the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
A quest for the biological sources of long chain alkyl diols in the western tropical North Atlantic Ocean
Julie Lattaud
Laura Villanueva
Sebastiaan W. Rampen
Corina P. D. Brussaard
Judith van Bleijswijk
Nicole Bale
Jaap S. Sinninghe Damsté
Stefan Schouten
Long chain alkyl diols (LCDs) are widespread in the marine water column and sediments, but their biological sources are mostly unknown. Here we combine lipid analyses with 18S rRNA gene amplicon sequencing on suspended particulate matter (SPM) collected in the photic zone of the western tropical North Atlantic Ocean at 24 stations to infer relationships between LCDs and potential LCD producers. The C30 1,15-diol was detected in all SPM samples and accounted for >95 % of the total LCDs, while minor proportions of C28 and C30 1,13-diols, C28 and C30 1,14-diols, as well as C32 1,15-diol were found. The concentration of the C30 and C32 diols was higher in the mixed layer of the water column compared to the deep chlorophyll maximum (DCM), whereas concentrations of C28 diols were comparable. Sequencing analyses revealed extremely low contributions (≈0.1 % of the 18S rRNA gene reads) of known LCD producers, but the contributions from two taxonomic classes with which known producers are affiliated, i.e. Dictyochophyceae and Chrysophyceae, followed a trend similar to that of the concentrations of C30 and C32 diols. Statistical analyses indicated that the abundance of 4 operational taxonomic units (OTUs) of the Chrysophyceae and Dictyochophyceae, along with 23 OTUs falling into other phylogenetic groups, were weakly (r≤0.6) but significantly (p value <0.01) correlated with C30 diol concentrations. It is not clear whether some of these OTUs might indeed correspond to C28−32 diol producers or whether these correlations are just indirect and the occurrence of C30 diols and specific OTUs in the same samples might be driven by other environmental conditions. Moreover, primer mismatches were unlikely, but cannot be excluded, and the variable number of rRNA gene copies within eukaryotes might have affected the analyses leading to LCD producers being undetected or undersampled. Furthermore, based on the average LCD content measured in cultivated LCD-producing algae, the detected concentrations of LCDs in SPM are too high to be explained by the abundances of the suspected LCD-producing OTUs. This is likely explained by the slower degradation of LCDs compared to DNA in the oxic water column and suggests that some of the LCDs found here were likely to be associated with suspended debris, while the DNA from the related LCD producers had been already fully degraded. This suggests that care should be taken in constraining biological sources of relatively stable biomarker lipids by quantitative comparisons of DNA and lipid abundances.
- Article
(2461 KB) - Full-text XML
-
Supplement
(668 KB) - BibTeX
- EndNote
Long chain alkyl diols (LCDs) are lipids that consist of a linear alkyl chain with 22–38 carbons, hydroxylated at both the terminal carbon atom and at an intermediate position, and usually saturated or monounsaturated. LCDs were identified for the first time in Black Sea sediments (de Leeuw et al., 1981) and have subsequently been found with widespread occurrence in both suspended particulate matter (SPM) and sediments from both coastal and off-shore sites throughout the world ocean (Jiang et al., 1994; Versteegh et al., 1997; Rampen et al., 2014b). LCDs can be preserved in marine sediments for long periods of time and their distribution can reflect the environmental conditions at the time they were produced.
The most abundant LCDs in seawater are the saturated C28 and C30 1,13-diols, C28 and C30 1,14-diols, and C30 and C32 1,15-diols (Rampen et al., 2014b), which are all likely produced by phytoplankton. However, the marine biological sources of LCDs are still not fully clear because, in contrast with the widespread occurrence of LCDs in the sediment, few marine taxa have been shown to contain these lipids. Eustigmatophyceae contain C30 1,13-, C30 1,15-, and C32 1,15-diols (Volkman et al., 1992; Rampen et al., 2014a), but they comprise mostly freshwater species, and only a few rare marine representatives from the genus Nannochloropsis are known (Andersen et al., 1998; Fawley and Fawley, 2007). Furthermore, the distribution of LCDs in the marine environment does not match that of LCDs of marine Eustigmatophyceae (Volkman et al., 1992; Rampen et al., 2012). Species of the diatom genus Proboscia and the dictyochophycean Apedinella radians contain C28−32 1,14-diols (Sinninghe Damsté et al., 2003; Rampen et al., 2009, 2011), with the former accounting for significant proportions of marine biomass mostly in upwelling regions (Moita et al., 2003; Lassiter et al., 2006), whereas the latter has been occasionally observed in estuarine environments (Seoane et al., 2005; Bergesch et al., 2008). Few other marine species from classes genetically related to diatoms and Eustigmatophyceae have been recently shown to produce LCDs (Table S1 in the Supplement). All the known LCD-producing phytoplankters belong to the eukaryotic supergroup Heterokontophyta, a division which includes, among others, diatoms and brown seaweeds. The widespread occurrence of LCDs in the marine environment, despite the restricted abundance and distribution of known marine LCD producers, suggests that these compounds may be produced by unknown phytoplankton species. In addition, LCD in the marine environment might also derive from vegetal debris of terrestrial or riverine origin. For example, C30−36 diols functionalized at the 1- and the ω18 or ω20 positions have previously been reported to occur in ferns (Jetter and Riederer, 1999; Speelman et al., 2009; Mao et al., 2017) and suggested to be part of the leaf cuticular waxes. Similarly, C26−32 diols have been occasionally detected in other plants (Buschhaus et al., 2013). This suggests that vegetal debris may in principle also source LCDs in seawater.
Several indices, based on ratios between the different diols, have been proposed for the reconstruction of past environmental conditions. The Diol Index, reflecting the proportion of C28 and C30 1,14-diols over the sum of C28 and C30 1,14-diols and the C30 1,15-diol, has been proposed to track ancient upwelling conditions since the 1,14-diols are believed to be mostly produced by upwelling diatoms of the genus Proboscia (Rampen et al., 2008). Another index, the long chain diol index (LDI), which is based on the proportion of the C30 1,15-diol over the C28 and C30 1,13-diols, shows a strong correlation with sea surface temperature (SST) and is used to determine past SST (Rampen et al., 2012; Plancq et al., 2014; Rodrigo-Gámiz et al., 2015). In addition, since the C32 1,15-diol is the major component of the LCDs of freshwater Eustigmatophyceae (Volkman et al., 1992; Rampen et al., 2014a), the fractional abundance of the C32 1,15-diol has been suggested to be a marker of riverine input in seawater (de Bar et al., 2016; Lattaud et al., 2017a, b). Other markers for riverine inputs in seawater are the C30−36 1,ω20-diols which are produced by the freshwater fern Azolla (Speelman et al., 2009; Mao et al., 2017). However, application of these proxies in the marine realm remains uncertain. For example, the growth of Proboscia spp. is typically promoted under low concentrations of dissolved silica, whereas other diatoms dominate the upwelling area under higher silica concentrations (Koning et al., 2001), making the Diol Index ineffective in predicting upwelling conditions when communities are dominated by other diatoms. In addition, the sources of the marine C28−32 1,13- and 1,15-diols are unknown, complicating the application of the LDI as a proxy.
A way of assessing the sources of biomarker lipids is to compare the abundance of lipids in environmental samples with the composition of the microbial community, as determined by genetic methods. For example, Villanueva et al. (2014) analysed both LCDs and eustigmatophycean 18S rRNA gene sequences in a tropical freshwater lake and found five clades of uncultured Eustigmatophyceae in the top 25 m of the water column of the lake, where LCDs were also abundant. Abundance determination by quantitative polymerase chain reaction (qPCR) highlighted that the number of eustigmatophycean 18S rRNA gene copies peaked at the same depth as the LCDs, suggesting that Eustigmatophyceae are a primary source for LCDs in freshwater (Villanueva et al., 2014). However, one of the limitations of this approach is that it relies on specific eustigmatophycean primers designed based on the sequences available in the genetic databases, which could be biased and not target all the existing LCD biological sources. To compensate for this limitation high throughput amplicon sequencing of the 18S rRNA gene allows the exploration of the total marine microbial communities in great detail (Stoeck et al., 2009; Logares et al., 2012; Christaki et al., 2014; Balzano et al., 2015; de Vargas et al., 2015; Massana et al., 2015). The combination of these analyses with lipid composition may potentially assist in identifying the main LCD producers in marine settings.
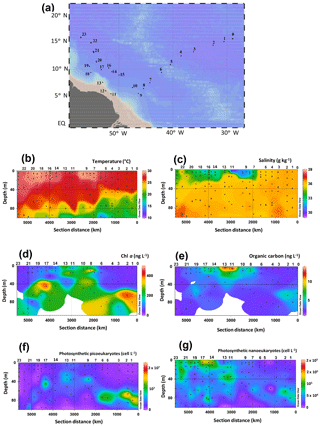
Figure 1HCC cruise track in the western tropical North Atlantic Ocean, physical seawater properties, and biological parameters. (a) Map of the sampling stations. Spatial distribution of (b) temperature, (c) salinity, the concentration of (d) Chl a, (e) organic carbon concentrations, and the abundance of photosynthetic (f) picoeukaryotes and (g) nanoeukaryotes. Temperature, salinity, as well as the concentrations of Chl a and organic carbon have also been published by Bale et al. (2018). Data were plotted using ODV software using kriging for interpolation between data points (Schlitzer, 2002). Dots represent the depth at which SPM was collected.
In the present study, we quantitatively analysed the composition and abundance of LCDs in suspended particulate matter (SPM) collected along the tropical North Atlantic (Fig. 1a) at different depths in the photic zone (surface, deep chlorophyll maximum (DCM), and bottom of the wind mixed layer (BWML); see also Bale et al., 2018). The 18S rRNA gene abundance and composition of the SPM was also analysed by quantitative PCR (qPCR) and high throughput amplicon sequencing to infer the taxonomic composition and to compare the abundance of the different taxa with that of the LCDs, in order to identify the potential marine biological sources of LCDs.
2.1 Cruise transect, ancillary data, and SPM collection
Samples were taken during the Heterocystous Cyanobacteria Cruise (HCC) (64PE393), which took place from 24 August to 21 September 2014 along a transect on the tropical North Atlantic Ocean (see Bale et al., 2018, for details). The transect was from Mindelo (Cape Verde) to a location about 500 km from the Amazon River mouth and then westwards along the coast towards Barbados (Fig. 1a). Temperature, salinity, and nutrient data have previously been reported in Bale et al. (2018).
