the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Unexpected scarcity of ANME archaea in hydrocarbon seeps within Monterey Bay
Amanda C. Semler
Anne E. Dekas
Marine hydrocarbon seeps typically harbor a relatively predictable microbiome, including anaerobic methanotrophic (ANME) archaea. Here, we sampled two cold seeps in Monterey Bay, CA – Clam Field and Extrovert Cliff – which have been known for decades but never characterized microbiologically. Many aspects of these seeps were typical of seeps worldwide, including elevated methane and sulfide concentrations, 13C-depleted dissolved inorganic carbon, and the presence of characteristic macrofauna. However, we observed atypical microbial communities: extremely few ANME sequences were detected in either 16S rRNA or mcrA gene surveys at Clam Field (< 0.1 % of total community reads), even after 6 months of incubation with methane in the laboratory, and only slightly more ANME sequences were recovered from Extrovert Cliff (< 0.3 % of total community reads). At Clam Field, a lack of ANME mcrA transcription, a lack of methane-dependent sulfate reduction, and a linear porewater methane profile were consistent with low or absent methanotrophy. Although the reason for the scarcity of ANME archaea is still unclear, we postulate that non-methane hydrocarbon release excludes anaerobic methanotrophs directly or indirectly (e.g., through competitive interactions with hydrocarbon-degrading bacteria). Our findings highlight the potential for hydrocarbon seeps without this critical biofilter and therefore greater methane emissions from sediments.
- Article
(5006 KB) - Full-text XML
-
Supplement
(4905 KB) - BibTeX
- EndNote
Monterey Bay is a well-studied region of the California coastline located within a tectonically active transform boundary. Right-lateral, strike-slip motion between the Pacific Plate to the west and the North American Plate to the east produces movement in the bay along two major fault zones (the San Gregorio and Monterey Bay fault zones) and causes extensive sedimentary compression and compaction (Clark, 1981; Orange et al., 1999). These forces drive fluid flow through the organic-rich, hydrocarbon-bearing sediments underlying Monterey Bay (Orange et al., 1999; Stakes et al., 1999), creating large networks of cold seeps – highly productive chemosynthetic ecosystems on the seafloor typically characterized by methane- and sulfide-rich fluids. In Monterey Bay, cold seeps are concentrated especially around high-porosity sediment layers and permeable fractures within the fault zones (Moore et al., 1991; Greene et al., 1999; Orange et al., 1999), as well as sites of recent erosion such as canyon walls (Paull et al., 2005).
Cold seeps have been surveyed and studied in Monterey Bay for more than 3 decades (Barry et al., 1996; Orange et al., 1999; Lorenson et al., 2002) – almost from the time cold seeps were first discovered (Paull et al., 1984; Suess et al., 1985). Investigations of Monterey Bay seeps have been particularly focused on their fluid chemistry and macrofaunal communities. Previous chemical analyses have demonstrated that fluids at most Monterey Bay seeps are enriched in sulfide and methane, and the 13C-depleted dissolved inorganic carbon (DIC) in pore fluids and in authigenic carbonates surrounding the seeps suggests the original methane is of a largely microbial origin (Martin et al., 1997). However, methane at some sites also has a distinct thermogenic isotope imprint, with potential input from deep fluids flowing through the organic-rich Monterey Formation (Martin et al., 1997; Rathburn et al., 2003; Füri et al., 2009). Non-methane hydrocarbons – including ethane, propane, and butane, as well as visible oil – have also been discovered in the seep fluids (Lorenson et al., 2002), especially at seeps located within the Monterey Bay fault zone. Utilizing the reduced compounds in Monterey Bay, seep fluids (either directly, or indirectly through a symbiont) have high numbers of vesicomyid clams, thiotrophic Beggiatoa mats, and, more rarely, vestimentiferan and pogonophoran tube worms (Fisher and Childress, 1992; Greene et al., 1994; Orange et al., 1994; Barry et al., 1996, 1997) – an overall community which, at broad, family-level taxonomic scales, mirrors the macrofauna found at cold seeps across the Pacific Basin (Barry et al., 1996). Species-level differences between macrofaunal taxa in Monterey Bay have been largely attributed to the differing sulfide concentrations at each individual seep (Barry et al., 1996).
However, while much is known about the geology, geochemistry, and macrofauna of Monterey Bay cold seeps, few investigations have targeted microbial communities here. Seep sediment has been previously retrieved from one Monterey Bay seep – Extrovert Cliff – for incubations in bioreactors (Girguis et al., 2005, 2003), and archaeal-specific 16S rRNA primers were utilized to generate clone libraries before and after enrichment. Clones belonging to characteristic seep microbial taxa were recovered – specifically, clones of anaerobic methanotrophic (ANME) archaea subgroups ANME-2b and ANME-2c. ANME archaea are core microbial taxa at cold seeps, as they couple the anaerobic oxidation of methane (AOM) to sulfate reduction with the help of syntrophic sulfate-reducing bacterial (SRB) partners (Boetius et al., 2000; Orphan et al., 2001). ANME archaea are comprised of three distinct polyphyletic clusters in the phylum Halobacterota (ANME-1 (Methanophagales), ANME-2 (Methanocomedenaceae, Methanogasteraceae, and Methanoperedenaceae in the Methanosarcinales), and ANME-3 (Methanovorans in the Methanosarcinales); following the naming scheme of Chadwick et al., 2022), and they associate and share electrons with a variety of SRB partners (Seep-SRB1, Seep-SRB2, Seep-SRB4, and thermophilic Hot-Seep1) typically in tight cell aggregates (in the case of ANME-2 and ANME-3). Both ANME archaea and their syntrophs are key components of the “seep microbiome” (Ruff et al., 2015) – the core set of microbial groups that dominate seeps globally. However, it has been noted that quantitative surveys for ANME archaea using fluorescence in situ hybridization (FISH) have been largely unsuccessful in Monterey Bay sediments, and cells with the typical ANME aggregate morphology are rare, even at Extrovert Cliff, where ANME-2 clones were recovered (Girguis et al., 2003).
Here, we investigated the microbial communities and sediment porewater geochemistry at two cold seeps in Monterey Bay: Clam Field (895–909 m b.s.l.) and Extrovert Cliff (965–990 m b.s.l.). Using deep amplicon sequencing of both the 16S rRNA and methyl coenzyme-M reductase (mcrA) genes, we characterized archaeal and bacterial community composition (DNA) and potential activity (RNA) in 20 cm sediment cores collected along 100 m transects from the center of each seep to “background” sediment. Using droplet digital PCR (ddPCR), we also investigated the abundance of mcrA genes and transcripts inside and outside each seep. Sediment from the Clam Field site was also collected and incubated under a variety of methane headspace concentrations for 6 months to enrich methanotrophic taxa and their sulfate-reducing syntrophs. Our goals were to (i) characterize the community of microorganisms within these well-known cold seeps and (ii) evaluate the impact of Monterey Bay's unusual and complex hydrocarbon geochemistry on seep communities. This comparative analysis provides the first deep-sequencing perspective of Monterey Bay cold seeps, and, more broadly, helps us understand how local geochemistry impacts methane oxidation potential.
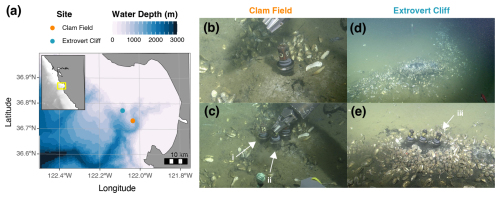
Figure 1(a) Bathymetric map of Monterey Bay and the location of the two sampling sites: Clam Field (CF – 895–909 m b.s.l.) and Extrovert Cliff (EC – 965–990 m b.s.l.). Inset shows the central California coast, with Monterey Bay indicated by a yellow box. ROV Doc Ricketts images of seep surface expression before (b) and during (c) sampling at Clam Field and before (d) and during (e) sampling at Extrovert Cliff. Push cores collected for in situ measurements (i; PC 75) and incubations (ii; PC 54) at the Clam Field “Seep” location and for in situ measurements at the Extrovert Cliff “Seep” location (iii; PC 64), indicated by white arrows. Bathymetric data from the General Bathymetric Chart of the Oceans (GEBCO) 2021 grid (GEBCO Compilation Group, 2021).
2.1 Site description and geochemical conditions
The two Monterey Bay cold seep sites investigated in this study were Clam Field and Extrovert Cliff (Fig. 1a). The Clam Field site is located on a sedimented apron of the Monterey Canyon wall and is constituted by a broad band of seepage parallel to the canyon. Dense fields of live clams are found across the main band of seepage, along with patchy microbial mats (Fig. 1b–c). Clam Field is located within the Monterey Bay fault zone (Orange et al., 1999), suggesting that fluid seepage there is tectonically influenced and likely driven by artesian flow through the highly fractured, hydrocarbon-rich shale of the Monterey Formation (Barry et al., 1996; Lorenson et al., 2002; LaBonte et al., 2007; Füri et al., 2009). Previously measured δ13C values of methane at this site (−50 ‰ to −55 ‰) are indicative of a high degree of thermogenic input – more so than at many other Monterey Bay seeps (Lorenson et al., 2002). The Extrovert Cliff site is located on the slope of a slide scar between the San Gregorio and Monterey Bay fault zones. The site is characterized by distinct, concentric rings of seepage covered by thick microbial mats and bordered by live clams (Fig. 1d–e). Fluid flow rates at Extrovert Cliff are temporally variable and tidally influenced, suggesting fluid conduits from an overpressurized aquifer (LaBonte et al., 2007; Füri et al., 2009).
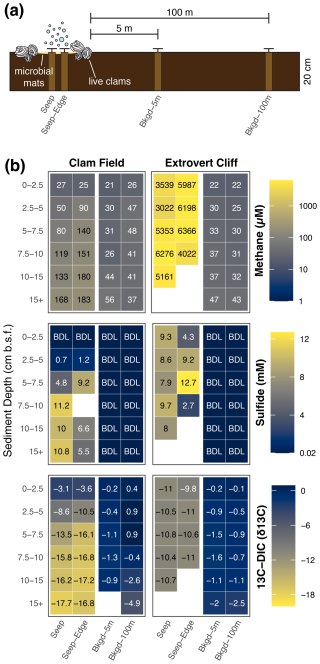
Figure 2(a) Diagram (not to scale) of core sampling scheme at both Clam Field and Extrovert Cliff. (b) Methane and sulfide concentration and δ13C–DIC values with sediment depth in all sampled cores. Blank values indicate no measurement was made. BDL indicates a value below the detection limit of the assay; detection limits were 1 µM and 0.02 mM for methane and sulfide, respectively.
2.2 Sample collection and processing
Sediment push cores were collected from both seep sites in April 2019 on the R/V Western Flyer, using ROV Doc Ricketts. At each site, two push cores up to 20 cm long were collected from each of four locations: (1) the center of the cold seep, (2) the inner edge of the cold seep, (3) 5 m outside the seep boundary, and (4) 100 m outside the seep boundary (Fig. 2a). These were categorized as “Seep,” “Seep-Edge”, “Background-5m,” and “Background-100m” cores, respectively (Table S1 in the Supplement). Seep boundaries were delineated by the sudden termination of white, filamentous microbial mats and live clam beds on the sediment surface. These boundaries were validated by the sulfidic smell of the Seep and Seep-Edge cores from each site once cores were recovered.
