the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Influence of carbon source and iron oxide minerals on methane production and magnetic mineral formation in salt marsh sediments
Kaleigh R. Block
Amy Arbetman
Sarah P. Slotznick
Thomas E. Hanson
George W. Luther III
Salt marshes can emit significant methane to the atmosphere. These emissions are highly variable, but the cause of this variability is not well understood. Although methanogenesis should be inhibited by sulfate reduction where sulfate is present due to its thermodynamic unfavorableness, methane emissions are not well predicted by sulfate concentrations; thus, other controls on methane production must be active. One hypothesis is that where sulfate is present, salt marsh methanogens are fueled by methylated carbon substrates that sulfate reducers do not compete for. It has also been shown that crystalline iron minerals can facilitate increased methane production in low-salinity wetlands, but this has not been explored in salt marshes. This study documents how different organic carbon sources (monomethylamine and ethanol) and Fe(III) minerals (ferrihydrite, magnetite, and hematite) influence methane production by microbial communities from a polyhaline tidal marsh creek in the Great Marsh Preserve, DE, USA. Carbon source had a strong influence on microbial community composition by the end of the incubations. More methane was produced with monomethylamine amendment than with ethanol, and the highest methane production rates were in incubations supplied with both monomethylamine and magnetite or hematite. This increased methane production in the presence of (semi)conductive iron minerals could indicate that interspecies electron transfer was active in some of our treatments. However, instead of the more commonly described syntrophic partners, this interaction appears to be between methylotrophic methanogens belonging to Methanococcoides and an unidentified iron-reducing bacterial group, possibly Candidatus Omnitrophus. Much less methane was measured with ethanol and ferrihydrite amendments. However, in ethanol-amended incubations, a small proportion of anaerobic methane oxidizers was detected, which suggests that both methane production and re-oxidation may have occurred, leading to low measured methane production. Although some iron reduction and Fe2+ production was observed in all treatments, significant transformation of ferrihydrite to magnetite was observed only with ethanol amendment. If microbially mediated magnetite formation occurs in salt marsh sediment, our observations indicate that the resulting magnetite could enhance methane production by methylotrophic methanogens. This study highlights the importance of methylated compounds to salt marsh methane production as well as the potential importance of iron mineral composition for predicting methane production and iron reduction rates.
- Article
(4028 KB) - Full-text XML
- BibTeX
- EndNote
Wetlands are the primary natural source of methane emissions to the atmosphere (Saunois et al., 2016; Zhang et al., 2017). However, only a fraction of these emissions is expected to come from high-salinity tidal wetlands, such as salt marshes, due to the influx of oxygen and sulfate during regular inundation (Bartlett et al., 1987; Hartman et al., 2024; Holmquist et al., 2023; Poffenbarger et al., 2011). According to the textbook paradigm, this is because thermodynamic considerations determine the dominance of microbial metabolisms. Organic carbon degraders are expected to follow the redox ladder, sequentially depleting oxygen, nitrate, manganese, and iron oxide minerals, and sulfate before methanogenesis is favored (Jørgensen, 2006; Jørgensen and Kasten, 2006; Sundby, 2006). In salt marshes, oxygen is often depleted within millimeters of the sediment surface, and sulfate reduction is responsible for a large fraction of organic carbon remineralization (Howarth, 1984; Howarth and Giblin, 1983; Howarth and Teal, 1979) as well as the oxidation of methane (Krause and Treude, 2021; Segarra et al., 2013). However, in more oxygenated marsh regions like creek banks and bioturbated zones where pore waters are more efficiently flushed by the tides, iron reduction can be the dominant microbial metabolism (Antler et al., 2019; Gribsholt et al., 2003; Kostka et al., 2002a, b; Krause et al., 2025). Iron oxides may also fuel anaerobic methane oxidation in salt marshes (Krause and Treude, 2021; Liu et al., 2025; Segarra et al., 2013). Moreover, salt marsh sediments can have high iron concentrations with seasonally fluctuating mineralogy (Kostka et al., 2002a, b; Kostka and Luther, 1994, 1995; Taillefert et al., 2007). Based on energy yield, the presence of measurable iron oxide minerals and sulfate should inhibit methanogenesis and facilitate the anaerobic oxidation of methane produced at deeper depths before it is released to marsh creeks and the atmosphere.
At the ecosystem scale, methane emission data from wetlands do follow a general decreasing trend with increasing sulfate concentration (Al-Haj and Fulweiler, 2020; Chmura et al., 2011; Hartman et al., 2024; Poffenbarger et al., 2011), but significant variability in methane emissions across salinity gradients has also been reported, indicating the importance of other controls (Al-Haj and Fulweiler, 2020; Bridgham et al., 2006; Chmura et al., 2011; Hartman et al., 2024; Rosentreter et al., 2021). In the mid-Atlantic region of the USA, high concentrations of methane and indications of active methanogenesis were found to coexist with sulfate reduction in marsh sediments (Seyfferth et al., 2020). This finding indicates that thermodynamics does not entirely predict the activity of microbial metabolisms and biogeochemical processes in salt marshes. Methanogens have been shown to coexist with sulfate reducers, although with lower activity (Sela-Adler et al., 2017; Senior et al., 1982). They have also been shown to circumvent the redox ladder via the utilization of organic substrates, such as methylated compounds, that are not as available to sulfate-reducing bacteria (Buckley et al., 2008; Oremland et al., 1982; Parkes et al., 2012a; Winfrey and Ward, 1983; Yuan et al., 2019). In the eastern USA, the low marsh is dominated by the native marsh grass Spartina alterniflora, which releases trimethylamine at depth through root exudates and root degradation (Wang and Lee, 1994a). Trimethylamine, dimethylamine, and monomethylamine have all been detected in the pore waters and sediments of salt marshes (Fitzsimons et al., 1997; Parkes et al., 2012b; Summons et al., 1998; Wang and Lee, 1994b). As sulfate-reducing bacteria are not known to utilize methylamines (Mausz and Chen, 2019), this supply of carbon substrate from S. alterniflora allows methanogenesis to proceed simultaneously with sulfate reduction (Seyfferth et al., 2020; Yuan et al., 2014, 2019).
Methanogens can also skip rungs of the redox ladder via participation in syntrophic relationships with iron-reducing bacteria (Cruz Viggi et al., 2014; Kato et al., 2012; Rotaru et al., 2014, 2021; Xu et al., 2019; Zhuang et al., 2019). Although this has not yet been observed in salt marsh microbial communities, methanogens can obtain their reducing equivalents through interspecies electron transfer (IET) facilitated by conductive iron minerals (Lovley, 2017a, b; Rotaru et al., 2021). Although the majority of iron in shallow marsh sediments undergoes a seasonal alternation between amorphous Fe(III) minerals and iron sulfides at shallow depths (Koretsky et al., 2003; Kostka and Luther, 1994), crystalline Fe(III) minerals with (semi)conductive properties that could facilitate IET have been shown to persist year-round, even at anoxic and sulfidic depths (Kostka and Luther, 1994, 1995; Taillefert et al., 2007). It has also been shown that ferrihydrite, an amorphous Fe(III) mineral, can be converted to the secondary conductive mineral magnetite over time in anoxic incubations of microbial communities from rice paddy soils, eventually enhancing methane production through IET (Tang et al., 2016; Zhuang et al., 2015, 2019). Although they have not been as extensively studied as sulfate reducers, iron-reducing bacteria have been reported in salt marsh sediments (Hyun et al., 2007; Koretsky et al., 2003, 2005; Lowe et al., 2000). These populations have the potential to cooperate with methanogens through IET or to outcompete them and transform Fe(III) minerals into aqueous Fe2+, mixed-valent minerals like magnetite, or Fe(II) minerals like pyrite. Salt marshes, with their naturally variable redox conditions, have also been found to host greigite-producing magnetotactic bacteria, often in sediments at the transition between oxic and anoxic conditions (Bazylinski et al., 1991; Blakemore, 1975; Demitrack, 1985; Simmons and Edwards, 2007a, b; Sparks et al., 1989). Moreover, both magnetite- and greigite-producing magnetotactic bacteria have been found in salt ponds neighboring salt marshes (Simmons et al., 2004; Simmons and Edwards, 2007b).
In this study, we investigate how specific carbon sources and Fe(III) minerals influence the production of methane and magnetic minerals in anoxic, sulfate-free incubations of marsh creek sediments from a polyhaline salt marsh in the mid-Atlantic region of the eastern USA. We studied non-vegetated marsh creek sediments adjacent to a marsh platform dominated by S. alterniflora to avoid the multiple and confounding influences of marsh grass stems and roots on redox conditions and methane emissions (Määttä and Malhotra, 2024). We investigated the effect of adding organic carbon substrates for methylotrophic and acetoclastic methanogenesis combined with iron minerals that supply Fe(III) with a range of reactivity and electrical conductivity values. Here, we compare methane production from monomethylamine (MMA), which represents methylated methanogenic substrates (Krause and Treude, 2021), to methane production from ethanol (EtOH), which is oxidized to acetate through fermentative pathways (Schink, 1997) and which has been used in many studies investigating the potential for magnetite and hematite to promote electric syntrophy between methanogens and bacteria in soil and sediment enrichments (Kato et al., 2012; Rotaru et al., 2019, 2018). MMA and EtOH amendments were combined with additions of magnetite, hematite, and ferrihydrite to determine the influence of these iron minerals on methane production and iron reduction by salt marsh microbial communities.
2.1 Study site description and sediment sampling
Sediments for this study were sampled from the Great Marsh Preserve in Lewes, DE, USA. At this location, low marsh is dominated by the marsh grass S. alterniflora, and it is dissected by a grid of marsh creeks originally dug as mosquito ditches in the 1930s but left to return to natural conditions by the 1970s. The marsh creeks and low marsh platform regularly experience inundation at high tide with water from the lower Delaware Bay via the Roosevelt Inlet and Canary Creek. High-tide salinity averages 30 PSU (where PSU denotes practical salinity units) during the month of June (2016–2020 average; Delaware Water Quality Portal Station BR40, 2024), and the tidal amplitude averages 1.2 m (Dewitt and Daiber, 1973; Guider et al., 2024). Sediments for our study were collected by push core from a mudflat adjacent to the low marsh platform within a marsh creek (38.78926° N, 75.16968° W) while it was covered by shallow water at low tide. The mudflat was selected to avoid living S. alterniflora stems and roots. Two sediment cores of 8.5 cm diameter and 22.5 cm length were taken on 29 June 2023. They were processed for geochemistry within 5 h of collection.
2.2 Downcore profiling of redox conditions
Before the sediment core was extruded in the laboratory, cyclic voltammetry (CV) was used to determine concentrations of dissolved O2, ΣH2S, ΣS2−, Fe2+, Fe3+, and FeSaq molecular clusters on the intact core as well as qualitative concentrations of solid Fe(III) nanoparticles and organic–Fe(III) aggregates. CV measurements were made with a three-electrode system composed of a solid-state gold amalgam () working microelectrode encased in glass, a silver–silver chloride () reference electrode, and a 0.5 mm platinum wire counter electrode (Brendel and Luther, 1995). Electrodes were coupled to a model DLK-60 electrochemical analyzer (Analytical Instrument Systems Inc., Flemington, NJ, USA). The reference and counter electrodes were secured to the core barrel such that they were suspended in marsh creek water, approximately 2 cm of which was retained above the sediment in the core barrel. The working electrode was lowered into the core at fine vertical resolution by a micromanipulator (Luther et al., 2008). Concentrations of ΣS2− and O2 were calculated from current using published parameters (Luther et al., 2008; Waite et al., 2006). Between the sediment–water interface and 0.35 cm depth, measurements were taken every 0.05 cm. Below 0.35 cm, measurements were taken every 0.1 cm until a depth of 1.2 cm. As the redox transitions were detected at shallower depths, CV measurements were discontinued at a depth of 1.2 cm. All scans were made in triplicate.
2.3 Ferrihydrite synthesis
Ferrihydrite was synthesized following the methods of (Smith et al., 2012). Ferric nitrate nonahydrate (20.25 g) and ammonium bicarbonate (11.91 g) were ground together with a mortar and pestle. The product of this reaction was dried overnight, rinsed the following day via vacuum filtration with small aliquots of deionized water, and dried again overnight. The final product was ground again with a mortar and pestle and used for ferrihydrite amendments in incubations.
2.4 Incubations
Although we observed a redox transition at ∼ 0.5 cm, sediments were unconsolidated at this depth. To avoid mixing soluble oxidants (e.g., oxygen, nitrate, and sulfate) from shallower depths into sediments used for incubations from below the redox transition, the top 2.5 cm of the core was discarded following CV measurements (Fig. 1a). Sediments between 2.5 cm and 16.5 cm depth provided sufficient sediment volume for our incubations, and this interval of sediment was extruded and homogenized in a 0.07 mm thickness, low-density polyethylene (LDPE) zip-top bag. The small amount of dead plant material within the core was removed by handpicking during extrusion and transfer to the bag. Homogenized sediments were refrigerated at 4 °C for ∼ 16 h prior to mixing sediments into a slurry and assembling the incubation bottles. The slurry was made up of homogenized marsh sediment and artificial sulfate-free seawater. Sulfate was not included in our artificial seawater to disadvantage sulfate reduction in favor of other microbial metabolisms and allow investigation of controls on methane production other than competition with sulfate-reducing bacteria. Artificial sulfate-free seawater contained 26.4 g of NaCl, 11.2 g of MgCl2⋅6H2O, 1.5 g of CaCl2⋅2H2O, and 0.7 g of KCl L−1 (Aromokeye et al., 2018) as well as 1 mM of an iron-free trace metal solution modified from DSMZ trace element solution SL-10 (Koblitz et al., 2023): 70.0 mg of ZnCl2, 100 mg of MnCl2⋅4H2O, 6 mg of H3BO3, 190 mg of CoCl2⋅6H2O, 2 mg of CuCl2⋅2H2O, 24 mg of NiCl2⋅6H2O, and 36 mg of Na2MoO4⋅2H2O L−1. Anaerobic incubations were performed in 125 mL serum bottles containing 50 mL of slurry and 75 mL of headspace. The headspace was purged of O2 by flushing for 2 min with () prior to sealing the bottles with an () headspace. Incubations were supplemented with 20 mM of organic carbon, either ethanol (EtOH) or monomethylamine hydrochloride (MMA). Incubations were also supplemented with 20 mM of powdered iron minerals, either synthesized ferrihydrite, magnetite, or hematite, or no iron. Powdered magnetite was purchased from Sigma-Aldrich (St. Louis, MO, USA), and powdered hematite was purchased from Strem Chemicals Inc. (Newburyport, MA, USA). In total, 24 bottles were prepared with the following treatment groups in triplicate: EtOH + no iron, EtOH + ferrihydrite, EtOH + magnetite, EtOH + hematite, MMA + no iron, MMA + ferrihydrite, MMA + magnetite, and MMA + hematite. Incubations were maintained at 30 °C under static conditions in the dark for 34 d.
