the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Potential for phenol biodegradation in cloud waters
Audrey Lallement
Ludovic Besaury
Elise Tixier
Martine Sancelme
Pierre Amato
Virginie Vinatier
Isabelle Canet
Olga V. Polyakova
Viatcheslay B. Artaev
Albert T. Lebedev
Laurent Deguillaume
Gilles Mailhot
Anne-Marie Delort
Phenol is toxic and can be found in many environments, in particular in the atmosphere due to its high volatility. It can be emitted directly from manufacturing processes or natural sources, and it can also result from benzene oxidation. Although phenol biodegradation by microorganisms has been studied in many environments, the cloud medium has not been investigated yet as the discovery of active microorganisms in cloud is rather recent.
The main objective of this work was to evaluate the potential degradation of phenol by cloud microorganisms. Phenol concentrations were measured by GC-MS on two cloud samples collected at the PUY station (summit of Puy de Dôme, 1465 m a.s.l., France): they ranged from 0.15 to 0.21 µg L−1.
The strategy for investigating its potential biodegradation involved a metatranscriptomic analysis and metabolic screening of bacterial strains from cloud water collected at the PUY station for phenol degradation capabilities (from the 145 tested strains, 33 were isolated for this work).
Among prokaryotic messenger RNA-enriched metatranscriptomes obtained from three cloud water samples, which were different from those used for phenol quantification, we detected transcripts of genes coding for enzymes involved in phenol degradation (phenol monooxygenases and phenol hydroxylases) and its main degradation product, catechol (catechol 1,2-dioxygenases). These enzymes were likely from Gammaproteobacteria, a dominant class in clouds, more specifically the genera Acinetobacter and Pseudomonas.
Bacterial isolates from cloud water samples (Pseudomonas spp., Rhodococcus spp., and strains from the Moraxellaceae family) were screened for their ability to degrade phenol: 93 % of the 145 strains tested were positive. These findings highlight the possibility of phenol degradation by microorganisms in clouds.
Metatranscriptomic analysis suggested that phenol could be biodegraded in clouds, while 93 % of 145 bacterial strains isolated from clouds were able to degrade phenol.
- Article
(795 KB) - Full-text XML
-
Supplement
(3438 KB) - BibTeX
- EndNote
Due to its toxicity, phenol is one of the main pollutants listed by the US Environmental Protection Agency (US EPA list) and its concentration in drinking water is inspected and regulated (Michalowicz and Duda, 2007). In France, the phenol concentration limit in drinking water is 0.5 µg L−1. Phenol is issued from natural sources such as organic matter decomposition and biomass burning (Schauer et al., 2001), but it mainly results from industrial processes. For instance phenol is involved in the production of oils, xylene, plastics, drugs, explosives, dyes, and pesticides; it is also present in oil refining and wood and leather preservatives (Gami et al., 2014; Schummer et al., 2009). Annual phenol production exceeded 10.7 million tons worldwide in 2016 (Merchant Research & Consulting Ltd.). Phenol has an environmental impact, particularly on aquatic biota (microorganisms, protozoa, invertebrates, and vertebrates; Babich and Davis, 1981; Duan et al., 2018). Phenol also represents a risk for human beings because it can be rapidly absorbed through the skin and by inhalation through the lungs. In particular, it provokes cutaneous exfoliation and cardiac arrhythmias; it is also toxic to the liver and kidneys (Babich and Davis, 1981; Lober, 1987; National Library of Medicine HSDB Database: https://toxnet.nlm.nih.gov/cgi-bin/sis/search/a?dbs+hsdb:@term+@DOCNO+113, last access: 4 August 2018).
Phenol can be found in all environmental compartments (soil, water), including the atmosphere (Atkinson et al., 1992; Rubio et al., 2012). Even if its volatility is low (≤7 % at 25 ∘C; National Institute for Occupational Safety and Health, NIOSH), phenol is present in the gas phase, but this polar compound can also be transferred to the aqueous phases of the atmosphere (rain, snow, clouds) thanks to its solubility described by the Henry's law constant ( M atm−1 at 298 K and mass accommodation = at 283 K; Harrison et al., 2002; Heal et al., 1995). Phenol can also be formed by the oxidation of precursors such as benzene directly in the atmosphere both in the gas and the aqueous phase (Grosjean, 1991; Harrison et al., 2005; Herrmann et al., 2015; Vione et al., 2004). The production of phenol by aqueous-phase reactivity is expected to be less efficient than in the gas phase. Indeed, benzene is a precursor of phenol but it will not accumulate in the droplet in significant amounts due to its relatively low Henry's law constant ( M atm−1). Phenol concentration ranges from 2.8 to 8.9 µg L−1 (0.03 to 0.09 µM) in cloud waters and it reaches up to 91.3 µg L−1 (0.97 µM) in rain (Harrison et al., 2005; Schummer et al., 2009).
In the gas phase, phenol is transformed into nitrophenols either in the presence of HO• and (during the day) or in the presence of and (during the night; Atkinson et al., 1992; Olariu, 2001; Olariu et al., 2002). In the aqueous phase, phenol can undergo transformations that should be much faster than in the gas phase, leading to the formation of nitrophenols (Vione et al., 2004). Recent studies show that direct photolysis should be competitive with the radical-driven one for phenol (Rayne et al., 2009) and that phenol exposed to atmospherically relevant photochemical conditions leads to the production of low-volatile compounds such as light-absorbing molecules (HULIS). In-cloud processing of phenol can therefore be a source of secondary organic aerosol (SOA; Gilardoni et al., 2016; Sun et al., 2010).
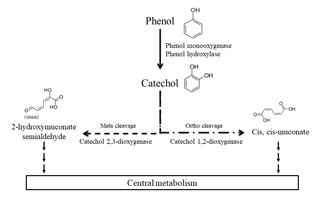
Figure 1Main phenol biodegradation pathways described for aerobic microorganisms as referred to in the KEGG database.
A great number of studies have been conducted to assess the biodegradation of phenol by microorganisms including bacteria, fungi, yeast, and algae in the context of environmental and water treatment chemistry (Michalowicz and Duda, 2007). Most of those microorganisms were isolated from soils (including the rhizosphere) and waters (fresh and marine waters, wastewater, and sediments) in which contamination by phenol has been studied (Basha et al., 2010; Kafilzadeh et al., 2010; Michalowicz and Duda, 2007; Mishra and Kumar, 2017; Sandhu et al., 2009; Sridevi et al., 2012; Tian et al., 2017). Only one team has focused on atmospheric phenol uptake by microorganisms (Sandhu et al., 2007, 2009). They studied a microbial community on leaves directly in contact with phenol in the air and found that they were able to degrade it. Many studies are based on direct measurement of the biodegradation activity of microbial isolates, in particular for biotechnological application in industrial effluent decontamination (Basha et al., 2010; Michalowicz and Duda, 2007; Mishra and Kumar, 2017; Sridevi et al., 2012). Alternatively, others used molecular-based approaches and reported microbial genes of phenol- or catechol-degrading enzymes (Brennerova et al., 2009; Fang et al., 2013; Sandhu et al., 2009; Sharma et al., 2012; Silva et al., 2013; Suenaga et al., 2009) or gained knowledge from metatranscriptomic analyses of microbial communities (Auffret et al., 2015). Ajaz et al. (2004) have identified 30 bacterial strains resistant to phenol in garden soil and Padmanabhan et al. (2003) have done DNA-SIP with 13C-labeled phenol to identify six phenol-degrading populations in soil thanks to 16S rRNA gene analysis. The main bacterial genera able to biodegrade phenol are Pseudomonas, Rhodococcus, Acinetobacter, and Bacillus; other genera are also described such as Arthrobacter, Alcalinogenes, Burkholderia, Thauera, etc. (Basha et al., 2010; Fang et al., 2013; Jadeja et al., 2014; Michalowicz and Duda, 2007; Padmanabhan et al., 2003; Silva et al., 2013). Major biodegradation pathways for aerobic bacteria have been established (Fig. 1). First, phenol can be oxidized into catechol by phenol hydroxylases or phenol monooxygenases, then the ring cleavage can be catalyzed by dioxygenases: catechol 1,2-dioxygenase produces cis, cis-muconate (“ortho” pathway), while catechol 2,3-dioxygenase leads to 2-hydroxymuconate semialdehyde (“meta” pathway). Finally, these products are integrated into the central metabolism of the bacteria and end up in CO2 production (Basha et al., 2010). Alternative pathways have been described with anaerobic microorganisms. In these cases, phenol is carboxylated by a carboxylase in the para position to produce 4-hydroxybenzoate, and this metabolite is further metabolized in benzoyl-CoA via anaerobic routes before its ring-opening step (Basha et al., 2010).