Seawater was collected from two or three depths at each station to measure the concentration of chlorophyll a (Chl a) and the abundances of photosynthetic picoeukaryotes and nanoeukaryotes. Seawater was collected during the up cast using Niskin bottles mounted on a CTD frame. The sampling depths were determined based on the evaluation of the vertical profiles of temperature, salinity, and chlorophyll fluorescence after the down cast of the CTD deployment. The depth of the BWML and the DCM were determined based on the lowest position of the mixed layer and the depth at which the highest values of chlorophyll fluorescence were observed. For Chl a determination, seawater was collected from the Niskin bottles and filtered through 0.7 µm pore-size glass-fibre (Whatman GF/F) filters, followed by frozen storage. Chl a was extracted with methanol buffered with 0.5 M ammonium acetate, homogenized for 15 s, and analysed by high-performance liquid chromatography.
Photosynthetic picoeukaryotes and nanoeukaryotes were enumerated by flow cytometry according to the protocol of Marie et al. (2005). In short, 1 mL samples were counted fresh using a Becton-Dickinson FACSCalibur (Erembodegem, Belgium) flow cytometer equipped with an air-cooled Argon laser (488 nm, 15 mW). Phytoplankton were discriminated based on their chlorophyll autofluorescence and scatter signature. Cyanobacteria, i.e. Synechococcus and Prochlorococcus, were not included in the current study. Size fractionation was performed by gravity filtration with >3 µm average cell diameter phytoplankton groups classified as nanoeukaryotic and those with <3 µm average cell diameter as picoeukaryotic phytoplankton.
Three McLane in situ pumps (McLane Laboratories Inc., Falmouth) were used to collect SPM from the water column for the analysis of both lipids and microbial communities. As with the collection of seawater with Niskin bottles for Chl a and flow cytometry analyses, the in situ pumps were deployed at the surface (3–5 m depth), the BWML, and the DCM (Table S2). Between 100 and 400 L of seawater was pumped and the SPM was collected on pre-combusted 0.7 µm GF/F filters (Pall Corporation, Washington) and immediately frozen at . For the determination of the organic carbon concentrations, SPM was freeze dried and analysis was carried out using a Flash 2000 series Elemental Analyzer (Thermo Scientific) equipped with a thermal conductivity detector.
2.2 Lipid extraction and analyses of LCDs
Lipids were extracted from the GF/F filters as described previously (Lattaud et al., 2017b). Briefly, 1∕4 of the filters were dried using a LyoQuest (Telstart, Life Sciences) freeze-dryer and lipids were extracted using base and acid hydrolysis. The base hydrolysis was achieved with 12 mL of a 1 M KOH in methanol solution by refluxing for 1 h. Subsequently, the pH was adjusted to 4 with 2 M HCl:CH3OH (1:1, v∕v) and the extract was transferred into a separatory funnel. The residues were further extracted once with CH3OH:H2O (1:1, v∕v), twice with CH3OH, and three times with dichloromethane (DCM). The extracts were combined in the separatory funnel and bidistilled water (6 mL) was added. The combined solutions were mixed, shaken, and separated into a CH3OH:H2O and a DCM phase; the DCM phase was removed and collected in a centrifuge tube. The aqueous layer was re-extracted twice with 3 mL DCM. The pooled DCM layers were dried over a sodium sulfate column and the DCM was evaporated under a stream of nitrogen. The extract was then acid hydrolysed with 2 mL of 1.5 M HCl in CH3OH solution under reflux for 2 h. The pH was adjusted to 4 by adding 2 M KOH:CH3OH; 2 mL of DCM and 2 mL of bidistilled water were added to the hydrolysed extract, mixed, and shaken and, after phase separation, the DCM layer was transferred into another centrifuge tube. The remaining aqueous layer was washed twice with 2 mL DCM. The combined DCM layers were dried over a sodium sulfate column, the DCM was evaporated under a stream of nitrogen, and a C22 5,17-diol was added to the extract as an internal standard. The extract was separated on an activated aluminium oxide column into three fractions using the following solvents: hexane:DCM (9:1, v∕v), hexane:DCM (1:1, v∕v), and DCM:CH3OH (1:1, v∕v). The latter (polar) fraction containing the diols was dried under a gentle nitrogen stream. Diols were derivatized by silylating an aliquot of the polar fraction with 10 µL N,O-bis(trimethylsilyl) trifluoroacetamide (BSTFA) and 10 µL pyridine, heating for 30 min at 60 ∘C and adding 30 µL of ethyl acetate. The analysis of diols was performed by gas chromatography-mass spectrometry (GC-MS) using an Agilent 7990B GC gas chromatograph, equipped with a fused silica capillary column (25 m×320 µm) coated with CP Sil-5 (film thickness 0.12 µm), coupled to an Agilent 5977A MSD mass spectrometer. The temperature regime for the oven was the same as that used by Lattaud et al. (2017b): held at 70 ∘C for 1 min, increased to 130 ∘C at a rate of 20 , increased to 320 ∘C at a rate of 4 , held at 320 ∘C for 25 min. The flow was held constant at 2 mL min−1. The MS source temperature was held at 250 ∘C and the MS quadrupole at 150 ∘C. The diols were identified and quantified via single ion monitoring (SIM) of the (C28 1,14-diol), 313.3 (C28 1,13-diol, C30 1,15-diol), 327.3 (C30 1,14-diol), and 341.3 (C30 1,13-diol, C32 1,15-diol) ions (Versteegh et al., 1997; Rampen et al., 2012). Surface samples, which contained the highest concentrations of LCDs, were also analysed by full scan to evaluate the presence of other eustigmatophycean biomarkers such as long chain alkenols and long chain hydroxy fatty acids. Absolute concentrations were calculated using the peak area of the internal standard as a reference.
2.3 DNA extraction, PCR, qPCR, and 18S rRNA gene sequencing
On ice a small portion of the GF/F filters, corresponding to 1 / 16 of their initial size, hence containing SPM from ca. 25 L of seawater, was cut into small pieces using sterile scissors and tweezers. Filter pieces were then transferred into 2 mL microtubes and the DNA was extracted using a MOBIO powersoil DNA isolation kit (Qiagen) following manufacturer instructions. We amplified the hypervariable V4 region of the 18S rRNA which is considered the best genetic marker for the identification of microbial eukaryotes (Logares et al., 2012; Massana et al., 2015). The V4 is located in a central region (565–584 to 964–981 bp for Saccharomyces cerevisiae) of the 18S rRNA and it was amplified from the genomic DNA by PCR using the universal eukaryotic primers TAReuk454FWD1 (5′-CCAGCASCYGCGGTAATTCC-3′) and TAReuk454REV3 (5′-ACTTTCGTTCTTGATYRA-3′) (Stoeck et al., 2010). Primers were modified for multiplex sequencing on a Roche 454 GS FLX system: a 454-adapter A (CCATCTCATCCCTGCGTGTCTCCGACTCAG), a key (TCAG), and a 10 bp sample-specific Multiple Identifier (MID, Table S3) were bound to the 5′ end of the forward primer, whereas a 454-adapter 2 (CCTATCCCCTGTGTGCCTTGGCAGTCTCAG) and a unique MID (CGTGTCA) were bound to the 5′ end of the reverse primer for all the samples. The PCR mixture included 25 µL Phusion Flash High-Fidelity PCR Master Mix (ThermoFisher Scientific) 19.1 µL deionized water, 1.5 µL dimethyl sulfoxide, 1.7 µL from each primer, and 25 ng genomic DNA, and the V4 region was amplified using the same thermal cycling as described by Logares et al. (2012). Amplicons were visualized on a 1 % agarose gel, V4 bands were excised and subsequently purified using a QIAquick Gel Extraction Kit (Qiagen), and DNA concentration was measured by Qubit fluorometric quantitation (ThermoFisher Scientific). For each sequencing run, 20 samples were pooled in equimolar amounts and sequenced using a 454 GS-FLX Plus (Macrogen Korea). Some samples yielded a low number of reads and were re-sequenced; overall 77 samples were sequenced in five sequencing runs.
To determine the concentration of total 18S rRNA genes within the seawater sampled, we carried out qPCR using the same primers and the same cycling conditions as described above. qPCR analysis was performed on a Biorad CFX96TM Real-Time System/C1000 Thermal cycler equipped with CFX Manager™ Software. Abundance of 18S rRNA gene sequences was determined with the same primer pair (TAReuk454FWD1/TAReuk454REV3) used for the 18S rRNA gene diversity analysis. Each reaction contained 12.5 µL MasterMix phusion, 8.25 µL deionized nuclease-free water, 0.75 µL DMSO, 1 µL from each primer and 0.5 µL Sybr green, and 1 µL of DNA template. Reactions were performed in iCycler iQTM 96-well plates (Bio-Rad). A mixture of V4 18S rRNA gene amplicons obtained as described above was used to prepare standard solutions. All qPCR reactions were performed in triplicate with standard curves from 6.4×103 to 6.4×109 V4 molecules per microlitre. Specificity of the qPCR was verified with melting curve analyses (50 to 95 ∘C).
2.4 Bioinformatic analyses
Bioinformatic analyses were carried out using the python-based bioinformatic pipeline quantitative insight in microbial ecology (QIIME) (Caporaso et al., 2010). Overall, we obtained 372 107 raw sequences; reads with a length comprised between 250 and 500 bp, less than 8 homopolymers, and a phred quality ≥25 over 50 bp sliding windows were kept for downstream analyses. Chimeric sequences were then identified by comparison with the Protist Ribosomal Database 2 (PR2) (Guillou et al., 2013) using the Uchime algorithm (Edgar et al., 2011) and removed from the dataset along with singletons (i.e. reads not sharing 100 % identity with at least one other read).
A total of 238 564 reads remaining after quality filtering were clustered into 2457 operational taxonomic units (OTUs) based on 95 % sequence identity using Uclust (Edgar, 2010). Samples containing less than 1000 sequencing reads were removed from the dataset. The taxonomic affiliation of the OTUs was then inferred by comparison with the PR2 (Guillou et al., 2013) using BLAST (Altschul et al., 1990) within the QIIME pipeline. Reads from metazoa and multicellular fungi were removed from the dataset, which finally contained 1871 OTUs and 184 279 reads. A representative set of sequences from the OTUs used here has been submitted to the GenBank with the accession numbers MH913521–MH915389. The abundances of the different taxa in each sample were estimated by multiplying the percentage of reads by the concentration of V4 copies measured by qPCR. Taxa containing C28−32 diol producers were extracted from the dataset and plotted using Ocean Data View (ODV) (Schlitzer, 2002).