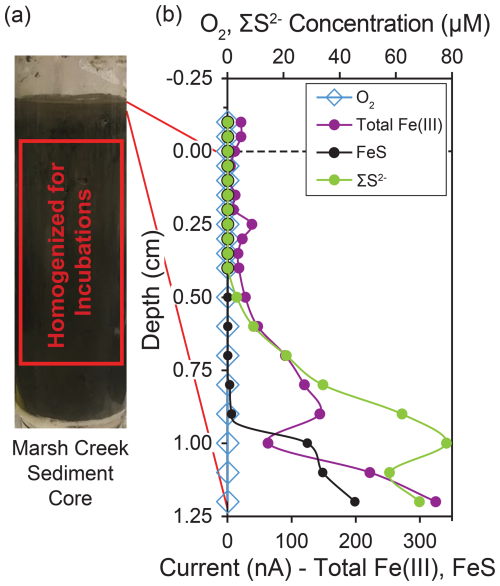
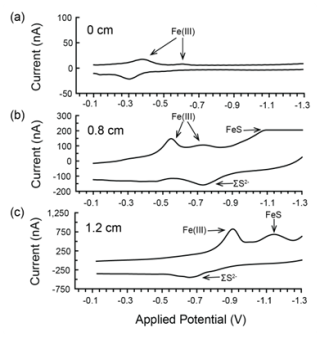
Figure 1Redox conditions and sampling depths in the marsh creek sediment core. (a) Photo of the 22.5 cm sediment core before extrusion with the interval of sediment used for incubations highlighted. (b) Microelectrode profiles of oxygen, iron, and sulfur species with depth in the sediment core as measured by cyclic voltammetry. Total Fe(III) is the summed peak areas for Fe(III)(aq), Fe(III)(s) nanoparticles of multiple sizes, and Fe(III) with organic ligands. Total Fe(III) and FeS are plotted with relative units. Traces of FeS were detected beginning at 0.5 cm. O2, Fe2+, and HS− were not detected in the sediment core or overlying water. The dashed line at 0 cm represents the sediment–water interface, while negative depths represent the height in the overlying water.
2.5 Methane and ferrous iron measurements
Headspace methane concentrations were measured twice weekly by gas chromatography with a flame-ionization detector (Agilent 7890B, Wilmington, DE, USA). Methane was separated isothermally on a HP-Plot Q column (Agilent, 30 m × 0.530 µm inner diameter, 40 µm film thickness). Headspace samples were manually injected with a gastight syringe in split mode with a split ratio of 5:1. The injector temperature was 250 °C, oven temperature was 60 °C, and detector temperature was 250 °C. Methane peaks were integrated using Agilent ChemStation data analysis software version F.01.03. A parallel set of 125 mL serum bottles were prepared as methane standards with the same ratio of distilled water to headspace as incubation bottles to account for dissolved methane. Although the solubility of methane in our slurries will not be the same as in distilled water, we expect these differences to be smaller than other sources of uncertainty and variability in our incubations. The headspace of the standard bottles was filled with and varying amounts of methane. Six standards with total methane concentrations ranging from 0.004 to 0.4 mmol were injected with each set of headspace methane measurements to obtain calibration curves. Methane concentrations and calculated methane production rates in incubations (Figs. 2, 3) were determined from a calibration curve of equilibrated standards and the ideal gas law.
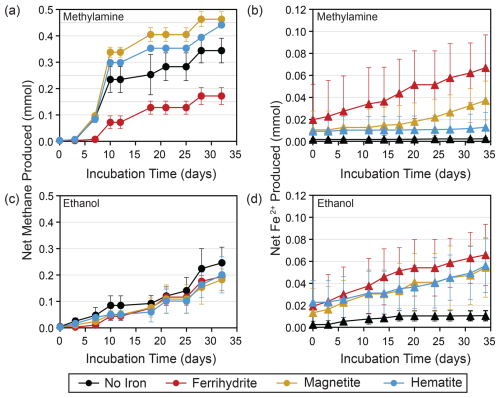
Figure 2(a) Net methane and (b) Fe2+ produced with methylamine amendments and all iron treatments averaged over three replicates. Similar plots for (c) net methane and (d) Fe2+ produced with ethanol amendments are also averaged over three replicates. Error bars represent the standard deviation from the replicates' average value.
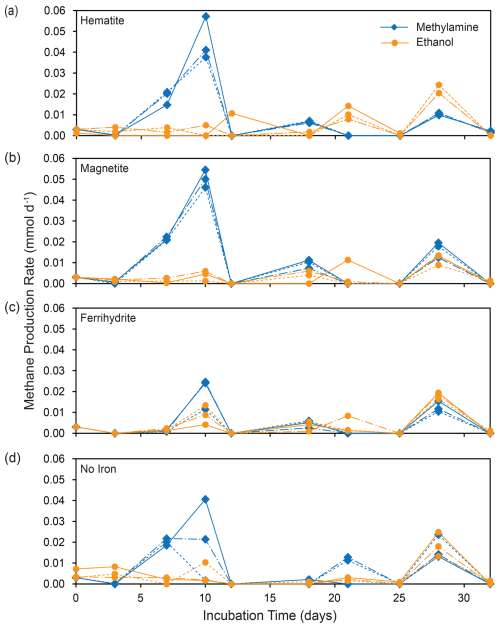
Figure 3Methane production rates for each replicate incubation bottle: shown for MMA incubations (blue) and EtOH incubations (orange) separated by iron treatment.
Measurements of dissolved ferrous iron were made twice weekly. Ferrous iron was quantified utilizing the ferrozine assay method developed by Stookey (1970). From each incubation, 0.5 mL of slurry was sampled from the bottles, filtered at 0.45 µm, and stabilized in ferrozine solution. Absorbance at 562 nm was measured with a HP-8452A diode array spectrophotometer (Hewlett-Packard/OLIS Inc., Athens, GA, USA).
2.6 DNA extraction, PCR, quantification, and sequencing
At the end of the incubation period, slurries were homogenized, and subsamples were then taken for DNA extraction. One bottle was selected at random from each treatment group for amplicon sequencing. DNA was extracted from the slurry using the DNeasy PowerSoil Pro Kit (QIAGEN, Germantown, MD, USA). Concentrations of DNA were quantified with a Qubit BR DNA assay (Invitrogen, Eugene, OR, USA) using a DeNovix DS-11 fluorometer (Wilmington, DE, USA). For all extractions, the total amount of DNA recovered was ≥ 6 ng. Amplification of extracted DNA was performed with a modified two-step barcoding method (Herbold et al., 2015; Tadley et al., 2023). Amplicons of the V4 region of the 16S rRNA gene were produced with the primers 515F (Parada et al., 2016) and 806R (Apprill et al., 2015) modified with the head sequence: 5'-GCTATGCGCGAGCTGC-3' (Herbold et al., 2015). The second PCR barcoding primer was paired to the head sequence. Each PCR reaction contained 1.0 µM forward and reverse primers in Invitrogen SuperFi Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) with denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 60 s for a total of 30 cycles in the first PCR and 5 cycles in the second PCR. During the first PCR, reactions were completed in triplicate, and PCR products were screened by gel electrophoresis. The barcoded amplicons were purified with sparQ PureMag beads (QuantaBio, Beverly, MA, USA). Sequencing on an Illumina MiSeq platform (MiSeq Reagent Kit v2, 2×250 bp, San Diego, CA, USA) was performed at the University of Delaware Sequencing and Genotyping Center.
Amplicon sequences were analyzed as described by Tadley et al. (2023). Sequences were sequentially processed with Python (3.10.10) and QIIME 2 (2021.2; Bolyen et al., 2019) for demultiplexing, as described by Herbold et al. (2015). Amplicon sequence variants were identified with DADA2 (per pipeline tutorial v1.16, DADA2 v1.21.0; Callahan et al., 2016) with quality trimming at Q30 per sample as determined by the FASTX-Toolkit (v0.0.14; Gordon et al., 2010). Taxonomy for all amplicons was assigned with the RDP classifier using a classifier based on the Silva 138 SSU Ref NR 99 database for 16S rRNA (Robeson et al., 2021). Relative abundance plots for taxa were generated in R using the FeatureTable package (version 0.0.11; Moore, 2020). Cluster dendrograms were generated using the pvclust R package (version 2.2.0; Suzuki et al., 2019) after double-square-root transformation to reduce artifacts from the large number of amplicon sequence variants (ASVs) that were individually relatively rare. Indicator analyses were completed using the indicspecies package in R (version 1.7.15; De Cáceres et al., 2010, 2012).
2.7 Magnetic measurements
Magnetic mineral measurements were made on pure iron oxide minerals used for amendments in incubations, on initial homogenized sediment prior to incubation before addition of iron amendments, and on sediments recovered at the end of the incubations. At the end of incubations, following subsampling for DNA, sediment was recovered from slurries by centrifugation. The supernatant was discarded, and sediments were frozen at −80 °C under an argon headspace until they were processed further. One bottle from the three replicates was selected at random from each treatment for magnetic measurements. Initial and post-incubation sediments were freeze-dried, pulverized with a mortar and pestle, and stored under an argon headspace until the next step of preparation. Powdered minerals and sediments were prepared for analysis by packing them into gel capsules with quartz wool in an anaerobic glove box. Magnetic analysis of minerals and sediments was performed on a Lakeshore 8604 vibrating sample magnetometer (VSM). The VSM was used to obtain hysteresis loops and direct current demagnetization at room temperature. Data from these VSM measurements are used to characterize and quantify the mineralogy, abundance, and grain size of magnetic minerals (Fig. 4). Hysteresis loops were slope-corrected to remove antiferromagnetic, paramagnetic, and diamagnetic contributions, and then saturation magnetization and coercivity were extracted for the ferromagnetic (sensu lato) components (Jackson and Solheid, 2010). The direct current demagnetization data were used to find the coercivity of remanence. Its first derivative also produced coercivity spectra, which were deconvolved using the MAX UnMix software program (Maxbauer et al., 2016).
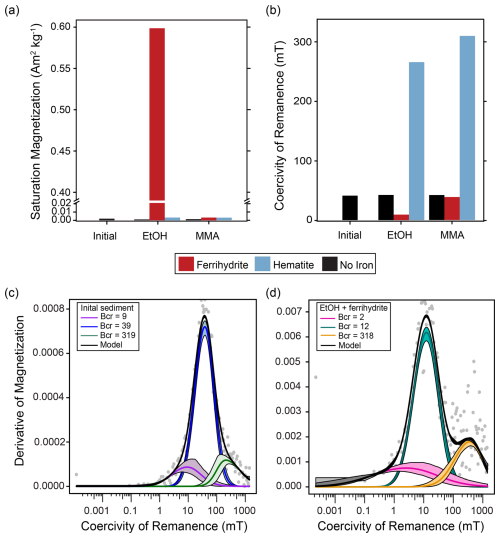
Figure 4Ferromagnetic mineral measurements of initial homogenized sediment used in all incubations and sediment recovered from all amended incubations at the end of the incubation period. (a) Abundance of ferromagnetic minerals as indicated by saturation magnetization (Ms; Am2 kg−1 of dried sediment sample). (b) Mineral identification summarized by coercivity of remanence (Bcr; mT). Examples of MAX UnMix data for (c) initial sediment with no iron mineral amendment and (d) EtOH + ferrihydrite sediment at the end of the incubation period. Data from sediments amended with magnetite are not shown, as magnetite amendment itself overwhelmed any signals of magnetite formation or loss.
3.1 Concentrations of redox-active species by cyclic voltammetry
By the time cyclic voltammetry measurements were made, 2 h after the core was collected, no dissolved oxygen was detected throughout the core, even at the sediment–water interface. However, at least one form of Fe(III) was detected throughout the measured interval of the sediment core, the upper 1.2 cm (Figs. 1b, A1). We detected multiple forms of Fe(III), including organic–Fe(III) aggregates and nanoparticles of different sizes (Fig. A1). The Fe(III) response generally shifted to more negative applied potential with depth in the core (Fig. A1), suggesting increasingly larger particles and an aging process (Taillefert et al., 2000). Reduced sulfur species (ΣS2−) were detected at 0.5 cm and deeper in the core, increasing in concentration to 78 µM at 1.0 cm. FeS was also detected at 0.5 cm depth and increased in concentration until the end of measurements at 1.2 cm (Fig. 1b). Thus, marsh creek sediments themselves supplied our incubations with iron and reduced sulfur in multiple forms and multiple oxidation states.
3.2 Methane production in incubations
Methane production was observed in all incubation bottles. The most methane, along with the highest and most consistent methane production rates, were produced in bottles amended with MMA and magnetite or hematite, followed by MMA without iron amendment (Figs. 2a and 3a and b). All other treatments produced similar quantities of methane (Fig. 2a, c). The highest production rates overall were observed within the first 10 d, followed by lower sporadic pulses (Fig. 3). MMA + ferrihydrite took longer to ramp up methane production compared to MMA with other iron treatments (Figs. 2a, 3c). On average, MMA + no iron bottles produced more methane than EtOH + no iron (Fig. 2a, c); however, methane production in no-iron incubations was more variable than it was with an iron amendment for both MMA and EtOH treatments (Fig. 3d). Bottles amended with EtOH produced less methane than MMA-amended bottles with the same iron treatment in all cases except with ferrihydrite amendment, in which methane production was comparable. There was also no significant difference between EtOH incubations with different iron treatments (Fig. 2c). In all EtOH-amended incubations, the highest methane production rates were observed as sporadic pulses late in the incubation period (Fig. 3). However, after the first 10 d, the trajectory of methane production was approximately the same regardless of the organic carbon or iron treatment. For example, all treatments had a moderate peak in methane production on day 28 (Fig. 3). This similarity suggests that methane production rates were no longer related to the initial organic carbon amendments by the final 2 weeks of the incubation period.
As Fe2+ concentrations reflect both production by microbial iron reduction and loss from iron mineral formation, they are less sensitive as a tracer of microbial activity than methane concentrations. Therefore, we treat Fe2+ concentrations as an indicator of iron reduction kinetics, following the approach of previous studies (Aromokeye et al., 2018). The most net Fe2+ production occurred in bottles with ferrihydrite amendments regardless of organic carbon treatment (Fig. 2a, c), suggesting that ferrihydrite yielded the highest iron reduction rates. In EtOH bottles, magnetite and hematite yielded almost as much Fe2+ as ferrihydrite, with a similar production rate in the first 10 d (Fig. 2d). With the MMA amendment, on the other hand, iron reduction was mineral-specific. Incubations with magnetite did not ramp up Fe2+ production until the final 2 weeks, and MMA + hematite incubations did not show evidence of significant iron reduction at any point during the incubation period (Fig. 2b).
3.3 Magnetic mineral formation
In bottles amended with EtOH and ferrihydrite, a progressive shift from reddish ferrihydrite to a dark-gray magnetic mineral was observed over the course of the incubation (Fig. C1). To further characterize magnetic mineral formation, magnetic measurements were made. However, as such an extensive transformation of ferrihydrite was not anticipated, we did not preserve subsamples of initial slurry with iron amendments for magnetic mineral characterization. Instead, initial sediments without iron amendments and each powdered iron mineral were characterized individually and used to contextualize magnetic measurements made on sediments recovered at the end of the incubations. Ferromagnetic minerals were identified and further characterized by their remanent coercivities, and saturation magnetization was used to measure ferromagnetic mineral abundance. Magnetite-amended incubations are not discussed further in the main text, as the amendment itself overwhelmed any detectable change in magnetic minerals that may have occurred during the incubation (Fig. C2, Table C1).