Although phenol is present in clouds, to our knowledge its transformation by microorganisms in these specific environments has never been assessed. Bacterial density usually ranges from 104 to 105 cells per mL of cloud water (Vaïtilingom et al., 2012). In spite of the numerous atmospheric stresses, it has been shown that microorganisms can survive in clouds, maintain metabolic activity, and degrade organic compounds (Delort et al., 2010, 2017; Hill et al., 2007; Sattler et al., 2001; Vaïtilingom et al., 2013). Among the bacteria known for phenol degradation, Pseudomonas (Gammaproteobacteria) and Rhodococcus (Actinobacteria) are frequently found viable and potentially active in clouds (Amato et al., 2017a, b).
The aim of this work was to explore the potential for phenol biodegradation in clouds. First, phenol concentration was quantified in atmospheric waters, and cloud water metatranscriptomes were checked for the presence of transcripts of phenol-degrading genes; second, bacterial strains isolated from cloud water were screened for phenol biodegradation ability.
2.1 Chemical reagents
Phenol (>99 %) and hydrogen peroxide (30 %) were obtained from Fluka, and sodium chloride (>99 %), dichloromethane (>99.8 %), and sulfuric acid (>95 %–97 %) were from Sigma Aldrich; acetonitrile (>99.9 %) was from VWR Chemicals, NaOH (99 %) from Merck, and MgSO4 (>98 %) from Carlo Erba Reagents (SDS).
2.2 Cloud water analysis
Cloud sampling. Cloud waters have been sampled at the PUY station (summit of the Puy de Dôme, 1465 m a.s.l.; 45∘46′ N, 2∘57′ E; France), which is part of the atmospheric survey networks EMEP (the European Monitoring and Evaluation Programme), GAW (Global Atmosphere Watch), and ACTRIS (Aerosols, Clouds, and Trace gases Research Infrastructure). The sampling site is fully described in Deguillaume et al. (2014). The global meteorological context was examined through 120 h back trajectories of the air masses sampled using the HYSPLIT model (HYbrid Single-Particle Lagrangian Inte-grated Trajectory). Two cloud water samples collected in 2016 (21 and 26 October) were analyzed in this work for phenol quantification by GC-MS. Three other samples were previously collected and analyzed in 2013 (5 November), 2014 (27 June), and 2016 (16 February; Lebedev et al., 2018). Samples were collected using a sterilized cloud droplet impactor and immediately filtered through a Minisart® PES filter (0.22 µm porosity; Sartorius, Germany) under sterilized conditions; these have been stored at −25 ∘C.
GC-MS analysis. Sample preparation was carried out according to US EPA 8270 method. Prior to utilization, all the glassware was cleaned with a piranha reagent composed of 6 mL of sulfuric acid mixed with 2 mL of hydrogen peroxide. The reagents were kept in the glassware for one night and after all the glasses were washed two times with ultrapure water and two times with dichloromethane. With clean dishes, cloud waters kept frozen were melted at room temperature and the pH adjusted to 2 and 11. Organic compounds were extracted three times with dichloromethane (keeping the ratio 10 mL of water for 1 mL of CH2Cl2). All the dichloromethane fractions were then dried with MgSO4 and evaporated to 1 mL using a rotary evaporator under reduced pressure; the temperature of the water bath was 20 ∘C. Samples were kept at 4 ∘C until analysis
All analyses related to cloud samples collected on 21 and 26 October 2016 were performed at Saint Joseph, MI, at LECO Corporation (USA). Accurate GC-MS measurements were performed with the high-resolution time-of-flight mass spectrometer Pegasus® GC-HRT in GC mode (software ChromaTOF-HRT). The obtained EI mass spectra were used for phenol identification by utilizing high mass accuracy data and retention time (Lebedev et al., 2013). Phenol concentrations were measured using naphthalene D8 as an internal standard. The response factor (0.7) was calculated using a standard solution of phenol. Phenol concentrations measured in cloud water samples collected on 5 November 2013, 27 June 2014, and 16 February 2016 are extracted from Lebedev et al. (2018).
2.3 Analysis of metatranscriptomes
Transcriptomic analysis. Cloud water samples were collected on 17 November 2014 for three consecutive periods of 5 h. The cloud air mass origin remained stable over the duration of sampling as attested by air mass backward trajectories (Fig. S1). The cloud droplets collected by impaction were immediately transferred by gravity into sterile collection bottles (Nalgene, Rochester, USA) through sterile (autoclaved) silicone tubing. Before cloud sampling operations started, control samples were made by pouring 200 mL of sterile water into the collection device and through the tubing and by processing it in parallel with the cloud water samples, including sequencing and data treatment. These controls were clearly distinct from samples: based on their contribution to identified ribosome sequences, these contained mostly Enterobacteriaceae (66 %), Dikarya (9.2 %), Streptococcaceae (5.4 %), Vibrionaceae (2.8 %), and Micrococcaceae (1.2 %), i.e., not the taxa of interest here. Conservatively, the sequences present in controls were further removed from sample files (BWA-MEM; Li, 2013). Immediately after collection, water samples were filtered (MO BIO 14880-50-WF) within a UV-sterilized laminar flow hood installed at the sampling site. The filters were then put into ∼5 mL of RNA Later solution (Sigma, Steinheim, Germany) and stored at −80 ∘C until further processing. Briefly, total RNA was extracted from filter halves using a MO BIO PowerWater RNA Kit, and bacterial ribosomal RNA was depleted using a MICROBExpress Bacterial mRNA Enrichment Kit (Life Technologies). Metatranscriptomes of the messenger RNAs were then obtained by multiple displacement amplification using a REPLI-g WGA & WTA kit.; shotgun libraries were sequenced on Illumina MiSeq paired-end 2×300 bp. Sequencing reads were quality checked (FastQC; Andrews, 2010) and trimmed (PRINSEQ-Lite; Schmieder and Edwards, 2011) before assembling the mate pairs using PANDA-SEQ (Masella et al., 2012). Annotations were made against the UNIPROTKB database (Leinonen et al., 2006), including protein sequences for bacteria, archaea, and fungi using BLASTX software (best hits with e value ). All steps were performed using custom scripts. The sequence files have been deposited to the European Nucleotide Archive (ENA) under the study accession number PRJEB25802.