2.5 Statistical analyses
Linear regression analyses between the concentrations of the different LCDs were performed to assess whether some of the LCDs were likely to derive from a common source. To investigate relationships between LCDs and environmental conditions we calculated the Spearman rank correlation coefficient (r) using the vegan R package (Dixon, 2003). The environmental data used were temperature, salinity, TOC, nutrients (nitrate, nitrite, ammonium, phosphate, and silica), as well the concentration of Chl a and the abundance of photosynthetic picoeukaryotes and nanoeukaryotes. Samples containing missing data and outliers were removed from the dataset before the calculations. Both correlation coefficients and p values were calculated and the latter were corrected for false discovery rates (Benjamini and Hochberg, 1995). Correlations were considered significant for p values <0.01.
To investigate the relationships between lipids and microbial taxa, we also calculated Spearman's rank correlation coefficient between the LCD concentrations and the abundance of the different taxa at both OTU and class levels. To this end, taxonomic data were normalized based on the number of V4 copies in the different samples measured by qPCR.
Comparisons at class level provide the advantage of pooling distribution data from several closely related OTUs, thus reducing the number of zeros (samples where a given OTU is absent), which complicate statistical analyses of biological distributions (Legendre and Gallagher, 2001). However, pooling OTUs at higher taxonomic levels likely leads to combining of species able and unable to produce LCDs falling into the same taxonomic level. We thus removed OTUs that were observed in fewer than 19 samples (25 %) and compared the resulting OTU table with the LCD concentrations. These analyses were performed using the qiime script observation_metadata_correlation.py (Caporaso et al., 2010) and the p values were corrected for false discovery rates (Benjamini and Hochberg, 1995).
3.1 Ancillary data
The HCC cruise sailed across tropical Atlantic waters (Fig. 1a) in late summer and was targeted at SPM from the photic zone collected at the surface, the BWML, and the DCM. The extent of the photic zone as well as the depths of both the BWML and DCM at each station were assessed based on the vertical profiles of temperature, salinity, and chlorophyll fluorescence. The temperature of photic zone waters ranged from 15 to 29 ∘C (Fig. 1b, Table S2), the BWML depth comprised between 9 and 40 m, whereas the depth of the DCM ranged from 45 to 105 m. Temperatures varied at the DCM, increasing westwards, whereas they were relatively constant at the surface and BWML. Salinity varied between 29 and 36.5 g kg−1 (Fig. 1c, Table S2) at the surface, whereas it was fairly constant in the DCM (36 to 37). The concentration of Chl a varied from 34 to 470 ng L−1 (Fig. 1d, Table S2), with the lowest values measured at the surface of the easternmost (1 to 6) and westernmost (21 to 23) stations and the relatively higher concentrations in surface waters of the shallowest stations (11 to 13) located above the continental shelf and about 500 km off the Amazon River mouth (Fig. 1a, Table S2). The POC concentration ranged from 0.6 to 13 mg L−1 and also peaked at the surface for the shallowest stations (Fig. 1e, Table S2).
Photosynthetic picoeukaryotes, quantified by flow cytometry, were more abundant at the DCM compared to the surface and BWML (Fig. 1f). Their abundance peaked at the DCM of Stations 1 and 2 ( cell L−1), whereas for surface waters the highest values were measured at Stations 11 to 13. In contrast, photosynthetic nanoeukaryotes did not vary substantially through the water column and their abundance peaked at the surface of Station 17, reaching a density of 1.4×105 cell L−1 (Fig. 1g).
3.2 Long chain alkyl diols
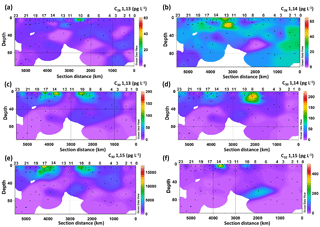
Six LCDs were detected, the C28 and C30 1,13-diols, C28 and C30 1,14-diols, and C30 and C32 1,15-diols (Fig. 2, Table S2). The C30 1,15-diol dominated all samples, accounting for >95 % of the total LCDs, and its concentration ranged from 100 to 1600 pg L−1. The concentration of the C28 1,13-diol ranged from 0 (i.e. undetectable) to 55 pg L−1, whereas the highest concentration measured for the C28 1,14-diol was 64 pg L−1. The other minor diols were usually more abundant than the C28 diols, reaching concentrations of up to 190 pg L−1 for the C30 1,13-diol, 240 pg L−1 for the C30 1,14-diol, and 480 pg L−1 for the C32 1,15-diol (Fig. 2). The concentration of the C28 1,13-diol peaked in the surface waters of Station 10, but it was below the detection limit in 19 samples from different depths and stations (Fig. 2a). The C28 1,14-diol reached its highest concentrations at the DCM of Station 12 (64 pg L−1) and at the surface of Station 13 (45 pg L−1) and tended to be more abundant in the waters of the eastern stations (Fig. 2b). The concentrations of both C28 1,13- and C28 1,14-diols did not vary significantly with depth (t-test, p value >0.1), while those of the C30 1,13-, C30 1,14-, and C30 1,15-diols were higher in the mixed layer (surface and BWML) compared to the DCM (p value <0.01).
The concentration of the C30 1,13-diol peaked at the surface of Stations 10 and 14 (Fig. 2c), while that of the C30 1,14-diol reached its maximum at the BWML of Stations 7 and 8 (Fig. 2d). The highest concentration of the C30 1,15-diol was measured at the surface of Station 17 (16 ng L−1, Fig. 2e). The concentration of the C32 1,15-diol peaked in the surface waters of Stations 10 and 14 and at the DCM of Station 7 (Fig. 2f) and its concentration did not vary significantly with depth. The concentrations of both the C30 and C32 diols peaked in the mixed layer of Stations 7–10 and 14–17, which are located in close proximity to the Amazon Shelf (Fig. 2c–f).
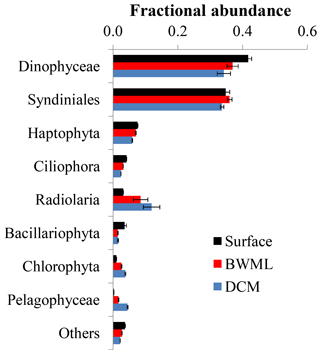
Figure 3Average fractional abundance of the reads obtained by 18S rRNA gene sequencing of SPM from the western tropical Atlantic Ocean over the various classes of eukaryotes. The V4 fragment of the 18S rRNA gene was sequenced using universal eukaryotic primers. Samples were pooled according to depth and the average contribution from each group at the different depth is shown. Error bars represent the standard deviation in the data from the various stations.
3.3 Eukaryotic 18S rRNA gene diversity analysis
Sequencing of the hypervariable V4 region of the 18S rRNA gene of 68 SPM samples resulted in 238 564 reads with an average of 4987 reads per sample (Table S2). Reads were clustered based on 95 % sequence identity and, after removal of reads of Metazoa and multicellular fungi, we obtained 1871 OTUs. Rarefaction analyses indicate that >90 % of the genetic diversity was captured (Fig. S1 in the Supplement), suggesting that no sample was undersequenced. Most (>90 %) reads sequenced here were assigned to Dinophyceae, Syndiniales, Haptophyta, and Radiolaria (Fig. 3). Samples were grouped according to the depth layer (surface, BWML, and DCM) and analysis of similarity (anosim) revealed that the average variance between samples from different groups was higher than the average variance between samples from the same group (p value ≈0.001), indicating that the eukaryotic community was mostly influenced by the water depth rather than the geographic location. The proportion of reads from Dinophyceae, Syndiniales, and Haptophyta was slightly higher in the mixed layer compared to the DCM, whereas Radiolaria and Pelagophyceae tended to be slightly more abundant in deeper waters (Fig. 3). All samples except surface waters from Station 12, the BWML from Station 11, and the DCM from Station 22 exhibited high contributions (>50 %) from Dinophyceae and Syndiniales (Fig. S2). Radiolaria dominated the DCM at Station 22, diatoms were relatively abundant (≈10–20 %) at the surface of Stations 12–14 and the BWML of Station 12, and the contribution of diatom reads was <5 % for all the other samples.
Table 1Distribution of the 18S rRNA gene reads associated with known LCD producers.
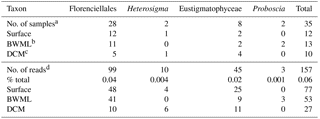
a Number of samples where 18S rRNA gene reads from
C28−32 diol producers were found. Overall, 68 samples were screened
for the
presence of 18S rRNA genes affiliated with LCD producers.
b Bottom wind mixed layer.
c Deep chlorophyll maximum.
d Number or proportion of 18S rRNA gene reads associated with
C28−32 diol producers.
18S rRNA gene reads of only four taxa containing known LCD producers were detected within our dataset: Proboscia spp., Florenciellales, Heterosigma spp., and Eustigmatophyceae (Table 1). In 33 out of 68 SPM samples we did not detect any 18S rRNA gene read from known LCD producers, whereas reads from these taxa accounted for <0.1 % of the total 18S rRNA reads in 24 samples, 0.1 % to 0.5 % in 8 samples, 0.5 % to 1 % in 2 samples, and 1.5 % in 1 sample (Station 20, BWML). The 18S rRNA gene reads from putative LCD producers were mostly recovered from the mixed layer (Table 1). Florenciellales was the most abundant taxon among the known LCD producers since it exhibited the highest number of reads (99) and was present in 28 out of 68 samples. The other taxa of putative LCD producers were detected only in 8 (Eustigmatophyceae) or 2 (Proboscia sp. and Heterosigma akashiwo) samples (Table 1) accounting for 3 (Proboscia) to 45 (Eustigmatophyceae) reads. Eustigmatophyceae (mostly affiliated with Nannochloropsis oculata) were found at the surface for Stations 11, 12, and 13, as well as at the DCM of Station 20 (Fig. 4a).