In initial homogenized sediments, a small but measurable saturation magnetization indicated the presence of magnetic minerals (Fig. 4a). This concentration was typical of magnetic mineral concentrations measured in sediments across a wide range of conditions (Kreisler, 2022). The presence of a ferromagnetic mineral with mid-coercivity (Bcr=39 mT), interpreted to be magnetite, and a minor high coercivity component, interpreted to be hematite, were both detected (Fig. 4c). At the end of the incubation period, the saturation magnetization of sediments recovered from the iron-free treatments looked similar to initial measurements. Small increases in most other treatments are hard to deconvolute from the addition of ferromagnetic mineral amendments (Fig. 4a; hematite incubations described further in Appendix C). The iron-free, hematite, and MMA + ferrihydrite treatments all had remanent coercivities and coercivity spectra similar to the initial sediment slurry (Figs. 4b, C2). However, the EtOH + ferrihydrite treatment showed a large increase in saturation magnetization (Fig. 4a). This signal could not have resulted simply from the addition of ferrihydrite to initial sediments, as ferrihydrite itself is antiferromagnetic to weakly ferromagnetic. Although the synthesized ferrihydrite analyzed contained a small ferromagnetic component, its saturation magnetization was similar to that of the hematite standard, and an amended bottle with no further mineralogical changes would be comparable. This indicates that the EtOH + ferrihydrite treatment was the only one that produced significant quantities of measurable ferromagnetic minerals. A large decrease in bulk remanent coercivity was noted with the EtOH + ferrihydrite treatment as well (Fig. 4b); coercivity spectra show that the predominant phase produced in the EtOH + ferrihydrite treatment is within the range for magnetite (Bcr∼ 9 mT) (Fig. 4d; Peters and Dekkers, 2003). Coercivity is not a definitive mineral identifier, and other ferromagnetic minerals have overlapping coercivity ranges, specifically pyrrhotite and greigite, but the low abundance of sulfide suggests that it is not either of these phases. Notably, the ferromagnetic mineral produced during EtOH + ferrihydrite incubations has a lower coercivity than the magnetite in initial sediments (Fig. 4c, d). In addition to this predominant mid-coercivity phase, there appears to be a slight increase in the abundance of high-coercivity phases as well (Bcr= 318), interpreted as hematite. A similar change is not seen in the MMA + ferrihydrite treatment, suggesting this is not simply due to abiotic aging of ferrihydrite but could also be related to microbial iron oxidation of magnetite instead of de novo production.
3.4 Microbial community composition
Microbial community composition at the end of the incubations from 16S rRNA sequence data is shown in Fig. 5 for families that represented > 5 % of total sequences. To facilitate more detailed comparisons, archaeal and bacterial community composition is also shown at the taxonomic level of genus in Fig. 6 for genera representing > 5 % of sequences from each domain. Hierarchical clustering of 16S rRNA sequences from the end of incubations (Fig. 5a) showed a clear pattern that was aligned with carbon substrate. The same pattern was observed when just bacterial or archaeal sequences were used (Fig. B1). In all three cases, clusters are supported by bootstrap values of 100, indicating that the organic carbon source was the major control on both the archaeal and bacterial community composition. Archaea were the dominant populations observed in all incubations. Bathyarchaeia made up the largest fraction of sequences in MMA incubations, which was distinct from EtOH incubations that were dominated by Methanosarcinaceae (Fig. 5b).
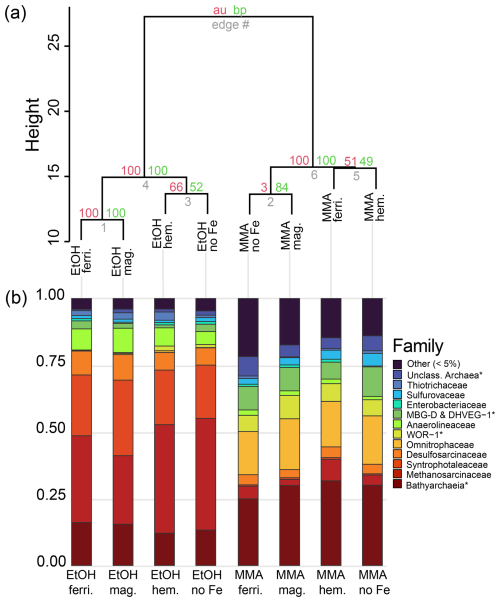
Figure 5Microbial community composition from 16S rRNA gene diversity at the end of the incubation period. (a) Cluster dendrogram of all gene sequences recovered with a universal 16S rRNA primer. Approximately unbiased probability (AU, red) and bootstrap probability (BP, green) are shown as percentages at nodes. (b) Relative abundances of the most abundant microbial groups classified at the level of family. ASVs with less than 10 counts were grouped into the “Other” category. Archaea and bacteria that could only be classified at higher taxonomic levels are denoted using an asterisk (*).
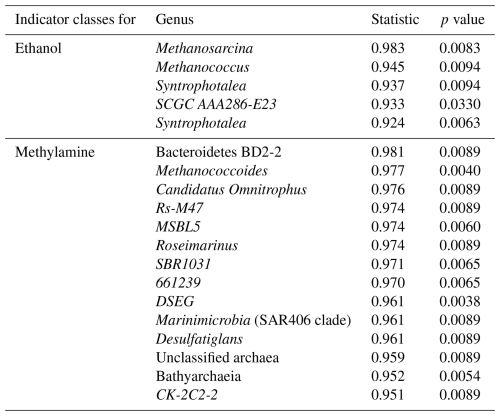
MMA-amended incubations had Shannon diversity index values that were consistently higher than EtOH-amended incubations (Fig. B2), indicating that MMA amendment resulted in a more diverse microbial community. Indicator species analysis performed at the taxonomic level of genus pointed to 68 bacterial and archaeal groups significantly associated (p<0.01) with MMA at high statistic values (> 0.95 %). The EtOH treatments had only one indicator archaeal genus with a similarly high statistic (0.983) and p value (0.008): Methanosarcina (Table B1). Iron was not found to be a strong driver of community structure, with no indicator species identified for any iron treatment. In MMA incubations, three genera comprised > 50 % of the sequences regardless of iron treatment: Bathyarchaeia, Candidatus Omnitrophus, and Marine Benthic Group D/Deep Hydrothermal Vent Euryarchaeotal Group 1 (MBG-D/DHVEG-1). In EtOH incubations, on the other hand, the top three genera, Methanosarcina, Syntrophotalea, and Bathyarchaeia, represented almost 75 % of recovered 16S rRNA sequences, in agreement with the lower diversity estimates in these treatments. We also looked for trends within archaea and bacteria genera individually (Fig. 6). Two different genera of the family Methanosarcinaceae were found to be indicators for carbon treatment (Fig. 6a). Methanosarcina was found to be an indicator for EtOH (statistic = 0.983; p value = 0.008), while Methanococcoides was an indicator for MMA (statistic = 0.977; p value = 0.004). Methanosarcina alone represented > 50 % of archaeal sequences in EtOH + hematite and EtOH + no iron treatments, and they were the largest fraction in the other iron treatments with EtOH amendment as well (Fig. 6a). The only other archaeal genera with significant representation in EtOH incubations were Bathyarchaeia and Methanococcus (Fig. 6a). However, Bathyarchaeia was found to be an indicator for MMA (statistic = 0.952; p value = 0.005), not EtOH. Individuals of this genus represented up to 51 % of archaeal sequences in MMA incubations. MBG-D/DHVEG-1 was also a larger fraction of the archaeal population in MMA treatments. A group of unclassified archaea was consistently recovered from incubations with MMA treatments as well (Figs. 5b, 6b). A BLAST search indicated that these unclassified archaea were most related to Ca. Thorarchaeota in the Asgard superphylum (94 %–96 % sequence similarity). None of these archaeal groups that represented the majority of archaeal sequences in MMA bottles are of the “classical” orders of methanogens (Evans et al., 2019). This was unexpected, as MMA incubations produced significantly more measured methane than EtOH incubations. Classical methanogens were found at very low relative abundance, e.g., Methanococcoides (mean of 1.9 %) and Methanofastidiosales (mean of 0.7 %).
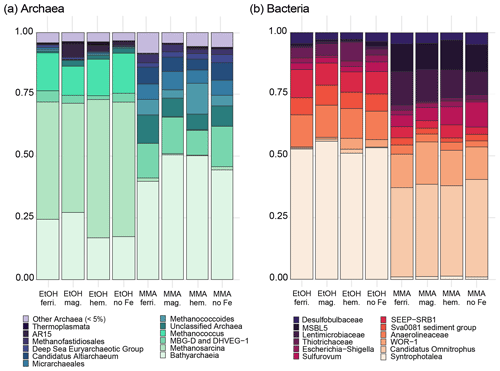
Figure 6Most abundant bacterial (a) and archaeal (b) genera from each incubation group from the universal 16S primer dataset. Bacterial data include ASVs with 10 or more counts, whereas archaeal data include ASVs that had 2 or more counts. Cluster dendrograms of archaeal and bacterial sequences are included separately in Appendix Fig. B1.
Bacterial assemblages were more distinct between carbon treatments than archaeal assemblages (Fig. 6b). MMA treatments resulted in high relative abundances (up to 30 %) of Ca. Omnitrophus followed by WOR-1 (also referred to as Saganbacteria; (Matheus Carnevali et al., 2019), Lentimicrobiaceae, MSBL5 (Mediterranean Sea Brine Lake Group 5), and Sulfurovum (Fig. 6b). The indicator with the highest statistic for MMA treatment, a member of the Bacteroidetes, was not highly abundant (Table B1). But Ca. Omnitrophus was also a strong indicator of MMA treatment (statistic = 0.976, p value = 0.009). In contrast, bacteria in EtOH incubations were dominated by Syntrophotalea, which made up ≥ 50 % of bacterial sequences across all iron treatments (Fig. 6b). Anaerolineaceae was also a substantial proportion of the community in EtOH bottles. Significant populations of Sva0081 sediment group and SEEP-SRB-1 were also found (Fig. 6b), both of which are sulfur reducers in the family Desulfosarcinaceae (Dyksma et al., 2018; Skennerton et al., 2017). Only Syntrophotalea, however, was found to be a strong indicator genus for EtOH treatment (statistic = 0.945, p value = 0.009).
4.1 Carbon source exerts a primary control on methane production
4.1.1 More methane production from noncompetitive methylamine than ethanol
Our experimental design included two carbon treatments to investigate the effect of substrate competition on methane production from salt marsh microbial communities. We found that the addition of a noncompetitive, methylated organic carbon substrate (MMA) yields more measurable methane than a substrate that is fermented to acetate (EtOH) (Fig. 2a, c). This likely reflects an adaptation of the natural microbial assemblage to the ready availability of methylated substrates in salt marshes. Methylamines are degradation products of common osmolytes synthesized by marsh grasses and other marsh organisms, such as glycine betaine, proline betaine, choline, and dimethylsulfoniopropionate (DMSP) (Cavalieri and Huang, 1981; Glob and Sørensen, 1987; King, 1988; Mulholland and Otte, 2002; Wang and Lee, 1994a). As such, methylamines are abundantly available in the sediment of salt marshes (Fitzsimons et al., 1997; Gibb et al., 1999; Lee and Olson, 1984; Oremland et al., 1982; Wang and Lee, 1994a; Yang et al., 1994). S. alterniflora has been shown to release methylamine to sediments (Wang and Lee, 1994a), and, consistent with our results, active methylotrophic methanogens have previously been found in North American salt marshes where S. alterniflora is native (Buckley et al., 2008; Cai et al., 2022; Franklin et al., 1988; Krause and Treude, 2021; Oremland et al., 1982; Oremland and Polcin, 1982; Parkes et al., 2012a). Our results are also consistent with the shift from hydrogenotrophic to methylotrophic methanogenesis that was observed with the invasion of S. alterniflora into China's coastal salt marshes where it is not native (Yuan et al., 2016, 2019).
Although our 16S rRNA sequence data reflect the microbial community at the end of the incubations, rather than at the time point of highest methane production for MMA incubations (Fig. 2a, c), these bottles continued to produce methane throughout the incubation period. It was, therefore, somewhat surprising to find that methylotrophic methanogens were not among the more abundant genera recovered in 16S rRNA sequences in bottles incubated with MMA amendments (Fig. 5b). In fact, all representatives of the classical orders of euryarchaeotal methanogens averaged only 0.6 % of recovered sequences in MMA incubations across the four iron treatments. Instead, Bathyarchaeia was the most represented among the archaea by far (averaged 13 % of all sequence reads in MMA incubations). As a class, Bathyarchaeia are found abundantly in anoxic marine sediments and terrestrial soils (Fillol et al., 2016; Zhou et al., 2018), where they are thought to be important methylotrophic acetogens that degrade a variety of organic carbon substrates (Farag et al., 2020; Hou et al., 2023). It has been proposed that methylotrophic methanogenesis may also be possible among the Bathyarchaeia based on genomic data (Evans et al., 2015); however, this has not been shown experimentally, and a recent study of existing metagenomes indicated that methanogenesis would not be widespread among them (Hou et al., 2023). Therefore, it is not likely that Bathyarchaeia produce methane in MMA-amended incubations, although it is possible.
A more likely source of the methane production in MMA incubations is a population of classical methylotrophic methanogens that is highly active, despite representing a small proportion of the sequences recovered at the end of incubations. A small population of methanogens with high activity has been described previously in salt marsh sediments (Cai et al., 2022). Of the sequences that we recovered from established methanogens, members of the genus Methanococcoides were most abundant (Fig. 6a). Methanococcoides have been identified in other studies of salt marsh sediments (Jameson et al., 2019; Jones et al., 2019; Munson et al., 1997), and they are known to utilize methylamines and other methylated compounds, such as choline and glycine betaine, but not H2 or acetate (L'Haridon et al., 2014; Liang et al., 2022; Singh et al., 2005; Watkins et al., 2014). A similarly high relative abundance of Bathyarchaeia and smaller abundance of classical methanogens, specifically Methanococcoides, have also been reported in seagrass meadow sediment, where methylated compounds fuel methane production via methylotrophic methanogenesis (Hall et al., 2024; Schorn et al., 2022).