Bioinformatics treatment. Known enzymes involved in phenol degradation were found in the KEGG database (see Fig. 1). We only focused on aerobic metabolism as the cloud environment is highly oxidative. Four nucleotide sequence databases were created from NCBI corresponding to phenol hydroxylases (69 sequences), phenol monooxygenases (29 sequences), catechol (regrouping catechol 1,2-dioxygenases and catechol 2,3-dioxygenases; 145 sequences), and a fourth database including genes coding for putative phenol degradation enzymes (38 sequences). The sequences from the cloud metatranscriptomes corresponding to the different created databases were then extracted using bowtie2 (very sensitive option; Langmead and Salzberg, 2012). The affiliation of the extracted sequences was determined using BLASTN on a local server (e value = 0.00001; Camacho et al., 2009).
2.4 Biodegradation of phenol by bacterial strains from cloud waters
Bacterial strains. Bacterial strains were isolated from cloud waters sampled at the PUY station and identified as previously described in Vaïtilingom et al. (2012). From our lab strain collection, we choose all the potential bacteria that could biodegrade phenol. From the 145 strains tested, 33 of the strains were isolated for this work; the others were published earlier (see Table S1 in the Supplement).
119 Pseudomonas, 24 Rhodococcus strains, and 2 strains form the Moraxella family were selected for the screening of phenol degradation (see Table S1). More precisely, Pseudomonas strains included 4 P. fluorescens, 10 P. graminis, 1 P. grimontii, 2 P. poae, 1 P. reactans, 1 P. reinekii, 3 P. rhizosphaerae, 35 P. syringae, 2 P. trivialis, 2 P. veronii, 1 P. viridiflava, and 57 Pseudomonas spp. Rhodococcus strains included 1 R. erythropolis, 1 R. enclensis, and 22 Rhodococcus spp. Moraxella family strains were one Moraxella sp. and one Psychrobacter sp.
Pseudomonas and Rhodococcus strains represent 20.4 % and 4.10 % of the 584 strains of our cloud bacterial collection. From our experience, at the genus level, Pseudomonas and Rhodococcus are among the most frequent bacteria in clouds: Pseudomonas strains in particular have been frequently isolated by culture (Vaïtilingom et al., 2012; Joly et al., 2013), and both targeted and untargeted molecular analyses (and metagenomes, respectively) demonstrated high occurrence in the bacterial communities. These represented 0.1 % to >2 % of the prokaryotes ribosome sequences in amplicon sequencing investigations (Amato et al., 2017a). Based on the biomass in clouds (∼104 bacteria cells mL−1; Vaïtilingom et al., 2012) and assuming even ribosome amplification between bacterial groups, we can infer the presence of ∼103 Pseudomonas mL−1 and ∼102 Rhodococcus mL−1 of cloud water.

Figure 2Relative abundance of transcripts in cloud waters encoding for enzymes involved in phenol degradation pathways. The absolute total number of hits for each sample is indicated (n).
Incubations. All the strains were grown in 25 mL of R2A medium for 48 h at 17 ∘C and 130 rpm (Reasoner and Geldreich, 1985). Then cultures were centrifuged at 4000 rpm for 15 min. Bacteria pellets were rinsed first with 5 mL of NaCl 0.8 % and after with Volvic® mineral water previously sterilized by filtration under sterile conditions using a 0.22 µm PES filter. Cells were resuspended in 5 mL of 0.1 mM phenol solution, prepared in Volvic® mineral water, and incubated at 17 ∘C and 130 rpm agitation during 5 days in the dark. To identify the concentration, the optical density (OD) for each strain was taken during the experiment. Strain concentrations were around 109 cells mL−1. The ratio number of bacterial cells ∕ phenol concentration was kept to that measured in cloud waters, and all the concentrations were multiplied by a factor of 104. Indeed, the mean bacteria concentration is around 105 cells mL−1 of cloud water, while that of phenol can reach 0.008 µM (0.74 µg L−1; see the Results section) in clouds collected at the PUY station. We showed in the past that when the cell ∕ substrate ratios are kept constant the rates of biodegradation are constant (Vaïtilingom et al., 2010). The temperature (17 ∘C) corresponds to the average temperature at the PUY station in summer under cloud conditions. It is well known that under culture conditions in a laboratory, a lag time can be observed before bacteria starts to biodegrade phenol that corresponds to the induction period of the gene expression (Al-Khalid and El-Naas, 2012).
Before sampling, the evaporation of water has been compensated for by adding Volvic® mineral water. A control experiment was performed by incubating phenol without bacteria; the phenol concentration remained stable with time (0.1 mM of phenol was obtained at the end of the experiment). For phenol quantification over time in the incubation experiments, 600 µL samples were centrifuged at 12 500 rpm for 3 min and the supernatants were kept frozen until HPLC analysis.
Phenol HPLC analysis. Before analysis, all the samples were filtered on an H-PTFE filter (pore size at 0.2 µm and diameter of 13 mm from Macherey-Nagel, Germany). Phenol detection was done on an HPLC VWR Hitachi Chromaster apparatus fitted with a DAD detector and driven by Chromaster software. Isocratic mode was used with a reverse-phase end-capped column (LiChrospher® RP-18, 150 mm × 4.6 mm, 5 µm, 100 Å). The mobile phase was composed of acetonitrile and filtered water (Durapore® membrane filters, 0.45 µm HVLP type, Ireland) in a 25 ∕ 75 ratio with a flow rate of 1.2 mL min−1 (adapted from Zhai, 2012). The sample injection volume was 50 µL, spectra were recorded at 272 nm, and the run time was 10 min.
Table 1Phenol concentrations measured by GC-MS in the five cloud waters sampled at the PUY station.

a From Lebedev et al. (2018). b This work.
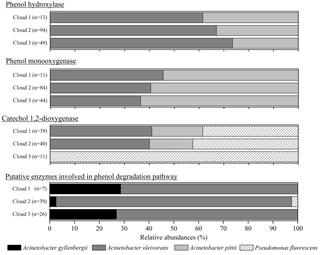
Figure 3Relative abundance of the putative taxonomic affiliation of the microorganisms involved in phenol degradation. For the five databases, microorganisms associated with a matching sequence with cloud transcripts are plotted here; the absolute total number of hits for each sample is indicated (n).
Phenol degradation. The percentage of phenol degradation was calculated by using the following equation.
The limit of phenol quantification was 3.8 µM. Strains are not considered active below 5 % of phenol degradation, corresponding to 5 µM.
Comparison of strain phenol degradation abilities was done using a nonparametric Kruskal–Wallis test (p value < 0.05) with Past software.
3.1 Phenol quantification in cloud waters
The objective of this paper was to explore the ability of microorganisms isolated or present in cloud waters collected at the PUY station to degrade phenol. From the 145 tested strains, 33 were isolated in this work; the others were already published (Table S1). We first checked for its presence in cloud waters sampled at the PUY station by performing GC-MS analysis. Figure S2 presents the back trajectories of the air masses corresponding to the five cloud events at the PUY station. The air mass origins of the five cloud samples determined from these back trajectories were classified as described in Deguillaume et al. (2014) and are reported in Table 1.
The GC-MS analysis performed on cloud samples allowed for the reliable identification and quantification of phenol in all samples (Table 1); the measured phenol concentrations ranged from 0.15 to 0.74 µg L−1. Figure S3a represents, as a typical example, the total ion current chromatogram of the cloud sample collected on 16 February 2016; the corresponding mass chromatogram based on the ion 94 current (characteristic for phenol) is represented in Fig. S3b. Quantification was done using similar mass chromatograms of all samples and the identification was proven by the correct retention time and accurate mass measurements (calculated: 94.0413; experimental: 94.0414).