On board, cores were kept at 4 °C until extruded from push core liners and sectioned within 8 h of collection. One core of each pair was sectioned into 2.5 cm (0–10 cm b.s.f.) or 5 cm (10+ cmbsf) horizons and subsampled for molecular and geochemical analyses, while the other was sectioned into 5 cm horizons and preserved anaerobically within Whirl-Paks sealed in Mylar bags at 4 °C for incubation experiments. Several 1 mL subsamples of each depth horizon in the molecular/geochemical cores were immediately flash-frozen in liquid nitrogen and preserved at −80 °C for later DNA and RNA extraction. Subsamples of 3 mL were transferred into 25 mL butyl-rubber-sealed vials filled with 5 mL of 5 M sodium hydroxide solution for methane analysis, 3–5 mL subsamples were scooped onto a pre-weighed sheet of aluminum foil for dehydration and porosity measurements, and 0.5 mL subsamples were transferred into 2 mL epitubes pre-loaded with 1 mL of 4 % PFA for microscopy. Porewater was squeezed from sediments using a porewater pressing bench (KC Denmark Research Equipment, Silkeborg, Denmark) under a stream of argon gas immediately after sectioning, and porewater was filtered with 0.2 µm Durapore® PVDF membrane filters (EMD Millipore, Burlington, MA, USA). A total of 0.5 mL of porewater was fixed with 0.5 mL of 0.5 M zinc acetate and stored at 4 °C for sulfide measurements, and 2 mL of porewater was added to a 12 mL Exetainer® (Labco Limited, Ceredigion, UK) pre-loaded with 1 mL 85 % phosphoric acid, evacuated, and N2-flushed for δ13C analysis of dissolved inorganic carbon (DIC). The remaining porewater was stored at −20 °C.
2.3 Incubation setup
Immediately after returning from the field, preserved sediment from all four Clam Field locations was anaerobically incubated and subsampled according to the method described in Dekas et al. (2009, 2014, 2016). Clam Field was chosen for incubations because its surface expression was similar to that of seeps sampled and characterized in previous studies (McVeigh et al., 2018; Seabrook et al., 2018; Semler et al., 2022), and thus responses of characteristic seep microbial communities to varying methane headspace concentration could be tested. The top two horizons (0–5 and 5–10 cm) from the Seep and Seep-Edge cores and the top three horizons (0–5, 5–10, and 10–15 cm) from the Background-5m and Background-100m cores (nhorizons=10) were incubated with varying concentrations (0–2 atm) of methane to evaluate community responses to methane addition; 15NH was also added to all incubations at a final concentration of 100 µM to measure total anabolic activity (99 at. % 15N; Cambridge Isotopes, NLM-467-1). The methane was a 4:1 mixture of natural abundance 13C 12C and 99 at. % 13C methane (Sigma-Aldrich, 490229-1L). Table S2 lists each of five methane treatments per horizon; all treatments were performed in triplicate (nincubations=150). Incubations were subsampled at 0-, 0.5-, 1-, 3-, and 6-month time points.
2.4 Porewater geochemistry
Headspace methane concentrations were measured from butyl-rubber-sealed vials (see above) using gas chromatography coupled to flame-ionization detection (Shimadzu GC-2014, Stanford University, Stanford, CA, USA) and back-calculated to porewater concentrations using porosity values determined via weight loss after dehydration (and assuming a sediment density of 2.65 g cm−3). Sulfide concentrations were measured in triplicate and determined colorimetrically from the zinc acetate-preserved porewater samples using the methylene blue method (Cline, 1969), with a detection limit of 0.02 mM. All other assays were performed without replication due to limitations on porewater volume.
2.5 Nucleic acid extraction, amplification, and sequencing of 16S rRNA and mcrA genes and transcripts
DNA and RNA were extracted from flash-frozen sediments using the RNeasy PowerSoil Total RNA Isolation Kit (RNA) and the RNA PowerSoil DNA Elution Accessory Kit (DNA; Mo Bio Laboratories, Carlsbad, CA, USA). The protocol was modified from the manufacturer's instructions to include a bead-beating step of 60 s at a speed of 5.5 m s−1 on a FastPrep instrument (MP Biomedicals, Santa Ana, CA, USA) to facilitate archaeal cell lysis as in Semler et al. (2022). RNA extracts were cleaned with the Ambion TURBO DNA-free Kit (Thermo Fisher Scientific, Waltham, MA, USA), and reverse transcription of RNA to cDNA was completed using Superscript III First Strand Synthesis Supermix (Thermo Fisher Scientific, Waltham, MA, USA).
DNA and cDNA were concentration normalized and amplified using a two-step PCR plan for Illumina amplicon sequencing. In the first step, universal primers 515F-Y/926R (Parada et al., 2016) were used to target the V4-V5 region of the 16S rRNA gene sequence, and mcrA_F/mcrA_R (Luton et al., 2002; Dekas et al., 2016) was used to target mcrA genes and transcripts. Both sets of primers included an extension complementary to the primers used in the second PCR. The gene-targeting regions of the primer sequences are listed in Table S3; 25 µL PCR reactions were performed containing 0.5 µL of forward and 0.5 µL of reverse primers (10 µM concentration), 10 µL 5PRIME HotMasterMix (2.5x, Quanta-Bio, Beverly, MA, USA), 13 µL DNase-free water, and 1 µL DNA or cDNA template. For 515F-Y/926R primers, the thermal cycling conditions were as follows: initial denaturing at 95 °C for 180 s; 28 cycles of 95 °C for 45 s, 50 °C for 45 s, and 68 °C for 90 s; a final elongation step at 68 °C for 300 s; and refrigeration at 4 °C until removal and storage. For mcrA_F/mcrA_R primers, the thermal cycling conditions were as follows: initial denaturing at 95 °C for 120 s; 35 cycles of 95 °C for 60 s, 50 °C for 60 s, and 72 °C for 60 s; a final elongation step at 72 °C for 300 s; and refrigeration at 4 °C until removal and storage. The 16S rRNA genes and mcrA genes were successfully amplified from all 45 sediment horizons, while 16S rRNA was successfully amplified from 42 of 45 sediment horizons, and mcrA transcripts were successfully amplified from 11 of 45 sediment horizons (Table S4).
In the second step, Illumina adaptors, barcodes, and indices were added to the amplicons. The same PCR reaction mix was used with custom primers targeting the primer extension in the first PCR. The thermal cycling conditions were as follows: initial denaturing at 95 °C for 180 s; 8 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s; a final elongation step at 72 °C for 300 s; and refrigeration at 4 °C until removal and storage. Amplicons were cleaned with 0.7x AMPure XP magnetic beads (Beckman Coulter, Brea, CA, USA), pooled, and quantified before being sent to the UC Davis DNA Technologies Core Facility (Davis, CA, USA) for Illumina MiSeq 2×250 base pair (bp) (16S rRNA) or 2x300 bp (mcrA) sequencing. Ten 16S rRNA and four mcrA samples were randomly chosen for duplicate amplification. The average weighted UniFrac distance between duplicate 16S rRNA samples was 0.067. Negative (molecular grade water) and positive (mock communities of known composition) controls were processed and sequenced in parallel with the samples. Lack of DNA contamination in the RNA extracts was confirmed by processing RNA extracts (without reverse transcription) in parallel and seeing no visible amplification on a gel after the second PCR.
2.6 Phylogenetic analysis of 16S rRNA and mcrA genes and transcripts
Demultiplexed sequences were trimmed with cutadapt (v. 2.10; Martin, 2011) and then filtered and processed using the R (v. 4.2.1) package DADA2 (v. 1.26.0; Callahan et al., 2016). Reads were trimmed to 216 (16S rRNA) or 260 and 230 (mcrA; forward and reverse reads, respectively) base pairs, with those containing more than two expected sequencing errors removed. Amplicon sequence variants (ASVs) were then inferred from filtered and merged reads. The majority of paired ends were merged for both genes (an average of 76 % for 16S rRNA reads in non-incubated samples and 95 % of mcrA reads), and the overlap length was roughly 30 bp for 16S rRNA and 50 bp for mcrA. Phylogenetic classification of 16S rRNA ASVs was based on the SILVA SSU database (v. 132; Quast et al., 2013). Classification of mcrA ASVs was determined manually based on placement on a reference tree (detailed below). On average, 937 16S rRNA reads were recovered per blank sample, while 36 372 16S rRNA genes and 4644 16S rRNA reads were recovered per in situ sample. The minimum number of reads recovered per 16S rRNA gene sample (DNA) was 7377, while the minimum number of reads recovered per 16S rRNA sample (RNA) was 71. On average, 4 mcrA reads were recovered per blank sample, while 46 941 mcrA gene and 11 066 mcrA transcript reads were recovered per in situ sample.
2.7 Classification of mcrA ASVs
To classify mcrA sequences, the tool EPA-ng (v 0.3.8) was used to place the ASVs onto a manually compiled reference tree. Reference tree sequences included published sequences from cultured methanogens or ANME archaea, as well as six of the ASVs themselves (those that represented > 10 % of any sample's total sequences and had no cultured match above 90 % similarity in NCBI and would therefore be represented poorly on the reference tree). Reference sequences were aligned with MAFFT (v. 7.490) and incorporated into a RAxML (v. 8.2.12) best-scoring maximum likelihood (ML) reference tree with the GTR+G+I substitution model and 100 bootstraps. After ASV placement with EPA-ng, relative abundances of those ASVs in each sample were displayed in a heatmap using ggtree (v. 3.6.2). Relative abundances of ASVs placed on internal tree nodes were divided among tip nodes associated with that internal node.
2.8 Sequence analysis and statistical methods
Non-metric multidimensional scaling (NMDS) of microbial communities was carried out based on the weighted UniFrac distance metric (Lozupone et al., 2007) using the R package vegan (v. 2.5-7; Oksanen et al., 2020). Analysis of similarity (ANOSIM) was used to determine the significance of microbial community differences between groups of samples, also based on the weighted UniFrac distance metric. The R package DESeq2 (v. 1.38.3; Love et al., 2014) was used to test whether ASVs were significantly enriched in abundance with time (0 months vs. 6 months) across Clam Field seep incubations. Raw data from four US Atlantic Margin (USAM) seep sites (characterized in Semler et al. 2022) were simultaneously processed using the same packages for comparison with Monterey Bay sites.
2.9 Droplet digital PCR
Droplet digital PCR (ddPCR) was used to quantify abundances of mcrA genes and transcripts in Seep and Background-5m cores from Clam Field and Extrovert Cliff using the primer pair mcrA_F/mcrA_R (Luton et al., 2002; Dekas et al., 2016). For comparison, mcrA genes and transcripts were also quantified in a seep and a background core from an alternative seep site (New England seep) on the northern US Atlantic Margin; 25 µL PCR reactions were performed containing 1 µL of forward and 1 µL of reverse primers (5 µM concentration), 12.5 µL EvaGreen Supermix (2x, Bio-Rad Laboratories, Hercules, CA, USA), 9 µL DNase-free water, 0.5 µL bovine serum albumin (2.5 µg µL−1), and 1 µL diluted DNA or cDNA template. Droplets were generated on a QX200 Droplet Generator (Bio-Rad) at the Stanford Functional Genomics Facility (Stanford, CA, USA) using droplet generation oil for EvaGreen (Bio-Rad). Thermal cycling was performed immediately afterward on a C1000 Touch Thermal Cycler, with thermal cycling conditions as follows: initial denaturing at 95 °C for 300 s; 45 cycles of 95 °C for 60 s, 52 °C for 90 s, and 72 °C for 75 s; signal stabilization steps at 4 °C for 300 s and 90 °C for 300 s; and a final 10 °C hold overnight. The overall ramp rate was set at 1 °C s−1.
Droplets were read with the QX200 Droplet Reader (Bio-Rad). Threshold fluorescence values were initially inspected using QuantaSoft software (Bio-Rad), but the values were later adjusted using the minimum density clustering method, which better separated droplet clusters upon manual inspection. Amplicon copy numbers per well were then converted to copies per gram of dry sediment. Technical replicates were run for a randomly selected half of the samples to confirm the precision of our assay. Results of the replicate runs are shown in Fig. S1 in the Supplement.