After Methanococcoide, Methanofastidiosales was the next most abundant of the established methanogens in MMA-amended incubations (Fig. 6a). Methanofastidiosales individuals are restricted to methanogenesis through methylated thiol reduction (Nobu et al., 2016). Methanethiol, which can be formed as a degradation product of DMSP and methionine or by methylation of sulfide (Lomans et al., 2002), is also commonly available in salt marsh sediments (Carrión et al., 2019; Dévai and Delaune, 1995; Kiene and Visscher, 1987) and was likely to have been naturally present in the sediments used for our incubations. Notably, Methanofastidiosales was found along with Methanococcoides and Bathyarchaeia in methanic sediments from seagrass meadows (Schorn et al., 2022). While the proportion of methanogen 16S rRNA sequences was lower overall in MMA treatments compared to EtOH treatments at the end of the incubations (Figs. 5b, 6a), the higher methane production rates observed with MMA likely indicate that a small population of salt marsh methanogens had high potential activity. They could have been primed to use noncompetitive methylated substrates, such as methylamines and methanethiol, that are not accessible to sulfate- or iron-reducing bacteria. Methanogens in our incubations could also have been supported by the products of organic matter degradation by Bathyarchaeia, as has been reported previously in other iron-rich, sulfate-poor sediments (Ruiz-Blas et al., 2024).
4.1.2 Ethanol yields a larger share of methanogens and potential concurrent methane oxidation
Incubations amended with EtOH also produced methane throughout the incubations, although less and later than with MMA amendments (Figs. 2, 3). EtOH would not directly support methanogens, but it can be fermented to methanogenic substrates by other members of the microbial community (Du et al., 2021; Schink, 1997). The lag in methane production in EtOH incubations relative to MMA incubations was likely related to the requirement that ethanol was converted to acetate or H2 by bacteria before it could be utilized. The two most abundant bacteria in incubations amended with EtOH were Syntrophotaleaceae and Anaerolineaceae (Figs. 5b, 6b). Both of these bacterial genera are likely involved in organic carbon fermentation (Hackmann, 2024; McIlroy et al., 2017). Syntrophotalea was the most abundant bacterial genus by far (Fig. 6b) and was the second most abundant among all taxa recovered from EtOH incubations (12 %–17 %). Isolates of the genus Syntrophotalea (formerly Pelobacter) have been shown to ferment ethanol in the presence of a H2-scavenging methanogen and to produce acetate among their fermentation products (Eichler and Schink, 1985; Schink, 1984, 2006). More recently, a member of Syntrophotalea was shown to transfer formate to a Methanococcus isolate in a mutualistic, methane-producing relationship (Day et al., 2022). Anaerolineaceae, although not as abundant, also likely contributed to fermentation in EtOH incubations. They have been identified as primary fermenters capable of degrading carbohydrates, proteinaceous material, or alkanes in anaerobic digesters (Bovio-Winkler et al., 2020; Liang et al., 2015; McIlroy et al., 2017). In a few instances, they have also been found to be associated with acetoclastic methanogens from the Methanothrix (formerly Methanosaeta) genus (Liang et al., 2015; McIlroy et al., 2017).
Even though (1) measured methane production was lower overall (Fig. 2c) and (2) acetate and H2 are competitive substrates that can be consumed by both methanogen and non-methanogen members of the microbial community, classical methanogens represented a much larger share of the 16S rRNA sequences recovered at the end of EtOH incubations (Fig. 5b). Methanosarcina was the dominant microbial genus, representing 15 % to 25 % of sequences recovered from EtOH incubations. Although Methanosarcina individuals are capable of hydrogenotrophic, acetoclastic, and methylotrophic methanogenesis, their ability to use acetate at high concentrations (Stams et al., 2019) was likely related to their proliferation in EtOH-amended incubations. Methanococcus, a genus of hydrogenotrophic methanogen, represented an additional 4 % to 8 % of total methanogen sequences in our EtOH incubations (Fig. 6a). Their abundance allows for the possibility that a mutualistic relationship, such as the one reported for Syntrophotalea and Methanococcus in an earlier study (Day et al., 2022), may have supported Methanococcus in our incubations as well. Although we did not find acetoclastic Methanothrix among the methanogens in our EtOH incubations, it is possible that a similar cooperative interaction that was described between Anaerolineaceae and Methanothrix (Liang et al., 2015; McIlroy et al., 2017) formed between Anaerolineaceae and an acetoclastic Methanosarcina in our incubations.
The larger proportion of methanogens but lower methane production observed in EtOH incubations relative to MMA incubations could result from a less active population of methanogens than in the MMA-amended bottles. However, an alternative explanation is that our measurements of methane concentrations underestimated methane production due to methane oxidation in EtOH incubations. Concurrent methane production and oxidation has been reported in salt marsh sediments (Krause et al., 2023, 2025; Krause and Treude, 2021; Segarra et al., 2013) as well in other coastal settings (Krause et al., 2023; Xiao et al., 2017, 2018). Methane oxidation is suggested by our identification of small numbers of ANME-3. Although ANME-3 comprised a small minority of sequences (included in “Other” in Fig. 6a), these individuals were most represented in EtOH incubations with ferrihydrite, magnetite, and hematite (1 %, 0.3 % and 0.8 %, respectively). We also detected the presence of the sulfate-reducing partner of anaerobic methanotrophs, SEEP-SRB1 (Schreiber et al., 2010; Skennerton et al., 2017), in all EtOH-amended incubations (Fig. 6b). Although sulfate-free artificial seawater was used to make up our sediment slurry, natural reduced sulfur of multiple forms was identified in the marsh creek sediments (Fig. 1b). Even though we did not measure sulfur species during the incubations and reduced sulfur native to the sediments would have been diluted by our use of sulfate-free artificial seawater, sulfur cycling is suggested by the presence of sulfur oxidizing Thiotrichaceae (Ravin et al., 2023) in addition to the sulfate-reducing Desulfosarcinaceae (Fig. 5b). It is also possible that sulfate-reducing bacteria were reducing Fe(III) in our incubations (Coleman et al., 1993; Lovley et al., 1993) or that anaerobic methane oxidation could be coupled to Fe(III) reduction (Cai et al., 2018; Ettwig et al., 2016; Yu et al., 2022), which has been observed previously in salt marsh sediments (Krause and Treude, 2021; Segarra et al., 2013). Overall, a picture emerges of a complex web of interspecies interactions mediated by multiple indirect electron shuttles and redox-active species in EtOH incubations.
4.2 Conductive minerals promote increased methylotrophic methane production
We expected EtOH incubations to reveal the potential for IET to enhance methane production by salt marsh microbial communities based on the results of previous studies. Many studies of co-cultures and enrichments in which IET was found use ethanol to enrich the bacterial and methanogen partners and to support methane production (Kato et al., 2012; Rotaru et al., 2019, 2014, 2018; Tang et al., 2016). Transfer of electrons between methanogens and electroactive bacteria has been shown through both magnetite and hematite (Aromokeye et al., 2018; Kato et al., 2012; Zhou et al., 2014). However, (semi)conductive iron minerals have also been shown to enhance the activity of acetoclastic methanogens themselves, without a bacterial partner (Fu et al., 2019; Wang et al., 2020; Xiao et al., 2021). The typical methanogen involved in either pathway that accelerates methane production is from the metabolically versatile genus Methanosarcina or the acetoclastic genus Methanothrix (Rotaru et al., 2021; Xiao et al., 2021). Although we did not detect Methanothrix, Methanosarcina was abundantly represented in our EtOH incubations (Fig. 5b). However, we did not observe any definitive increase in methane production in the presence of either magnetite or hematite relative to no iron amendment in EtOH incubations (Fig. 2d). This suggests that IET was not active in EtOH incubations. Genera of well-known electroactive bacteria (Garber et al., 2024; Lovley and Holmes, 2022) were also not detected in EtOH incubations.
Instead, the most methane produced and the highest methane production rates that we observed were found in MMA + magnetite incubations followed by MMA + hematite incubations in the first 10 d (Figs. 2, 3). Although methane production with no iron amendment was variable, the average among replicates was lower than that with magnetite or hematite amendments, and methane production with MMA + ferrihydrite was significantly lower. This pattern of higher methane production rates with magnetite and hematite than with ferrihydrite or no iron has been reported previously in rice paddy soil microbial enrichments (Kato et al., 2012; Zhou et al., 2014). However, the most abundant methanogens in MMA incubations with these iron treatments were neither Methanosarcina nor Methanothrix. A few other methanogenic genera have been found capable of participating in IET (e.g., Methanobacterium; Zheng et al., 2020, 2021), suggesting that the full diversity of IET-capable methanogens is not yet known. Methylotrophic methanogens, such as those that were most abundant in our MMA incubations (Fig. 6a), have not been identified as capable of IET to date. Still, Methanococcoides are a member of Methanosarcinales, an order with other IET-capable genera. Some Methanococcoides species have been found with cytochromes (Jussofie and Gottschalk, 1986; Saunders et al., 2005) that could facilitate extracellular electron transfer.
The bacterial partner in IET interactions is more variable and, therefore, more difficult to predict. In studies of IET-active co-cultures and enrichments, Geobacter is often the syntrophic partner of methanogens and can be found in high abundance, although not always (Deng et al., 2023; Holmes et al., 2017; Kato et al., 2012; Liu et al., 2015; Rotaru et al., 2014; Zhou et al., 2014). We did not recover 16S rRNA sequences belonging to Geobacter or other bacterial genera that are found in syntrophic association with methanogens (e.g., Syntrophomonas: Yuan et al., 2020; Zhao et al., 2018; or Rhodoferax: Yee and Rotaru, 2020; Zhao et al., 2020). Instead, the most abundant bacterial genus was Ca. Omnitrophus (Fig. 6b). Ca. Omnitrophus has a widespread distribution in the environment, but these individuals have remained relatively uncharacterized. Members of the Ca. Omnitrophus genus have been associated with metal-contaminated groundwater (Bärenstrauch et al., 2022) and magnetite production in the form of magnetosomes (Kolinko et al., 2016; Lin et al., 2018). In the class Omnitrophia, metagenomic evidence indicates the potential to produce cytochromes for use in dissimilatory metal reduction and conductive pili (Perez-Molphe-Montoya et al., 2022; Seymour et al., 2023). These features allow for the possibility that Ca. Omnitrophus individuals are the electroactive syntrophic partner of Methanococcoides in our MMA incubations. However, Ca. Omnitrophus was also found abundantly in MMA incubations with no iron and ferrihydrite treatments (Fig. 6b), and these incubations did not appear to have active IET. The other bacterial genera with the highest abundances, WOR-1, or Saganbacteria; Lentimicrobiaceae; MSBL5; and Sulfurovum (Fig. 6b) are less likely to be the electroactive bacterial partner. Lentimicrobiaceae individuals are thought to be fermenters (Sun et al., 2016), Sulfurovum comprises sulfur-reducing bacteria (Li et al., 2023; Wang et al., 2023), and metabolic capabilities and biogeochemical roles of Saganbacteria and MBSL5 are largely unknown. Like Ca. Omnitrophus, the abundances of these genera are also not distinctly different between bottles with magnetite and hematite amendments vs. bottles with ferrihydrite and no iron amendments. Despite the distinctly higher methane production rates early on, the microbial communities in MMA + magnetite and MMA + hematite incubations were not significantly different from MMA + no iron and MMA + ferrihydrite by the end of the incubations. The similar bacterial assemblages in MMA incubations may reflect similar methane production rates at the end of the incubations (Fig. 3) or they could be explained by the microbial community responding to the small amount of magnetite naturally present in salt marsh sediments (Fig. 4).
4.3 Magnetite formation from ferrihydrite with ethanol supplementation
Despite general similarities in methane production and microbial community composition in all incubations with the same organic carbon amendment, large quantities of magnetic minerals were produced in only one of our incubations: EtOH + ferrihydrite (Figs. 4, C1). Coercivity, remanent coercivity, and coercivity spectra suggest that the mineral is magnetite and that the magnetite produced in EtOH + ferrihydrite incubations was different from that natively present in marsh creek sediments (Fig. 4c, d). The low coercivity suggests very fine grained magnetite (above 25 nm but probably not over 80 nm (Dunlop, 1973; Dunlop and Ozdemir, 2011; Enkin and Dunlop, 1987). It is also lower coercivity than seen in magnetosomes, suggesting extracellular precipitation (Bai et al., 2022; Egli, 2004; Xue et al., 2024). Such a transformation of ferrihydrite is not surprising considering that iron-reducing bacteria are known to facilitate the conversion of poorly crystalline Fe(III) minerals to more crystalline, iron-bearing minerals such as magnetite (Benner et al., 2002; Hansel et al., 2003; Zachara et al., 2002). However, no detectable magnetite was formed from ferrihydrite during incubation with MMA (Fig. 4a), even though Fe2+ concentrations were elevated during the incubation, indicating that iron reduction occurred (Fig. 2b), and there was a small delay in methane production, indicating that iron reduction was competing with methanogenesis early in the incubations (Fig. 2a). Although the mineral fate of Fe(II) produced by iron reducers has been tied to the rate of iron reduction (Kappler et al., 2023), incubations with EtOH + ferrihydrite, MMA + ferrihydrite, and EtOH + magnetite treatments all produced similar amounts of net Fe2+ (Fig. 2b, d), suggesting similar iron reduction rates. Microbial community composition did not provide strong clues as to the identity of the bacteria responsible for magnetite formation either. There was no significant difference between the bacterial community in the EtOH + ferrihydrite incubation and other EtOH treatments (Fig. 6b). No well-known dissimilatory iron reducers (or magnetotactic bacteria) were among the bacterial genera that we recovered from EtOH incubations (Bazylinski et al., 2013; Kappler et al., 2021; Simmons and Edwards, 2007a; Zhao et al., 2025), with the exception of Ca. Omnitrophus. In contrast to MMA incubations, however, only a small fraction of sequences in EtOH incubations were from Ca. Omnitrophus. Although the genus Ca. Omnitrophus does include magnetotactic bacteria (Kolinko et al., 2016; Perez-Molphe-Montoya et al., 2022), they were much less abundant in EtOH incubations than in MMA incubations. There is a possibility that bacteria of the genus Methanosarcina, which were highly abundant in EtOH incubations, are directly involved in iron reduction and ferromagnetic mineral formation, as cultures of Methanosarcina barkeri have been shown to be capable of ferrihydrite reduction (Bond and Lovley, 2002; Sivan et al., 2016; Yu et al., 2022).
In rice paddy soils, the transformation of ferrihydrite to magnetite results in an eventual enhancement of methane production (Tang et al., 2016; Zhuang et al., 2015, 2019), but the impact of magnetite formation on methane production has not been characterized yet in salt marshes. We did not observe a similar increase in methane production following magnetite formation in EtOH + ferrihydrite incubations to that observed in rice paddy incubations, at least on the timescale of our 34 d incubations. However, this may result from a mismatch between the more prolific methylotrophic methane producers in our salt marsh incubations and the formation of magnetite, which only occurred in EtOH incubations. Although we did not detect magnetite formation in MMA + ferrihydrite incubations, the widespread presence of crystalline iron minerals at anoxic and euxinic depths resulting from the transformation of amorphous iron minerals has been reported previously (Kostka and Luther, 1995; Taillefert et al., 2007). Separate populations of iron-reducing bacteria accessing different carbon substrates may be involved in magnetite formation and IET in salt marshes. The activity of both would influence the rates and quantities of methane production by methylotrophic methanogens.
