contribution to bacterial diversity, while bacterial diversity had a strong positive influence on the primed soil C (Table 2). For instance, salinity properties of EC had a directly negative influence on the bacterial diversity but positive influence on the primed soil C. Meanwhile, pH was negatively correlated with bacterial diversity and positively correlated with substrate-derived C.
4.1 Soil priming effects along salty gradients
Understanding soil C dynamics along salinity gradients is crucial to predict C sequestration in salty soils. In the early stage of the incubation, we observed that the cumulative substrate-derived CO2 in the soils with lower salinity was significantly higher than in soils with higher salinity (Fig. 1), which can be possibly explained by that high-salinity-inhibited microbial activity. Many studies have reported the influence of soil salinity on organic matter decomposition; the decomposition of organic matter is mostly decreased by salinity (Wichern et al., 2006; Ghollarata and Raiesi, 2007; Tripathi et al., 2007; Setia et al., 2012; Lu et al., 2019). Yet, the response of microbial communities to the increasing levels of salinity and consequent effects on soil priming effects remains largely unknown.
Here, we found soil priming effects were gradually changed from a negative to a positive priming effect (Fig. 1). The early pattern of the dynamics of the priming effect in this study was similar to other studies showing preferential utilization of labile C substances. The first phase of negative priming effects was likely to be caused by microbial assimilation of substrate. The soil microbes started to use the new added substrate and thus used less of the original SOC. This was attributed to “preferential substrate utilization” (Perelo et al., 2005).
Soil-microbial-biomass-related growth predominating in the first phase was most likely to utilize SOC, leading to a positive priming effect after the substrate largely vanished. The magnitude of priming effects depends on soil microbial biomass size (Schneckenberger et al., 2008). It was found that the amount of easily available organic C added is beyond 50 % of microbial biomass C (Blagodatskaya et al., 2008). Namely, the second phase of positive priming effects (PEs) probably was due to increased biomass size and enhanced demand on SOC. Secondly, C that was assimilated into microbial biomass in the first stage may also be mineralized in the second stage due to the turnover of microbial biomass (Shahbaz et al., 2017; Perelo et al., 2005).
4.2 Microbial community along salt gradients
Previous studies concerning the impact of salinity on soil microbial communities used different soils with a range of salt levels. In the present study we investigated the influence of soil salinity on microbial communities in soils from the closed area covering a range of salt content. Similarly, Rousk et al. (2011) also used agricultural soils from the same area representing a range of soil salinity. Here, we found microbial diversity (α diversity) decreased with increasing salinity (Fig. 3). The negative impact on microbial diversity can be explained by the accumulation of large amounts of salt in the soil raising the extracellular osmotic concentration (Rath and Rousk, 2015; Oren, 2010). The high osmotic pressures made it difficult for many microorganisms to adapt and thus reduce their biological activity. The changes in soil microbial community structure were also explained by salinity (Herlemann et al., 2011; Campbell and Kirchman, 2012). We found that Bacteroidetes, Firmicutes, Acidobacteria, and Deinococcus-Thermus were dominant in these soils (Fig. 4). These results are supported by previous findings that Firmicutes possess high salinity resistance. Other studies also found that Bacteroidetes is a dominant taxon in alkaline saline soil because of its resistant to salt (Valenzuela-Encinas et al., 2009; Keshri et al., 2013). Other study shows that the dominant phyla are Bacteroidetes and then Proteobacteria in the haloalkaline soil (Keshri et al., 2012). These results are consistent with the estuarine or marine environments despite some studies suggesting that soil salinity is not found to be a decisive factor for bacterial communities and their growth (Rousk et al., 2011).
Table 3The variable influence projection (VIP) value and Spearman's correlation between the relative abundances of genera and C dynamics. Note: , .