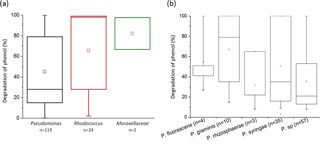
Figure 4Biodegradation of phenol by bacterial strains isolated from cloud waters. Results are expressed as the percentage of phenol biodegradation measured by HPLC after 5 days of incubation at 17 ∘C. (a) Results obtained for the 119 Pseudomonas strains, the 24 Rhodococcus strains, and the 2 strains from the Moraxellaceae family. (b) Focus on the Pseudomonas species. Only species groups with a minimum of three strains are plotted here.
3.2 Possibility of in-cloud phenol degradation by the cloud microbiome using a metatranscriptomic analysis
The presence of transcripts involved in the biodegradation of phenol (Fig. 1) was investigated from prokaryotic messenger RNA-enriched metatranscriptomes obtained from three cloud water samples. Sequence data were searched for the presence of transcripts of genes involved in phenol biodegradation among the 281 sequences included in our database (more details about the affiliation of the sequences are given in Table S2).
Gene transcripts were detected for all the enzymes, except the catechol 2,3-dioxygenase, showing a possible implication of the microorganisms in the degradation of phenol in cloud (Fig. 2). However, the number of hits and the relative abundance of the transcripts coding for the different enzymes varied according to the cloud samples. A total of 257 hits (sequence homology) could be counted for cloud 2, with only 70 in cloud 1 and 130 in cloud 3. Transcripts corresponding to the enzyme involved in the first step of oxidation of phenol leading to catechols (phenol hydroxylases and phenol monooxygenases) were the most abundant in clouds 2 and 3, while those corresponding to the cleavage of the catechol ring (catechol 1,2-dioxygenase) were dominant in cloud 1. For all the samples the transcripts corresponding to putative phenol degradation enzyme pathways (i.e., none explicitly described enzymes) remained low. However, the slight differences observed between the three cloud samples are not significant when analyzed by a nonparametric Kruskal–Wallis test.
Figure 3 presents the relative abundance of the putative taxonomic affiliation of microorganisms involved in phenol biodegradation based on the information associated with sequences in the databases. All the sequences were affiliated with Gammaproteobacteria from only two genera, namely Pseudomonas and Acinetobacter, corresponding to only four species (P. fluorescens, A. gyllenbergii, A. oleivorans, and A. pitii) matched with cloud transcripts, among a total of 50 (Table S2). This very low diversity was unexpected considering that sequences from 50 bacterial genera, including 109 species, were used for our search in databases. In addition, the relative abundance of sequences affiliated with a bacterial species varied a lot with the considered enzymes and clouds (Fig. 3).
Gammaproteobacteria were found to contribute up to 21 % of the ribosome sequences identified in bacteria in targeted sequencing investigations. Pseudomonas in particular was highlighted as one of the most represented genera (contributing alone up to 2 % of the ribosome sequences) and the most active genus based on its representation in transcriptomes and consecutively high ribosomal cDNA : DNA ratio (Amato et al., 2017a; Fig. S4). Acinetobacter and Rhodococcus were much less represented (<0.1 % of the ribosome sequences) but also accounted for groups of interest regarding potential metabolic activity.
Rhodococcus were previously isolated from clouds at the PUY station (Vaïtilingom et al., 2012), but genes for phenol degradation affiliated with this genus were not detected here.
3.3 Screening of bacterial strains isolated from cloud waters for their ability to biodegrade phenol
From our strain collection of 826 culturable microorganisms isolated from clouds collected at the PUY station between March 2003 and June 2016, we selected strains belonging to genera of interest concerning their potential ability for phenol biodegradation. We choose to test specifically Pseudomonas and Acinetobacter strains as they were detected in our metatranscriptomic analysis. As no Acinetobacter was available in our bacterial collection we choose closely related genera, namely two strains of Moraxella and Psychrobacter. In addition, Rhodococcus is well known to biodegrade phenol in the literature (as well as Pseudomonas and Acinetobacter). Pseudomonas and Rhodococcus are also the most frequently found genera in culturable bacteria from clouds (Renard et al., 2016; Vaïtilingom et al., 2012). Altogether 145 bacterial strains were tested (Table S1). The percentage of phenol degradation measured by HPLC after 5 days of incubation at 17 ∘C is reported in Fig. 4 and in Table S1. As the objective of the work was to perform a large screening with different types of cells, an incubation duration of 5 days was chosen to be sure that the induction period necessary for laboratory experiments was long enough to be able to detect biodegradation ability for all the tested cells. This time is quite long for a cloud but the objective here is not to evaluate a rate of biodegradation but to investigate the potential of biodegradation of microorganisms present in cloud waters.
We found that 93.1 % of the 145 tested strains were able to degrade phenol after 5 days of incubation. Globally, in our experimental conditions, all the families tested were very good phenol degraders (see Fig. 4a). No significant difference was found in the capacity of phenol degradation between Pseudomonas, Rhodococcus, and Moraxellaceae strains. A focus on the Pseudomonas strains according to their species is presented in Fig. 4b. The mean capacity of phenol degradation varied between 31 % and 67 % (for Pseudomonas rhizosphaerae and Pseudomonas graminis, respectively); however, no significant difference was observed between the species according to the Kruskal–Wallis test. Considering specifically Pseudomonas syringae strains, which are the most abundant species among cultural strains present in cloud waters (Renard et al., 2016), only 2 out of 35 were not capable of degrading phenol (strains PDD-32b-31and PDD-69b-20; see Table S1).
Phenol was present in the cloud water samples at concentrations ranging from 0.15 to 0.74 µg L−1; these values are within the range usually measured in atmospheric waters at remote sampling sites (3.0 to 5.4 µg L−1; Harrison et al., 2005), but globally the concentrations measured at the PUY station are rather in the lower range of values. Although the concentration of phenol remains within the same order of magnitude in the five cloud samples, it seems that the origin of the air masses had an impact on this concentration; it was 3 times higher in non-polluted air masses (west) than in polluted ones (northwest–north).
The results of combining a molecular approach and biodegradation assays involving culturable bacteria indicate that phenol-degrading microorganisms are present in clouds. The molecular approach allowed for the detection of transcripts belonging to Pseudomonas and Acinetobacter strains but not for the sequences of the other strains present in Table S2. It was surprising not to find Rhodococcus sequences as this genus is well known to degrade phenol as reported in the literature. In parallel, Rhodococcus strains isolated from clouds were very active phenol degraders but no Acinetobacter have been isolated from clouds. This difference reflects the complementarity but also the bias of each approach (molecular vs. cultural). Metatranscriptomic analysis can be biased by technical issues (extraction, sequencing, etc.) or by the creation of an incomplete database. In the future, the database for phenol degradation could be improved by integrating more sequences, especially considering data banks other than NCBI. For instance, the catechol operon sequences of Pseudomonas synringae (Berge et al., 2014) could be added to the database. We recently published the genome sequence of Pseudomonas syringae 32b-74, Pseudomonas graminis 13b-3, and Rhodococcus enclensis 23b-28, which are degrading phenol (Table S1; Besaury et al., 2017a, b; Lallement et al., 2017); they could be used to implement the database. Finally, in the future the genome of many phenol degraders (Table S1) could be also sequenced and integrated.