2.10 DAPI staining and microscopy
To visualize putative ANME aggregates under the microscope, cells were fixed on board in 4 % paraformaldehyde according to the protocol of Dekas et al. (2009). Fixed sediment was diluted 1:20 in phosphate-buffered saline (PBS), sonicated 3×10 s at an amplitude of 30 on a Q500 sonicator (Qsonica, Newtown, CT, USA), and floated on top of a preestablished Percoll–PBS gradient (protocol described in Orphan et al., 2002, and Dekas and Orphan, 2011). In total, a 1 mL sonicated sample was added to 9 mL Percoll–PBS. After floating the sample, the mixture was centrifuged at 4780 rpm for 15 min at 4 °C. The supernatant was then filtered through a 25 mm 3 µm polycarbonate filter (Millipore, no. TSTP02500) backed with a glass microfiber filter (Cytiva Whatman, GF/F, no. 1825-025) under low (< 5 psi) vacuum and washed with 1 mL PBS and then 1 mL 100 % EtOH. Each filter was sectioned with a razor blade and stained with 4′,6-diamidino-2-phenylindole (DAPI; no. D9542, Sigma-Aldrich, Darmstadt, Germany) before visualization on an inverted fluorescence microscope (Nikon Eclipse Ti). The number of aggregates in 100 random fields of view at 400× magnification was counted, corresponding to a detection limit (< 1 aggregate per 100 fields of view) of 8.58×104 aggregates (g−1 dry sediment).
2.11 Methane diffusive flux calculations
The amount of methane potentially diffusing out of sediments in the absence of biological consumption was estimated according to Fick's laws of diffusion (Boudreau, 1997). Equation (1) calculates the diffusion rate (J) in mmol m−2 yr−1:
where ϕ is the sediment porosity, Ds is the sediment diffusion coefficient (in m2 yr−1), and ddz is the methane concentration gradient over sediment depth (in mmol m−3 m−1). The sediment diffusion coefficient, Ds, can be calculated via Eq. (2):
where cm2 s−1 (the initial diffusion coefficient of methane at 20 °C; Boudreau, 1997), n=3 (the lithology factor of silty clay), and ϕ is the sediment porosity.
3.1 Geochemical environment
Both cold seep sites contained moderate to high concentrations of porewater methane in all cores collected within the putative seep boundaries (i.e., both the Seep and Seep-Edge cores; Fig. 2b). Methane concentrations were 2 orders of magnitude higher at Extrovert Cliff (ranging from ∼ 3000 to ∼6400 µM) than at Clam Field (ranging from 25 to 183 µM), but even Clam Field contained methane concentrations roughly 4 times higher than the surrounding background sediment. At Clam Field, methane concentrations increased with depth in the sediment; methane concentrations at Extrovert Cliff, in contrast, showed no trend with depth but were consistently high throughout the sediment core. At both sites, methane concentrations in background sediments were measurable and ranged from 21 to 56 µM.
Sulfide concentrations at both cold seep sites were also elevated within the seep boundaries (Fig. 2b). At Extrovert Cliff, sulfide concentrations were elevated at all sediment depths (ranging from 2.7 to 12.7 mM), with no depth trend. At Clam Field, sulfide concentrations were below detection at the surface and increased with sediment depth (up to 11.2 mM).
With sediment depth, δ13C–DIC decreased at Clam Field (Fig. 2b) from −3.1 ‰ to −17.7 ‰, with the lowest values found in the deepest depths (Fig. 2). At Extrovert Cliff, δ13C–DIC was consistent with sediment depth (from −9.8 ‰ to −11 ‰), though at both sites, δ13C–DIC was more negative in cores collected within the seep than in cores collected in background sediments (from −0.1 ‰ to −4.9 ‰).
3.2 Quantification of mcrA genes and transcripts
To evaluate methane cycling both on and off each cold seep and to compare those results with methane and sulfide concentrations, we quantified mcrA genes and transcripts with droplet digital PCR (ddPCR). mcrA encodes the alpha subunit of methyl coenzyme-M reductase and is a marker gene for both methanogens and anaerobic methanotrophs (Luton et al., 2002; Hallam et al., 2003; Krüger et al., 2003); therefore, quantification of the gene cannot distinguish between the two metabolisms. mcrA gene copies (g−1 dry sediment) were highest in Extrovert Cliff seep sediments and peaked between 2.5–5 cmbsf at 5.3×107 copies (g−1 dry sediment) – roughly the same sediment depth where methane concentrations began to decrease up the core (Fig. 3). Gene copies of mcrA were an order of magnitude less abundant at Clam Field seep – peaking in the uppermost sediment horizons at concentrations of 4.2×106 copies (g−1 dry sediment). However, gene copy numbers at Clam Field seep were still above the average of 7.4×105 mcrA gene copies (g−1 dry sediment) in background sediments from either site. Transcript copies of mcrA (per g dry sediment) were elevated in Extrovert Cliff seep sediments compared to background sediments (by a factor of 2) but were equivalent between Clam Field seep and background sediments.
3.3 Community composition at Monterey Bay cold seeps via 16S rRNA gene and 16S rRNA amplicon sequencing
To characterize the microbial communities at these sites, we performed 16S rRNA gene and 16S rRNA amplicon sequencing in the seep (Seep and Seep-Edge) and background samples (5 and 100 m from seepage) from both sites. In the 21 seep samples, we recovered 6402 unique 16S rRNA ASVs. The most abundant of these were members of the phyla Bacteroidota, Campylobacterota, Desulfobacterota, Chloroflexi, Proteobacteria (mainly Gammaproteobacteria), and Verrucomicrobiota. The relative abundances of Campylobacteria and Chloroflexi tended to increase with sediment depth in a core (by an average of 141 % and 422 %, respectively), while the relative abundance of Bacteroidota decreased (by an average of 53.4 %) with sediment depth (Fig. 4a). Across 24 background samples, we recovered 12 731 unique 16S rRNA ASVs. Microbial communities in the background sediments from both sites were similar to one another at the phylum level (ANOSIM: p value = 0.066) with Bacteroidota, Chloroflexi, Desulfobacterota, and Proteobacteria as the most abundant groups (Fig. 4a). There was no significant difference between microbial communities in Background-5m (5 m from seepage) and Background-100m cores (100 m from seepage) – neither at the phylum level (ANOSIM: p value = 0.102) nor at the ASV level (ANOSIM: p value = 0.084). Consistent with the typical “seep microbiome” (Ruff et al., 2015; Semler et al., 2022), Caldatribacteriota, Campylobacterota, and Verrucomicrobiota were enriched in seep cores in comparison to background cores (> 3 times greater) at both sites, while Desulfobacterota and Proteobacteria (mainly Gammaproteobacteria) had particularly high relative abundances in background cores in comparison to seep cores (roughly 2 times greater; Fig. 4a). While relative potential activity of many groups (as inferred by the presence of 16S rRNA (cDNA)) mirrored many of the trends seen in the DNA, reads from several phyla, including Halobacterota and Desulfobacterota, were more than twice as abundant in the cDNA fraction of the seep core at Extrovert Cliff; Halobacterota was 40 times more abundant in the cDNA analysis of the seep core at Extrovert Cliff.
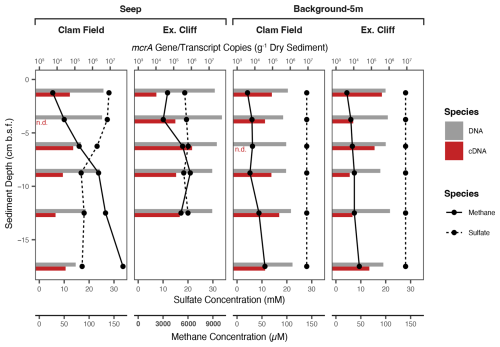
Figure 3Methane and calculated sulfate (28 mM – sulfide) concentration (lines) and mcrA gene (DNA) and transcript (cDNA) concentration (bars) with sediment depth in Seep and Background-5m cores from both study sites. (mcrA gene and transcript concentrations were not measured in Seep-Edge nor Background-100m cores.) Note the difference in x-axis values for methane concentration in the Extrovert Cliff Seep panel (axes labels in bold). n.d. – measured, but not detected in a given sample.
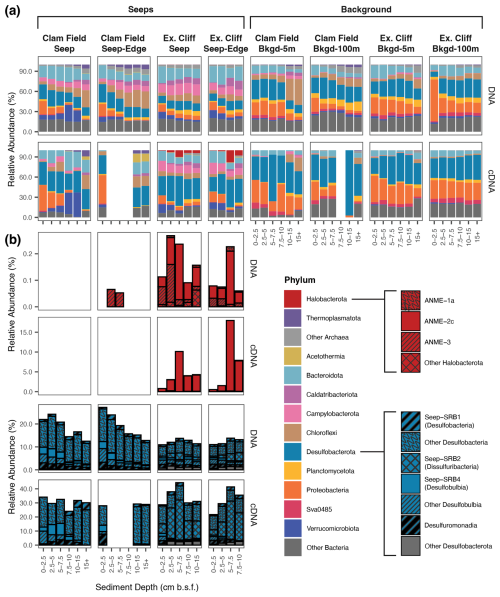
Figure 4Relative abundance (%) of Archaea and Bacteria phyla (a) and of methane-cycling and sulfate-reducing subgroups within Halobacterota and Desulfobacterota, respectively (b), across sampled sediment horizons from Seep, Seep-Edge, Background-5m, and Background-100m cores, as inferred by 16S rRNA gene (DNA) and 16S rRNA (cDNA) sequencing. Boxes surround each individual core, with sediment depth in each core increasing from left to right; 16S rRNA was not successfully amplified from blank samples in (a). Note the variable relative abundance percentages on the y axis.
Halobacterota – the phylum containing the majority of methanogenic and anaerobic methanotrophic taxa (including ANME archaea) – were present in very low relative abundances in the 16S rRNA gene dataset. At Clam Field, these organisms were not detected in seep or background sediments, with the exception of extremely low (< 0.1 %) abundances of ANME-3 in the two upper sediment horizons of the Seep-Edge core (Fig. 4b). No Halobacterota reads were detected in cDNA at Clam Field. At Extrovert Cliff, all seep sediment horizons contained low levels of Halobacterota (< 0.3 % of the community), with the majority assigned to ANME-2c or ANME-3. A small number of methanogenic Methanomicrobia were also present in the lowest sediment depths of the Extrovert Cliff Seep core. While Halobacterota were not highly abundant at Extrovert Cliff, their relative potential activity was high – up to ∼ 18 % of total community cDNA in some cores. This transcriptional activity was almost entirely associated with ANME-2c and was highest in the 5–7.5 cmbsf depth horizons, consistent with the location of the peak in mcrA transcripts in the ddPCR data. Relative abundance of rRNA is a not precise proxy for relative activity between taxa, but it can indicate which taxa are likely translationally active (Blazewicz et al., 2013). We refer to the detection of rRNA as reflecting “potential” activity to emphasize the limitations of this proxy.
Desulfobacterota – the phylum comprising known ANME syntrophs as well as other free-living sulfate reducers common at cold seeps – comprised roughly 20 % of the Clam Field microbial community and roughly 10 % of the Extrovert Cliff microbial community (Fig. 4b). The most common subgroups within the Desulfobacterota were Desulfobacteria (including Seep-SRB1), Seep-SRB2 (at Extrovert Cliff only), and Seep-SRB4 (particularly at Clam Field). Seep-SRB1, the group containing many known obligate ANME syntrophs (Knittel et al., 2003; Schreiber et al., 2010; Skennerton et al., 2017; Metcalfe et al., 2021), were found in low abundances at both seep sites, ranging from 0.70 %–2.04 % of sequences in Clam Field seep cores and from 0.73 %–1.26 % of sequences at Extrovert Cliff. Also present – particularly at Clam Field – were Desulfomonadia, a group associated with sulfur- and iron-reducing capabilities (Ravenschlag et al., 1999; Wunder et al., 2021). At Clam Field, the cDNA profiles of Desulfobacterota generally mirrored the DNA profiles, with Desulfobacteria dominating the reads overall, and Desulfobulbia, Seep-SRB4, and Seep-SRB1 showing relative potential activity peaks in the top, middle, and bottom portions of the cores, respectively. At Extrovert Cliff, Desulfobacterota comprised roughly 3 times more of the cDNA reads than they did for the DNA, mirroring the high relative potential activity observed for the Halobacteria there. The increase was largely due to Seep-SRB2, which alone comprised up to 30 % of the cDNA reads in the 5–7.5 cmbsf depth horizon in both cores. This is consistent with the previously described association of members of ANME-2c and Seep-SRB2 (Krukenberg et al., 2018; Kleindienst et al., 2012) and suggests active anaerobic methanotrophy by these groups at this depth.