The difference in microbial community structure is affected by many soil variables, and pH and EC were the most important ones (Fig. 3; Table 2). Our results showed that the value of soil pH and EC would significantly affect the microbial community structure, and the combined contribution rate of these two variables to microbial community was 43 % (Table 2). At high levels of salt and under arid alkaline conditions, soil pH has also been shown to have a very powerful influence on soil bacterial community structures (Bååth and Anderson, 2003; Fierer and Jackson, 2006; Rousk et al., 2010). Meanwhile, as a consequence it is unlikely that soil pH differences between the studied soils obscured the influence of salt (Rousk et al., 2011). Salinity has been identified as one of the most potent environmental factors that determine the assembly of microbiomes. Salinity has been regarded as playing a vital role in shaping microbial communities in different ecosystems. This, despite the clear evidence from aquatic microbial ecology (Lozupone and Knight, 2007), shows a potential for salt to affect soil microbial communities apart from that of pH (Rath and Rousk, 2015).
4.3 The core microbial taxa regulating C decomposition along salinity gradient
The correlation of microbial taxa and SOC decomposition (priming) was found according to the results of O2PLS and SEM (Table 3; Fig. S4). Here we showed that Streptomyces (Actinobacteria), Glycomyces (branch of Actinobacteria), Agromyces (branch of Actinobacteria), and Sphingomonas (branch of Proteobacteria) at the genus level were significantly correlated with the C process, particularly primed-soil-derived C. Most of these functional taxa belonged to Actinobacteria and Proteobacteria. In a recent study, Ren et al. (2018) found that Actinobacteria had a negative impact on SOC mineralization across land-use change (Fierer et al., 2007; Goldfarb et al., 2011) and Proteobacteria drove the positive soil respiration (He et al., 2012), indicating that the balance of soil C dynamics was largely regulated by these two phyla. We found a similar result for Streptomyces (branch of Actinobacteria) in that it had a negative correlation with primed soil CO2. Actinobacteria are able to grow preferentially on the C-rich refractory materials and relatively easily decompose the cellulose and lignocellulose (Khodadad et al., 2011), indicating these microorganisms preferentially use the C source that is used partially by others.
Although some studies suggest soil salinity may not be a vital factor for C decomposers (Rousk et al., 2010), the composition of a microbial community is considered to play a decisive role in determining dynamic C processes in response to salt stress (Ramsey et al., 2005; Schimel et al., 2007; Nottingham et al., 2009). Here, the analysis showed that soil pH and EC in salted soils reduced microbial diversity and thus limited the utilization of SOC by microbial communities. It was reported that high pH and salinity are the major determinants of soil microbial activity and community structure (Kamble et al., 2014).
Cottonseed meal is a kind of organic material with high nitrogen content, and adding cottonseed meal to salinized soil can stimulate and promote the release of soil nutrients. The microorganisms mainly use the organic matter in the cottonseed meal in the pre-culture period, so the soil carbon excitation is negative excitation. The soil priming effect turned from negative to positive at the later stage of incubation (day 28) because microorganisms started to decompose SOC from the labile substrate. With the increase in salinity, the diversity of microbial community decreased. The soil microbial community was mainly controlled by soil pH and EC. By O2PLS, we found that Actinobacteria and Proteobacteria (Luteimonas, Hoeflea, and Stenotrophomonas), dominant in these soils, were the core microbial taxa that affected the process of organic C mineralization, particularly primed soil CO2.
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
The supplement related to this article is available online at: https://doi.org/10.5194/bg-21-1-2024-supplement.
KW conceptualized and conducted the experiment. HZ and DC conducted the data analysis, conducted the indoor experiment, and wrote the manuscript. CM and ZZ assisted in conducting the experiment. All authors reviewed the manuscript. All authors contributed to the manuscript and approved the submitted version.
The contact author has declared that none of the authors has any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
This study was supported by the Special Fund for the Science and Technology Department of Xinjiang Uygur Autonomous Region (grant no. 2022B02021-3-1) and the Ningbo Key Research and Development Program (2021Z101).
This paper was edited by Luo Yu and reviewed by two anonymous referees.