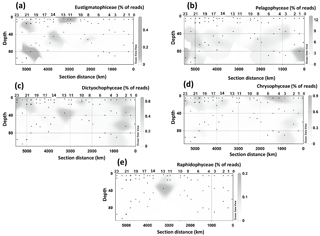
Figure 4Spatial distribution of the 18S rRNA gene fragments related to taxa containing LCD producers at different stations and depth. (a) Pelagophyceae, (b) Chrysophyceae, (c) Dictyochophyceae, (d) radial centric diatoms, (e) Eustigmatophyceae, and (f) Raphidophyceae. Data were plotted using ODV software using kriging for interpolation between data points (Schlitzer, 2002).
Since species genetically related to cultivated microalgae known to produce LCDs may also contain LCDs, we expanded our community composition analyses to groups at a higher taxonomic level and focused on those classes or divisions that contain LCD producers (Table S1). Specifically we investigated the distribution of Eustigmatophyceae, since they are the most well-known class of LCD producers, Pelagophyceae and Chrysophyceae, which include the LCD producers Sarcinochrysis marina and Chrysosphaera parvula, respectively (Table S1), Dictyochophyceae, which include Apedinella radians (Rampen et al., 2011), and Raphidophyceae, which include two LCD producers, H. akashiwo and Haramonas dimorpha. We did not detect any representative of Pinguiophyceae, a class which includes the LCD producer Phaeomonas parva (Table S1). Reads associated with Pelagophyceae, and mostly (97 %) affiliated with Pelagomonas calceolata, were recovered more frequently as they were present in 55 samples with an average abundance of 85 reads (2 % of total reads) per sample and a maximum value of 935 reads (12 % of total) in the DCM of Station 23 (Fig. 4b). Pelagophyceae reads were mostly detected in the DCM and were particularly abundant at the three westernmost stations investigated, where they comprised 8 % of total reads (Fig. 4b).
Chrysophyceae and Dictyochophyceae were also detected in most samples (54 and 57 samples, respectively) and their reads were recovered more frequently at the surface and BWML of the westernmost part of the transect (Stations 20–23) and at the surface of Stations 3–4 (Fig. 4c and d). Their 18S rRNA gene reads reached abundances of up to 55 and 41 reads (0.4 % and 0.6 % of the total, respectively), for Chrysophyceae and Dictyochophyceae, respectively, in the BWML of Station 20 (Table S4). Raphidophyceae were present only in three samples from Stations 11, 12, and 13 (Fig. 4e).
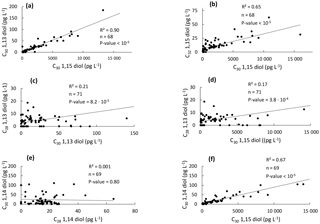
Figure 5Scatter plots of the concentrations of the different LCDs in the western tropical Atlantic Ocean. (a) C30 1,13-diol vs. C30 1,15-diol, (b) C32 1,15-diol vs. C30 1,15-diol, (c) C28 1,13-diol vs. C30 1,13-diol, (d) C28 1,13-diol vs. C30 1,15-diol, (e) C30 1,14-diol vs. C28 1,14- diol, and (f) C30 1,14-diol vs. C30 1,15-diol.
4.1 Comparison of diol distributions
In general, it is thought that 1,13- and 1,15-diols derive from a different source than 1,14-diols in the marine realm (Sinninghe Damsté et al., 2003; Rampen et al., 2007, 2011). Indeed, linear regressions showed that the concentration of the C30 1,15-diol is significantly correlated with those of the C30 1,13- and C32 1,15-diols (Fig. 5a–b). We did not observe any significant correlation between the concentrations of the C28 1,13- and C30 1,13- or C30 1,15-diols (Fig. 5c–d), which might be due to the fact that the C28 1,13-diol was below the detection limit in 19 out of 71 samples and its distribution could be compared to that of the widespread C30−32 diols only for the remaining 52 samples. This low abundance of the C28 1,13-diol is consistent with the relatively high temperatures observed for the tropical Atlantic Ocean (Fig. 1b), since the LCD core top calibration study has revealed that the fractional abundance of the C30 1,15-diol is high and that of the C28 1,13-diol is low when SST is relatively high (Rampen et al., 2012).
The concentration of the C28 1,14-diol was not correlated with that of the C30 1,14-diol (Fig. 5e), potentially suggesting a different origin for the C28 and C30 1,14-diols. However, the concentration of the C30 1,14-diol was significantly correlated with the C30 1,15-diol (Fig. 5f). This is quite surprising as the 1,14-diols in seawater have been suggested to derive from Proboscia spp. (Sinninghe Damsté et al., 2003; Rampen et al., 2009), and to a lesser extent from A. radians (Rampen et al., 2011), whereas the 1,13- and 1,15-diols are thought to be associated with Eustigmatophyceae (Rampen et al., 2014a, and references cited therein). Previous studies highlighted indeed good correlations in the fluxes of C28 and C30 1,14-diols in the water column of the Arabian Sea (Rampen et al., 2007) and the northwestern Indian Ocean (Rampen et al., 2008). Proboscia spp. also contain unsaturated 1,14-diols which were not found here; specifically the warm water species Proboscia indica is dominated by C28:1 and C30:1 1,14-diols (Rampen et al., 2007), suggesting that the 1,14-diols found here do not derive from Proboscia spp. This is confirmed by the absence or very low proportions of 18S rRNA gene reads from the major producers of C28−30 1,14-diols, i.e. Proboscia spp. and A. radians (Table 1). This suggests different sources for the C28 and C30 1,14-diols. Since the C30 1,15-diol accounted for >95 % of the C28−32 diols, it is possible that the C30 1,14-diol was biosynthesized in low amounts, along with C30 1,13-diol, by the producers of the C30 1,15-diol. This is supported by the fact that Eustigmatophyceae can contain small amounts of 1,14-diols along with large quantities of 1,15-diols (Rampen et al., 2014a); specifically, the C28 1,14-diol accounts for up to 15 % of the total LCDs in Pseudostarastrum enorme, and lower proportions (1 %–5 %) of C30 1,14-diols were previously found in Vischeria punctata and Eustigmatos vischeri (Rampen et al., 2014a).
It has been reported that the distributions of LCDs can be affected by riverine input, which is reflected by elevated amounts of the C32 1,15-diol (>10 %, de Bar et al., 2016; Lattaud et al., 2017b). However, the fractional abundance of the C32 1,15-diol in the SPM is low (0 % to 4 %, data not shown), far lower than the values typically measured in river-influenced ecosystems such as the Iberian Atlantic Margin (de Bar et al., 2016), the Kara Sea (Lattaud et al., 2017b), or the Congo River plume (Versteegh et al., 2000). We did not detect other eustigmatophycean biomarkers such as C32 alkenols or C30−32 hydroxy fatty acids (Volkman et al., 1992; Gelin et al., 1997b), suggesting that riverine or marine Eustigmatophyceae were unlikely to source the C28−32 diols found here. The HCC cruise took place in a period of the year (August/September) when the water discharge from the Amazon River is typically low (Molleri et al., 2010), thus leading to low inputs of riverine organic matter into the sea. The distribution of LCDs in the sampled SPM is thus likely not impacted by terrestrial input of LCDs.
Beyond Heterokontophyta, LCDs may also be produced by lower (Speelman et al., 2009) and higher (Wen and Jetter, 2007; Racovita and Jetter, 2016) plants. However, only four reads from our dataset were associated with a plant species, i.e. Panax ginseng (Table S4), which is not known to contain LCDs. The near absence of 18S rRNA gene reads from higher plants confirms the low riverine input of organic matter in the SPM of the tropical North Atlantic waters analysed here.
Table 2Spearman rank correlation coefficients between LCD and environmental variablesa.
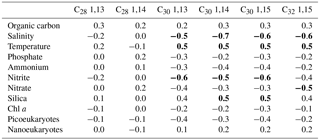
a Significant (p value <0.01) correlation values are in bold.
We explored the variations in the concentrations of the LCDs with respect to environmental data. The C28 1,13- and 1,14-diols, both occurring in low abundance, did not exhibit significant correlations with any of the environmental data measured here (Table 2). In contrast, the concentrations of C30 1,13-, 1,14-, and 1,15-diols exhibited significant but weak positive correlations with temperature and dissolved silica and weak negative correlations with salinity and nitrite. The concentration of the C32 1,15-diol revealed a correlation with the same environmental variables as the C30 diols, except for dissolved silica and nitrite, and exhibited a weak negative correlation with the concentration of nitrate. The correlations found here are likely simply due to different water masses: the mixed layer, where the highest proportions of LCDs were measured, exhibited indeed higher temperatures and lower salinities compared to the DCM. We repeated the analyses after excluding DCM samples and did not find strong positive or negative correlations between LCDs and environmental variables (data not shown). Thus, there does not seem to be a major control of environmental conditions on the concentrations of LCDs.
4.2 Comparison with eukaryotic abundance and diversity
Although C28−32 diols are likely produced by phytoplankton, the variability in LCD abundance is not correlated with that of Chl a concentration, or photosynthetic picoeukaryote and nanoeukaryote abundances (Table 2). This lack of correlation suggests that the LCD producers accounted for only a small proportion of phytoplankton. The high proportion of Dinophyceae, Syndiniales, and Radiolaria revealed by our genetic libraries agrees with previous studies on marine microbial communities based on 18S rRNA gene sequencing in different environments (Comeau et al., 2011; Christaki et al., 2014; de Vargas et al., 2015). However, these taxa do not necessarily dominate marine microbial communities, and so our results are likely due to a relatively high number of rRNA gene copies per cell (Zhu et al., 2005). Larger-sized dinoflagellates such as Prorocentrum minimum and Amphidinium carterae can contain up to 1000 gene copies per cell compared to <10 of rRNA gene copies for smaller-sized (<3 µm) species of Chlorophyta, Pelagophyceae, and Haptophyta (Zhu et al., 2005).