3.4 Community composition at Monterey Bay cold seeps via mcrA amplicon sequencing
To provide greater insight into the diversity of methane-cycling microorganisms at these sites, and specifically increase our ability to detect low-abundance methanotrophs, if present, we sequenced mcrA genes at both sites. Sequencing the mcrA gene specifically increased our detection limit of methane-cycling organisms by nearly 3 orders of magnitude, as we recovered on average 46 941 reads per sample using mcrA gene sequencing versus 61 reads of putative methane-cycling organisms per sample with 16S rRNA gene sequencing. Within Extrovert Cliff seep samples, the mcrA gene results were generally consistent with the 16S rRNA results, with mcrA sequences affiliated with ANME-3 detected in shallow-depth horizons and ANME-2c and methanogenic archaea appearing deeper in the cores. Interestingly, within the two seep cores at Extrovert Cliff (both Seep and Seep-Edge), a single ASV affiliated with the ANME-2c group comprised ∼ 50 % of the total mcrA reads, peaking between 5 and 10 cmbsf (Fig. 5). Similar observations of local dominance of individual ANME ASVs had been made at other seep sites previously (Semler et al., 2022).
The mcrA sequences from Clam Field revealed a more nuanced perspective of methane cycling than the 16S rRNA sequences, with seep samples containing ANME-1 as well as a variety of putatively methanogenic archaea. Notably, most of these, including the ANME-1 reads, were more relatively abundant in the background samples at Clam Field than the seep samples, indicating they were not enriched by increasing methane concentrations. The one exception was ASV.2, which was the most abundant ASV at Clam Field seeps (alone comprising roughly 45 % of seep ASVs) and was almost absent from background sediments (0.1 % of background ASVs). ASV.2 has no cultured relative above 90 % similarity in NCBI but clusters with sequences from Methanohalophilus halophilus and Methanomethylovorans hollandica, anaerobes involved in methylotrophic methanogenesis (Lomans et al., 1999). The most highly abundant ASV (ASV.3) in Clam Field and Extrovert Cliff background sediments is most closely related to strain MO-MCD, also a methylotrophic methanogen, belonging to the genus Methanococcoides (Singh et al., 2005). Although this organism is also present within the Clam Field seep, the clear difference in distribution between ASV.2 and ASV.3 across the seep boundary highlights niche separation between putatively similar methylotrophic methanogens at these sites. In general, while ANME-affiliated sequences dominated the mcrA dataset from seep samples at Extrovert Cliff (85.6 % of mcrA reads), reads associated with methylotrophic methanogens dominated both Clam Field (> 80 % of mcrA reads; only 5.6 % were affiliated with ANME archaea) and the background sediments from each site (> 75 % and > 60 % of reads from background cores at Extrovert Cliff and Clam Field, respectively). We also attempted to sequence mcrA transcripts at Clam Field (Table S4), but mcrA expression was below the limit of detection (no visible amplification of the cDNA on a gel).
By multiplying the mcrA copy number in samples (as determined by ddPCR) by the proportion of ANME-affiliated mcrA reads in that sample (as determined by the amplicon sequencing analysis using the same primer set) – and by assuming a single mcrA gene copy per cell – ANME cell numbers in each sample were estimated (Table S5). In total, ANME archaea were not enriched in Clam Field seep sediments (4.5×102 to 8.2×104 ANME cells g−1 dry sediment) relative to background sediments (8.3×103 to 6.9×105 ANME cells g−1 dry sediment), and ANME mcrA genes at Clam Field were attributed primarily to ANME-1 (Fig. 5). ANME cell numbers were estimated to be roughly 4 orders of magnitude higher at Extrovert Cliff seep (8.2×106 to 4.7×107 ANME cells g−1 dry sediment) than at Clam Field seep (Table S5), with ANME mcrA genes attributed primarily to ANME-2c and ANME-3 (Fig. 5). The assumption of one mcrA copy per cell is imperfect, but only one or two copies of mcrA have been found in sequenced methanogen genomes (Alvarado et al., 2014), including that of Methanosarcina mazei – a close relative of ANME-2 (Deppenmeier et al., 2002; Nunoura et al., 2006). The ANME-1 genome also contains a single operon for MCR (Krukenberg et al., 2018; Chadwick et al., 2022; Laso-Pérez et al., 2023). As a result, the difference of roughly 4 orders of magnitude in ANME cell numbers between Clam Field and Extrovert Cliff would not be significantly affected by likely variation in ANME mcrA copy number.
3.5 Visualization of ANME aggregates
To visualize and quantify potential aggregates of ANME archaea and sulfate-reducing bacteria from these seeps, we examined DAPI-stained cells from methane and sulfate replete sediments at Clam Field (7.5–10 cmbsf) and Extrovert Cliff (5–7.5 cmbsf) seeps. ANME-2 and ANME-3 typically form tight associations with their syntrophic partners, resulting in cellular aggregates of characteristic morphologies (Boetius et al., 2000; Orphan et al., 2002). While not a taxa-specific assay, quantifying DAPI-stained cell aggregates typical of the ANME–SRB morphology provides independent support for the presence or absence of aggregate-forming ANME-2 and ANME-3 archaea in the molecular data and has been used previously to approximate potential ANME–SRB aggregate abundances (Dekas et al., 2009; Zhang et al., 2011). In sediments from Extrovert Cliff, where ANME-2 and ANME-3 were found in the DNA (0.2 % of 16S rRNA gene reads) and cDNA analyses (10.1 % of 16S rRNA reads), we detected putative ANME cell aggregates at a concentration of 4.35×106 aggregates (g−1 dry sediment) (Fig. S2). Considering ANME aggregates can contain anywhere from tens to thousands of cells, this value is roughly consistent with the estimate of ANME density derived from the molecular data. Cell aggregates were not found at Clam Field, consistent with the lack of ANME-2 and dearth of ANME-3 sequences detected at this site. Our aggregate detection limit corresponded to 8.58×104 aggregates (g−1 dry sediment).
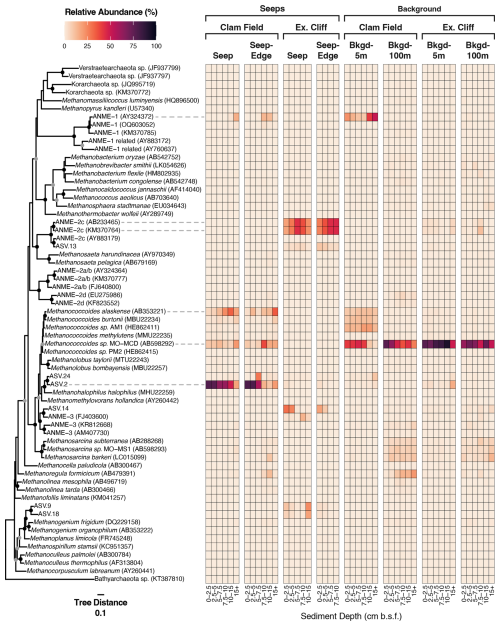
Figure 5Relative abundance (%) of all ASVs inferred from mcrA gene sequencing (DNA) at Clam Field and Extrovert Cliff and their distribution on an mcrA reference tree. Sequences on the tree were aligned with MAFFT (v. 7.490) and incorporated into a RAxML (v. 8.2.12) best-scoring ML reference tree with 100 bootstraps. Heatmap values were calculated by adding the relative abundance of all ASVs assigned to each tip node by EPA-ng (v. 0.3.8). (ASVs assigned to internal nodes were evenly divided among all tip nodes associated with that internal node. See methods.) Tree was rooted with a member of Bathyarchaeota (KT387810). The scale bar indicates the average number of amino acid substitutions per site, and the filled circles signal nodes with at least 60 % (grey) or 80 % (black) bootstrap support.
3.6 Changes in microbial community and geochemistry with methane addition
To investigate whether sulfate-coupled methane oxidation was occurring at Clam Field and determine whether canonical ANME archaea could be enriched there, Clam Field sediments were incubated under varying concentrations of methane. Sulfide concentrations increased continuously over 6 months in sediment from the seep center, up to 10 mM, indicating active sulfate reduction (Fig. S3). A smaller increase (∼ 1–2 mM) was observed in sediments from the Seep-Edge core. However, there was not a statistically significant difference in sulfide production with or without methane in either core, indicating that sulfate reduction was not methane-dependent. We therefore did not see evidence of sulfate-coupled methane oxidation in these sediments, in contrast to previous observations at other seeps (e.g., Hydrate Ridge (Nauhaus et al., 2002, 2007), Eel River basin (Dekas et al., 2009), and the Costa Rica margin (Dekas et al., 2014)).
Consistent with a lack of methane oxidation, ANME archaea were not enriched during the 6-month incubation, as assessed by 16S rRNA sequencing (DNA and cDNA; Fig. S4). Taxa that were significantly enriched after 6 months with methane included several ASVs within the Desulfobacterota, specifically Desulfovibrio, Desulfobulbus, Desulfuromonas, and Seep-SRB4 (Fig. S5a). A putatively methylotrophic methanogen in the genus Methanococcoides was also significantly enriched relative to the pre-incubation community. Aerobic sulfide-oxidizing groups, including Candidatus Maribeggiatoa, Colwellia, and Thiomargarita, significantly decreased in abundance over 6 months with methane. Without methane, none of the above taxa were significantly enriched or unenriched after 6 months of incubation time (Fig. S5b). A single Verrucomicrobia ASV had significantly increased in abundance, and a single Proteobacteria ASV (from the family Beggiatoaceae) had significantly decreased in abundance. Furthermore, there were no ASVs that were significantly enriched or under-enriched when comparing incubations with and without methane after 6 months.
3.7 Comparison to previously characterized sites
To directly compare Monterey Bay seep microbial communities with canonical seep communities, we compared them to those of four seeps along the US Atlantic Margin (USAM), which were sampled and sequenced with the same methodologies. First, we compared the 16S rRNA gene profiles of the Monterey Bay seeps to those of the USAM seeps using non-metric multidimensional scaling (NMDS). Monterey Bay seep samples formed a separate cluster from the Atlantic samples (Fig. 6a), and communities from Extrovert Cliff were nested within those from Clam Field. While the primary axis of the NMDS plot was defined by sediment depth (Fig. 6b), the secondary axis was defined by geographic region. When including background samples from all sites, background samples from Monterey Bay clustered together within background sediment communities sequenced from the USAM (Fig. 6c). As in the seep-only community comparison, the primary axis of the plot was defined by sediment depth (Fig. 6d), though the secondary axis was instead defined by environment type (seep vs. background), rather than sampling region.
We also measured mcrA concentrations via ddPCR at New England seep – one of the four USAM seeps – to better contextualize the trends in mcrA abundance observed within Monterey Bay. At the New England seep, where ANME archaea had represented 13.4 % of the microbial community in 16S rRNA gene surveys on average (Semler et al., 2022), we found that mcrA gene copy numbers reached 2.5×108 copies (g−1 dry sediment) at the deepest sediment depths, outnumbering those at both Extrovert Cliff and Clam Field by 1 and 2 orders of magnitude, respectively (Fig. S1). However, mcrA transcript copy numbers at the New England seep were comparable to those at Extrovert Cliff, though they peaked at lower sediment depths (15+ cmbsf).