In this study, we hypothesized that the noncompetitive, methylated substrate MMA would yield more methane production than EtOH without the addition of iron oxide minerals but that the acetoclastic methanogens that we expected to find in EtOH incubations would respond to the presence of (semi)conductive iron oxides with increased methane production, whereas methylotrophic methanogens in MMA incubations would not. Instead, we found that a noncompetitive substrate and the presence of conductive minerals worked in concert to yield the highest methane production from salt marsh microbial communities. Therefore, methanogens in salt marshes can employ multiple simultaneous strategies to circumvent the redox ladder. Adding a methylated organic carbon substrate had a stronger effect on measured methane production and on the microbial community than the presence of conductive minerals. This suggests that it is not only the presence of sulfate in polyhaline salt marshes that controls methane production but also the contribution of methyl-containing osmolytes from marsh plants and animals that support methane production in higher-salinity coastal environments. The transformation of ferrihydrite to magnetite and the persistent presence of crystalline iron minerals in anoxic zones of salt marsh sediments could further enhance methane production in salt marshes. The importance of these multiple factors and their inevitable variability across space, depth, and seasons are likely to contribute to the heterogeneity that has been observed in methane emissions from salt marshes.

Figure B1Cluster dendrograms for DNA-based 16S rRNA gene diversity sorted only to the (a) domain Bacteria and the (b) domain Archaea. Approximately unbiased (AU) p values are in red (as a percentage), while bootstrap probability (BP) values are shown in green.
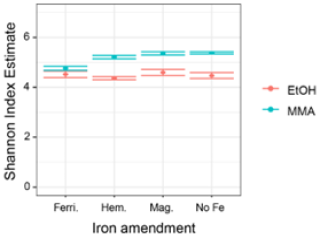
Figure B2Shannon diversity index values for microbial assemblages at the end of the incubation period.
Sediments from incubations amended with hematite
A higher saturation magnetization was observed in the MMA + hematite treatment compared to the EtOH + hematite treatment. Coercivity spectra are similar between the two treatments with two clear magnetic phases (hematite and magnetite), but the relative ratios of magnetite and hematite differ slightly. The ratio is 43:57 in the EtOH + hematite treatment vs. 37:63 in the MMA + hematite treatment. This is reflected in the slightly lower coercivity values in the EtOH + hematite treatment than the MMA + hematite treatment (Fig. 4b, Table C1).
Table C1Magnetic measurement results for iron minerals and all incubations at the beginning of the incubation periods (sediment + no amendments) and at the end of the incubation period (all other incubations).
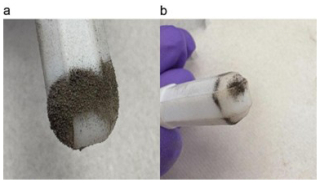
Figure C1Photo of ferromagnetic minerals pulled out of freeze-dried sediment with a magnetic stir bar. These sediments were recovered from EtOH + ferrihydrite incubations at the end of the incubation period. The magnetic mineral sticking to the stir bar is likely magnetite and/or greigite. (a) Stir bar with magnetic mineral and sediments. (b) Stir bar after sediments had been tapped off, more selectively leaving the magnetic mineral attached.
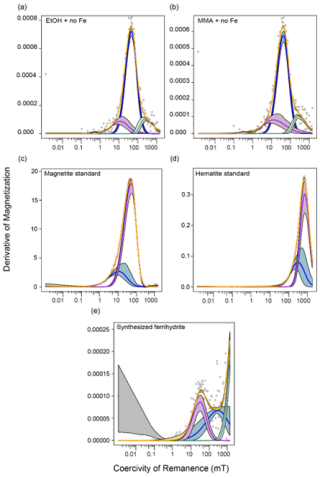
Figure C2MAX UnMix coercivity spectra of (a) EtOH with no iron amendment at the end of the incubation period, (b) MMA with no iron amendment at the end of the incubation period, (c) the magnetite standard used in all magnetite-containing incubations, (d) the hematite standard used in all hematite-containing incubations, and (e) the synthesized ferrihydrite used in all ferrihydrite-containing incubations. Because ferrihydrite is only weakly ferromagnetic, there is high noise in the data, making it difficult to deconvolve a peak.
An Rmd file used to generate all DNA figures and data tables can be found at https://hansonlabgit.dbi.udel.edu/hanson/block_et_al_manuscript.git (Block et al., 2025a).
Amplicon sequence data are available from https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1200716/ (Block et al., 2025b).
This project was conceptualized by SRSW, with input from KRB and AA. Development of the methodology and investigation was done by KRB, AA, and SRSW. All authors contributed to data curation and formal analyses. Data visualization was done by KRB, SRSW, TEH, and SPS. Original draft preparation was done by KB and SRSW. All authors contributed to reviewing and editing the manuscript. Funding acquisition, project administration, and supervision were done by SRSW.
The contact author has declared that none of the authors has any competing interests.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors. Views expressed in the text are those of the authors and do not necessarily reflect the views of the publisher.
Undergraduate interns participating in an NSF-supported Marine Sciences REU program over multiple years contributed to the development of this project: Sarah Bartoloni in 2019; Ethan Goulart in 2021; Samantha Reynolds in 2024; and an undergraduate intern participating in the UD Summer Scholars program, Matthew Hicks, in 2021. Particular thanks is owed to S. Reynolds for her contributions and discussions during the writing of the manuscript. We also thank lab members Justin Guider, Sawyer Hilt, and Leland Wood, for their assistance with field work; Andrew Wozniak, for the use of his GC-FID instrument; and Jennifer Biddle, for her advice and suggestions during the years of iterative improvement. Our manuscript was significantly improved by helpful discussions about cyclic voltammetry data interpretation with Mustafa Yücel.
The development of this project was supported by grant no. NSF OCE-2342979 to SRSW. The participation of AA (and previous undergraduate interns) was made possible by the UD Marine Sciences Research Experiences for Undergraduates (REU) program administered by Joanna York (grant no. NSF OCE-2051069). KB was supported by grant no. NSF OCE-2342979 as well as a University of Delaware Graduate Scholars Award.
This paper was edited by Robert Rhew and reviewed by William D. Orsi and one anonymous referee.
Al-Haj, A. N. and Fulweiler, R. W.: A synthesis of methane emissions from shallow vegetated coastal ecosystems, Glob. Chang. Biol., 26, 2988–3005, 2020.
Antler, G., Mills, J. V., Hutchings, A. M., Redeker, K. R., and Turchyn, A. V.: The Sedimentary Carbon-Sulfur-Iron Interplay – A Lesson From East Anglian Salt Marsh Sediments, Front Earth Sci., 7, 140, https://doi.org/10.3389/feart.2019.00140, 2019.
Apprill, A., McNally, S., Parsons, R., and Weber, L.: Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton, Aquat. Microb. Ecol., 75, 129–137, 2015.
Aromokeye, D. A., Richter-Heitmann, T., Oni, O. E., Kulkarni, A., Yin, X., Kasten, S., and Friedrich, M. W.: Temperature Controls Crystalline Iron Oxide Utilization by Microbial Communities in Methanic Ferruginous Marine Sediment Incubations, Front. Microbiol., 9, 2574, https://doi.org/10.3389/fmicb.2018.02574, 2018.
Bai, F., Chang, L., Pei, Z., Harrison, R. J., and Winklhofer, M.: Magnetic biosignatures of magnetosomal greigite from micromagnetic calculation, Geophys. Res. Lett., 49, https://doi.org/10.1029/2022gl098437, 2022.
Bärenstrauch, M., Vanhove, A. S., Allégra, S., Peuble, S., Gallice, F., Paran, F., Lavastre, V., and Girardot, F.: Microbial diversity and geochemistry of groundwater impacted by steel slag leachates, Sci. Total Environ., 843, 156987, https://doi.org/10.1016/j.scitotenv.2022.156987, 2022.
Bartlett, K. B., Bartlett, D. S., Harriss, R. C., and Sebacher, D. I.: Methane emissions along a salt marsh salinity gradient, Biogeochemistry, 4, 183–202, 1987.
Bazylinski, D. A., Frankel, R. B., Garratt-Reed, A. J., and Mann, S.: Biomineralization of Iron Sulfides in Magnetotactic Bacteria from Sulfidic Environments, in: Iron Biominerals, edited by: Frankel, R. B. and Blakemore, R. P., Springer US, Boston, MA, 239–255, https://doi.org/10.1007/978-1-4615-3810-3_17, 1991.
Bazylinski, D. A., Williams, T. J., Lefèvre, C. T., Trubitsyn, D., Fang, J., Beveridge, T. J., Moskowitz, B. M., Ward, B., Schübbe, S., Dubbels, B. L., and Simpson, B.: Magnetovibrio blakemorei gen. nov., sp. nov., a magnetotactic bacterium (Alphaproteobacteria: Rhodospirillaceae) isolated from a salt marsh, Int. J. Syst. Evol. Microbiol., 63, 1824–1833, 2013.
Benner, S. G., Hansel, C. M., Wielinga, B. W., Barber, T. M., and Fendorf, S.: Reductive dissolution and biomineralization of iron hydroxide under dynamic flow conditions, Environ. Sci. Technol., 36, 1705–1711, 2002.
Blakemore, R.: Magnetotactic bacteria, Science, 190, 377–379, 1975.
Block, K. R., Arbetman, A., Slotznick, S. P., Hanson, T. E., Luther III, G. W. and Shah Walter, S. R.: Block et al Manuscript Markdowns, Block_et_al_Manuscript, https://hansonlabgit.dbi.udel.edu/hanson/block_et_al_manuscript (last access: 10 February 2025), 2025a.
Block, K. R., Arbetman, A., Slotznick, S. P., Hanson, T. E., Luther III, G. W. and Shah Walter, S. R.: NCBI BioProject ID PRJNA1200716, Influence of Carbon Substrate and Iron Oxide Minerals on Methane Production and Magnetic Mineral Formation in Salt Marsh Sediments, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1200716/ (last access: 10 February 2025), 2025b.
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., Alexander, H., Alm, E. J., Arumugam, M., Asnicar, F., Bai, Y., Bisanz, J. E., Bittinger, K., Brejnrod, A., Brislawn, C. J., Brown, C. T., Callahan, B. J., Caraballo-Rodríguez, A. M., Chase, J., Cope, E. K., Da Silva, R., Diener, C., Dorrestein, P. C., Douglas, G. M., Durall, D. M., Duvallet, C., Edwardson, C. F., Ernst, M., Estaki, M., Fouquier, J., Gauglitz, J. M., Gibbons, S. M., Gibson, D. L., Gonzalez, A., Gorlick, K., Guo, J., Hillmann, B., Holmes, S., Holste, H., Huttenhower, C., Huttley, G. A., Janssen, S., Jarmusch, A. K., Jiang, L., Kaehler, B. D., Kang, K. B., Keefe, C. R., Keim, P., Kelley, S. T., Knights, D., Koester, I., Kosciolek, T., Kreps, J., Langille, M. G. I., Lee, J., Ley, R., Liu, Y.-X., Loftfield, E., Lozupone, C., Maher, M., Marotz, C., Martin, B. D., McDonald, D., McIver, L. J., Melnik, A. V., Metcalf, J. L., Morgan, S. C., Morton, J. T., Naimey, A. T., Navas-Molina, J. A., Nothias, L. F., Orchanian, S. B., Pearson, T., Peoples, S. L., Petras, D., Preuss, M. L., Pruesse, E., Rasmussen, L. B., Rivers, A., Robeson 2nd, M. S., Rosenthal, P., Segata, N., Shaffer, M., Shiffer, A., Sinha, R., Song, S. J., Spear, J. R., Swafford, A. D., Thompson, L. R., Torres, P. J., Trinh, P., Tripathi, A., Turnbaugh, P. J., Ul-Hasan, S., van der Hooft, J. J. J., Vargas, F., Vázquez-Baeza, Y., Vogtmann, E., von Hippel, M., Walters, W., Wan, Y., Wang, M., Warren, J., Weber, K. C., Williamson, C. H. D., Willis, A. D., Zech Xu, Z., Zaneveld, J. R., Zhang, Y., Zhu, Q., Knight, R. and Caporaso, J. G.: Author Correction: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 37, 1091, https://doi.org/10.1038/s41587-019-0252-6, 2019.
Bond, D. R. and Lovley, D. R.: Reduction of Fe(III) oxide by methanogens in the presence and absence of extracellular quinones, Environ. Microbiol., 4, 115–124, 2002.
Bovio-Winkler, P., Cabezas, A., and Etchebehere, C.: Database mining to unravel the ecology of the phylum Chloroflexi in methanogenic full scale bioreactors, Front. Microbiol., 11, 603234, https://doi.org/10.3389/fmicb.2020.603234, 2020.
Brendel, P. J. and Luther, G. W.: Development of a gold amalgam voltammetric microelectrode for the determination of dissolved Fe, Mn, O2, and S(-II) in porewaters of marine and freshwater sediments, Environ. Sci. Technol., 29, 751–761, 1995.
Bridgham, S. D., Megonigal, J. P., Keller, J. K., Bliss, N. B., and Trettin, C.: The carbon balance of North American wetlands, Wetlands, 26, 889–916, 2006.
Buckley, D. H., Baumgartner, L. K., and Visscher, P. T.: Vertical distribution of methane metabolism in microbial mats of the Great Sippewissett Salt Marsh, Environ. Microbiol., 10, 967–977, 2008.
Cai, C., Leu, A. O., Xie, G.-J., Guo, J., Feng, Y., Zhao, J.-X., Tyson, G. W., Yuan, Z., and Hu, S.: A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction, ISME J., 12, 1929–1939, 2018.
Cai, M., Yin, X., Tang, X., Zhang, C., Zheng, Q., and Li, M.: Metatranscriptomics reveals different features of methanogenic archaea among global vegetated coastal ecosystems, Sci. Total Environ., 802, 149848, https://doi.org/10.1016/j.scitotenv.2021.149848, 2022.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P.: DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 13, 581–583, 2016.
Carrión, O., Pratscher, J., Richa, K., Rostant, W. G., Farhan Ul Haque, M., Murrell, J. C., and Todd, J. D.: Methanethiol and dimethylsulfide cycling in stiffkey saltmarsh, Front. Microbiol., 10, 1040, https://doi.org/10.3389/fmicb.2019.01040, 2019.
Cavalieri, A. J. and Huang, A. H. C.: Accumulation of proline and glycinebetaine in Spartina alterniflora Loisel in response to NaCl and nitrogen in the marsh, Oecologia, 49, 224–228, 1981.
Chmura, G. L., Kellman, L., and Guntenspergen, G. R.: The greenhouse gas flux and potential global warming feedbacks of a northern macrotidaland microtidal salt marsh, Environ. Res. Lett., 6, 044016, https://doi.org/10.1088/1748-9326/6/4/044016, 2011.
Coleman, M. L., Hedrick, D. B., Lovley, D. R., White, D. C., and Pye, K.: Reduction of Fe(III) in sediments by sulphate-reducing bacteria, Nature, 361, 436–438, 1993.
Cruz Viggi, C., Rossetti, S., Fazi, S., Paiano, P., Majone, M., and Aulenta, F.: Magnetite particles triggering a faster and more robust syntrophic pathway of methanogenic propionate degradation, Environ. Sci. Technol., 48, 7536–7543, 2014.