On the other hand, it is well known that culturable microorganisms only represent 1 % or less than the total community, notably in clouds (Amann et al., 1995; Vaïtilingom et al., 2012). Strains of Acinetobacter, Pseudomonas, and Rhodococcus genera are known to degrade phenol in other environments (Basha et al., 2010; Gami et al., 2014; Michalowicz and Duda, 2007; Sandhu et al., 2007). The cloud microbiota, as described from a culturable approach, harbors species usually affiliated with the phyllosphere (Amato et al., 2017b; Vaïtilingom et al., 2012). Sandhu et al. (2007) explored the presence of phenol degraders among microbial communities on plant leaves. They did not find Pseudomonas, but they isolated Acinetobacter and Rhodococcus strains and noticed globally a low diversity of phenol degraders. Only the genes encoding for the ortho pathway for phenol degradation that involves the catechol 1,2-dioxygenase activity were present in both Proteobacteria and Actinobacteria. Similarly, we did not find transcripts of genes coding for catechol 2,3-dioxygenase but only those coding for phenol hydroxylase, phenol monooxygenase, and catechol 1,2-dioxygenase. In principle, bacteria can have either ortho or meta pathways or both, but their expression is dependent on phenol concentration. The enzyme catechol 1,2-dioxygenase is produced at low phenol concentration, while catechol 2,3-dioxygenase enzymes become dominant at high phenol concentrations (3 mM; Sandhu et al., 2009). This might explain why bacteria from clouds and the phyllosphere only produce catechol 1,2-dioxygenase as the phenol concentration in the atmosphere is much lower than in polluted surface water, for instance (in the range of a few µg L−1 versus 100 to 1000 µg L−1; Gami et al., 2014; Harrison et al., 2005; Schummer et al., 2009; Sturaro et al., 2010).
In our study we focused on Pseudomonas strains as they are the most frequent culturable strains (Vaïtilingom et al., 2012) and belong to the most active strains in cloud waters (Amato et al., 2017a). We observed that these strains likely issued from the phyllosphere; P. graminis P. syringae, P. fluorescence, P. poae, and P. viriflava were able to degrade phenol. In the literature, P. aeruginosa and P. putida are the most popular phenol degraders (Basha et al., 2010; Der Yang and Humphrey, 1975; Erhan et al., 2004; Gami et al., 2014; Kumar et al., 2005; Molin and Nilsson, 1985). Interestingly, Bartoli et al. (2015) showed that the genome of several P. syringae pathogens of woody plants contained a catechol operon, while it was not the case for other P. syringae strain pathogens of herbaceous plants. These results strongly suggested that the enzymes present in the catechol pathway could help the degradation of aromatics present in lignins. In addition, Berge et al. (2014) showed that some P. syringae strains from phylogroups 1 and 3 that were ice nuclei active (INA+) also contained the catechol operon. In our case we also measured the ice nucleation activity of the 35 Pseudomonas syringae strains as described in Joly et al. (2013). Figure S5 presents the strains that were INA+ ( ∘C) versus their phenol degradation ability. Among the phenol degraders, 57.6 % of the bacteria were INA+.
Clouds can be considered as a medium for microorganism transport, and INA+ bacteria are suspected to induce precipitation and thus participate in the water cycle (Morris et al., 2008). Consequently, the presence of Pseudomonas syringae in clouds combining ice nucleation and phenol degradation properties can be of major importance for the pathogenicity on woody plants in terms of epidemiology, dispersion of pathogens, and emergence of plant diseases.
We showed that microorganisms from clouds were able to degrade phenol. The question raised is what is the potential impact of this biotransformation on the fate of phenol in real clouds? First, the presence of transcripts of phenol-degrading enzymes measured directly in situ demonstrates the real in-cloud activity of microorganisms. However, these data do not give any exact quantitative contribution of the microbial activity to phenol transformation in real clouds. On the other hand, the large screening performed with selected cloud strains showed that they have the enzymatic equipment for phenol degradation. Future work should be conducted to evaluate this potential for phenol biodegradation in real clouds where a larger microbial diversity is present. In particular, precise biodegradation rates should be determined under “realistic cloud conditions” to evaluate its real impact. It will also be very important to compare the relative contribution of biological degradation versus radical chemistry, especially with photochemistry. It is well known that phenol can react with the •OH, , and radicals alone or in combination to give rise to catechol, 2-nitrophenol, 4-nitrophenol, and 2,4-dinitrophenol; these compounds can be further degraded in intermediates after the ring cleavage (Harrison et al., 2005).
In addition to examining the presence of phenol-degrading pathways, we also looked for biological pathways leading to the potential formation of phenol from benzene (Choi et al., 2013; Tao et al., 2004) involving toluene monooxygenases in eight species of Actinobacteria, Alphaproteobacteria, and Betaproteobacteria (Table S3). None of these sequences were found in the cloud prokaryote metatranscriptomes. This result should be confirmed by incubating cloud microorganisms directly with benzene to assess the real potential of cloud microorganisms to produce phenol under these conditions. Consequently, the origin of phenol in cloud waters could only result from mass transfer from the gas phase to the aqueous phase or production via radical processes in the atmosphere. For instance, the production of phenol in the gas phase can result from the reactivity of the •OH radical with benzene (Grosjean, 1991; Volkamer et al., 2002). Considering that benzene has a very low solubility in water, it is likely that the production of phenol mainly occurs in the gas phase and is transferred to the water phase. The contributions of the biotic or abiotic transformation of benzene into phenol in the water phase should remain minor processes.
In conclusion, this is the first report showing that cloud water is inhabited by microorganisms that have phenol degradation ability. The study was centered on bacteria present in cloud waters collected at the PUY station where phenol concentrations were measured by GC-MS and found in the range of 0.15 to 0.74 µg L−1. Metatranscriptomic analysis suggested that phenol could be biodegraded in clouds, while a large screening of isolated strains showed that the enzymatic equipment to degrade phenol was not rare. These two combined approaches suggested that Pseudomonas, Acinetobacter, and Rhodococcus strains were the major genera potentially involved in phenol biodegradation. Further work is needed to evaluate the relative contribution of this biological activity and radical chemistry (particularly photochemistry) to phenol transformation. For that, experiments will be set up to measure phenol biodegradation rates under realistic cloud conditions and compare them with abiotic degradation rates. This will produce valuable information to better describe the fate of this pollutant in the atmosphere. Since phenol is highly toxic and is one of the main pollutants listed by the US Environmental Protection Agency (US EPA no.440/5-80-066: https://nepis.epa.gov/Exe/ZyPDF.cgi/2000LNAI.PDF?Dockey=2000LNAI.PDF, last access: 5 September 2018), this work will help to better assess its impact on health and air quality. Most probably, microorganisms could participate in a natural remediation process of the atmosphere.
The sequence files have been deposited in the European Nucleotide Archive (ENA) under the study accession number PRJEB25802.
The supplement related to this article is available online at: https://doi.org/10.5194/bg-15-5733-2018-supplement.
Cloud sampling was supervised by LD and performed by PA, AL, LD, and LB. Phenol has been extracted by IC and AL, and it was identified and quantified by OVP, VBA, and ATL. Bioinformatics data have been analyzed by PA, LB, and AL. Strain incubations have been performed by AL, MS, ET, and VV. All this work was supervised by AMD and GM. The paper was written by AL, AMD, and LD.
The authors declare that they have no conflict of interest.
This work does not involve human or animal subjects. There is no ethical conflict.
This work was mainly funded by the French ANR program BIOCAP (ANR-13-BS06-0004), the ANR-DFG program CHLOROFILTER (ANR-DFG-14-CE35-005-02), and the CNRS EC2CO program FONCOMIC.