4.2.1 LCD producers
Although the primers used in this study have a perfect match with the 18S rRNA gene sequences of most eukaryotes (including all the classes containing LCD producers), and the rarefaction curves indicate that we sampled an appropriate (i.e. >90 %) proportion of the eukaryotic community, we cannot fully exclude that some species will remain undetected because of undersampling or primer mismatches. Moreover, the large number (100–1000) of rRNA gene copies per cell present within dinoflagellates and Radiolaria might have somehow affected the detection of LCD producers. In particular, Nannochloropsis salina has been shown to possess only one to two copies of the 18S rRNA gene (Zhu et al., 2005), and similarly, the other marine Nannochloropsis species, which do not differ greatly in size from N. salina (Fawley and Fawley, 2007), are also likely to have a low number of 18S rRNA gene copies. Known species of LCD producers were present in only 51 % of our SPM samples as revealed by sequencing data (Table 1), whereas the major LCD, the C30 1,15-diol, was present in all samples. This suggests either (1) that the LCDs found here were produced by other species which were not detected using the current methodology, (2) that the LCD producers were undersampled because of their low number of rRNA gene copies per cell, or (3) that the DNA of the LCD producers was no longer present in the SPM at the moment of sampling. Specifically, marine Eustigmatophyceae were represented by only two OTUs (denovo2075, Nannochloropsis oculata, and denovo229, uncultured Eustigmatophycea, Table S4) detected in only eight samples, confirming the hypothesis of Volkman et al. (1992) and Rampen et al. (2012) that they are not the major producers of LCDs in the marine environment. Even if we expand our analyses of LCD-related species to a higher taxonomic level, we do not find large proportions of 18S rRNA reads (generally <0.9 % of total reads) except for the class Pelagophyceae, which accounts for up to 12 % of total reads (Fig. 2a–e). However, Pelagophyceae are unlikely to be the source of any of the LCDs found here because their vertical distribution (i.e. mostly detected in the DCM, Figs. 3, 4b) does not correspond well to that of LCDs, which were either more abundant in the upper layers (C30 1,13-, 1,14-, and 1,15-diols and C32 1,15-diol) or did not vary greatly with depth (C28 diols, Fig. 2). Chrysophyceae and Dictyochophyceae were instead more abundant in the upper layers (Fig. 4b–c), and although none of the three known LCD producers from these classes produces the most abundant LCD detected in the SPM, i.e. C30 1,15-diol (Table S1), other species within the Chrysophyceae and Dictyochophyceae may possibly be a source for the C30 diols.
The C28 diols exhibited higher concentrations at the BWML of Station 12 and at the surface in Station 13 (Fig. 2a and b), and higher proportions of 18S rRNA gene reads were recovered from Pelagophyceae (2.4 %) and Eustigmatophyceae (0.5 %), at the surface of Stations 11–12 (Fig. 4d–f). The scattered occurrences of these groups and the mismatches in distributions when compared to the LCDs suggest that the LCDs in the tropical North Atlantic Ocean are unlikely to derive from Pelagophyceae, radial centric diatoms, Raphidophyceae, and/or Eustigmatophyceae.
Overall the abundance of known LCD producers is low and scattered and does not match the observed abundance patterns observed for the LCDs, suggesting that most of the LCDs measured here were not produced by any of these species.
4.2.2 Correlations between the abundance of OTUs and LCD concentration
Since LCDs have been shown to be present within two genetically distant eukaryotic supergroups, the Heterokontophyta and the Archaeplastida, the latter including plants as well as green and red algae, the genetic and enzymatic machinery required for the biosynthesis of LCDs might be present in other genera and classes, including uncultured species. We, therefore, also compared the concentration of LCDs with the composition of the entire eukaryotic microbial community, normalized with respect to the 18S rRNA gene abundance, at both class and OTU levels to identify co-occurrence patterns. No significant correlation was found at class level (data not shown), whereas the correlations at the OTU level were weak (r≤0.60) but significant (p value <0.01) for 27 OTUs affiliated with 11 different classes (Table 3). A reason behind the lack of correlation between taxonomic classes and LCDs can be that pooling OTUs at higher taxonomic levels likely leads to combining the LCD producers with species which are unable to produce LCDs but which fall into the same taxonomic level. The ability of microorganisms to biosynthesize LCDs can indeed vary, even between genetically related species; some genera include both LCD producers and species which do not contain LCDs (Table S1).
Table 3Correlation coefficient (r) for the OTUs, representing 95 % of the sequence identity, whose abundance was correlateda with the concentration of LCDs in SPM samples obtained in the HCC cruise.
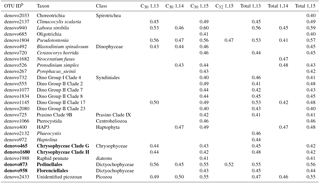
a Only significant (p value <0.01 after FDR correction)
correlations are shown.
b OTUs closely related to known LCD producers are in bold.
The C30 1,15-diol exhibited significant correlations (p<0.01) with 23 OTUs and, overall, 27 OTUs were significantly correlated with C30 or, to a lesser extent, C32 diols (Table 3). Of the 27 OTUs, 4 OTUs were affiliated with classes containing known LCD producers (Chrysophyceae and Dictyochophyceae, Table 3). The abundance of the two chrysophycean OTUs (denovo465 and denovo1680, Table 3) exhibited significant correlations with the concentrations of both C30 1,13- and 1,15-diols and accounted for 52 % of the total reads from this class and the only known LCD producer from this class (Chrysosphaera parvula) was found to contain C32 1,15-diol (Sebastiaan Rampen, unpublished results). The two OTUs affiliated with Dictyochophyceae (denovo873 and denovo958) and exhibiting positive correlation with C30−32 diols, cluster within Pedinellales and Florenciellales families, respectively, and are thus closely related to two known LCD producers, Florenciella parvula and Apedinella radians. However, F. parvula contains C24 1,13-, C24 1,14-, and C24 1,15-diols (Sebastiaan Rampen, unpublished results) and A. radians produces C28, C30, and C32 1,14-diols (Rampen et al., 2011), whereas the two dictyochophycean OTUs denovo873 and denovo958 exhibited a positive correlation with the C30 1,15-diol (Table 3).
The correlation values found here are nearly all low (r≈0.4–0.5), raising the question of whether these relationships reflect the ability of these species to produce LCDs or whether they are simply driven by other environmental conditions leading to similar spatial distributions of OTUs and LCDs. Other OTUs showing significant correlations with C30 1,15-diols represent species that are rare in the marine environment. For example, Centroheliozoa (OTU denovo1066) are mostly known as freshwater predators (Slapeta et al., 2005), and in seawater, they have only been sporadically detected in anoxic environments (Stock et al., 2009; Stoeck et al., 2009), suggesting that the centroheliozoan reads found here are unlikely to derive from active microorganisms. In contrast, the other OTUs include marine representatives commonly found in the photic zone of seawater and thus the reads found here might derive from living organisms: Syndiniales are intracellular parasites of other marine protists, and the genetic clades found here (Group I Clade 4, Group II Clades 2, 7, 8, 17, and 23) are commonly detected in the upper 100 m of the water column (Guillou et al., 2008). Spirotrichea include several heterotrophic and mixotrophic marine planktonic ciliates (Agatha et al., 2004; Santoferrara et al., 2017), whereas Phaeocystis is a widespread primary producer. The OTUs of uncultured classes exhibiting significant positive correlations with LCDs (Prasino Clade IX and the HAP-3 clade) are also commonly observed in the photic zone (Shi et al., 2009; Egge et al., 2015; Lopes dos Santos et al., 2016). However, cultivated representatives would be required in order to confirm whether species within these clades are capable of LCD synthesis.
4.3 Can 18S rRNA gene-based community composition analysis be used to determine LCD biological sources?
The lack of correlations of C28 diols with any OTUs as well as the low degree of correlation between OTUs and C30−32 diols and the trace abundance or near absence of known LCD producers suggest that the 18S rRNA genes from the microorganisms sourcing the LCDs were either absent or present below detection level in the seawater sampled. The fact that we sampled >90 % of the OTUs potentially present (Fig. S1) and the use of universal eukaryotic primers suggests that LCD producers have been unlikely to escape detection. However, the relatively low number of rRNA gene copies found for N. oculata (Zhu et al., 2005), and likewise also in other smaller-sized marine Eustigmatophyceae, suggests that LCD producers might have been undersampled with respect to larger-sized species which can contain up to 1000 rRNA copies per cell (Zhu et al., 2005).
It should be considered that both the LCDs and DNA in the SPM might derive not only from active or senescent cells, but also from detritus (Not et al., 2009). In addition, LCDs can persist in seawater for likely much longer periods than the DNA of the related LCD producers. Although the biological function of LCDs is unclear for most species, they have been shown to be the building blocks of cell wall polymers in Eustigmatophyceae, and likewise they might occur in other biopolymers of marine or terrestrial origin. In Nannochloropsis cell walls, LCDs and long chain alkenols are likely to be bound together through ester and ether bonds to form highly refractory polymers known as algaenans (Gelin et al., 1997a; Scholz et al., 2014). These biopolymers are thought to be quite persistent and accumulate in ancient sediments for millions of years (Tegelaar et al., 1989; Derenne and Largeau, 2001; de Leeuw et al., 2006). Indeed, LCDs are ubiquitous in recent surface sediments (Rampen et al., 2012) and ancient sediments of up to 65 million years old (Yamamoto et al., 1996) showing their recalcitrant nature.
Recent laboratory experiments highlighted that LCDs from dead biomass of Nannochloropsis oculata can persist in seawater for longer than 250 days under both anoxic (Grossi et al., 2001) and oxic conditions (Reiche et al., 2018). In contrast, much shorter turnover times (6 h to 2 months) are typically reported for extracellular DNA in the oxic water column (Nielsen et al., 2007). This suggests that the DNA from LCD producers likely reflects the living eukaryotic community (recently) present when seawater was sampled, while the LCDs probably represent an accumulation that occurred over longer periods of time (weeks to months or even years).
Because of this large difference in turnover rates between LCDs and the DNA from the LCD producers, 18S rRNA gene analysis of environmental samples may be unsuccessful for identifying LCD producers. This is seemingly in contrast to a previous study that showed that the LCD concentration in the upper 25 m of the freshwater Lake Challa (Tanzania) was related to the number of eustigmatophycean 18S rRNA gene copies (Villanueva et al., 2014). However, Villanueva et al. (2014) used Eustigmatophyceae-biased primers and, since this was a lake system, Eustigmatophyceae are likely to be the major source of LCDs in freshwater ecosystems. Importantly, they found a mismatch for the uppermost part of the water column (0–5 m), where high LCD abundance (38–46 ng L−1) coincided with few or no Eustigmatophyceae 18S rRNA gene copies. This pattern was explained by them to be caused by wind-driven and convective mixing of preserved LCDs, while phytoplankton adjusted its buoyancy at greater depth (Villanueva et al., 2014). The high salinity values (≥33 g kg−1) detected in most surface samples, the low proportions of both C32 1,15-diols (2.2 % over the total LCDs), and 18S rRNA gene reads associated with plants (4 out of 238 564), as well as the low input of freshwater from the Amazon River to the stations analysed here during the sampling period (Molleri et al., 2010) suggest that the LCDs found here are unlikely to have a freshwater origin.