Day, L. A., Kelsey, E. L., Fonseca, D. R., and Costa, K. C.: Interspecies Formate Exchange Drives Syntrophic Growth of Syntrophotalea carbinolica and Methanococcus maripaludis, Appl. Environ. Microbiol., 88, e0115922, https://doi.org/10.1128/aem.01159-22, 2022.
De Cáceres, M., Legendre, P., and Moretti, M.: Improving indicator species analysis by combining groups of sites, Oikos, 119, 1674–1684, 2010.
De Cáceres, M., Legendre, P., and Wiser, S. K.: Using species combinations in indicator value analyses, Methods Ecol. Evol., https://doi.org/10.1111/j.2041-210X.2012.00246.x, 2012.
Demitrack, A.: A Search for Bacterial Magnetite in the Sediments of Eel Marsh, Woods Hole, Massachusetts, in: Magnetite Biomineralization and Magnetoreception in Organisms: A New Biomagnetism, edited by: Kirschvink, J. L., Jones, D. S., and MacFadden, B. J., Springer US, Boston, MA, 625–645, https://doi.org/10.1007/978-1-4613-0313-8_35, 1985.
Deng, P., Wang, L., Li, X., Zhang, J., and Jiang, H.: Geobacter grbiciae – A new electron donor in the formation of co-cultures via direct interspecies electron transfer, Microbiol. Res. (Pavia), 14, 1774–1787, 2023.
Dévai, I. and Delaune, R.: Formation of volatile sulfur compounds in salt marsh sediment as influenced by soil redox condition, Organic Geochemistry, 23, 283–287, 1995.
Dewitt, P. and Daiber, F. C.: The hydrography of the Broadkill River estuary, Delaware, Chesapeake Science, 14, 28–40, 1973.
Du, J., Yin, Q., Gu, M., and Wu, G.: New insights into the effect of ethanol and volatile fatty acids proportions on methanogenic activities and pathways, Environ. Res., 194, 110644, https://doi.org/10.1016/j.envres.2020.110644, 2021.
Dunlop, D. J.: Superparamagnetic and single-domain threshold sizes in magnetite, J. Geophys. Res., 78, 1780–1793, 1973.
Dunlop, D. J. and Ozdemir, O.: Cambridge studies in magnetism: Rock magnetism: Fundamentals and frontiers series number 3, Cambridge University Press, Cambridge, England, https://doi.org/10.1017/CBO9780511612794, 2011.
Dyksma, S., Lenk, S., Sawicka, J. E., and Mußmann, M.: Uncultured Gammaproteobacteria and Desulfobacteraceae account for major acetate assimilation in a coastal marine sediment, Front. Microbiol., 9, 3124, https://doi.org/10.3389/fmicb.2018.03124, 2018.
Egli, R.: Characterization of individual rock magnetic components by analysis of remanence curves, 1. Unmixing natural sediments, Stud. Geophys. Geod., https://doi.org/10.1023/B:SGEG.0000020839.45304.6d, 2004.
Eichler, B. and Schink, B.: Fermentation of primary alcohols and diols and pure culture of syntrophically alcohol-oxidizing anaerobes, Arch. Microbiol., 143, 60–66, 1985.
Enkin, R. J. and Dunlop, D. J.: A micromagnetic study of pseudo single-domain remanence in magnetite, J. Geophys. Res., 92, 12726, https://doi.org/10.1029/JB092iB12p12726, 1987.
Ettwig, K. F., Zhu, B., Speth, D., Keltjens, J. T., Jetten, M. S. M., and Kartal, B.: Archaea catalyze iron-dependent anaerobic oxidation of methane, Proc. Natl. Acad. Sci. USA, 113, 12792–12796, 2016.
Evans, P. N., Parks, D. H., Chadwick, G. L., Robbins, S. J., Orphan, V. J., Golding, S. D., and Tyson, G. W.: Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics, Science, 350, 434–438, 2015.
Evans, P. N., Boyd, J. A., Leu, A. O., Woodcroft, B. J., Parks, D. H., Hugenholtz, P., and Tyson, G. W.: An evolving view of methane metabolism in the Archaea, Nat. Rev. Microbiol., 17, 219–232, 2019.
Farag, I. F., Biddle, J. F., Zhao, R., Martino, A. J., House, C. H., and León-Zayas, R. I.: Metabolic potentials of archaeal lineages resolved from metagenomes of deep Costa Rica sediments, ISME J., 14, 1345–1358, 2020.
Fillol, M., Auguet, J.-C., Casamayor, E. O., and Borrego, C. M.: Insights in the ecology and evolutionary history of the Miscellaneous Crenarchaeotic Group lineage, ISME J., 10, 665–677, 2016.
Fitzsimons, M. F., Jemmett, A. W., and Wolff, G. A.: A preliminary study of the geochemistry of methylamines in a salt marsh, Org. Geochem., 27, 15–24, 1997.
Franklin, M. J., Wiebe, W. J., and Whitman, W. B.: Populations of methanogenic bacteria in a georgia salt marsh, Appl. Environ. Microbiol., 54, 1151–1157, 1988.
Fu, L., Zhou, T., Wang, J., You, L., Lu, Y., Yu, L., and Zhou, S.: NanoFe3O4 as solid electron shuttles to accelerate acetotrophic methanogenesis by Methanosarcina barkeri, Front. Microbiol., 10, 388, https://doi.org/10.3389/fmicb.2019.00388, 2019.
Garber, A. I., Nealson, K. H., and Merino, N.: Large-scale prediction of outer-membrane multiheme cytochromes uncovers hidden diversity of electroactive bacteria and underlying pathways, Front. Microbiol., 15, 1448685, https://doi.org/10.3389/fmicb.2024.1448685, 2024.
Gibb, S. W., Mantoura, R. F. C., and Liss, P. S.: Ocean-atmosphere exchange and atmospheric speciation of ammonia and methylamines in the region of the NW Arabian Sea, Global Biogeochem. Cy., 13, 161–178, 1999.
Glob, E. and Sørensen, J.: Determination of dissolved and exchangeable trimethylamine pools in sediments, Journal of Microbiological Methods, 6, 347–355, 1987.
Gordon, A., Plessy, C., and K., S.: fastx_toolkit: FASTA/FASTQ pre-processing programs, Github, https://github.com/Debian/fastx-toolkit (last access: 10 February 2025), 2010.
Gribsholt, B., Kostka, J. E., and Kristensen, E.: Impact of fiddler crabs and plant roots on sediment biogeochemistry in a Georgia saltmarsh, Mar. Ecol. Prog. Ser., 259, 237–251, 2003.
Guider, J. T., Yoshimura, K. M., Block, K. R., Biddle, J. F., and Shah Walter, S. R.: Archaeal blooms and busts in an estuarine time series, Environ. Microbiol., 26, e16584, https://doi.org/10.1111/1462-2920.16584, 2024.
Hackmann, T. J.: The Vast Landscape of Carbohydrate Fermentation in Prokaryotes, FEMS Microbiology Reviews, 48, fuae016, https://doi.org/10.1093/femsre/fuae016, 2024.
Hall, N., Wong, W. W., Lappan, R., Ricci, F., Glud, R. N., Kawaichi, S., Rotaru, A.-E., Greening, C., and Cook, P. L. M.: Aerotolerant methanogens use seaweed and seagrass metabolites to drive marine methane emissions, bioRxiv [preprint], https://doi.org/10.1101/2024.10.14.618369, 2024.
Hansel, C. M., Benner, S. G., Neiss, J., Dohnalkova, A., Kukkadapu, R. K., and Fendorf, S.: Secondary mineralization pathways induced by dissimilatory iron reduction of ferrihydrite under advective flow, Geochim. Cosmochim. Acta, 67, 2977–2992, 2003.
Hartman, W. H., Bueno de Mesquita, C. P., Theroux, S. M., Morgan-Lang, C., Baldocchi, D. D., and Tringe, S. G.: Multiple microbial guilds mediate soil methane cycling along a wetland salinity gradient, mSystems, 9, e00936-23, https://doi.org/10.1128/msystems.00936-23, 2024.
Herbold, C. W., Pelikan, C., Kuzyk, O., Hausmann, B., Angel, R., Berry, D., and Loy, A.: A flexible and economical barcoding approach for highly multiplexed amplicon sequencing of diverse target genes, Front. Microbiol., 6, 731, https://doi.org/10.3389/fmicb.2015.00731, 2015.
Holmes, D. E., Shrestha, P. M., Walker, D. J. F., Dang, Y., Nevin, K. P., Woodard, T. L., and Lovley, D. R.: Metatranscriptomic evidence for direct interspecies electron transfer between Geobacter and Methanothrix species in methanogenic rice paddy soils, Appl. Environ. Microbiol., 83, https://doi.org/10.1128/AEM.00223-17, 2017.
Holmquist, J. R., Eagle, M., Molinari, R. L., Nick, S. K., Stachowicz, L. C., and Kroeger, K. D.: Mapping methane reduction potential of tidal wetland restoration in the United States, Communications Earth & Environment, 4, 1–11, 2023.
Hou, J., Wang, Y., Zhu, P., Yang, N., Liang, L., Yu, T., Niu, M., Konhauser, K., Woodcroft, B. J., and Wang, F.: Taxonomic and carbon metabolic diversification of Bathyarchaeia during its coevolution history with early Earth surface environment, Sci. Adv., 9, eadf5069, https://doi.org/10.1126/sciadv.adf5069, 2023.
Howarth, R. W.: The ecological significance of sulfur in the energy dynamics of salt marsh and coastal marine sediments, Biogeochemistry, 1, 5–27, 1984.
Howarth, R. W. and Giblin, A.: Sulfate reduction in the salt marshes at Sapelo Island, Georgia, Limnol. Oceanogr., 28, 70–82, 1983.
Howarth, R. W. and Teal, J. M.: Sulfate reduction in a New England salt marsh, Limnol. Oceanogr., 24, 999–1013, 1979.
Hyun, J.-H., Smith, A. C., and Kostka, J. E.: Relative contributions of sulfate- and iron(III) reduction to organic matter mineralization and process controls in contrasting habitats of the Georgia saltmarsh, Appl. Geochem., 22, 2637–2651, 2007.
Jackson, M. and Solheid, P.: On the quantitative analysis and evaluation of magnetic hysteresis data: HYSTERESIS LOOP EVALUATION AND ANALYSIS, Geochem. Geophys. Geosyst., 11, Q04Z15, https://doi.org/10.1029/2009GC002932, 2010.
Jameson, E., Stephenson, J., Jones, H., Millard, A., Kaster, A.-K., Purdy, K. J., Airs, R., Murrell, J. C., and Chen, Y.: Deltaproteobacteria (Pelobacter) and Methanococcoides are responsible for choline-dependent methanogenesis in a coastal saltmarsh sediment, ISME J., 13, 277–289, 2019.
Jones, H. J., Kröber, E., Stephenson, J., Mausz, M. A., Jameson, E., Millard, A., Purdy, K. J., and Chen, Y.: A new family of uncultivated bacteria involved in methanogenesis from the ubiquitous osmolyte glycine betaine in coastal saltmarsh sediments, Microbiome, 7, 120, https://doi.org/10.1186/s40168-019-0732-4, 2019.
Jørgensen, B. B.: Bacteria and Marine Biogeochemistry, in: Marine Geochemistry, edited by: Schulz, H. D. and Zabel, M., Springer Berlin Heidelberg, Berlin, Heidelberg, 169–206, https://doi.org/10.1007/3-540-32144-6_5, 2006.
Jørgensen, B. B. and Kasten, S.: Sulfur Cycling and Methane Oxidation, in: Marine Geochemistry, edited by: Schulz, H. D. and Zabel, M., Springer Berlin Heidelberg, Berlin, Heidelberg, 271–309, https://doi.org/10.1007/3-540-32144-6_8, 2006.
Jussofie, A. and Gottschalk, G.: Further studies on the distribution of cytochromes in methanogenic bacteria, FEMS Microbiol. Lett., 37, 15–18, 1986.
Kappler, A., Bryce, C., Mansor, M., Lueder, U., Byrne, J. M., and Swanner, E. D.: An evolving view on biogeochemical cycling of iron, Nat. Rev. Microbiol., 19, 360–374, 2021.
Kappler, A., Thompson, A., and Mansor, M.: Impact of biogenic magnetite formation and transformation on biogeochemical cycles, Elements (Que.), 19, 222–227, 2023.
Kato, S., Hashimoto, K., and Watanabe, K.: Methanogenesis facilitated by electric syntrophy via (semi)conductive iron-oxide minerals, Environ. Microbiol., 14, 1646–1654, 2012.
Kiene, R. P. and Visscher, P. T.: Production and fate of methylated sulfur compounds from methionine and dimethylsulfoniopropionate in anoxic salt marsh sediments, Appl. Environ. Microbiol., 53, 2426–2434, 1987.
King, G. M.: Methanogenesis from methylated amines in a hypersaline algal mat, Appl. Environ. Microbiol., 54, 130–136, 1988.
Koblitz, J., Halama, P., Spring, S., Thiel, V., Baschien, C., Hahnke, R. L., Pester, M., Overmann, J., and Reimer, L. C.: MediaDive: the expert-curated cultivation media database, Nucleic Acids Res., 51, D1531–D1538, 2023.
Kolinko, S., Richter, M., Glöckner, F.-O., Brachmann, A., and Schüler, D.: Single-cell genomics of uncultivated deep-branching magnetotactic bacteria reveals a conserved set of magnetosome genes: Genomic analysis of an uncultivated multicellular magnetotactic prokaryote, Environ. Microbiol., 18, 21–37, 2016.
Koretsky, C. M., Moore, C. M., Lowe, K. L., Meile, C., DiChristina, T. J., and Van Cappellen, P.: Seasonal oscillation of microbial iron and sulfate reduction in saltmarsh sediments (Sapelo Island, GA, USA), Biogeochemistry, 64, 179–203, 2003.
Koretsky, C. M., Van Cappellen, P., DiChristina, T. J., Kostka, J. E., Lowe, K. L., Moore, C. M., Roychoudhury, A. N., and Viollier, E.: Salt marsh pore water geochemistry does not correlate with microbial community structure, Estuar. Coast. Shelf Sci., 62, 233–251, 2005.
Kostka, J. E. and Luther, G. W.: Partitioning and speciation of solid phase iron in saltmarsh sediments, Geochim. Cosmochim. Acta, 58, 1701–1710, 1994.
Kostka, J. E. and Luther, G. W.: Seasonal cycling of Fe in saltmarsh sediments, Biogeochemistry, 29, 159–181, 1995.
Kostka, J. E., Roychoudhury, A., and Van Cappellen, P.: Rates and controls of anaerobic microbial respiration across spatial and temporal gradients in saltmarsh sediments, Biogeochemistry, 60, 49–76, 2002a.
Kostka, J. E., Gribsholt, B., Petrie, E., Dalton, D., Skelton, H., and Kristensen, E.: The rates and pathways of carbon oxidation in bioturbated saltmarsh sediments, Limnol. Oceanogr., 47, 230–240, 2002b.