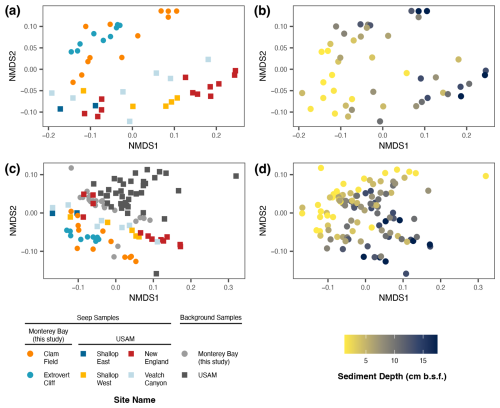
Figure 6Non-metric multi-dimensional scaling (NMDS) of seep samples only (a–b) and of both seep and background samples (c–d) from Monterey Bay (Clam Field and Extrovert Cliff; this study), as well as from US Atlantic Margin seep and background samples (Semler et al., 2022). Samples colored by site (a, c) and by sediment depth (b, d). NMDS was based on a weighted UniFrac distance metric and was inferred by 16S rRNA gene sequencing.
We also chose two USAM seeps (New England and Shallop Canyon East) at which to visualize putative ANME aggregates. Again, fixed sediments from methane and sulfate replete depths (0–3 cmbsf) at these two sites were examined under an epifluorescence microscope with a DAPI stain. In the chosen horizons, ANME archaea had comprised 4.8 % (New England) and 0.5 % (Shallop Canyon East) of the microbial community in 16S rRNA gene surveys (Semler et al., 2022). Consistent with their higher relative abundances in the molecular data compared to the Monterey seeps, we discovered higher concentrations of putative aggregates in these sediments by microscopy: 1.15×107 and 1.34×106 aggregates (g−1 dry sediment), respectively.
3.8 Methane diffusive flux
The porewater methane concentrations at Clam Field decreased approximately linearly up the core, indicative of diffusive flow between the methane-rich sediments and the methane-poor overlying water column. This is in contrast to a concave uptrend, which is indicative of biological methane consumption and therefore AOM (Reeburgh, 1976; Martens and Berner, 1977; Ward et al., 1987). The lack of methane-dependent sulfide production in sediments from this site, as well as the scarcity of ANME archaea, further supports a lack of significant biological methane consumption. Although we cannot exclude the possibility of low levels of biological oxidation, if we assume diffusion is the only mechanism removing methane from seep sediment, we can calculate the diffusive flux to determine the potential amount of methane released to the water column at this site. The methane concentration gradient over sediment depth (ddz) was determined from the average slope of the linear relationship between methane concentration and sediment depth (Fig. S6). Calculated fluxes were 17.8 and 18.5 mmol methane m−2 yr−1 for the Clam Field Seep and Seep-Edge cores, respectively (Table S6).
The scarcity of 16S rRNA and mcrA sequences belonging to ANME archaea in the Monterey Bay seeps, and particularly at Clam Field, is unexpected and intriguing. In typical methane seep sediments, a characteristic suite of microbial community members – termed the “seep microbiome” – are present and remarkably consistent despite geographical separation (Ruff et al., 2015; Semler et al., 2022). This community is primarily composed of ANME archaea (including ANME-1a, ANME-1ab; ANME-2a, ANME-2b, ANME-2c, ANME-2d; and ANME-3) and SRB partners (including members of the Seep-SRB1 and Seep-SRB2 in the Desulfobacterales, Desulfobulbus and Seep-SRB4 in the Desulfobulbales, and thermophilic Hot-Seep1). Sulfide-oxidizing and aerobic methane-oxidizing Gammaproteobacteria, as well as the putatively methanotrophic JS1 lineage of Atribacterota, are also abundant at seeps. While Monterey Bay is a somewhat geographically isolated environment, seep communities are thought to assemble primarily deterministically – based on local geochemical variables like methane, sulfate/sulfide, and ammonium concentrations – and do not appear to be limited by dispersal (Semler et al., 2022). It is therefore unlikely that the low levels of ANME archaea in Monterey Bay are a result of dispersal limitation.
At Extrovert Cliff, ANME–SRB consortia members are not highly abundant (< 0.3 % ANME archaea in any given sample), but they have high-potential relative activity, and active methane oxidation is likely. ANME archaea typically comprise a large portion of seep communities, for instance, 13.4 % at New England Seep on the USAM. Methane concentrations are extremely high at Extrovert Cliff, and most geochemical variables show no obvious depth-related trend in the top 15 cm (Fig. 2b). Other studies have noted the high, though temporally variable, fluid flux here (LaBonte et al., 2007; Füri et al., 2009), which potentially homogenizes the upper sediment layers and masks depth-related trends. mcrA gene copy numbers, which reached a maximum of 5.3×107 copies (g−1 dry sediment), were moderately low as compared to concentrations at other seeps, for instance, 109 mcrA gene copies (g−1 wet sediment) in a seep in the Nankai Trough (Nunoura et al., 2006), 108 copies (g−1 wet sediment) in a seep in the Kumano Knoll (Miyazaki et al., 2009), 107 copies (g−1 wet sediment) in a seep core from the South China Sea (Niu et al., 2017), and 2.5×108 copies (g−1 dry sediment) measured in this study for USAM seep sediment (Fig. S1). However, ANME-2c comprises > 10 % of 16S rRNA sequences (RNA fraction) in some Extrovert Cliff seep samples, coincident with likely syntrophic Seep-SRB2 comprising > 20 % (Fig. 4b). At Extrovert Cliff, it is therefore likely that a small number of extremely active ANME-SRB consortia (particularly ANME-2c and Seep-SRB2) perform substantial methane oxidation.
At Clam Field, ANME archaea were both low in abundance and low in potential relative activity, as ANME sequences were not detected in the cDNA from this site at all. While extraction, sequencing, and/or primer biases can cause artificial underestimates, our successful recovery of ANME 16S rRNA and mcrA genes/transcripts at Extrovert Cliff here and previously at US Atlantic Margin sites using the same protocols (Semler et al., 2022) suggest a technical artifact is unlikely. Furthermore, our use of both 16S rRNA and mcrA primer sets, as well as microscopy to detect ANME-typical morphologies, reduces the possibility that a diverged ANME lineage was overlooked by a particular primer set. Though 5.6 % of mcrA genes at Clam Field were affiliated with ANME-1, mcrA gene concentrations reached a maximum of 4.2×106 copies (g−1 dry sediment) throughout the seep, indicating that the overall abundance of ANME-1 was low. Additionally, the relative and inferred absolute abundances of the ANME-1 ASVs found within Clam Field seeps were actually higher in background cores, consistent with the possibility that this group is not exclusively methanotrophic (Lloyd et al., 2011; Kevorkian et al., 2021). While it is possible that biological methane oxidation is being performed by organisms not identified as methane oxidizers at this site – with mcrA ASV.2 the most likely candidate – the lack of evidence for active methane oxidation in the porewater methane profile and the sediment incubations over time makes this possibility unlikely.
A lack of ANME archaea is unexpected in methane-rich marine sediments. Methane concentrations at Clam Field are elevated far above background concentrations (up to 183 µM), and although they are lower than at Extrovert Cliff, they are higher than at three of the four USAM sites where ANME archaea are abundant (Semler et al., 2022). A dearth of ANME sequences in methanic sediments has been observed at just a few other methane-rich sites (Goffredi et al., 2008; Ruff et al., 2019; Thurber et al., 2020), but these observations have been made only in recently perturbed environments such as whale falls or young, newly emerged (< 1 year) seeps. Clam Field, in contrast, has a documented history of elevated methane concentrations and of characteristic seep macrofauna going back nearly 3 decades (Barry et al., 1996, 1997; Lorenson et al., 2002). Despite (1) the lack of ANME dispersal limitation globally (Ruff et al., 2015; Semler et al., 2022), (2) the success of these protocols at detecting ANME archaea at other seeps, and (3) the current and historically high concentrations of methane and sulfide at this site, we do not detect an active ANME population at Clam Field. Therefore, an external factor likely limits the presence of ANME archaea.
The location of Clam Field seep may be the key to its unique microbial assemblage, as historical data indicate that this site experiences non-methane hydrocarbon inputs in addition to methane. The site is situated within the Monterey Bay fault zone and, in particular, within sediments where the hydrocarbon-rich Monterey Formation crops out. Lorenson et al. (2002) recovered two oil-stained rocks at this site, roughly 20 cmbsf, which were later measured to contain n-chain alkanes from C16–C35, along with a complex mixture of unresolved hydrocarbons. Orange et al. (1999) and Stakes et al. (1999) described an authigenic carbonate sample with a distinct aromatic hydrocarbon odor. Furthermore, methane here has a distinct thermogenic fingerprint; reported 13C–CH4 values were heavier than at other measured cold seep sites – from −50 ‰ to −55 ‰ rather than from −70 ‰ to −85 ‰ (Orange et al., 1999; Lorenson et al., 2002).
The δ13C–DIC concentrations we measured at Clam Field (−3 ‰ to −18 ‰) are consistent with the oxidation of non-methane hydrocarbons. The values are heavier than at a typical cold seep with active methanotrophy (typically −40 ‰ to −30 ‰ (Paull et al., 2000; Liu et al., 2020; Sauer et al., 2021), even with a thermogenic methane source (Sauer et al., 2021)), though lighter than those in background sediments at this site (0.9 ‰ to −4.9 ‰; Fig. 2b). The oxidation of non-methane hydrocarbons, which have heavier isotopic signatures (−33 ‰ to −29 ‰ for oil) than methane (Joye, 2020), leads to DIC that is less 13C-depleted, as is observed at Clam Field. However, at Extrovert Cliff, the δ13C–DIC is also only moderately depleted (−9.8 ‰ to −11 ‰), despite signs of active methanotrophs. It is possible that the oxidation of hydrocarbons is common at both sites and dominates the δ13C–DIC signal despite the co-occurrence of methane oxidation at Extrovert Cliff. But a moderate δ13C–DIC signal derived from isotopically light methane-derived DIC mixed with seawater–DIC (consistent with turbulence at a higher flux seep system) could also result in the observed intermediate values at Extrovert Cliff.
The presence of oil and other non-methane hydrocarbons has been previously documented to play a role in shaping microbial communities at seeps (Orcutt et al., 2010; Vigneron et al., 2017), and some of the lineages responsible for their oxidation are abundant in our dataset. In Gulf of Mexico seep sediments, seeps with input of non-methane hydrocarbons were typically characterized by specific Desulfobacterota lineages (Vigneron et al., 2017), which can be involved in the anaerobic oxidation of non-methane hydrocarbons with sulfate as an electron acceptor (Kleindienst et al., 2014; Vigneron et al., 2017; Joye, 2020). The Desulfococcus/Desulfosarcina (DSS) clade comprises organisms that degrade short-chain alkanes, as well as degraders of mid-chain or long-chain alkanes and alkenes, or aromatic compounds (Aeckersberg et al., 1998; Harms et al., 1999; So and Young, 1999; Meckenstock et al., 2002). Hydrocarbon degradation genes, including assA and bssA, are also found in members of the non-DSS clades Desulfobacteraceae, Syntrophobacteraceae, and Desulfatiglans (Widdel and Grundmann, 2010; Vigneron et al., 2023), and members of Desulfatiglans, Seep-SRB1d, and Seep-SRB4 have been continually overrepresented in sediments affected by seepage of non-methane hydrocarbons (Kleindienst et al., 2014; Vigneron et al., 2017). Notably, ASVs affiliated with the Desulfobacterota groups Desulfatiglans, Desulfobacteraceae, and Seep-SRB4 were the 1st and 6th, 15th, and 17th most potentially relatively active (cDNA) ASVs in Clam Field seep sediments, respectively (Table S7).