Laboratory experiments carried out under different conditions of temperature, light irradiance, salinity, and nitrate concentrations revealed an average cellular LCD content of about 23 fg cell−1 (Balzano et al., 2017) for Nannochloropsis oceanica. The average LCD concentration in the SPM investigated was ca. 2.6 ng L−1, which would correspond to ca. 1.1×106 pico/nano algal cells L−1. We detected average phytoplankton abundances of 3.3×106 cell L−1 for picoeukaryotes and 3.6×104 cell L−1 for nanoeukaryotes. Although nanoplanktonic Eustigmatophyceae might produce larger amounts of LCDs than those measured in our previous study (Balzano et al., 2017), because of their larger cell size, the nanoplankton abundances measured here are 2 orders of magnitude lower than the densities required to source the LCDs (1.1×106 cell L−1). Therefore, if the LCDs measured here were biosynthesized by intact microorganisms in the water column, nanoplankton alone would not be able to source all the LCDs measured, and therefore in addition at least one-third of the picophytoplankton should be able to produce LCDs, which is unrealistic. This supports the idea that most of the LCDs detected here are of a fossil nature and not contained in living cells. The higher concentrations of LCDs found in the SPM from the mixed layer compared to the DCM suggest that LCDs were originally produced at a higher frequency in the mixed layer. Moreover, their possible fossil nature indicates that LCDs were likely to persist in the mixed layer for long periods, eventually associated with suspended particulate matter.
The combination of lipid and DNA analyses is often complicated by different turnover rates, especially for refractory compounds such as LCDs. Studies focused on more labile biomarker lipids such as fatty acids or intact polar lipids can be more successful, e.g. with short branched fatty acids (Balzano et al., 2011), cyanobacterial glycolipids (Bale et al., 2018), or archaeal phospholipids (Pitcher et al., 2011; Buckles et al., 2013). Therefore, care has to be taken in inferring sources of biomarker lipids by the quantitative comparison of DNA abundance with biomarker lipid concentrations. Analysis of intact polar lipids, rather than total lipids, might have facilitated the identification of diol producers.
The combination of lipid analyses and 18S rRNA gene amplicon sequencing revealed some weak correlations between the abundances of 27 OTUs and the concentration of C30 diols. Four of these OTUs are affiliated with classes that include few LCD-producing species (i.e. Chrysophyceae and Dictyochophyceae), whereas the remaining 23 OTUs belong to taxa in which the presence of LCDs has never been assessed. In both cases it remains unclear whether the correlation between these 27 OTUs and the C30 diols reflects novel LCD producers or is driven by other environmental conditions.
The abundances of photosynthetic picoeukaryotes and nanoeukaryotes measured here suggest that these microbial populations are highly unlikely to source all the LCDs found. Some of the LCDs found here might be associated with suspended debris rather than intact cells, with the DNA from their producers being already degraded at the time of sampling. DNA degradation rates in the oxygenated water column are indeed faster than those of most lipids, including LCDs. The freshness of the organic matter and the turnover rates of both lipids and DNA in a given environment should thus be considered when identifying the biological sources of a specific class of lipids through DNA sequencing. In addition, the extraction methods applied in our study did not discriminate between free and bound lipids and we thus do not know whether the compounds found here were originally present in seawater as free or ester-bound diols. Finally, the 18S rRNA gene amplicon sequencing can be suitable for tracking LCD sources (1) for simple ecosystems or laboratory/in situ mesocosms with high proportions of fresh organic matter and (2) for low oxygen/anoxic environments where extracellular DNA can persist for longer periods.
DNA sequences are on the Genbank under accession numbers MH913521–MH915389. All the other data are in the Supplement.
The supplement related to this article is available online at: https://doi.org/10.5194/bg-15-5951-2018-supplement.
SB extracted and amplified the DNA, carried out the bioinformatic analyses and wrote most of the paper. JL extracted and analysed the lipids and helped in data interpretation along with SWR. LV and JvB contributed in designing the best strategy for analysing DNA. CPDB analysed picoeukaryotes and nanoeukaryotes by flow cytometry, NB collected the samples used in this study, and gave useful suggestions for the analyses and the interpretation of lipid data, and JSSD and SS made major contributions in designing the experiments as well as in the writing of the manuscript. All the co-authors made their contribution to the writing and successful revisions of the manuscript.
The authors declare that they have no conflict of interest.
We thank the captains and the crew of the R/V Pelagia for their
support during the cruise. We thank Denise Dorhout for sample collection,
Sharyn Ossebaar for nutrient analysis, Harry Witte, Elda Panoto,
and Sanne Vreugdenhil for support
in molecular biology, Hans Malschaert for the bioinformatics, and Marc Besseling for helpful discussions. John Volkman and two
anonymous referees provided helpful comments on an earlier version of this
paper. This research was funded by the European Research Council (ERC) under
the European Union's Seventh Framework Program (FP7/2007-2013) ERC grant
agreement (339206). Stefan Schouten and Jaap S. Sinninghe Damsté receive
financial support from the Netherlands Earth System Science Centre (NESSC)
though a gravitation grant (NWO 024.002.001) from the Dutch Ministry for
Education, Culture and Science.
Edited by:
Silvio Pantoja
Reviewed by: John K. Volkman and two anonymous
referees
Agatha, S., Struder-Kypke, M. C., and Beran, A.: Morphologic and genetic variability in the marine planktonic ciliate Laboea strobila Lohmann, 1908 (Ciliophora, Oligotrichia), with notes on its ontogenesis, J. Eukaryot. Microbiol., 51, 267–281, https://doi.org/10.1111/j.1550-7408.2004.tb00567.x, 2004.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J.: Basic local alignment search tool, J. Mol. Bio., 215, 403–410, https://doi.org/10.1016/S0022-2836(05)80360-2, 1990.
Andersen, R. A., Brett, R. W., Potter, D., and Sexton, J. P.: Phylogeny of the Eustigmatophyceae based upon 18S rDNA, with emphasis on Nannochloropsis, Protist, 149, 61–74, https://doi.org/10.1016/S1434-4610(98)70010-0, 1998.
Bale, N. J., Villareal, T. A., Hopmans, E. C., Brussaard, C. P. D., Besseling, M., Dorhout, D., Sinninghe Damsté, J. S., and Schouten, S.: C5 glycolipids of heterocystous cyanobacteria track symbiont abundance in the diatom Hemiaulus hauckii across the tropical North Atlantic, Biogeosciences, 15, 1229–1241, https://doi.org/10.5194/bg-15-1229-2018, 2018.
Balzano, S., Pancost, R. D., Lloyd, J. R., and Statham, P. J.: Changes in fatty acid composition in degrading algal aggregates, Mar. Chem., 124, 2–13, https://doi.org/10.1016/j.marchem.2010.11.001, 2011.
Balzano, S., Abs, E., and Leterme, S. C.: Protist diversity along a salinity gradient in a coastal lagoon, Aquat. Microb. Ecol., 74, 263–277, https://doi.org/10.3354/ame01740, 2015.
Balzano, S., Villanueva, L., de Bar, M., Sinninghe Damsté, J. S., and Schouten, S.: Impact of culturing conditions on the abundance and composition of long chain alkyl diols in species of the genus Nannochloropsis, Org. Geochem., 108, 9–17, https://doi.org/10.1016/j.orggeochem.2017.02.006, 2017.
Benjamini, Y. and Hochberg, Y.: Controlling the false discovery rate. A pratical and powerful approach to multiple testing., J. Roy. Stat. Soc. B Met., 57, 289–300, 1995.
Bergesch, M., Odebrecht, C., and Moestrup, O.: Nanoflagellates from coastal waters of southern Brazil (32 degrees S), Bot. Mar., 51, 35–50, https://doi.org/10.1515/bot.2008.003, 2008.
Buckles, L. K., Villanueva, L., Weijers, J. W. H., Verschuren, D., and Sinninghe Damste, J. S.: Linking isoprenoidal GDGT membrane lipid distributions with gene abundances of ammonia-oxidizing Thaumarchaeota and uncultured crenarchaeotal groups in the water column of a tropical lake (Lake Challa, East Africa), Environ. Microbiol., 15, 2445–2462, https://doi.org/10.1111/1462-2920.12118, 2013.
Buschhaus, C., Peng, C., and Jetter, R.: Very-long-chain 1,2-and 1,3-bifunctional compounds from the cuticular wax of Cosmos bipinnatus petals, Phytochemistry, 91, 249–256, https://doi.org/10.1016/j.phytochem.2012.07.018, 2013.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., Fierer, N., Pena, A. G., Goodrich, J. K., Gordon, J. I., Huttley, G. A., Kelley, S. T., Knights, D., Koenig, J. E., Ley, R. E., Lozupone, C. A., McDonald, D., Muegge, B. D., Pirrung, M., Reeder, J., Sevinsky, J. R., Tumbaugh, P. J., Walters, W. A., Widmann, J., Yatsunenko, T., Zaneveld, J., and Knight, R.: QIIME allows analysis of high-throughput community sequencing data, Nat. Methods, 7, 335–336, https://doi.org/10.1038/nmeth.f.303, 2010.
Christaki, U., Kormas, K. A., Genitsaris, S., Georges, C., Sime-Ngando, T., Viscogliosi, E., and Monchy, S.: Winter-summer succession of unicellular eukaryotes in a meso-eutrophic coastal system, Microb. Ecol., 67, 13–23, https://doi.org/10.1007/s00248-013-0290-4, 2014.