The authors also acknowledge financial support from the Regional Council
of Auvergne, the Observatoire de Physique du Globe de Clermont-Ferrand
(OPGC), the Fédération de Recherche en Environnement through the
CPER Environnement founded by Région Auvergne-Rhône-Alpes, the French
ministry, and FEDER from the European Community.
Edited by: Tina Treude
Reviewed by: Odile
Berge and one anonymous referee
Ajaz, M., Noor, N., Rasool, S. A., and Khan, S. A.: Phenol resistant bacteria from soil: identification-characterization and genetical studies, Pak. J. Bot., 36, 415–424, 2004.
Al-Khalid, T. and El-Naas, M. H.: Aerobic biodegradation of phenols: a comprehensive review, Crit. Rev. Env. Sci. Tech., 42, 1631–1690, https://doi.org/10.1080/10643389.2011.569872, 2012.
Amann, R. I., Ludwig, W., and Schleifer, K. H.: Phylogenetic identification and in situ detection of individual microbial cells without cultivation, Microbiol. Rev., 59, 143–169, 1995.
Amato, P., Joly, M., Besaury, L., Oudart, A., Taib, N., Moné, A. I., Deguillaume, L., Delort, A.-M., and Debroas, D.: Active microorganisms thrive among extremely diverse communities in cloud water, PLoS One, 12, e0182869, https://doi.org/10.1371/journal.pone.0182869, 2017a.
Amato, P., Brisebois, E., Draghi, M., Duchaine, C., Fröhlich-Nowoisky, J., Huffman, J. A., Mainelis, G., Robine, E., and Thibaudon, M.: Main biological aerosols, specificities, abundance, and diversity, in: Microbiology of Aerosols, edited by: Delort, A. and Amato, P., 3–22, https://doi.org/10.1002/9781119132318.ch1a, 2017b.
Andrews, S.: FastQC: a quality control tool for high throughput sequence data, Babraham Bioinfomatics, available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (last access: 27 June 2017), 2010.
Atkinson, R., Aschmann, S. M., and Arey, J.: Reactions of OH and NO3 radicals with phenol, cresols, and 2-nitrophenol at 296±2 K, Environ. Sci. Technol., 26, 1397–1403, https://doi.org/10.1021/es00031a018, 1992.
Auffret, M. D., Yergeau, E., Labbé, D., Fayolle-Guichard, F., and Greer, C. W.: Importance of Rhodococcus strains in a bacterial consortium degrading a mixture of hydrocarbons, gasoline, and diesel oil additives revealed by metatranscriptomic analysis, Appl. Microbiol. Biot., 99, 2419–2430, https://doi.org/10.1007/s00253-014-6159-8, 2015.
Babich, H. and Davis, D. L.: Phenol: a review of environmental and health risks, Regul. Toxicol. Pharm., 1, 90–109, https://doi.org/10.1016/0273-2300(81)90071-4, 1981.
Bartoli, C., Lamichhane, J. R., Berge, O., Guilbaud, C., Varvaro, L., Balestra, G. M., Vinatzer, B. A., and Morris, C. E.: A framework to gauge the epidemic potential of plant pathogens in environmental reservoirs: the example of kiwifruit canker, Mol. Plant Pathol., 16, 137–149, https://doi.org/10.1111/mpp.12167, 2015.
Basha, K. M., Rajendran, A., and Thangavelu, V.: Recent advances in the biodegradation of phenol: a review, Asian Journal of Experimental Biological Sciences, 1, 219–234, 2010.
Berge, O., Monteil, C. L., Bartoli, C., Chandeysson, C., Guilbaud, C., Sands, D. C., and Morris, C. E.: A user's guide to a data base of the diversity of Pseudomonas syringae and its application to classifying strains in this phylogenetic complex, PLoS One, 9, e105547, https://doi.org/10.1371/journal.pone.0105547, 2014.
Besaury, L., Amato, P., Sancelme, M., and Delort, A. M.: Draft genome sequence of Pseudomonas syringae PDD-32b-74, a model strain for ice-nucleation studies in the atmosphere, Genome Announcements, 5, 28751406, https://doi.org/10.1128/genomeA.00742-17, 2017a.
Besaury, L., Amato, P., Wirgot, N., Sancelme, M., and Delort, A. M.: Draft genome sequence of Pseudomonas graminis PDD-13b-3, a model strain isolated from cloud water, Genome Announcements, 5, https://doi.org/10.1128/genomeA.00464-17, 2017b.
Brennerova, M. V., Josefiova, J., Brenner, V., Pieper, D. H., and Junca, H.: Metagenomics reveals diversity and abundance of meta-cleavage pathways in microbial communities from soil highly contaminated with jet fuel under air-sparging bioremediation, Environ. Microbiol., 11, 2216–2227, https://doi.org/10.1111/j.1462-2920.2009.01943.x, 2009.
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., and Madden, T. L.: BLAST+: architecture and applications, BMC Bioinformatics, 10, 421, https://doi.org/10.1186/1471-2105-10-421, 2009.
Choi, E. J., Jin, H. M., Lee, S. H., Math, R. K., Madsen, E. L., and Jeon, C. O.: Comparative genomic analysis and BTEX degradation pathways of Pseudoxanthomonas spadix BD-a59, Appl. Environ. Microbiol., 79, 663–671, https://doi.org/10.1128/AEM.02809-12, 2013.
Deguillaume, L., Charbouillot, T., Joly, M., Vaïtilingom, M., Parazols, M., Marinoni, A., Amato, P., Delort, A.-M., Vinatier, V., Flossmann, A., Chaumerliac, N., Pichon, J. M., Houdier, S., Laj, P., Sellegri, K., Colomb, A., Brigante, M., and Mailhot, G.: Classification of clouds sampled at the puy de Dôme (France) based on 10 yr of monitoring of their physicochemical properties, Atmos. Chem. Phys., 14, 1485–1506, https://doi.org/10.5194/acp-14-1485-2014, 2014.
Delort, A.-M., Vaïtilingom, M., Amato, P., Sancelme, M., Parazols, M., Mailhot, G., Laj, P., and Deguillaume, L.: A short overview of the microbial population in clouds: potential roles in atmospheric chemistry and nucleation processes, Atmos. Res., 98, 249–260, https://doi.org/10.1016/j.atmosres.2010.07.004, 2010.
Delort, A. M., Vaïtilingom, M., Joly, M., Amato, P., Wirgot, N., Lallement, A., Sancelme, M., Matulova, M., and Deguillaume, L.: Clouds: a transient and stressing habitat for microorganisms, in: Microbial Ecology of Extreme Environments, edited by: Chénard, C. and Lauro, F. M., 215–245, Springer International Publishing, https://doi.org/10.1007/978-3-319-51686-8_10, 2017.
Der Yang, R. and Humphrey, A. E.: Dynamic and steady state studies of phenol biodegradation in pure and mixed cultures, Biotechnol. Bioeng., 17, 1211–1235, https://doi.org/10.1002/bit.260170809, 1975.
Duan, W., Meng, F., Cui, H., Lin, Y., Wang, G., and Wu, J.: Ecotoxicity of phenol and cresols to aquatic organisms: a review, Ecotox. Environ. Safe., 157, 441–456, https://doi.org/10.1016/j.ecoenv.2018.03.089, 2018.
Erhan, E., Yer, E., Akay, G., Keskinler, B., and Keskinler, D.: Phenol degradation in a fixed-bed bioreactor using micro-cellular polymer-immobilized Pseudomonas syringae, J. Chem. Technol. Biotechnol., 79, 195–206, https://doi.org/10.1002/jctb.938, 2004.