Krause, S. J. E. and Treude, T.: Deciphering cryptic methane cycling: Coupling of methylotrophic methanogenesis and anaerobic oxidation of methane in hypersaline coastal wetland sediment, Geochim. Cosmochim. Acta, 302, 160–174, 2021.
Krause, S. J. E., Liu, J., Yousavich, D. J., Robinson, D., Hoyt, D. W., Qin, Q., Wenzhöfer, F., Janssen, F., Valentine, D. L., and Treude, T.: Evidence of cryptic methane cycling and non-methanogenic methylamine consumption in the sulfate-reducing zone of sediment in the Santa Barbara Basin, California, Biogeosciences, 20, 4377–4390, https://doi.org/10.5194/bg-20-4377-2023, 2023.
Krause, S. J. E., Wipfler, R., Liu, J., Yousavich, D. J., Robinson, D., Hoyt, D. W., Orphan, V. J., and Treude, T.: Spatial evidence of cryptic methane cycling and methylotrophic metabolisms along a land–ocean transect in salt marsh sediment, Geochim. Cosmochim. Acta, https://doi.org/10.1016/j.gca.2025.07.019, 2025.
Kreisler, J.: Magnetite preservation and diagenesis in marine sediments, Senior honors thesis, Department of Earth Sciences, Dartmouth College, 2022.
Lee, C. and Olson, B. L.: Dissolved, exchangeable and bound aliphatic amines in marine sediments: initial results, Organic Geochemistry, 6, 259–263, 1984.
L'Haridon, S., Chalopin, M., Colombo, D., and Toffin, L.: Methanococcoides vulcani sp. nov., a marine methylotrophic methanogen that uses betaine, choline and N,N-dimethylethanolamine for methanogenesis, isolated from a mud volcano, and emended description of the genus Methanococcoides, Int. J. Syst. Evol. Microbiol., 64, 1978–1983, 2014.
Li, Y., Wang, S., Ji, B., Yuan, Q., Wei, S., Lai, Q., Wu, K., Jiang, L., and Shao, Z.: Sulfurovum mangrovi sp. nov., an obligately chemolithoautotrophic, hydrogen-oxidizing bacterium isolated from coastal marine sediments, Int. J. Syst. Evol. Microbiol., 73, https://doi.org/10.1099/ijsem.0.006142, 2023.
Liang, B., Wang, L.-Y., Mbadinga, S. M., Liu, J.-F., Yang, S.-Z., Gu, J.-D., and Mu, B.-Z.: Anaerolineaceae and Methanosaeta turned to be the dominant microorganisms in alkanes-dependent methanogenic culture after long-term of incubation, AMB Express, 5, 117, https://doi.org/10.1186/s13568-015-0117-4, 2015.
Liang, L., Sun, Y., Dong, Y., Ahmad, T., Chen, Y., Wang, J., and Wang, F.: Methanococcoides orientis sp. nov., a methylotrophic methanogen isolated from sediment of the East China Sea, Int. J. Syst. Evol. Microbiol., 72, https://doi.org/10.1099/ijsem.0.005384, 2022.
Lin, W., Zhang, W., Zhao, X., Roberts, A. P., Paterson, G. A., Bazylinski, D. A., and Pan, Y.: Genomic expansion of magnetotactic bacteria reveals an early common origin of magnetotaxis with lineage-specific evolution, ISME J., 12, 1508–1519, 2018.
Liu, F., Rotaru, A.-E., Shrestha, P. M., Malvankar, N. S., Nevin, K. P., and Lovley, D. R.: Magnetite compensates for the lack of a pilin-associated c-type cytochrome in extracellular electron exchange: Magnetite compensates for the lack of OmcS, Environ. Microbiol., 17, 648–655, 2015.
Liu, J., Klonicki-Ference, E., Krause, S. J. E., and Treude, T.: Iron oxides fuel anaerobic oxidation of methane in the presence of sulfate in hypersaline coastal wetland sediment, Environ. Sci. Technol., 59, 513–522, 2025.
Lomans, B. P., van der Drift, C., Pol, A., and Op den Camp, H. J. M.: Microbial cycling of volatile organic sulfur compounds, Cell. Mol. Life Sci., 59, 575–588, 2002.
Lovley, D. R.: Happy together: microbial communities that hook up to swap electrons, ISME J., 11, 327–336, 2017a.
Lovley, D. R.: Syntrophy Goes Electric: Direct Interspecies Electron Transfer, Annu. Rev. Microbiol., 71, 643–664, 2017b.
Lovley, D. R. and Holmes, D. E.: Electromicrobiology: the ecophysiology of phylogenetically diverse electroactive microorganisms, Nat. Rev. Microbiol., 20, 5–19, 2022.
Lovley, D. R., Roden, E. E., Phillips, E. J. P., and Woodward, J. C.: Enzymatic iron and uranium reduction by sulfate-reducing bacteria, Mar. Geol., 113, 41–53, 1993.
Lowe, K. L., Dichristina, T. J., Roychoudhury, A. N., and Van Cappellen, P.: Microbiological and Geochemical Characterization of Microbial Fe(III) Reduction in Salt Marsh Sediments, Geomicrobiol. J., 17, 163–178, 2000.
Luther, G. W., III, Glazer, B. T., Ma, S., Trouwborst, R. E., Moore, T. S., Metzger, E., Kraiya, C., Waite, T. J., Druschel, G., Sundby, B., Taillefert, M., Nuzzio, D. B., Shank, T. M., Lewis, B. L., and Brendel, P. J.: Use of voltammetric solid-state (micro)electrodes for studying biogeochemical processes: Laboratory measurements to real time measurements with an in situ electrochemical analyzer (ISEA), Mar. Chem., 108, 221–235, 2008.
Määttä, T. and Malhotra, A.: The hidden roots of wetland methane emissions, Glob. Chang. Biol., 30, e17127, https://doi.org/10.1111/gcb.17127, 2024.
Matheus Carnevali, P. B., Schulz, F., Castelle, C. J., Kantor, R. S., Shih, P. M., Sharon, I., Santini, J. M., Olm, M. R., Amano, Y., Thomas, B. C., Anantharaman, K., Burstein, D., Becraft, E. D., Stepanauskas, R., Woyke, T., and Banfield, J. F.: Hydrogen-based metabolism as an ancestral trait in lineages sibling to the Cyanobacteria, Nat. Commun., 10, 463, https://doi.org/10.1038/s41467-018-08246-y, 2019.
Mausz, M. A. and Chen, Y.: Microbiology and ecology of methylated Amine metabolism in marine ecosystems, Curr. Issues Mol. Biol., 33, 133–148, 2019.
Maxbauer, D. P., Feinberg, J. M., and Fox, D. L.: MAX UnMix: A web application for unmixing magnetic coercivity distributions, Comput. Geosci., 95, 140–145, 2016.
McIlroy, S. J., Kirkegaard, R. H., Dueholm, M. S., Fernando, E., Karst, S. M., Albertsen, M., and Nielsen, P. H.: Culture-independent analyses reveal novel Anaerolineaceae as abundant primary fermenters in anaerobic digesters treating waste activated sludge, Front. Microbiol., 8, 1134, https://doi.org/10.3389/fmicb.2017.01134, 2017.
Moore, R.: FeatureTable – R6 and s3 classes for feature tables, data, and microbiome analysis, Github, https://github.com/mooreryan/featuretable (last access: 10 February 2025), 2020.
Mulholland, M. M. and Otte, M. L.: The effects of nitrogen supply and salinity on DMSP, glycine betaine and proline concentrations in leaves of Spartina anglica, Aquat. Bot., 72, 193–200, 2002.
Munson, M. A., Nedwell, D. B., and Embley, T. M.: Phylogenetic diversity of Archaea in sediment samples from a coastal salt marsh, Appl. Environ. Microbiol., 63, 4729–4733, 1997.
Nobu, M. K., Narihiro, T., Kuroda, K., Mei, R., and Liu, W.-T.: Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen, ISME J., 10, 2478–2487, 2016.
Oremland, R. S. and Polcin, S.: Methanogenesis and sulfate reduction: competitive and noncompetitive substrates in estuarine sediments, Appl. Environ. Microbiol., 44, 1270–1276, 1982.
Oremland, R. S., Marsh, L. M., and Polcin, S.: Methane production and simultaneous sulphate reduction in anoxic, salt marsh sediments, Nature, 296, 143–145, 1982.
Parada, A. E., Needham, D. M., and Fuhrman, J. A.: Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples, Environ. Microbiol., 18, 1403–1414, 2016.
Parkes, R. J., Brock, F., Banning, N., Hornibrook, E. R. C., Roussel, E. G., Weightman, A. J., and Fry, J. C.: Changes in methanogenic substrate utilization and communities with depth in a salt-marsh, creek sediment in southern England, Estuar. Coast. Shelf Sci., 96, 170–178, 2012a.
Parkes, R. J., Brock, F., Banning, N., Hornibrook, E. R. C., Roussel, E. G., Weightman, A. J., and Fry, J. C.: Changes in methanogenic substrate utilization and communities with depth in a salt-marsh, creek sediment in southern England, Estuar. Coast. Shelf Sci., 96, 170–178, 2012b.
Perez-Molphe-Montoya, E., Küsel, K., and Overholt, W. A.: Redefining the phylogenetic and metabolic diversity of phylum Omnitrophota, Environ. Microbiol., 24, 5437–5449, 2022.
Peters, C. and Dekkers, M. J.: Selected room temperature magnetic parameters as a function of mineralogy, concentration and grain size, Phys. Chem. Earth, 28, 659–667, 2003.
Poffenbarger, H. J., Needelman, B. A., and Megonigal, J. P.: Salinity influence on methane emissions from tidal marshes, Wetlands, 31, 831–842, 2011.
Ravin, N. V., Muntyan, M. S., Smolyakov, D. D., Rudenko, T. S., Beletsky, A. V., Mardanov, A. V., and Grabovich, M. Y.: Metagenomics Revealed a New Genus “Candidatus Thiocaldithrix dubininis” gen. nov., sp. nov. and a New Species “Candidatus Thiothrix putei” sp. nov. in the Family Thiotrichaceae, Some Members of Which Have Traits of Both Na+- and H+-Motive Energetics, Int. J. Mol. Sci., 24, 14199, https://doi.org/10.3390/ijms241814199, 2023.
Robeson, M. S., 2nd, O'Rourke, D. R., Kaehler, B. D., Ziemski, M., Dillon, M. R., Foster, J. T., and Bokulich, N. A.: RESCRIPt: Reproducible sequence taxonomy reference database management, PLoS Comput. Biol., 17, e1009581, https://doi.org/10.1371/journal.pcbi.1009581, 2021.
Rosentreter, J. A., Borges, A. V., Deemer, B. R., Holgerson, M. A., Liu, S., Song, C., Melack, J., Raymond, P. A., Duarte, C. M., Allen, G. H., Olefeldt, D., Poulter, B., Battin, T. I., and Eyre, B. D.: Half of global methane emissions come from highly variable aquatic ecosystem sources, Nat. Geosci., 14, 225–230, 2021.
Rotaru, A., Posth, N., Löscher, C., Miracle, M. R., Vincence, E., Cox, R. P., Thompson, J., Poulton, S., and Thamdrup, B.: Interspecies interactions mediated by conductive minerals in the sediments of the Iron rich Meromictic Lake La Cruz, Spain, Limnetica, 38, 21–40, 2019.
Rotaru, A.-E., Shrestha, P. M., Liu, F., Shrestha, M., Shrestha, D., Embree, M., Zengler, K., Wardman, C., Nevin, K. P., and Lovley, D. R.: A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane, Energy Environ. Sci., 7, 408–415, 2014.
Rotaru, A.-E., Calabrese, F., Stryhanyuk, H., Musat, F., Shrestha, P. M., Weber, H. S., Snoeyenbos-West, O. L. O., Hall, P. O. J., Richnow, H. H., Musat, N., and Thamdrup, B.: Conductive Particles Enable Syntrophic Acetate Oxidation between Geobacter and Methanosarcina from Coastal Sediments, MBio, 9, 00226–00218, 2018.
Rotaru, A.-E., Yee, M. O., and Musat, F.: Microbes trading electricity in consortia of environmental and biotechnological significance, Curr. Opin. Biotechnol., 67, 119–129, 2021.
Ruiz-Blas, F., Bartholomäus, A., Yang, S., Wagner, D., Henny, C., Russell, J. M., Kallmeyer, J., and Vuillemin, A.: Metabolic features that select for Bathyarchaeia in modern ferruginous lacustrine subsurface sediments, ISME Commun., 4, ycae112, https://doi.org/10.1093/ismeco/ycae112, 2024.
Saunders, N. F. W., Goodchild, A., Raftery, M., Guilhaus, M., Curmi, P. M. G., and Cavicchioli, R.: Predicted roles for hypothetical proteins in the low-temperature expressed proteome of the Antarctic archaeon Methanococcoides burtonii, J. Proteome Res., 4, 464–472, 2005.
Saunois, M., Bousquet, P., Poulter, B., Peregon, A., and Ciais, P.: Global Methane Budget 2000–2012 (V.1.0, issued June 2016 and V.1.1, issued December 2016), ESS-DIVE repository [data set], https://doi.org/10.3334/CDIAC/GLOBAL_METHANE_BUDGET_2016_V1.1, 2016.
Schink, B.: Fermentation of 2,3-butanediol by Pelobacter carbinolicus sp. nov. and Pelobacter propionicus sp. nov., and evidence for propionate formation from C2 compounds, Arch. Microbiol., 137, 33–41, 1984.
Schink, B.: Energetics of syntrophic cooperation in methanogenic degradation, Microbiol. Mol. Biol. Rev., 61, 262–280, 1997.
Schink, B.: The Genus Pelobacter, in: The Prokaryotes, Springer New York, New York, NY, 5–11, https://doi.org/10.1007/978-1-4757-2191-1_24, 2006.
Schorn, S., Ahmerkamp, S., Bullock, E., Weber, M., Lott, C., Liebeke, M., Lavik, G., Kuypers, M. M. M., Graf, J. S., and Milucka, J.: Diverse methylotrophic methanogenic archaea cause high methane emissions from seagrass meadows, Proc. Natl. Acad. Sci. USA, 119, e2106628119, https://doi.org/10.1073/pnas.2106628119, 2022.
Schreiber, L., Holler, T., Knittel, K., Meyerdierks, A., and Amann, R.: Identification of the dominant sulfate-reducing bacterial partner of anaerobic methanotrophs of the ANME-2 clade: SEEP-SRB1 associated with ANME-2, Environ. Microbiol., 12, 2327–2340, 2010.
Segarra, K. E. A., Comerford, C., Slaughter, J., and Joye, S. B.: Impact of electron acceptor availability on the anaerobic oxidation of methane in coastal freshwater and brackish wetland sediments, Geochim. Cosmochim. Acta, 115, 15–30, 2013.