Comeau, A. M., Li, W. K. W., Tremblay, J.-E., Carmack, E. C., and Lovejoy, C.: Arctic Ocean microbial community structure before and after the 2007 record sea ice minimum, Plos One, 6, e27492, https://doi.org/10.1371/journal.pone.0027492, 2011.
de Bar, M. W., Dorhout, D. J. C., Hopmans, E. C., Rampen, S. W., Sinninghe Damsté, J. S., and Schouten, S.: Constraints on the application of long chain diol proxies in the Iberian Atlantic margin, Org. Geochem., 101, 184–195, https://doi.org/10.1016/j.orggeochem.2016.09.005, 2016.
de Leeuw, J. W., Irene, W., Rijpstra, C., and Schenck, P. A.: The occurrence and identification of C30, C31 and C32 alkan-1, 15-diols and alkan-15-one-1-ols in Unit I and Unit II Black Sea sediments, Geochim. Cosmochim. Ac., 45, 2281–2285, https://doi.org/10.1016/0016-7037(81)90077-6, 1981.
de Leeuw, J. W., Versteegh, G. J. M., and van Bergen, P. F.: Biomacromolecules of algae and plants and their fossil analogues, Plant Ecol., 182, 209–233, https://doi.org/10.1007/s11258-005-9027-x, 2006.
Derenne, S. and Largeau, C.: A review of some important families of refractory macromolecules: Composition, origin, and fate in soils and sediments, Soil Sci., 166, 833–847, https://doi.org/10.1097/00010694-200111000-00008, 2001.
de Vargas, C., Audic, S., Henry, N., Decelle, J., Mahé, F., Logares, R., Lara, E., Berney, C., Le Bescot, N., Probert, I., Carmichael, M., Poulain, J., Romac, S., Colin, S., Aury, J.-M., Bittner, L., Chaffron, S., Dunthorn, M., Engelen, S., Flegontova, O., Guidi, L., Horák, A., Jaillon, O., Lima-Mendez, G., Lukeš, J., Malviya, S., Morard, R., Mulot, M., Scalco, E., Siano, R., Vincent, F., Zingone, A., Dimier, C., Picheral, M., Searson, S., Kandels-Lewis, S., Acinas, S. G., Bork, P., Bowler, C., Gorsky, G., Grimsley, N., Hingamp, P., Iudicone, D., Not, F., Ogata, H., Pesant, S., Raes, J., Sieracki, M. E., Speich, S., Stemmann, L., Sunagawa, S., Weissenbach, J., Wincker, P., and Karsenti, E.: Eukaryotic plankton diversity in the sunlit ocean, Science, 348, 1261605, https://doi.org/10.1126/science.1261605, 2015.
Dixon, P.: VEGAN, a package of R functions for community ecology, J. Veg. Sci., 14, 927–930, https://doi.org/10.1111/j.1654-1103.2003.tb02228.x, 2003.
Edgar, R. C.: Search and clustering orders of magnitude faster than BLAST, Bioinformatics, 26, 2460–2461, https://doi.org/10.1093/bioinformatics/btq461, 2010.
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R.: UCHIME improves sensitivity and speed of chimera detection, Bioinformatics, 27, 2194–2200, https://doi.org/10.1093/bioinformatics/btr381, 2011.
Egge, E. S., Johannessen, T. V., Andersen, T., Eikrem, W., Bittner, L., Larsen, A., Sandaa, R.-A., and Edvardsen, B.: Seasonal diversity and dynamics of haptophytes in the Skagerrak, Norway, explored by high-throughput sequencing, Mol. Ecol., 24, 3026–3042, https://doi.org/10.1111/mec.13160, 2015.
Fawley, K. P. and Fawley, M. W.: Observations on the diversity and ecology of freshwater Nannochloropsis (Eustigmatophyceae), with descriptions of new taxa, Protist, 158, 325–336, https://doi.org/10.1016/j.protis.2007.03.003, 2007.
Gelin, F., Boogers, I., Noordeloos, A. A. M., Sinninghe Damsté, J. S., Riegman, R., and De Leeuw, J. W.: Resistant biomacromolecules in marine microalgae of the classes Eustigmatophyceae and Chlorophyceae: geochemical implications, Org. Geochem., 26, 659–675, https://doi.org/10.1016/s0146-6380(97)00035-1, 1997a.
Gelin, F., Volkman, J. K., deLeeuw, J. W., and Sinninghe Damsté, J. S.: Mid-chain hydroxy long-chain fatty acids in microalgae from the genus Nannochloropsis, Phytochemistry, 45, 641–646, https://doi.org/10.1016/s0031-9422(97)00068-x, 1997b.
Grossi, V., Blokker, P., and Sinninghe Damste, J. S.: Anaerobic biodegradation of lipids of the marine microalga Nannochloropsis salina, Org. Geochem., 32, 795–808, https://doi.org/10.1016/s0146-6380(01)00040-7, 2001.
Guillou, L., Viprey, M., Chambouvet, A., Welsh, R. M., Kirkham, A. R., Massana, R., Scanlan, D. J., and Worden, A. Z.: Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata), Environ. Microbiol., 10, 3349–3365, https://doi.org/10.1111/j.1462-2920.2008.01731.x, 2008.
Guillou, L., Bachar, D., Audic, S., Bass, D., Berney, C., Bittner, L., Boutte, C., Burgaud, G., de Vargas, C., Decelle, J., del Campo, J., Dolan, J. R., Dunthorn, M., Edvardsen, B., Holzmann, M., Kooistra, W. H. C. F., Lara, E., Le Bescot, N., Logares, R., Mahé, F., Massana, R., Montresor, M., Morard, R., Not, F., Pawlowski, J., Probert, I., Sauvadet, A.-L., Siano, R., Stoeck, T., Vaulot, D., Zimmermann, P., and Christen, R.: The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy, Nucleic Acid Res., 41, 597–604, https://doi.org/10.1093/nar/gks1160, 2013.
Jetter, R. and Riederer, M.: Long-chain alkanediols, ketoaldehydes, ketoalcohols and ketoalkyl esters in the cuticular waxes of Osmunda regalis fronds, Phytochemistry, 52, 907–915, https://doi.org/10.1016/s0031-9422(99)00309-x, 1999.
Jiang, S. C., Oleary, T., Volkman, J. K., Zhang, H. Z., Jia, R. F., Yu, S. H., Wang, Y., Luan, Z. F., Sun, Z. Q., and Jiang, R. H.: Origins and simulated thermal alteration of sterols and keto-alcohols in deep-sea marine sediment of the Okinawa Trough., Org. Geochem., 21, 415–422, https://doi.org/10.1016/0146-6380(94)90203-8, 1994.
Koning, E., van Iperen, J. M., van Raaphorst, W., Helder, W., Brummer, G. J. A., and van Weering, T. C. E.: Selective preservation of upwelling-indicating diatoms in sediments off Somalia, NW Indian Ocean, Deep-Sea Res. Pt. I, 48, 2473–2495, https://doi.org/10.1016/s0967-0637(01)00019-x, 2001.
Lassiter, A. M., Wilkerson, F. P., Dugdale, R. C., and Hogue, V. E.: Phytoplankton assemblages in the CoOP-WEST coastal upwelling area, Deep-Sea Res. Pt. II, 53, 3063–3077, https://doi.org/10.1016/j.dsr2.2006.07.013, 2006.
Lattaud, J., Dorhout, D., Schulz, H., Casta neda, I. S., Schefuß, E., Sinninghe Damsté, J. S., and Schouten, S.: The C32 alkane-1,15-diol as a proxy of late Quaternary riverine input in coastal margins, Clim. Past, 13, 1049–1061, https://doi.org/10.5194/cp-13-1049-2017, 2017a.
Lattaud, J., Kim, J. H., De Jonge, C., Zell, C., Sinninghe Damste, J. S., and Schouten, S.: The C32 alkane-1,15-diol as a tracer for riverine input in coastal seas, Geochim. Cosmochim. Ac., 202, 146–158, https://doi.org/10.1016/j.gca.2016.12.030, 2017b.
Legendre, P. and Gallagher, E. D.: Ecologically meaningful transformations for ordination of species data, Oecologia, 129, 271–280, https://doi.org/10.1007/s004420100716, 2001.
Logares, R., Audic, S., Santini, S., Pernice, M. C., de Vargas, C., and Massana, R.: Diversity patterns and activity of uncultured marine heterotrophic flagellates unveiled with pyrosequencing, ISME J., 6, 1823–1833, https://doi.org/10.1038/ismej.2012.36, 2012.
Lopes dos Santos, A., Gourvil, P., Tragin, M., Noel, M.-H., Decelle, J., Romac, S., and Vaulot, D.: Diversity and oceanic distribution of prasinophytes clade VII, the dominant group of green algae in oceanic waters, ISME J., 11, 512–528, https://doi.org/10.1038/ismej.2016.120, 2016.
Mao, S. Y., Zhu, X. W., Wu, N. Y., Jia, G. D., Sun, Y. G., Guan, H. X., and Wu, D. D.: Alcohol compounds in Azolla imbricata and potential source implication for marine sediments, Sci. China Earth Sci., 60, 348–359, https://doi.org/10.1007/s11430-016-5177-6, 2017.
Marie, D., Simon, N., and Vaulot, D.: Phytoplankton cell counting by flow cytometry., in: Algal Culturing Techniques, edited by: RA, A., Elsevier Academic Press, Burlington, MA, 253–268, 2005.
Massana, R., Gobet, A., Audic, S., Bass, D., Bittner, L., Boutte, C., Chambouvet, A., Christen, R., Claverie, J. M., Decelle, J., Dolan, J. R., Dunthorn, M., Edvardsen, B., Forn, I., Forster, D., Guillou, L., Jaillon, O., Kooistra, W., Logares, R., Mahe, F., Not, F., Ogata, H., Pawlowski, J., Pernice, M. C., Probert, I., Romac, S., Richards, T., Santini, S., Shalchian-Tabrizi, K., Siano, R., Simon, N., Stoeck, T., Vaulot, D., Zingone, A., and de Vargas, C.: Marine protist diversity in European coastal waters and sediments as revealed by high-throughput sequencing, Environ. Microbiol., 17, 4035–4049, https://doi.org/10.1111/1462-2920.12955, 2015.
Moita, M. T., Oliveira, P. B., Mendes, J. C., and Palma, A. S.: Distribution of chlorophyll a and Gymnodinium catenatum associated with coastal upwelling plumes off central Portugal, Acta Oecol., 24, S125–S132, https://doi.org/10.1016/s1146-609x(03)00011-0, 2003.