Fang, H., Cai, L., Yu, Y., and Zhang, T.: Metagenomic analysis reveals the prevalence of biodegradation genes for organic pollutants in activated sludge, Bioresource Technol., 129, 209–218, https://doi.org/10.1016/j.biortech.2012.11.054, 2013.
Gami, A. A., Shukor, M. Y., Khalil, K. A., Dahalan, F. A., Khalid, A., and Ahmad, S. A.: Phenol and its toxicity, Journal of Environmental Microbiology and Toxicology, 2, 11–24, 2014.
Gilardoni, S., Massoli, P., Paglione, M., Giulianelli, L., Carbone, C., Rinaldi, M., Decesari, S., Sandrini, S., Costabile, F., Gobbi, G. P., Pietrogrande, M. C., Visentin, M., Scotto, F., Fuzzi, S., and Facchini, M. C.: Direct observation of aqueous secondary organic aerosol from biomass-burning emissions, P. Natl. Acad. Sci. USA, 113, 10013–10018, https://doi.org/10.1073/pnas.1602212113, 2016.
Grosjean, D.: Atmospheric fate of toxic aromatic compounds, Sci. Total Environ., 100, 367–414, https://doi.org/10.1016/0048-9697(91)90386-S, 1991.
Harrison, M. A. J., Cape, J. N., and Heal, M. R.: Experimentally determined Henry's law coefficients of phenol, 2-methylphenol and 2-nitrophenol in the temperature range 281–302 K, Atmos. Environ., 36, 1843–1851, https://doi.org/10.1016/S1352-2310(02)00137-1, 2002.
Harrison, M. A. J., Barra, S., Borghesi, D., Vione, D., Arsene, C., and Olariu, R. I.: Nitrated phenols in the atmosphere: a review, Atmos. Environ., 39, 231–248, https://doi.org/10.1016/j.atmosenv.2004.09.044, 2005.
Heal, M. R., Pilling, M. J., Titcombe, P. E., and Whitaker, B. J.: Mass accommodation of aniline, phenol and toluene on aqueous droplets, Geophys. Res. Lett., 22, 3043–3046, https://doi.org/10.1029/95GL02944, 1995.
Herrmann, H., Schaefer, T., Tilgner, A., Styler, S. A., Weller, C., Teich, M., and Otto, T.: Tropospheric aqueous-phase chemistry: kinetics, mechanisms, and its coupling to a changing gas phase, Chem. Rev., 115, 4259–4334, https://doi.org/10.1021/cr500447k, 2015.
Hill, K. A., Shepson, P. B., Galbavy, E. S., Anastasio, C., Kourtev, P. S., Konopka, A., and Stirm, B. H.: Processing of atmospheric nitrogen by clouds above a forest environment, J. Geophys. Res., 112, 1–16, https://doi.org/10.1029/2006JD008002, 2007.
Jadeja, N. B., More, R. P., Purohit, H. J., and Kapley, A.: Metagenomic analysis of oxygenases from activated sludge, Bioresource Technol., 165, 250–256, https://doi.org/10.1016/j.biortech.2014.02.045, 2014.
Joly, M., Attard, E., Sancelme, M., Deguillaume, L., Guilbaud, C., Morris, C. E., Amato, P., and Delort, A.-M.: Ice nucleation activity of bacteria isolated from cloud water, Atmos. Environ., 70, 392–400, https://doi.org/10.1016/j.atmosenv.2013.01.027, 2013.
Kafilzadeh, F., Farhangdoost, M.-S., and Tahery, Y.: Isolation and identification of phenol degrading bacteria from Lake Parishan and their growth kinetic assay, Afr. J. Biotechnol., 9, 6721–6726, 2010.
Kumar, A., Kumar, S., and Kumar, S.: Biodegradation kinetics of phenol and catechol using Pseudomonas putida MTCC 1194, Biochem. Eng. J., 22, 151–159, https://doi.org/10.1016/j.bej.2004.09.006, 2005.
Lallement, A., Besaury, L., Eyheraguibel, B., Amato, P., Sancelme, M., Mailhot, G., and Delort, A. M.: Draft genome sequence of Rhodococcus enclensis 23b-28, a model strain isolated from cloud water, Genome Announcements, 5, 29074669, https://doi.org/10.1128/genomeA.01199-17, 2017.
Langmead, B. and Salzberg, S. L.: Fast gapped-read alignment with Bowtie 2, Nature Methods, 9, 357–359, https://doi.org/10.1038/nmeth.1923, 2012.
Lebedev, A. T., Polyakova, O. V., Mazur, D. M., and Artaev, V. B.: The benefits of high resolution mass spectrometry in environmental analysis, Analyst, 138, 6946–6953, https://doi.org/10.1039/C3AN01237A, 2013.
Lebedev, A. T., Polyakova, O. V., Mazur, D. M., Artaev, V. B., Canet, I., Lallement, A., Vaïtilingom, M., Deguillaume, L., and Delort, A.-M.: Detection of semi-volatile compounds in cloud waters by GC × GC-TOF-MS. Evidence of phenols and phthalates as priority pollutants, Environ. Pollut., 241, 616–625, https://doi.org/10.1016/j.envpol.2018.05.089, 2018.
Leinonen, R., Nardone, F., Zhu, W., and Apweiler, R.: UniSave: the UniProtKB sequence/annotation version database, Bioinformatics, 22, 1284–1285, https://doi.org/10.1093/bioinformatics/btl105, 2006.
Li, H.: Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM, arXiv preprint arXiv:1303.3997, available at: https://arxiv.org/pdf/1303.3997.pdf (last access: 5 September 2018), 2013.
Lober, C. W.: Chemexfoliation – indications and cautions, J. Am. Acad. Dermatol., 17, 109–112, https://doi.org/10.1016/S0190-9622(87)70181-9, 1987.
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G., and Neufeld, J. D.: PANDAseq: paired-end assembler for illumina sequences, BMC Bioinformatics, 13, 31, https://doi.org/10.1186/1471-2105-13-31, 2012.
Michalowicz, J. and Duda, W.: Phenols transformations in the environment and living organisms, Current Topics in Biophysics, 30, 24–36, 2007.
Mishra, V. K. and Kumar, N.: Microbial degradation of phenol: a review, Journal of Water Pollution and Purification Research, 4, 17–22, 2017.
Molin, G. and Nilsson, I.: Degradation of phenol by Pseudomonas putida ATCC 11172 in continuous culture at different ratios of biofilm surface to culture volume, Appl. Environ. Microbiol., 50, 946–950, 1985.
Morris, C. E., Sands, D. C., Vinatzer, B. A., Glaux, C., Guilbaud, C., Buffiere, A., Yan, S., Dominguez, H., and Thompson, B. M.: The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle, ISME J., 2, 321–334, https://doi.org/10.1038/ismej.2007.113, 2008.
Olariu, R. I.: Atmospheric oxidation of selected aromatic hydrocarbons, Doctoral Thesis, Bergische Universitat Gesamthochschule Wuppertal, Wuppertal, 2001.
Olariu, R. I., Klotz, B., Barnes, I., Becker, K. H., and Mocanu, R.: FT–IR study of the ring-retaining products from the reaction of OH radicals with phenol, o-, m-, and p-cresol, Atmos. Environ., 36, 3685–3697, https://doi.org/10.1016/S1352-2310(02)00202-9, 2002.
Padmanabhan, P., Padmanabhan, S., DeRito, C., Gray, A., Gannon, D., Snape, J. R., Tsai, C. S., Park, W., Jeon, C., and Madsen, E. L.: Respiration of 13C-labeled substrates added to soil in the field and subsequent 16S rRNA gene analysis of 13C-labeled soil DNA, Appl. Environ. Microbiol., 69, 1614–1622, https://doi.org/10.1128/AEM.69.3.1614-1622.2003, 2003.