In seep sediments such as Clam Field where sulfate penetrates, sulfate reducers generally outcompete methanogens for hydrogen and acetate (Schönheit et al., 1982; Lovley and Klug, 1986; Reeburgh, 2007), leading to a local depletion in hydrogen and the possible stimulation of sulfate-dependent AOM (Hoehler et al., 1994; Lloyd et al., 2011; Kevorkian et al., 2021; Coon et al., 2023). However, certain methylated compounds like methylamines and methyl sulfides are more favorable for methylotrophic methanogens than sulfate reducers, allowing these methanogens to persist and be noncompetitive even in sediments where sulfate is not fully depleted (Oremland and Polcin, 1982; Winfrey and Ward, 1983) and where sulfate reducers are abundant. Methylated compounds can also be the degradation products of complex organic compounds (Yancey and Somero, 1980; Oremland et al., 1982; Alcolombri et al., 2015) and are likely ubiquitous in Clam Field sediments. Among mcrA-containing taxa, the dominance of methylotrophic methanogens in Clam Field seep sediments supports this possibility. mcrA ASV.2, clustering with methylotrophic methanogens Methanohalophilus halophilus and Methanomethylovorans hollandica was the most abundant at all sampled sediment depths within the Clam Field seep area, particularly at the shallowest sediment depths.
While the presence of non-methane hydrocarbons may explain the presence of particular microorganisms, it is difficult to explain the near-exclusion of ANME archaea. Compared to AOM, the oxidation of non-methane hydrocarbons provides a higher energy yield per molecule of sulfate reduced (Bowles et al., 2011; Adams et al., 2013), particularly when energy yields from AOM must be split between the two partner organisms. While coexistence between ANME archaea and other hydrocarbon oxidizers has been documented elsewhere (Orcutt et al., 2010; Vigneron et al., 2017), it is possible that in the particular conditions of Clam Field seep, coexistence is not viable. The precise mechanism of ANME exclusion – whether it be competition for essential nutrients, competition for sulfate as an electron acceptor, or inhibition by an unknown environmental factor – remains unknown.
The scarcity of known anaerobic methanotrophic populations at Clam Field has implications for estimated methane emissions from marine cold seeps. Our findings suggest a previously unknown sensitivity of ANME archaea to certain biogeochemical conditions and highlight the potential for hydrocarbon seeps without this critical methane biofilter. At Clam Field, this may result in the escape of 17.8–18.5 mmol methane m−2 yr−1. While aerobic methanotrophs in the water column can also prevent the release of methane to the atmosphere, and can thus curb its climate-warming impact, aerobic methanotrophy above seeps is poorly constrained due to potential variations in oxygen concentration, the depth of overlying water, total methane flux at the seep (which can affect the presence/size of methane bubbles), and microbial community response time to new methane inputs. While current estimates suggest that anaerobic methanotrophs in sediments oxidize 80 % of methane before it reaches the seafloor (Reeburgh, 2007), our data demonstrate large site-to-site variations in methanotrophic efficiency that are not reflected by the seep's surface expression or benthic macrofauna. Especially as new hydrocarbon seeps develop along continental margins worldwide due to warming bottom waters and dissociating methane hydrate reserves (Phrampus and Hornback, 2012; Skarke et al., 2014; Davies et al., 2024), direct observations of methanotrophs and methanotrophy will be necessary to confirm subsurface oxidation.
Nucleotide sequences from this study were deposited in the European Nucleotide Archive, project number PRJEB72125. Full sample metadata, including accession numbers, are available at https://doi.org/10.6084/m9.figshare.25769601 (Semler, 2025).
The supplement related to this article is available online at: https://doi.org/10.5194/bg-22-385-2025-supplement.
ACS and AED designed the experiments and sampling campaign; ACS collected the samples. ACS performed sequencing, geochemical measurements, and microscopy and analyzed the resulting data. ACS and AED interpreted the data together. ACS wrote the initial manuscript draft; AED reviewed and edited the manuscript.
The contact author has declared that neither of the authors has any competing interests.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
We thank the captain, crew, and science party of R/V Western Flyer MMV19, particularly Nicolette Meyer and Sebastian Sudek, as well as the pilots and engineers of ROV Doc Ricketts, for facilitating sample collection. We thank Alexandra Worden for the opportunity to collect samples on this expedition. We thank the members of the Dekas Geomicrobiology Lab for discussions and feedback, especially Steffen Buessecker for conversations about methane measurements.
Funding was provided by Stanford University, including a McGee/Levorsen Research Grant to Amanda C. Semler and through support to the Stanford Geomicrobiology Shared Laboratories Core Facility (RRID:SCR_025000).
This paper was edited by Andrew Thurber and reviewed by Cornelia U. Welte and one anonymous referee.
Adams, M. M., Hoarfrost, A. L., Bose, A., Joye, S. B., Girguis, P. R., Teske, A., and Huber, J. A.: Anaerobic oxidation of short-chain alkanes in hydrothermal sediments: potential influences on sulfur cycling and microbial diversity, Front Microbiol., 4, 110, https://doi.org/10.3389/fmicb.2013.00110, 2013.
Aeckersberg, F., Rainey, F. A., and Widdel, F.: Growth, natural relationships, cellular fatty acids and metabolic adaptation of sulfate-reducing bacteria that utilize long-chain alkanes under anoxic conditions, Arch. Microbiol., 170, 361–369, https://doi.org/10.1007/s002030050654, 1998.
Alcolombri, U., Ben-Dor, S., Feldmesser, E., Levin, Y., Tawfik, D. S., and Vardi, A.: Identification of the algal dimethyl sulfide–releasing enzyme: A missing link in the marine sulfur cycle, Science, 348, 1466–1469, https://doi.org/10.1126/science.aab1586, 2015.
Alvarado, A., Montañez-Hernández, L. E., Palacio-Molina, S. L., Oropeza-Navarro, R., Luévanos-Escareño, M. P., and Balagurusamy, N.: Microbial trophic interactions and mcrA gene expression in monitoring of anaerobic digesters, Front. Microbiol., 5, 597, https://doi.org/10.3389/fmicb.2014.00597, 2014.
Barry, J. P., Greene, H. G., Orange, D. L., Baxter, C. H., Robison, B. H., Kochevar, R. E., Nybakken, J. W., Reed, D. L., and McHugh, C. M.: Biologic and geologic characteristics of cold seeps in Monterey Bay, California, Deep-Sea Res. Pt. I, 43, 1739–1762, https://doi.org/10.1016/S0967-0637(96)00075-1, 1996.
Barry, J. P., Kochevar, R. E., and Baxter, C. H.: The influence of pore-water chemistry and physiology on the distribution of vesicomyid clams at cold seeps in Monterey Bay: Implications for patterns of chemosynthetic community organization, Limnol. Oceanogr., 42, 318–328, https://doi.org/10.4319/LO.1997.42.2.0318, 1997.
Blazewicz, S. J., Barnard, R. L., Daly, R. A., and Firestone, M. K.: Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses, ISME J., 7, 2061–2068, https://doi.org/10.1038/ismej.2013.102, 2013.
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gleeske, A., Amann, R., Jorgensen, B. B., Witte, U., and Pfannkuche, O.: A marine microbial consortium apparently mediating AOM., Nature, 407, 623–626, 2000.
Boudreau, B. P.: Diagenetic Models and Their Implementation, Springer, Berlin, https://doi.org/10.1007/97S-3-642-60421-S, 1997.
Bowles, M. W., Samarkin, V. A., Bowles, K. M., and Joye, S. B.: Weak coupling between sulfate reduction and the anaerobic oxidation of methane in methane-rich seafloor sediments during ex situ incubation, Geochim. Cosmochim. Ac., 75, 500–519, https://doi.org/10.1016/J.GCA.2010.09.043, 2011.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P.: DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 13, 581–583, https://doi.org/10.1038/nmeth.3869, 2016.
Chadwick, G. L., Skennerton, C. T., Laso-Pérez, R., Leu, A. O., Speth, D. R., Yu, H., Morgan-Lang, C., Hatzenpichler, R., Goudeau, D., Malmstrom, R., Brazelton, W. J., Woyke, T., Hallam, S. J., Tyson, G. W., Wegener, G., Boetius, A., and Orphan, V. J.: Comparative genomics reveals electron transfer and syntrophic mechanisms differentiating methanotrophic and methanogenic archaea, PLOS Biol., 20, e3001508, https://doi.org/10.1371/journal.pbio.3001508, 2022.
Clark, J.: Stratigraphy, paleontology, and geology of the central Santa Cruz Mountains, California Coast Ranges: U.S. Geological Survey Professional Paper 1168, Professional Paper, https://doi.org/10.3133/pp1168, 1981.
Cline, J. D.: Spectrophotometric determination of hydrogen sulfide in natural waters, Limnol. Oceanogr., 14, 454–458, https://doi.org/10.4319/lo.1969.14.3.0454, 1969.
Coon, G., Duesing, P., Paul, R., Baily, J., and Lloyd, K. G.: Biological methane production and accumulation under sulfate-rich conditions at Cape Lookout Bight, NC, Front Microbiol., 14, 1268361, https://doi.org/10.3389/fmicb.2023.1268361, 2023.
Davies, R. J., Yang, J., Ireland, M. T., Berndt, C., Ángel, M., Maqueda, M., and Huuse, M.: Long-distance migration and venting of methane from the base of the hydrate stability zone, Nat. Geosci., 17, 32–37, https://doi.org/10.1038/s41561-023-01333-w, 2024.
Dekas, A. E. and Orphan, V. J.: Chapter Twelve – Identification of Diazotrophic Microorganisms in Marine Sediment via Fluorescence In Situ Hybridization Coupled to Nanoscale Secondary Ion Mass Spectrometry (FISH-NanoSIMS), in: Research on Nitrification and Related Processes, Part A, edited by: Klotz, M. G., vol. 486, Academic Press, 281–305, https://doi.org/10.1016/B978-0-12-381294-0.00012-2, 2011.
Dekas, A. E., Poretsky, R. S., and Orphan, V. J.: Deep-Sea archaea fix and share nitrogen in methane-consuming microbial consortia, Science, 326, 422–426, https://doi.org/10.1126/science.1178223, 2009.
Dekas, A. E., Chadwick, G. L., Bowles, M. W., Joye, S. B., and Orphan, V. J.: Spatial distribution of nitrogen fixation in methane seep sediment and the role of the ANME archaea, Environ. Microbiol., 16, 3012–3029, https://doi.org/10.1111/1462-2920.12247, 2014.
Dekas, A. E., Connon, S. A., Chadwick, G. L., Trembath-Reichert, E., and Orphan, V. J.: Activity and interactions of methane seep microorganisms assessed by parallel transcription and FISH-NanoSIMS analyses, ISME J., 10, 678–692, https://doi.org/10.1038/ismej.2015.145, 2016.
Deppenmeier, U., Johann, A., Hartsch, T., Merkl, R., Schmitz, R. A., Martinez-Arias, R., Henne, A., Wiezer, A., Bäumer, S., Jacobi, C., Brüggemann, H., Lienard, T., Christmann, A., Bömeke, M., Steckel, S., Bhattacharyya, A., Lykidis, A., Overbeek, R., Klenk, H.-P., Gunsalus, R. P., Fritz, H.-J., and Gottschalk, G.: The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea, J. Mol. Microbiol. Biotechnol., 4, 453–461, 2002.
Fisher, C. and Childress, J.: Organic Carbon Transfer from Methanotrophic Symbionts to the Host Hydrocarbon-seep Mussel, Symbiosis, 12, 221–235, 1992.
Füri, E., Hilton, D. R., Brown, K. M., and Tryon, M. D.: Helium systematics of cold seep fluids at Monterey Bay, Geochem. Geophy. Geosy, 10, 8013, https://doi.org/10.1029/2009GC002557, 2009.
GEBCO Compilation Group: GEBCO 2021 Grid, https://doi.org/10.5285/c6612cbe-50b3-0cff-e053-6c86abc09f8f, 2021.
Girguis, P. R., Orphan, V. J., Hallam, S. J., and DeLong, E. F.: Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor, Appl. Environ. Microbiol., 69, 5472–5482, https://doi.org/10.1128/AEM.69.9.5472-5482.2003, 2003.