Ali, N., Khan, S., Li, Y., Zheng, N., and Yao, H.: Influence of biochars on the accessibility of organochlorine pesticides and microbial community in contaminated soils, Sci. Total. Environ., 647, 551–560, https://doi.org/10.1016/j.scitotenv.2018.07.425, 2018.
Aoyama, M., Angers, D. A., N'Dayegamiye, A., and Bissonnette, N.: Metabolism of 13C-labeled glucose in aggregates from soils with manure application, Soil Biol. Biochem., 32, 295–300, https://doi.org/10.1016/S0038-0717(99)00152-2, 2000.
Asghar, H. N., Setia, R., and Marschner, P.: Community composition and activity of microbes from saline soils and non-saline soils respond similarly to changes in salinity, Soil Biol. Biochem., 47, 175–178, https://doi.org/10.1016/j.soilbio.2012.01.002, 2012.
Bååth, E. and Anderson, T. H.: Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques, Soil Biol. Biochem., 35, 955–963, https://doi.org/10.1016/s0038-0717(03)00154-8, 2003.
Bao, S. D.: Soil and Agricultural Chemistry Analysis, 3rd Edn., China Agriculture Press, Beijing, 9787109066441, 2000.
Blagodatskaya, Å. and Kuzyakov, Y.: Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: critical review, Biol. Fert. Soils, 45, 115–131, https://doi.org/10.1007/s00374-008-0334-y, 2008.
Bird, J. A., Herman, D. J., and Firestone, M. K.: Rhizosphere priming of soil organic matter by bacterial groups in a grassland soil, Soil Biol. Biochem., 43, 718–725, https://doi.org/10.1016/j.soilbio.2010.08.010, 2010.
Campbell, B. J. and Kirchman, D. L.: Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient, ISME. J., 7, 210–220, https://doi.org/10.1038/ismej.2012.93, 2012.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., Fierer, N., Peña, A. G., Goodrich, J. K., Gordon, J. I., Huttley, G. A., Kelley, S. T., Knights, D., Koenig, J. E., Ley, R. E., Lozupone, C. A., McDonald, D., Muegge, B. D., Pirrung, M., Reeder, J., Sevinsky, J. R., Turnbaugh, P. J., Walters, W. A., Widmann, J., Yatsunenko, T., Zaneveld, J., and Knight, R.: QIIME allows analysis of high-throughput community sequencing data, Nat. Methods., 7, 335–336, https://doi.org/10.1038/nmeth.f.303, 2012.
Dai, H., Chen, Y., Yang, X., Cui, J., and Sui, P.: The effect of different organic materials amendment on soil bacteria communities in barren sandy loam soil, Environ. Sci. Pollut. R., 24, 1–10, https://doi.org/10.1007/s11356-017-0031-1, 2017.
Edgar, R. C.: Search and clustering orders of magnitude faster than BLAST, Bioinformatics, 26, 2460–2461, https://doi.org/10.1093/bioinformatics/btq461, 2010.
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R.: UCHIME improves sensitivity and speed of chimera detection, Bioinformatics, 27, 194–220, https://doi.org/10.1093/bioinformatics/btr381, 2011.
Faith, D. P.: Conservation evaluation and phylogenetic diversity, Biol. Conserv., 61, 1–10, https://doi.org/10.1016/0006-3207(92)91201-3, 1992.
Fierer, N. and Jackson, R. B.: The diversity and biogeography of soil bacterial communities, P. Natl. Acad. Sci. USA, 103, 626–631, https://doi.org/10.1073/pnas.0507535103, 2006.
Fierer, N., Bradford, M., and Jackson, R.: Toward an ecological classification of soil bacteria, Ecology, 88, 1354–1364, https://doi.org/10.1890/05-1839, 2007.
Filley, T. R. and Boutton, T. W.: Ecosystems in flux: molecular and stable isotope assessments of soil organic matter storage and dynamics, Soil Biol. Biochem., 38, 3181–3183, https://doi.org/10.1016/j.soilbio.2006.05.006, 2006.