Rayne, S., Forest, K., and Friesen, K. J.: Mechanistic aspects regarding the direct aqueous environmental photochemistry of phenol and its simple halogenated derivatives. A review, Environ. Int., 35, 425–437, https://doi.org/10.1016/j.envint.2008.09.004, 2009.
Reasoner, D. J. and Geldreich, E. E.: A new medium for the enumeration and subculture of bacteria from potable water, Appl. Environ. Microb., 49, 1–7, 1985.
Renard, P., Canet, I., Sancelme, M., Wirgot, N., Deguillaume, L., and Delort, A.-M.: Screening of cloud microorganisms isolated at the Puy de Dôme (France) station for the production of biosurfactants, Atmos. Chem. Phys., 16, 12347–12358, https://doi.org/10.5194/acp-16-12347-2016, 2016.
Rubio, M. A., Lissi, E., Herrera, N., Pérez, V., and Fuentes, N.: Phenol and nitrophenols in the air and dew waters of Santiago de Chile, Chemosphere, 86, 1035–1039, https://doi.org/10.1016/j.chemosphere.2011.11.046, 2012.
Sandhu, A., Halverson, L. J., and Beattie, G. A.: Bacterial degradation of airborne phenol in the phyllosphere, Environ. Microbiol., 9, 383–392, https://doi.org/10.1111/j.1462-2920.2006.01149.x, 2007.
Sandhu, A., Halverson, L. J., and Beattie, G. A.: Identification and genetic characterization of phenol-degrading bacteria from leaf microbial communities, Microb. Ecol., 57, 276–285, https://doi.org/10.1007/s00248-008-9473-9, 2009.
Sattler, B., Puxbaum, H., and Psenner, R.: Bacterial growth in supercooled cloud droplets, Geophys. Res. Lett., 28, 239–242, https://doi.org/10.1029/2000GL011684, 2001.
Schauer, J. J., Kleeman, M. J., Cass, G. R., and Simoneit, B. R. T.: Measurement of emissions from air pollution sources. 3. C1-C29 organic compounds from fireplace combustion of wood, Environ. Sci. Technol., 35, 1716–1728, https://doi.org/10.1021/es001331e, 2001.
Schmieder, R. and Edwards, R.: Quality control and preprocessing of metagenomic datasets, Bioinformatics, 27, 863–864, https://doi.org/10.1093/bioinformatics/btr026, 2011.
Schummer, C., Groff, C., Al Chami, J., Jaber, F., and Millet, M.: Analysis of phenols and nitrophenols in rainwater collected simultaneously on an urban and rural site in east of France, Sci. Total Environ., 407, 5637–5643, https://doi.org/10.1016/j.scitotenv.2009.06.051, 2009.
Sharma, N., Tanksale, H., Kapley, A., and Purohit, H. J.: Mining the metagenome of activated biomass of an industrial wastewater treatment plant by a novel method, Indian J. Microbiol., 52, 538–543, https://doi.org/10.1007/s12088-012-0263-1, 2012.
Silva, C. C., Hayden, H., Sawbridge, T., Mele, P., De Paula, S. O., Silva, L. C. F., Vidigal, P. M. P., Vicentini, R., Sousa, M. P., Torres, A. P. R., Santiago, V. M. J., and Oliveira, V. M.: Identification of genes and pathways related to phenol degradation in metagenomic libraries from petroleum refinery wastewater, PLoS One, 8, e61811, https://doi.org/10.1371/journal.pone.0061811, 2013.
Sridevi, V., Chandana Lakshmi, M. V. V., Manasa, M., and Sravani, M.: Metabolic pathways for the biodegradation of phenol, International Journal of Engineering Science and Advanced Technology, 2, 695–705, 2012.
Sturaro, A., Rella, R., Parvoli, G., and Ferrara, D.: Long-term phenol, cresols and BTEX monitoring in urban air, Environ. Monit. Assess., 164, 93–100, https://doi.org/10.1007/s10661-009-0877-x, 2010.
Suenaga, H., Koyama, Y., Miyakoshi, M., Miyazaki, R., Yano, H., Sota, M., Ohtsubo, Y., Tsuda, M., and Miyazaki, K.: Novel organization of aromatic degradation pathway genes in a microbial community as revealed by metagenomic analysis, ISME J., 3, 1335–1348, https://doi.org/10.1038/ismej.2009.76, 2009.
Sun, Y. L., Zhang, Q., Anastasio, C., and Sun, J.: Insights into secondary organic aerosol formed via aqueous-phase reactions of phenolic compounds based on high resolution mass spectrometry, Atmos. Chem. Phys., 10, 4809–4822, https://doi.org/10.5194/acp-10-4809-2010, 2010.
Tao, Y., Fishman, A., Bentley, W. E., and Wood, T. K.: Oxidation of benzene to phenol, catechol, and 1, 2, 3-trihydroxybenzene by toluene 4-monooxygenase of Pseudomonas mendocina KR1 and toluene 3-monooxygenase of Ralstonia pickettii PKO1, Appl. Environ. Microbiol., 70, 3814–3820, https://doi.org/10.1128/AEM.70.7.3814-3820.2004, 2004.
Tian, M., Du, D., Zhou, W., Zeng, X., and Cheng, G.: Phenol degradation and genotypic analysis of dioxygenase genes in bacteria isolated from sediments, Braz. J. Microbiol., 48, 305–313, https://doi.org/10.1016/j.bjm.2016.12.002, 2017.
Vaïtilingom, M., Amato, P., Sancelme, M., Laj, P., Leriche, M., and Delort, A.-M.: Contribution of microbial activity to carbon chemistry in clouds, Appl. Environ. Microbiol., 76, 23–29, https://doi.org/10.1128/AEM.01127-09, 2010.
Vaïtilingom, M., Attard, E., Gaiani, N., Sancelme, M., Deguillaume, L., Flossmann, A. I., Amato, P., and Delort, A.-M.: Long-term features of cloud microbiology at the puy de Dôme (France), Atmos. Environ., 56, 88–100, https://doi.org/10.1016/j.atmosenv.2012.03.072, 2012.
Vaïtilingom, M., Deguillaume, L., Vinatier, V., Sancelme, M., Amato, P., Chaumerliac, N., and Delort, A.-M.: Potential impact of microbial activity on the oxidant capacity and organic carbon budget in clouds, P. Natl. Acad. Sci. USA, 110, 559–564, https://doi.org/10.1073/pnas.1205743110, 2013.
Vione, D., Barra, S., de Gennaro, G., de Rienzo, M., Gilardoni, S., Perrone, M. G., and Pozzoli, L.: Polycyclic aromatic hydrocarbons in the atmosphere: monitoring, sources, sinks and fate. II: sinks and fate, Ann. Chim. Banner, 94, 257–268, https://doi.org/10.1002/adic.200490031, 2004.
Volkamer, R., Klotz, B., Barnes, I., Imamura, T., Wirtz, K., Washida, N., Heinz Becker, K., and Platt, U.: OH-initiated oxidation of benzene Part I. Phenol formation under atmospheric conditions, Phys. Chem. Chem. Phys., 4, 1598–1610, https://doi.org/10.1039/B108747A, 2002.
Zhai, A.: Determination of phenols in drinking water with agilent bond elut plexa SPE and HPLC, Agilent, available at: https://www.agilent.com/cs/library/applications/5990-9730EN.pdf (last access: 10 October 2017), 2012.