Girguis, P. R., Cozen, A. E., and DeLong, E. F.: Growth and population dynamics of anaerobic methane-oxidizing archaea and sulfate-reducing bacteria in a continuous-flow bioreactor, Appl. Environ. Microbiol., 71, 3725–3733, https://doi.org/10.1128/AEM.71.7.3725-3733.2005, 2005.
Goffredi, S. K., Wilpiszeski, R., Lee, R., and Orphan, V. J.: Temporal evolution of methane cycling and phylogenetic diversity of archaea in sediments from a deep-sea whale-fall in Monterey Canyon, California, ISME J., 2, 204–220, https://doi.org/10.1038/ismej.2007.103, 2008.
Greene, H. G., Yoklavich, M. M., Barry, J. P., Orange, D. L., Sullivan, D. E., and Cailliet, G. M.: Geology and related benthic habitats of Monterey canyon, central California, EOS Transactions of the American Geophysical Union Supplement, 75, 203, 1994.
Greene, H. G., Maher, N., Haehr, T. H., and Orange, D. L.: Fluid flow in the offshore Monterey Bay region, in: Late Cenozoic Fluid Seeps and Tectonics Along the San Gregorio Fault Zone in the Monterey Bay Region, California, vol. GB-76, edited by: Garrison, R. E., Aiello, I. W., and Moore, C. E., The Pacific Section American Association of Petroleum Geologists, 1–19, https://doi.org/10.32375/1999-GB76, 1999.
Hallam, S. J., Girguis, P. R., Preston, C. M., Richardson, P. M., and DeLong, E. F.: Identification of Methyl Coenzyme M Reductase A (mcrA) Genes Associated with Methane-Oxidizing Archaea, Appl. Environ. Microbiol., 69, 5483–5491, https://doi.org/10.1128/AEM.69.9.5483-5491.2003, 2003.
Harms, G., Rabus, R., and Widdel, F.: Anaerobic oxidation of the aromatic plant hydrocarbon p-cymene by newly isolated denitrifying bacteria, Arch. Microbiol., 172, 303–312, https://doi.org/10.1007/s002030050784, 1999.
Hoehler, T. M., Alperin, M. J., Albert, D. B., and Martens, C. S.: Field and laboratory studies of methane oxidation in an anoxic marine sediment: Evidence for a methanogen-sulfate reducer consortium, Global Biogeochem. Cy., 8, 451–463, https://doi.org/10.1029/94GB01800, 1994.
Joye, S. B.: The Geology and Biogeochemistry of Hydrocarbon Seeps, Annu. Rev. Earth Pl. Sci., 48, 205–231, https://doi.org/10.1146/annurev-earth-063016-020052, 2020.
Kevorkian, R. T., Callahan, S., Winstead, R., and Lloyd, K. G.: ANME-1 archaea may drive methane accumulation and removal in estuarine sediments, Environ. Microbiol. Rep., 13, 185–194, https://doi.org/10.1111/1758-2229.12926, 2021.
Kleindienst, S., Ramette, A., Amann, R., and Knittel, K.: Distribution and in situ abundance of sulfate-reducing bacteria in diverse marine hydrocarbon seep sediments, Environ. Microbiol., 14, 2689–2710, https://doi.org/10.1111/j.1462-2920.2012.02832.x, 2012.
Kleindienst, S., Herbst, F.-A., Stagars, M., von Netzer, F., von Bergen, M., Seifert, J., Peplies, J., Amann, R., Musat, F., Lueders, T., and Knittel, K.: Diverse sulfate-reducing bacteria of the Desulfosarcina/Desulfococcus clade are the key alkane degraders at marine seeps, ISME J., 8, 2029–2044, https://doi.org/10.1038/ismej.2014.51, 2014.
Knittel, K., Boetius, A., Lemke, A., Eilers, H., Lochte, K., Pfannkuche, O., Linke, P., and Amann, R.: Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrate (Cascadia margin, Oregon), Geomicrobiol. J., 20, 269–294, https://doi.org/10.1080/01490450303896, 2003.
Krüger, M., Meyerdierks, A., Glöckner, F. O., Amann, R., Widdel, F., Kube, M., Reinhardt, R., Kahnt, J., Böcher, R., Thauer, R. K., and Shima, S.: A conspicuous nickel protein in microbial mats that oxidize methane anaerobically, Nature, 426, 878–881, https://doi.org/10.1038/nature02207, 2003.
Krukenberg, V., Riedel, D., Gruber-Vodicka, H. R., Buttigieg, P. L., Tegetmeyer, H. E., Boetius, A., and Wegener, G.: Gene expression and ultrastructure of meso- and thermophilic methanotrophic consortia, Environ. Microbiol., 20, 1651–1666, https://doi.org/10.1111/1462-2920.14077, 2018.
LaBonte, A. L., Brown, K. M., and Tryon, M. D.: Monitoring periodic and episodic flow events at Monterey Bay seeps using a new optical flow meter, J. Geophys. Res.-Sol. Ea., 112, B02105, https://doi.org/10.1029/2006JB004410, 2007.
Laso-Pérez, R., Wu, F., Crémière, A., Speth, D. R., Magyar, J. S., Zhao, K., Krupovic, M., and Orphan, V. J.: Evolutionary diversification of methanotrophic ANME-1 archaea and their expansive virome, Nat. Microbiol., 8, 231–245, https://doi.org/10.1038/s41564-022-01297-4, 2023.
Liu, W., Wu, Z., Xu, S., Wei, J., Peng, X., Li, J., and Wang, Y.: Pore-water dissolved inorganic carbon sources and cycling in the shallow sediments of the Haima cold seeps, South China Sea, J. Asian Earth Sci., 201, 104495, https://doi.org/10.1016/J.JSEAES.2020.104495, 2020.
Lloyd, K. G., Alperin, M. J., and Teske, A.: Environmental evidence for net methane production and oxidation in putative ANaerobic MEthanotrophic (ANME) archaea, Environ. Microbiol., 13, 2548–2564, https://doi.org/10.1111/j.1462-2920.2011.02526.x, 2011.
Lomans, B. P., Maas, R., Luderer, R., Op Den Camp, H. J. M., Pol, A., Van Der Drift, C., and Vogels, G. D.: Isolation and characterization of Methanomethylovorans hollandica gen. nov., sp. nov., isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol, Appl. Environ. Microbiol., 65, 3641–3650, https://doi.org/10.1128/AEM.65.8.3641-3650.1999, 1999.
Lorenson, T. D., Kvenvolden, K. A., Hostettler, F. D., Rosenbauer, R. J., Orange, D. L., and Martin, J. B.: Hydrocarbon geochemistry of cold seeps in the Monterey Bay National Marine Sanctuary, Mar. Geol., 181, 285–304, https://doi.org/10.1016/S0025-3227(01)00272-9, 2002.
Love, M. I., Huber, W., and Anders, S.: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 15, 550–550, https://doi.org/10.1186/s13059-014-0550-8, 2014.
Lovley, D. R. and Klug, M. J.: Model for the distribution of sulfate reduction and methanogenesis in freshwater sediments, Geochim. Cosmochim. Ac., 50, 11–18, https://doi.org/10.1016/0016-7037(86)90043-8, 1986.
Lozupone, C. A., Hamady, M., Kelley, S. T., and Knight, R.: Quantitative and Qualitative β Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities, Appl. Environ. Microbiol., 73, 1576 LP–1585, https://doi.org/10.1128/AEM.01996-06, 2007.
Luton, P. E., Wayne, J. M., Sharp, R. J., and Riley, P. W.: The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill, Microbiol., 148, 3521–3530, https://doi.org/10.1099/00221287-148-11-3521, 2002.
Martens, C. S. and Berner, R. A.: Interstitial water chemistry of anoxic Long Island Sound sediments. 1. Dissolved gases1, Limnol. Oceanogr., 22, 10–25, https://doi.org/10.4319/lo.1977.22.1.0010, 1977.
Martin, J. B., Orange, D. L., Lorenson, T. D., and Kvenvolden, K. A.: Chemical and isotopic evidence of gas-influenced flow at a transform plate boundary: Monterey Bay, California, J. Geophys. Res. Sol.-Ea., 102, 24903–24915, https://doi.org/10.1029/97JB02154, 1997.
Martin, M.: Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet.journal, 17, 1, https://doi.org/10.14806/ej.17.1.200, 2011.
McVeigh, D., Skarke, A., Dekas, A. E., Borrelli, C., Hong, W.-L., Marlow, J. J., Pasulka, A., Jungbluth, S. P., Barco, R. A., and Djurhuus, A.: Characterization of benthic biogeochemistry and ecology at three methane seep sites on the Northern U.S. Atlantic margin, Deep-Sea Res. Pt. II, 150, 41–56, https://doi.org/10.1016/j.dsr2.2018.03.001, 2018.
Meckenstock, R. U., Morasch, B., Kästner, M., Vieth, A., and Richnow, H. H.: Assessment of Bacterial Degradation of Aromatic Hydrocarbons in the Environment by Analysis of Stable Carbon Isotope Fractionation, Water Air Soil Poll. Focus, 2, 141–152, https://doi.org/10.1023/A:1019999528315, 2002.
Metcalfe, K. S., Murali, R., Mullin, S. W., Connon, S. A., and Orphan, V. J.: Experimentally-validated correlation analysis reveals new anaerobic methane oxidation partnerships with consortium-level heterogeneity in diazotrophy, ISME J., 15, 377–396, https://doi.org/10.1038/s41396-020-00757-1, 2021.
Miyazaki, J., Higa, R., Toki, T., Ashi, J., Tsunogai, U., Nunoura, T., Imachi, H., and Takai, K.: Molecular characterization of potential nitrogen fixation by anaerobic methane-oxidizing archaea in the methane seep sediments at the number 8 Kumano Knoll in the Kumano Basin, offshore of Japan, Appl. Environ. Microbiol., 75, 7153–7162, https://doi.org/10.1128/AEM.01184-09, 2009.
Moore, J. C., Brown, K. M., Horath, F., Cochrane, G., MacKay, M., and Moore, G.: Plumbing Accretionary Prisms: Effects of Permeability Variations, Philos. T. Phys. Sci. Eng., 335, 275–288, 1991.
Nauhaus, K., Boetius, A., Kruger, M., and Widdel, F.: In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area, Environ. Microbiol., 4, 296–305, 2002.
Nauhaus, K., Albrecht, M., Elvert, M., Boetius, A., and Widdel, F.: In vitro cell growth of marine archaeal-bacterial consortia during anaerobic oxidation of methane with sulfate, Environ. Microbiol., 9, 187–196, https://doi.org/10.1111/j.1462-2920.2006.01127.x, 2007.
Niu, M., Fan, X., Zhuang, G., Liang, Q., and Wang, F.: Methane-metabolizing microbial communities in sediments of the Haima cold seep area, northwest slope of the South China Sea, FEMS Microbiol. Ecol., 93, 1–13, https://doi.org/10.1093/femsec/fix101, 2017.
Nunoura, T., Oida, H., Toki, T., Ashi, J., Takai, K., and Horikoshi, K.: Quantification of mcrA by quantitative fluorescent PCR in sediments from methane seep of the Nankai Trough, FEMS Microbiol. Ecol., 57, 149–157, https://doi.org/10.1111/j.1574-6941.2006.00101.x, 2006.
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., Minchin, P. R., O'Hara, B., Simpson, G. L., Solymos, P., Stevens, M. H. H., Szoecs, E., and Wagner, H.: vegan: Community Ecology Package. R package version 2.5-7. https://CRAN.R-project.org/package=vegan (last access: 21 September 2024), 2020.
Orange, D. L., Greene, H. G., Barry, J. P., and Kochevar, R.: ROV investigations of cold seeps along fault zones and mud volcanoes in Monterey Bay, T. Am. Geophys. Un., 75, 1994.