Fontaine, S., Henault, C., Aamor, A., Bdioui, N., Bloor, J. M. G., Maire, V., Mary, B., Revaillot, S., and Maron, P. A.: Fungi mediate long term sequestration of carbon and nitrogen in soil through their priming effect, Soil Biol. Biochem., 43, 86–96, https://doi.org/10.1016/j.soilbio.2010.09.017, 2003.
Fontaine, S., Mariotti, A., and Abbadie, L.: The priming effect of organic matter: A question of microbial competition?, Soil Biol. Biochem., 35, 837–843, https://doi.org/10.1016/s0038-0717(03)00123-8, 2011.
Ghollarata, M. and Raiesi, F.: The adverse effects of soil salinization on the growth of trifolium alexandrinum l. and associated microbial and biochemical properties in a soil from iran, Soil Biol. Biochem., 39, 1699–1702, https://doi.org/10.3389/fmicb.2011.00094, 2011.
Goldfarb, K., Karaoz, U., Hanson, C., Santee, C., Bradford, M., Treseder, K., Wallenstein, M., and Brodie, E.: Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance, Front. Microbiol., 2, 94, https://doi.org/10.3389/fmicb.2011.00094, 2011.
Hartmann, M., Frey, B., Mayer, J., and Widmer, F.: Distinct soil microbial diversity under long-term organic and conventional farming, ISME J., 9, 1177–1194, https://doi.org/10.1038/ismej.2014.210, 2014.
Hagemann, S.: Vulnerability of permafrost carbon to climate change: Implications for the global carbon cycle, Bioscience, 58, 701–714, https://doi.org/10.1641/b580807, 2008.
He, Z., Piceno, Y., Deng, Y., Xu, M., Lu, Z., DeSantis, T., Andersen, G., Hobbie, S. E., Reich, P. B., and Zhou, J.: The phylogenetic composition and structure of soil microbial communities shifts in response to elevated carbon dioxide, ISME J., 6, 259–272, https://doi.org/10.1038/ismej.2011.99, 2012.
Herlemann, D. P., Labrenz, M., Jürgens, K., Bertilsson, S., Waniek, J. J., and Andersson, A. F.: Transitions in bacterial communities along the 2000 km salinity gradient of the baltic sea, ISME J., 5, 1571–1579, https://doi.org/10.1038/ismej.2011.41, 2011.
Kamble, P. N., and Bååth, E.: Induced N-limitation of bacterial growth in soil: Effect of carbon loading and N status in soil, Soil Biol. Biochem., 70, 11–20, https://doi.org/10.1016/j.soilbio.2014.02.015, 2014.
Keshri, J., Mody, K., and Jha, B.: Bacterial community structure in a semi-arid haloalkaline soil using culture independent method, Geomicrobiol. J., 30, 517–529, https://doi.org/10.1080/01490451.2012.737092, 2012.
Khodadad, C. L. M., Zimmerman, A. R., Green, S. J., Uthandi, S., and Foster, J. S.: Taxa-specific changes in soil microbial community composition induced by pyrogenic carbon amendments, Soil Biol. Biochem., 43, 385–392, https://doi.org/10.1016/j.soilbio.2010.11.005, 2010.
Kuzyakov, Y., Friedel, J. K., and Stahr, K.: Review of mechanisms and quantification of priming effects, Soil Biol. Biochem., 32, 1485–1498, https://doi.org/10.1016/s0038-0717(00)00084-5, 2000.
Laura, R. D.: Effects of neutral salts on carbon and nitrogen mineralisation of organic matter in soil, Plant Soil., 41, 113–127, https://doi.org/10.1007/bf00017949, 1974.
Lavelle, P., Bignell, D., Lepage, M., Wolters, V., Roger, P., Ineson, P., Heal, O. W., and Dhillion, S.: Soil function in a changing world: the role of invertebrate ecoystem engineers, Eur. J. Soil. Biol., 33, 159–193, https://doi.org/10.1016/j.ejsobi.2006.10.002, 2006.