Sela-Adler, M., Ronen, Z., Herut, B., Antler, G., Vigderovich, H., Eckert, W., and Sivan, O.: Co-existence of Methanogenesis and Sulfate Reduction with Common Substrates in Sulfate-Rich Estuarine Sediments, Front. Microbiol., 8, 00766, https://doi.org/10.3389/fmicb.2017.00766, 2017.
Senior, E., Lindström, E. B., Banat, I. M., and Nedwell, D. B.: Sulfate reduction and methanogenesis in the sediment of a saltmarsh on the East coast of the United kingdom, Appl. Environ. Microbiol., 43, 987–996, 1982.
Seyfferth, A. L., Bothfeld, F., Vargas, R., Stuckey, J. W., Wang, J., Kearns, K., Michael, H. A., Guimond, J., Yu, X., and Sparks, D. L.: Spatial and temporal heterogeneity of geochemical controls on carbon cycling in a tidal salt marsh, Geochim. Cosmochim. Acta, 282, 1–18, 2020.
Seymour, C. O., Palmer, M., Becraft, E. D., Stepanauskas, R., Friel, A. D., Schulz, F., Woyke, T., Eloe-Fadrosh, E., Lai, D., Jiao, J.-Y., Hua, Z.-S., Liu, L., Lian, Z.-H., Li, W.-J., Chuvochina, M., Finley, B. K., Koch, B. J., Schwartz, E., Dijkstra, P., Moser, D. P., Hungate, B. A., and Hedlund, B. P.: Hyperactive nanobacteria with host-dependent traits pervade Omnitrophota, Nat. Microbiol., 8, 727–744, 2023.
Simmons, S. L. and Edwards, K. J.: Geobiology of Magnetotactic Bacteria, in: Magnetoreception and Magnetosomes in Bacteria, edited by: Schüler, D., Springer Berlin Heidelberg, Berlin, Heidelberg, 77–102, https://doi.org/10.1007/7171_039, 2007a.
Simmons, S. L. and Edwards, K. J.: Unexpected diversity in populations of the many-celled magnetotactic prokaryote, Environ. Microbiol., 9, 206–215, 2007b.
Simmons, S. L., Sievert, S. M., Frankel, R. B., Bazylinski, D. A., and Edwards, K. J.: Spatiotemporal distribution of marine magnetotactic bacteria in a seasonally stratified coastal salt pond, Appl. Environ. Microbiol., 70, 6230–6239, 2004.
Singh, N., Kendall, M. M., Liu, Y., and Boone, D. R.: Isolation and characterization of methylotrophic methanogens from anoxic marine sediments in Skan Bay, Alaska: description of Methanococcoides alaskense sp. nov., and emended description of Methanosarcina baltica, Int. J. Syst. Evol. Microbiol., 55, 2531–2538, 2005.
Sivan, O., Shusta, S. S., and Valentine, D. L.: Methanogens rapidly transition from methane production to iron reduction, Geobiology, 14, 190–203, 2016.
Skennerton, C. T., Chourey, K., Iyer, R., Hettich, R. L., Tyson, G. W., and Orphan, V. J.: Methane-fueled syntrophy through extracellular electron transfer: Uncovering the genomic traits conserved within diverse bacterial partners of anaerobic methanotrophic Archaea, MBio, 8, https://doi.org/10.1128/mBio.00530-17, 2017.
Smith, S. J., Page, K., Kim, H., Campbell, B. J., Boerio-Goates, J., and Woodfield, B. F.: Novel synthesis and structural analysis of ferrihydrite, Inorg. Chem., 51, 6421–6424, 2012.
Sparks, N. H. C., Lloyd, J., and Board, R. G: Saltmarsh ponds – a preferred habitat for magnetotactic bacteria?, Lett. Appl. Microbiol., 8, 109–111, 1989.
Stams, A. J. M., Teusink, B., and Sousa, D. Z.: Ecophysiology of Acetoclastic Methanogens, in: Biogenesis of Hydrocarbons, Springer International Publishing, Cham, 109–121, https://doi.org/10.1007/978-3-319-78108-2_21, 2019.
Stookey, L. L.: Ferrozine – a new spectrophotometric reagent for iron, Anal. Chem., 42, 779–781, 1970.
Summons, R. E., Franzmann, P. D., and Nichols, P. D.: Carbon isotopic fractionation associated with methylotrophic methanogenesis, Org. Geochem., 28, 465–475, 1998.
Sundby, B.: Transient state diagenesis in continental margin muds, Mar. Chem., 102, 2–12, 2006.
Sun, L., Toyonaga, M., Ohashi, A., Tourlousse, D. M., Matsuura, N., Meng, X.-Y., Tamaki, H., Hanada, S., Cruz, R., Yamaguchi, T., and Sekiguchi, Y.: Lentimicrobium saccharophilum gen. nov., sp. nov., a strictly anaerobic bacterium representing a new family in the phylum Bacteroidetes, and proposal of Lentimicrobiaceae fam. nov, Int. J. Syst. Evol. Microbiol., 66, 2635–2642, 2016.
Suzuki, R., Terada, Y., and Shimodaira, H.: pvclust: An R package for hierarchical clustering with p-values, Github, https://github.com/shimo-lab/pvclust (last access: 10 February 2025), 2019.
Tadley, P., Bennett, A., Hansen, K., Hanson, T., Colosi, J., and Goudsouzian, L. K.: Identification of Bacterial Taxa Present in a Concrete Pennsylvania Bridge, Microbiol. Resour. Announc., 12, e0021123, https://doi.org/10.1128/mra.00211-23, 2023.
Taillefert, M., Bono, A. B., and Luther, G. W.: Reactivity of freshly formed Fe(III) in synthetic solutions and (pore)waters: Voltammetric evidence of an aging process, Environ. Sci. Technol., 34, 2169–2177, 2000.
Taillefert, M., Neuhuber, S., and Bristow, G.: The effect of tidal forcing on biogeochemical processes in intertidal salt marsh sediments, Geochem. Trans., 8, https://doi.org/10.1186/1467-4866-8-6, 2007.
Tang, J., Zhuang, L., Ma, J., Tang, Z., Yu, Z., and Zhou, S.: Secondary mineralization of ferrihydrite affects microbial methanogenesis in Geobacter-Methanosarcina cocultures, Appl. Environ. Microbiol., 82, 5869–5877, 2016.
Waite, T. J., Kraiya, C., Trouwborst, R. E., Ma, S., and Luther III, G. W.: An investigation into the suitability of bismuth as an alternative to gold-amalgam as a working electrode for the in situ determination of chemical redox species in the natural environment, Electroanalysis, 18, 1167–1172, 2006.
Wang, H., Byrne, J. M., Liu, P., Liu, J., Dong, X., and Lu, Y.: Redox cycling of Fe(II) and Fe(III) in magnetite accelerates aceticlastic methanogenesis by Methanosarcina mazei, Environ. Microbiol. Rep., 12, 97–109, 2020.
Wang, J., Zheng, Q., Wang, S., Zeng, J., Yuan, Q., Zhong, Y., Jiang, L., and Shao, Z.: Characterization of two novel chemolithoautotrophic bacteria of Sulfurovum from marine coastal environments and further comparative genomic analyses revealed species differentiation among deep-sea hydrothermal vent and non-vent origins, Front. Mar. Sci., 10, https://doi.org/10.3389/fmars.2023.1222526, 2023.
Wang, X.-C. and Lee, C.: Sources and distribution of aliphatic amines in salt marsh sediment, Org. Geochem., 22, 1005–1021, 1994a.
Wang, X.-C. and Lee, C.: Sources and distribution of aliphatic amines in salt marsh sediment, Org. Geochem., 22, 1005–1021, 1994b.
Watkins, A. J., Roussel, E. G., Parkes, R. J., and Sass, H.: Glycine betaine as a direct substrate for methanogens (Methanococcoides spp.), Appl. Environ. Microbiol., 80, 289–293, 2014.
Winfrey, M. R. and Ward, D. M.: Substrates for sulfate reduction and methane production in intertidal sediments, Appl. Environ. Microbiol., 45, 193–199, 1983.
Xiao, K.-Q., Beulig, F., Kjeldsen, K. U., Jørgensen, B. B., and Risgaard-Petersen, N.: Concurrent methane production and oxidation in surface sediment from Aarhus Bay, Denmark, Front. Microbiol., 8, 1198, https://doi.org/10.3389/fmicb.2017.01198, 2017.
Xiao, K. Q., Beulig, F., Røy, H., Jørgensen, B. B., and Risgaard-Petersen, N.: Methylotrophic methanogenesis fuels cryptic methane cycling in marine surface sediment, Limnol. Oceanogr., 63, 1519–1527, 2018.
Xiao, L., Lichtfouse, E., and Senthil Kumar, P.: Advantage of conductive materials on interspecies electron transfer-independent acetoclastic methanogenesis: A critical review, Fuel (Lond.), 305, 121577, https://doi.org/10.1016/j.fuel.2021.121577, 2021.
Xu, H., Chang, J., Wang, H., Liu, Y., Zhang, X., Liang, P., and Huang, X.: Enhancing direct interspecies electron transfer in syntrophic-methanogenic associations with (semi)conductive iron oxides: Effects and mechanisms, Sci. Total Environ., 695, 133876, https://doi.org/10.1016/j.scitotenv.2019.133876, 2019.
Xue, P., Chang, L., Pei, Z., and Harrison, R. J.: The origin of magnetofossil coercivity components: Constraints from coupled experimental observations and micromagnetic calculations, J. Geophys. Res. Solid Earth, 129, https://doi.org/10.1029/2023jb028501, 2024.
Yang, X. H., Scranton, M. I., and Lee, C.: Seasonal variations in concentration and microbial uptake of methylamines in estuarine waters, Marine Ecology Progress Series, 108, 303–312, 1994.
Yee, M. O. and Rotaru, A.-E.: Extracellular electron uptake in Methanosarcinales is independent of multiheme c-type cytochromes, Sci. Rep., 10, 372, https://doi.org/10.1038/s41598-019-57206-z, 2020.
Yu, L., He, D., Yang, L., Rensing, C., Zeng, R. J., and Zhou, S.: Anaerobic methane oxidation coupled to ferrihydrite reduction by Methanosarcina barkeri, Sci. Total Environ., 844, 157235, https://doi.org/10.1016/j.scitotenv.2022.157235, 2022.
Yuan, J., Ding, W., Liu, D., Xiang, J., and Lin, Y.: Methane production potential and methanogenic archaea community dynamics along the Spartina alterniflora invasion chronosequence in a coastal salt marsh, Appl. Microbiol. Biotechnol., 98, 1817–1829, 2014.
Yuan, J., Ding, W., Liu, D., Kang, H., Xiang, J., and Lin, Y.: Shifts in methanogen community structure and function across a coastal marsh transect: effects of exotic Spartina alterniflora invasion, Sci. Rep., 6, 18777, https://doi.org/10.1038/srep18777, 2016.
Yuan, J., Liu, D., Ji, Y., Xiang, J., Lin, Y., Wu, M., and Ding, W.: Spartina alterniflora invasion drastically increases methane production potential by shifting methanogenesis from hydrogenotrophic to methylotrophic pathway in a coastal marsh, J. Ecol., 107, 2436–2450, 2019.
Yuan, T., Ko, J. H., Zhou, L., Gao, X., Liu, Y., Shi, X., and Xu, Q.: Iron oxide alleviates acids stress by facilitating syntrophic metabolism between Syntrophomonas and methanogens, Chemosphere, 247, 125866, https://doi.org/10.1016/j.chemosphere.2020.125866, 2020.
Zachara, J. M., Kukkadapu, R. K., Fredrickson, J. K., Gorby, Y. A., and Smith, S. C.: Biomineralization of poorly crystalline Fe(III) oxides by dissimilatory metal reducing bacteria (DMRB), Geomicrobiol. J., 19, 179–207, 2002.
Zhang, Z., Zimmermann, N. E., Stenke, A., Li, X., Hodson, E. L., Zhu, G., Huang, C., and Poulter, B.: Emerging role of wetland methane emissions in driving 21st century climate change, Proc. Natl. Acad. Sci. USA, 114, 9647–9652, 2017.
Zhao, N., Ding, H., Zhou, X., Guillemot, T., Zhang, Z., Zhou, N., and Wang, H.: Dissimilatory iron-reducing microorganisms: The phylogeny, physiology, applications and outlook, Crit. Rev. Environ. Sci. Technol., 55, 73–98, 2025.
Zhao, Z., Li, Y., Yu, Q., and Zhang, Y.: Ferroferric oxide triggered possible direct interspecies electron transfer between Syntrophomonas and Methanosaeta to enhance waste activated sludge anaerobic digestion, Bioresour. Technol., 250, 79–85, 2018.
Zhao, Z., Sun, C., Li, Y., Peng, H., and Zhang, Y.: Upgrading current method of anaerobic co-digestion of waste activated sludge for high-efficiency methanogenesis: Establishing direct interspecies electron transfer via ethanol-type fermentation, Renew. Energy, 148, 523–533, 2020.
Zheng, S., Liu, F., Wang, B., Zhang, Y., and Lovley, D. R.: Methanobacterium capable of direct interspecies electron transfer, Environ. Sci. Technol., 54, 15347–15354, 2020.
Zheng, S., Li, M., Liu, Y., and Liu, F.: Desulfovibrio feeding Methanobacterium with electrons in conductive methanogenic aggregates from coastal zones, Water Res., 202, 117490, 2021.
Zhou, S., Xu, J., Yang, G., and Zhuang, L.: Methanogenesis affected by the co-occurrence of iron(III) oxides and humic substances, FEMS Microbiol. Ecol., 88, 107–120, 2014.
Zhou, Z., Pan, J., Wang, F., Gu, J.-D., and Li, M.: Bathyarchaeota: globally distributed metabolic generalists in anoxic environments, FEMS Microbiol. Rev., 42, 639–655, 2018.
Zhuang, L., Xu, J., Tang, J., and Zhou, S.: Effect of ferrihydrite biomineralization on methanogenesis in an anaerobic incubation from paddy soil, J. Geophys. Res. Biogeosci., 120, 876–886, 2015.
Zhuang, L., Tang, Z., Yu, Z., Li, J., and Tang, J.: Methanogenic Activity and Microbial Community Structure in Response to Different Mineralization Pathways of Ferrihydrite in Paddy Soil, Front Earth Sci., 7, 00325, https://doi.org/10.3389/feart.2019.00325, 2019.
- Abstract
- Introduction
- Materials and methods
- Results
- Discussion
- Conclusions
- Appendix A: Extended cyclic voltammetry results
- Appendix B: Extended results from DNA analysis
- Appendix C: Extended results and discussion of magnetic analyses
- Code availability
- Data availability
- Author contributions
- Competing interests
- Disclaimer
- Acknowledgements
- Financial support
- Review statement
- References
- Abstract
- Introduction
- Materials and methods
- Results
- Discussion
- Conclusions
- Appendix A: Extended cyclic voltammetry results
- Appendix B: Extended results from DNA analysis
- Appendix C: Extended results and discussion of magnetic analyses
- Code availability
- Data availability
- Author contributions
- Competing interests
- Disclaimer
- Acknowledgements
- Financial support
- Review statement
- References