Orange, D. L., Greene, H. G., Reed, D., Martin, J. B., McHugh, C. M., Ryan, W. B. F., Maher, N., Stakes, D., and Barry, J.: Widespread fluid expulsion on a translational continental margin: Mud volcanoes, fault zones, headless canyons, and organic-rich substrate in Monterey Bay, California, GSA Bull., 111, 992–1009, https://doi.org/10.1130/0016-7606(1999)111<0992:WFEOAT>2.3.CO;2, 1999.
Orcutt, B. N., Joye, S. B., Kleindienst, S., Knittel, K., Ramette, A., Reitz, A., Samarkin, V., Treude, T., and Boetius, A.: Impact of natural oil and higher hydrocarbons on microbial diversity, distribution, and activity in Gulf of Mexico cold-seep sediments, Deep-Sea Res. Pt. II, 57, 2008–2021, https://doi.org/10.1016/j.dsr2.2010.05.014, 2010.
Oremland, R. S. and Polcin, S.: Methanogenesis and Sulfate Reduction: Competitive and Noncompetitive Substrates in Estuarine Sediments, Appl. Environ. Microbiol., 44, 1270–1276, https://doi.org/10.1128/aem.44.6.1270-1276.1982, 1982.
Oremland, R. S., Marsh, L. M., and Polcin, S.: Methane production and simultaneous sulphate reduction in anoxic, salt marsh sediments, Nature, 296, 143–145, https://doi.org/10.1038/296143a0, 1982.
Orphan, V. J., House, C. H., Hinrichs, K. U., McKeegan, K. D., and DeLong, E. F.: Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis, Science, 293, 484–487, https://doi.org/10.1126/science.1061338, 2001.
Orphan, V. J., House, C. H., Hinrichs, K.-U., McKeegan, K. D., and DeLong, E. F.: Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments, P. Natl. Acad. Sci. USA, 99, 7663–7668, https://doi.org/10.1073/pnas.072210299, 2002.
Parada, A. E., Needham, D. M., and Fuhrman, J. A.: Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples, Environ. Microbiol., 18, 1403–1414, https://doi.org/10.1111/1462-2920.13023, 2016.
Paull, C. K., Hecker, B., Commeau, R., Freeman-Lynde, R. P., Neumann, C., Corso, W. P., Golubic, S., Hook, J. E., Sikes, E., and Curray, J.: Biological Communities at the Florida Escarpment Resemble Hydrothermal Vent Taxa, Science), 226, 965–967, https://doi.org/10.1126/SCIENCE.226.4677.965, 1984.
Paull, C. K., Borowski, W., Paull, C., Lorenson, T., Borowski, W., Ussler III, W., Olsen, K., and Rodriguez, N.: Isotopic composition of CH4, CO2 species, and sedimentary organic matter within samples from the Blake Ridge: gas source implications, Scientific Results, 164, Ms 164SR-207, https://doi.org/10.2973/odp.proc.sr.164.207.2000, 2000.
Paull, C. K., Schlining, B., Ussler, I., Paduan, J. B., Caress, D., and Greene, H. G.: Distribution of chemosynthetic biological communities in Monterey Bay, California, Geology, 33, 85–88, https://doi.org/10.1130/G20927.1, 2005.
Phrampus, B. J. and Hornbach, M. J.: Recent changes to the Gulf Stream causing widespread gas hydrate destabilization, Nature, 490, 527–530, https://doi.org/10.1038/nature11528, 2012.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glöckner, F. O.: The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 41, D590–D596, https://doi.org/10.1093/nar/gks1219, 2013.
Rathburn, A. E., Martin, J. B., Day, S. A., Mahn, C., Gieskes, J., Ziebis, W., Williams, D., Bahls, A., Rathburn, A. E., Pérez, M. E., Martin, J. B., Day, S. A., Mahn, C., Gieskes, J., Ziebis, W., Williams, D., and Bahls, A.: Relationships between the distribution and stable isotopic composition of living benthic foraminifera and cold methane seep biogeochemistry in Monterey Bay, California, Geochem. Geophy. Geosy., 4, 1106, https://doi.org/10.1029/2003GC000595, 2003.
Ravenschlag, K., Sahm, K., Jakob, P. J., and Rudolf, A. R.: High Bacterial Diversity in Permanently Cold Marine Sediments, Appl. Environ. Microbiol., 65, 3982–3989, https://doi.org/10.1128/AEM.65.9.3982-3989.1999, 1999.
Reeburgh, W. S.: Methane consumption in Cariaco Trench waters and sediments, Earth Planet. Sc. Lett., 28, 337–344, https://doi.org/10.1016/0012-821X(76)90195-3, 1976.
Reeburgh, W. S.: Oceanic Methane Biogeochemistry, ChemInform, 38, 486–513, https://doi.org/10.1002/chin.200720267, 2007.
Ruff, S. E., Biddle, J. F., Teske, A. P., Knittel, K., Boetius, A., and Ramette, A.: Global dispersion and local diversification of the methane seep microbiome, P. Natl. Acad. Sci. USA, 112, 4015–4020, https://doi.org/10.1073/pnas.1421865112, 2015.
Ruff, S. E., Felden, J., Gruber-Vodicka, H. R., Marcon, Y., Knittel, K., Ramette, A., and Boetius, A.: In situ development of a methanotrophic microbiome in deep-sea sediments, ISME J., 13, 197–213, https://doi.org/10.1038/s41396-018-0263-1, 2019.
Sauer, S., Hong, W. L., Yao, H., Lepland, A., Klug, M., Eichinger, F., Himmler, T., Crémière, A., Panieri, G., Schubert, C. J., and Knies, J.: Methane transport and sources in an Arctic deep-water cold seep offshore NW Svalbard (Vestnesa Ridge, 79° N), Deep-Sea Res. Pt. I, 167, 103430, https://doi.org/10.1016/J.DSR.2020.103430, 2021.
Schönheit, P., Kristjansson, J. K., and Thauer, R. K.: Kinetic mechanism for the ability of sulfate reducers to out-compete methanogens for acetate, Arch. Microbiol., 132, 285–288, https://doi.org/10.1007/BF00407967, 1982.
Schreiber, L., Holler, T., Knittel, K., Meyerdierks, A., and Amann, R.: Identification of the dominant sulfate-reducing bacterial partner of anaerobic methanotrophs of the ANME-2 clade, Environ. Microbiol., 12, 2327–2340, https://doi.org/10.1111/j.1462-2920.2010.02275.x, 2010.
Seabrook, S., C. De Leo, F., Baumberger, T., Raineault, N., and Thurber, A. R.: Heterogeneity of methane seep biomes in the Northeast Pacific, Deep-Sea Res. Pt. II, 150, 195–209, https://doi.org/10.1016/j.dsr2.2017.10.016, 2018.
Semler, A.: Supplementary Dataset.xlsx, figshare [data set], https://doi.org/10.6084/m9.figshare.25769601.v1, 2025.
Semler, A. C., Fortney, J. L., Fulweiler, R. W., and Dekas, A. E.: Cold Seeps on the Passive Northern U.S. Atlantic Margin Host Globally Representative Members of the Seep Microbiome with Locally Dominant Strains of Archaea, Appl. Environ. Microbiol., 88, e00468-22, https://doi.org/10.1128/aem.00468-22, 2022.
Singh, N., Kendall, M. M., Liu, Y., and Boone, D. R.: Isolation and characterization of methylotrophic methanogens from anoxic marine sediments in Skan Bay, Alaska: Description of Methanococcoides alaskense sp. nov., and emended description of Methanosarcina baltica, Int. J. Syst. Evol. Microbiol., 55, 2531–2538, https://doi.org/10.1099/ijs.0.63886-0, 2005.
Skarke, A., Ruppel, C., Kodis, M., Brothers, D., and Lobecker, E.: Widespread methane leakage from the sea floor on the northern US Atlantic margin, Nat. Geosci., 7, 657–661, https://doi.org/10.1038/ngeo2232, 2014.
Skennerton, C. T., Chourey, K., Iyer, R., Hettich, R. L., Tyson, G. W., and Orphan, V. J.: Methane-Fueled Syntrophy through Extracellular Electron Transfer: Uncovering the Genomic Traits Conserved within Diverse Bacterial Partners of Anaerobic Methanotrophic Archaea, mBio, 8, 4, https://doi.org/10.1128/mBio.00530-17, 2017.
So, C. M. and Young, L. Y.: Initial reactions in anaerobic alkane degradation by a sulfate reducer, strain AK-01, Appl. Environ. Microbiol., 65, 5532–5540, https://doi.org/10.1128/aem.65.12.5532-5540.1999, 1999.
Stakes, D. S., Orange, D., Paduan, J. B., Salamy, K. A., and Maher, N.: Cold-seeps and authigenic carbonate formation in Monterey Bay, California, Mar. Geol., 159, 93–109, https://doi.org/10.1016/S0025-3227(98)00200-X, 1999.
Suess, E., Carson, B., Ritger, S. D., Moore, J. C., Jones, M. L., Kulm, L. D., and Cochrane, G. R.: Biological communities at vent sites along the subduction zone off Oregon, Bull. Biol. Soc. Wash., 6, 475–484, 1985.
Thurber, A. R., Seabrook, S., and Welsh, R. M.: Riddles in the cold: Antarctic endemism and microbial succession impact methane cycling in the Southern Ocean, P. Roy. Soc. B, 287, 20201134, https://doi.org/10.1098/rspb.2020.1134, 2020.
Vigneron, A., Alsop, E. B., Cruaud, P., Philibert, G., King, B., Baksmaty, L., Lavallée, D., Lomans, B. P., Kyrpides, N. C., Head, I. M., and Tsesmetzis, N.: Comparative metagenomics of hydrocarbon and methane seeps of the Gulf of Mexico, Sci. Rep., 7, 1–12, https://doi.org/10.1038/s41598-017-16375-5, 2017.
Vigneron, A., Cruaud, P., Lovejoy, C., and Vincent, W. F.: Genomic insights into cryptic cycles of microbial hydrocarbon production and degradation in contiguous freshwater and marine microbiomes, Microbiome, 11, 104, https://doi.org/10.1186/s40168-023-01537-7, 2023.
Ward, B. B., Kilpatrick, K. A., Novelli, P. C., and Scranton, M. I.: Methane oxidation and methane fluxes in the ocean surface layer and deep anoxic waters, Nature, 327, 226–229, https://doi.org/10.1038/327226a0, 1987.
Widdel, F. and Grundmann, O.: Biochemistry of the Anaerobic Degradation of Non-Methane Alkanes, in: Handbook of Hydrocarbon and Lipid Microbiology, edited by: Timmis, K. N., Springer Berlin Heidelberg, Berlin, Heidelberg, 909–924, https://doi.org/10.1007/978-3-540-77587-4_64, 2010.
Winfrey, M. R. and Ward, D. M.: Substrates for Sulfate Reduction and Methane Production in Intertidal Sediments, Appl. Environ. Microbiol., 45, 193–199, https://doi.org/10.1128/aem.45.1.193-199.1983, 1983.
Wunder, L. C., Aromokeye, D. A., Yin, X., Richter-Heitmann, T., Willis-Poratti, G., Schnakenberg, A., Otersen, C., Dohrmann, I., Römer, M., Bohrmann, G., Kasten, S., and Friedrich, M. W.: Iron and sulfate reduction structure microbial communities in (sub-)Antarctic sediments, ISME J., 15, 3587–3604, https://doi.org/10.1038/s41396-021-01014-9, 2021.
Yancey, P. H. and Somero, G. N.: Methylamine osmoregulatory solutes of elasmobranch fishes counteract urea inhibition of enzymes, J. Exp. Zoo., 212, 205–213, https://doi.org/10.1002/JEZ.1402120207, 1980.
Zhang, Y., Maignien, L., Zhao, X., Wang, F., and Boon, N.: Enrichment of a microbial community performing anaerobic oxidation of methane in a continuous high-pressure bioreactor, BMC Microbiol., 11, 137, https://doi.org/10.1186/1471-2180-11-137, 2011.