Lehmann, J. and Kleber, M.: The contentious nature of soil organic matter, Nature, 528, 60–68, https://doi.org/10.1038/nature16069, 2015.
Li, X., Chen, Q. L., He, C., Shi, Q., Chen, S. C., Reid, B. J., Zhu, Y. G., and Sun, G. X.: Organic Carbon Amendments Affect the Chemodiversity of Soil Dissolved Organic Matter and Its Associations with Soil Microbial Communities, Environ. Sci. Technol., 53, 50–59, https://doi.org/10.1021/acs.est.8b04673, 2018.
Ling, L., Fu, Y. Y., Jeewani, P. H., Tang, C. X., Pan, S. T., Reid, B. J., Gunina, A., Li, Y. F., Li, Y. C., Cai, Y. J., Kuzyakov, Y., Li, Y., Su, W. Q., Singh, B. P., Luo, Y., and Xu, J. M.: Organic matter chemistry and bacterial community structure regulate decomposition processes in post-fire forest soils, Soil Biol. Biochem., 160, 108311, https://doi.org/10.1016/j.soilbio.2021.108311, 2021.
Ling, L., Luo, Y., Jiang, B., Lv, J. T., Meng, C. M., Liao, Y. H., Reid, B. J., Ding, F., Lu, Z. J., Kuzyakov, Y., and Xu, J. M.: Biochar induces mineralization of soil recalcitrant components by activation of biochar responsive bacteria groups, Soil Biol. Biochem., 172, 108778, https://doi.org/10.1016/j.soilbio.2022.108778, 2022.
Löhnis, F.: Nitrogen abailability of green manures, Soil Sci., 22, 253–290, 1926.
Lozupone, C. A. and Knight, R.: Global patterns in bacterial diversity, P. Natl. Acad. Sci. USA, 104, 11436–11440, https://doi.org/10.1073/pnas.0611525104, 2007.
Lozupone, C. A., Hamady, M., Kelley, S. T., and Knight, R.: Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities, Appl. Environ. Microb., 73, 1576–1585, https://doi.org/10.1128/aem.01996-06, 2007.
Lu, X. H., Li, Y. F., Wang, H. L., Singh, B. P., Hu, S. D., Luo, Y., Li, J. W., Xiao, Y. H., Cai, X. Q., and Li, Y. C.: Responses of soil greenhouse gas emissions to different application rates of biochar in a subtropical Chinese chestnut plantation, Agr. Forest Meteorol., 271, 168–179, https://doi.org/10.1016/j.agrformet.2019.03.001, 2019.
Luo, Y., Zang, H., Yu, Z., Chen, Z., Gunina, A., Kuzyakov, Y., Xu, J., Zhang, K., and Brookes, P. C.: Priming effects in biochar enriched soils using a three-source-partitioning approach: 14C labelling and 13C natural abundance, Soil Biol. Biochem., 106, 28–35, https://doi.org/10.1016/j.soilbio.2016.12.006, 2016.
Martin, W., Livia, U., Hobley, E., Lang, B., von Ltzow, M., Marin-Spiotta, E., Van Wesemael, B., Rabot, E., Lie, M., Garcia-Franco, N., Wollschlger, U., Vogel, H. J., and Kgel-Knabner, I.: Soil organic carbon storage as a key function of soils – A review of drivers and indicators at various scales, Geoderma, 333, 149–162, https://doi.org/10.1016/j.geoderma.2018.07.026, 2018.
Nottingham, A. T., Griffiths, H., Chamberlain, P. M., Stott, A. W., and Tanner, E. V. J.: Soil priming by sugar and leaf-litter substrates: a link to microbial groups, Appl. Soil Ecol., 42, 183–190, https://doi.org/10.1016/j.apsoil.2009.03.003, 2009.
Oren, A.: Thermodynamic limits to microbial life at high salt concentrations, Environ. Microbiol., 13, 1908–1923, https://doi.org/10.1111/j.1462-2920.2010.02365.x, 2010.
Pathak, H. and Rao, D. L. N.: Carbon and nitrogen mineralization from added organic matter in saline and alkaline soils, Soil Biol. Biochem., 30, 695–702, https://doi.org/10.1016/S0038-0717(97)00208-3, 1998.
Perelo, L. W., Jimenez, M., and Munch, J. C.: Microbial immobilisation and turnover of 15N labelled substrates in two arable soils under field and laboratory conditions, Soil Biol. Biochem., 38, 912–922, https://doi.org/10.1016/j.soilbio.2005.02.039, 2005.
Ramsey, P. W., Rillig, M. C., Feris, K. P., Gordon, N. S., Moore, J. N., Holben, W. E., and Gannon, J. E.: Relationship between communities and processes; new insights from a field study of a contaminated ecosystem, Ecol. Lett., 8, 1201–1210, https://doi.org/10.1111/j.1461-0248.2005.00821.x, 2005.
Rath, K. M. and Rousk, J.: Salt effects on the soil microbial decomposer community and their role in organic carbon cycling: A review, Soil Biol. Biochem., 81, 108–123, https://doi.org/10.1016/j.soilbio.2014.11.001, 2014.
Ren, C., Wang, T., Xu, Y., Deng, J., Zhao, F., Yang, G., Han, X., Feng, Y., and Ren, G.: Differential soil microbial community responses to the linkage of soil organic carbon fractions with respiration across land-use changes, Forest Ecol. Manag., 12, 170–178, https://doi.org/10.1016/j.foreco.2017.11.011, 2018.
Rietz, D. N. and Haynes, R. J.: Effects of irrigation-induced salinity and sodicity on soil microbial activity, Soil Biol. Biochem., 35, 845–854, https://doi.org/10.1016/s0038-0717(03)00125-1, 2003.
Rousk, J., Bååth, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., Knight, R., and Fierer, N.: Soil bacterial and fungal communities across a pH gradient in an arable soil, ISME J., 4, 1340–1351, https://doi.org/10.1038/ismej.2010.58, 2010.
Rousk, J., Elyaagubi, F. K., Jones, D. L., and Godbold, D. L.: Bacterial salt tolerance is unrelated to soil salinity across an arid agroecosystem salinity gradient, Soil Biol. Biochem., 43, 1881–1887, https://doi.org/10.1016/j.soilbio.2011.05.007, 2011.
Sardinha, M., Müller, T., Schmeisky, H., and Joergensen, R. G.: Microbial performance in soils along a salinity gradient under acidic conditions, Appl. Soil Ecol., 23, 237–244, https://doi.org/10.1016/s0929-1393(03)00027-1, 2003.
Schneckenberger, K., Demin, D., Stahr, K., and Kuzyakov, Y.: Microbial utilization and mineralization of 14C glucose added in six orders of concentration to soil, Soil Biol. Biochem., 40, 1981–1988, https://doi.org/10.1016/j.soilbio.2008.02.020, 2008.
Scheu, S. and Parkinson, D.: Effects of earthworms on nutrient dynamics, carbon turnover and microorganisms in soils from cool temperate forests of the Canadian Rocky Mountains – laboratory studies, Appl. Soil Ecol., 5, 23–27, https://doi.org/10.1016/0929-1393(94)90031-0, 1994.
Schimel, J., Balser, T. C., and Wallenstein, M.: Microbial stress-response physiology and its implications for ecosystem function, Ecology, 88, 1386–1394, https://doi.org/10.1890/06-0219, 2007.
Setia, R., Setia, D., and Marschner, P.: Short-term carbon mineralization in saline-sodic soils, Biol. Fert. Soils, 48, 475–479, https://doi.org/10.1007/s00374-011-0643-4, 2012.
Shahbaz, M., Kuzyakov, Y., Sanaullah, M., Heitkamp, F., Zelenev, V., Kumar, A., and Blagodatskaya, E.: Microbial decomposition of soil organic matter is mediated by quality and quantity of crop residues: mechanisms and thresholds, Biol. Fert. Soils, 53, 287–301, https://doi.org/10.1007/s00374-016-1174-9, 2017.
Soina, V. S., Mergelov, N. S., Kudinova, A. G., Lysak, L. V., Demkina, E. V., Vorobyova, E. A., Dolgikh, A. V., and Shorkunov, I. G.: Microbial Communities of Soils and Soil-like Bodies in Extreme Conditions of East Antarctica, Paleontol. J., 52, 1186–1195, https://doi.org/10.1134/s0031030118100143, 2018.
Stevenson, B. S., Eichorst, S. A., Wertz, J. T., Schmidt, T. M., and Breznak, J. A.: New strategies for cultivation and detection of previously uncultured microbes, Appl. Environ. Microb., 70, 4748–4755, https://doi.org/10.1128/aem.70.8.4748-4755.2004, 2004.
Su, J. Q., An, X. L., Li, B., Chen, Q. L., Gillings, M. R., and Chen, H.: Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in china, Microbiome, 5, 84–85, https://doi.org/10.1186/s40168-017-0298-y, 2017.
Sullivan, B. W. and Hart, S. C.: Evaluation of mechanisms controlling the priming of soil carbon along a substrate age gradient, Soil Biol. Biochem., 58, 293–301, https://doi.org/10.1016/j.soilbio.2012.12.007, 2013.
Tripathi, S., Chakraborty, A., Chakrabarti, K., and Bandyopadhyay, B, K.: Enzyme activities and microbial biomass in coastal soils of India, Soil Biol. Biochem., 39, 2840–2848, https://doi.org/10.1016/j.soilbio.2007.05.027, 2007.
Valenzuela-Encinas, C., Neria-González, I., Alcántara-Hernández, R. J., Estrada-Alvarado, I., Dendooven, L., and Marsch, R.: Changes in the bacterial populations of the highly alkaline saline soil of the former lake Texcoco (Mexico) following flooding, Extremophiles, 13, 609–621, https://doi.org/10.1007/s00792-009-0244-4, 2009.
Vance, E. D., Brookes, P. C., and Jenkinson, D. S.: An extraction method for measuring soil microbial biomass C, Soil Biol. Biochem., 19, 703–707, https://doi.org/10.1016/0038-0717(87)90052-6, 1987.
Wagner, B. and Raquel, G.: Impacts of sewage sludge in tropical soil: a case study in brazil, Applied and Environmental Soil Science, 2011, 212807, https://doi.org/10.1155/2011/212807, 2011.
Wang, Z. M., Lu, Z. M., Shi, J. S., and Xu, Z. H.: Exploring flavour-producing core microbiota in multispecies solid-state fermentation of traditional Chinese vinegar, Sci. Rep.-UK, 31, 26818, https://doi.org/10.1038/srep26818, 2016.
Wichern, J., Wichern, F., and Joergensen, R. G.: Impact of salinity on soil microbial communities and the decomposition of maize in acidic soils, Geoderma, 137, 100–108, https://doi.org/10.1016/j.geoderma.2006.08.001, 2006.
Wu, J., Joergensen, R. G., and Pommerening, B.: Measurement of soil microbial biomass C by fumigation-extraction: an automated procedure, Soil Biol. Biochem., 22, 1167–1169, https://doi.org/10.1016/0038-0717(90)90046-3, 1990.
Xun, W., Zhao, J., Xue, C., Zhang, G., Ran, W., Wang, B., Shen, Q., and Zhang, R.: Significant alteration of soil bacterialcommunities and organic carbon decomposition by different longterm fertilization management conditions of extremely lowproductivity arable soil in South China, Environ. Microbiol., 18, 1907–1917, https://doi.org/10.1111/1462-2920.13098, 2016.