Molleri, G. S. F., Novo, E., and Kampel, M.: Space-time variability of the Amazon River plume based on satellite ocean color, Cont. Shelf Res., 30, 342–352, https://doi.org/10.1016/j.csr.2009.11.015, 2010.
Nielsen, K. M., Johnsen, P. J., Bensasson, D., and Daffonchio, D.: Release and persistence of extracellular DNA in the environment, Environmental Biosafety Research, 6, 37–53, https://doi.org/10.1051/ebr:2007031, 2007.
Not, F., del Campo, J., Balague, V., de Vargas, C., and Massana, R.: New Insights into the Diversity of Marine Picoeukaryotes, Plos One, 4, e7143, https://doi.org/10.1371/journal.pone.0007143, 2009.
Pitcher, A., Villanueva, L., Hopmans, E. C., Schouten, S., Reichart, G. J., and Sinninghe Damste, J. S.: Niche segregation of ammonia-oxidizing archaea and anammox bacteria in the Arabian Sea oxygen minimum zone, ISME J., 5, 1896–1904, https://doi.org/10.1038/ismej.2011.60, 2011.
Plancq, J., Mattioli, E., Pittet, B., Simon, L., and Grossi, V.: Productivity and sea-surface temperature changes recorded during the late Eocene-early Oligocene at DSDP Site 511 (South Atlantic), Palaeogeogr. Palaeocl., 407, 34–44, https://doi.org/10.1016/j.palaeo.2014.04.016, 2014.
Racovita, R. C. and Jetter, R.: Identification of In-Chain-Functionalized Compounds and Methyl-Branched Alkanes in Cuticular Waxes of Triticum aestivum cv. Bethlehem, Plos One, 11, 25, https://doi.org/10.1371/journal.pone.0165827, 2016.
Rampen, S. W., Schouten, S., Wakeham, S. G., and Sinninghe Damste, J. S.: Seasonal and spatial variation in the sources and fluxes of long chain diols and mid-chain hydroxy methyl alkanoates in the Arabian Sea, Org. Geochem., 38, 165–179, https://doi.org/10.1016/j.orggepchem.2006.10.008, 2007.
Rampen, S. W., Schouten, S., Koning, E., Brummer, G.-J. A., and Sinninghe Damsté, J. S.: A 90 kyr upwelling record from the northwestern Indian Ocean using a novel long-chain diol index, Earth Planet. Sc. Lett., 276, 207–213, https://doi.org/10.1016/j.epsl.2008.09.022, 2008.
Rampen, S. W., Schouten, S., Schefuss, E., and Sinninghe Damsté, J. S.: Impact of temperature on long chain diol and mid-chain hydroxy methyl alkanoate composition in Proboscia diatoms: results from culture and field studies, Org. Geochem., 40, 1124–1131, https://doi.org/10.1016/j.orggeochem.2009.08.005, 2009.
Rampen, S. W., Schouten, S., and Sinninghe Damsté, J. S.: Occurrence of long chain 1,14-diols in Apedinella radians, Org. Geochem., 42, 572–574, https://doi.org/10.1016/j.orggeochem.2011.03.009, 2011.
Rampen, S. W., Willmott, V., Kim, J.-H., Uliana, E., Mollenhauer, G., Schefuss, E., Sinninghe Damsté, J. S., and Schouten, S.: Long chain 1,13-and 1,15-diols as a potential proxy for palaeotemperature reconstruction, Geochim. Cosmochim. Ac., 84, 204–216, https://doi.org/10.1016/j.gca.2012.01.024, 2012.
Rampen, S. W., Datema, M., Rodrigo-Gamiz, M., Schouten, S., Reichart, G.-J., and Sinninghe Damsté, J. S.: Sources and proxy potential of long chain alkyl diols in lacustrine environments, Geochim. Cosmochim. Ac., 144, 59–71, https://doi.org/10.1016/j.gca.2014.08.033, 2014a.
Rampen, S. W., Willmott, V., Kim, J.-H., Rodrigo-Gamiz, M., Uliana, E., Mollenhauer, G., Schefuss, E., Sinninghe Damsté, J. S., and Schouten, S.: Evaluation of long chain 1,14-alkyl diols in marine sediments as indicators for upwelling and temperature, Org. Geochem., 76, 39–47, https://doi.org/10.1016/j.orggeochem.2014.07.012, 2014b.
Reiche, S., Rampen, S., Dorhout, D., Sinninghe Damste, J. S., and Schouten, S.: The impact of oxygen exposure on long-chain alkyl diols and the long chain diol index (LDI) – a long-term incubation study, Biogeosciences Discuss., in review, 2018.
Rodrigo-Gámiz, M., Rampen, S. W., de Haas, H., Baas, M., Schouten, S., and Sinninghe Damsté, J. S.: Constraints on the applicability of the organic temperature proxies , TEX86 and LDI in the subpolar region around Iceland, Biogeosciences, 12, 6573–6590, https://doi.org/10.5194/bg-12-6573-2015, 2015.
Santoferrara, L. F., Alder, V. V., and McManus, G. B.: Phylogeny, classification and diversity of Choreotrichia and Oligotrichia (Ciliophora, Spirotrichea), Mol. Phylogenet. Evol., 112, 12–22, https://doi.org/10.1016/j.ympev.2017.03.010, 2017.
Schlitzer, R.: Interactive analysis and visualization of geoscience data with Ocean Data View, Comput. Geosci., 28, 1211–1218, https://doi.org/10.1016/s0098-3004(02)00040-7, 2002.
Scholz, M. J., Weiss, T. L., Jinkerson, R. E., Jing, J., Roth, R., Goodenough, U., Posewitz, M. C., and Gerken, H. G.: Ultrastructure and composition of the Nannochloropsis gaditana cell wall, Eukaryot. Cell, 13, 1450–1464, https://doi.org/10.1128/ec.00183-14, 2014.
Seoane, S., Laza, A., Urrutxurtu, M., and Orive, E.: Phytoplankton assemblages and their dominant pigments in the Nervion River estuary, Hydrobiologia, 549, 1–13, https://doi.org/10.1007/s10750-005-1736-6, 2005.
Shi, X. L., Marie, D., Jardillier, L., Scanlan, D. J., and Vaulot, D.: Groups without Cultured Representatives Dominate Eukaryotic Picophytoplankton in the Oligotrophic South East Pacific Ocean, Plos One, 4, e7657, https://doi.org/10.1371/journal.pone.0007657, 2009.
Sinninghe Damsté, J. S., Rampen, S., Irene, W., Rupstra, C., Abbas, B., Muyzer, G., and Schouten, S.: A diatomaceous origin for long-chain diols and mid-chain hydroxy methyl alkanoates widely occurring in Quaternary marine sediments: Indicators for high-nutrient conditions, Geochim. Cosmochim. Ac., 67, 1339–1348, https://doi.org/10.1016/s0016-7037(02)01225-5, 2003.
Slapeta, J., Moreira, D., and Lopez-Garcia, P.: The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes, P. Roy. Soc. B-Biol. Sci., 272, 2073–2081, https://doi.org/10.1098/rspb.2005.3195, 2005.
Speelman, E. N., Reichart, G. J., de Leeuw, J. W., Rijpstra, W. I. C., and Sinninghe Damste, J. S.: Biomarker lipids of the freshwater fern Azolla and its fossil counterpart from the Eocene Arctic Ocean, Org. Geochem., 40, 628–637, https://doi.org/10.1016/j.orggeochem.2009.02.001, 2009.
Stock, A., Juergens, K., Bunge, J., and Stoeck, T.: Protistan diversity in suboxic and anoxic waters of the Gotland Deep (Baltic Sea) as revealed by 18S rRNA clone libraries, Aquat. Microb. Ecol., 55, 267–284, https://doi.org/10.3354/ame01301, 2009.
Stoeck, T., Behnke, A., Christen, R., Amaral-Zettler, L., Rodriguez-Mora, M. J., Chistoserdov, A., Orsi, W., and Edgcomb, V. P.: Massively parallel tag sequencing reveals the complexity of anaerobic marine protistan communities, BMC Biol., 7, 72, https://doi.org/10.1186/1741-7007-7-72, 2009.
Stoeck, T., Bass, D., Nebel, M., Christen, R., Jones, M. D. M., Breiner, H. W., and Richards, T. A.: Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water, Mol. Ecol., 19, 21–31, https://doi.org/10.1111/j.1365-294X.2009.04480.x, 2010.
Tegelaar, E. W., Deleeuw, J. W., Derenne, S., and Largeau, C.: A reappraisal of kerogen formation, Geochim. Cosmochim. Ac., 53, 3103–3106, https://doi.org/10.1016/0016-7037(89)90191-9, 1989.
Versteegh, G. J. M., Bosch, H. J., and De Leeuw, J. W.: Potential palaeoenvironmental information of C-24 to C-36 mid-chain diols, keto-ols and mid-chain hydroxy fatty acids; a critical review, Org. Geochem., 27, 1–13, https://doi.org/10.1016/s0146-6380(97)00063-6, 1997.
Versteegh, G. J. M., Jansen, J. H. F., De Leeuw, J. W., and Schneider, R. R.: Mid-chain diols and keto-ols in SE Atlantic sediments: A new tool for tracing past sea surface water masses?, Geochim. Cosmochim. Ac., 64, 1879–1892, https://doi.org/10.1016/s0016-7037(99)00398-1, 2000.
Villanueva, L., Besseling, M., Rodrigo-Gamiz, M., Rampen, S. W., Verschuren, D., and Sinninghe Damsté, J. S.: Potential biological sources of long chain alkyl diols in a lacustrine system, Org. Geochem., 68, 27–30, https://doi.org/10.1016/j.orggeochem.2014.01.001, 2014.
Volkman, J. K., Barrett, S. M., Dunstan, G. A., and Jeffrey, S. W.: C-30-C-32 Alkyl diols and unsaturated alcohols in microalgae of the class Eustigmatophyceae, Org. Geochem., 18, 131–138, https://doi.org/10.1016/0146-6380(92)90150-v, 1992.
Wen, M. and Jetter, R.: Very-long-chain hydroxyaldehydes from the cuticular wax of Taxus baccata needles, Phytochemistry, 68, 2563–2569, https://doi.org/10.1016/j.phytochem.2007.05.029, 2007.
Yamamoto, M., Ficken, K., Baas, M., Bosch, H. J., and de Leeuw, J. W.: Molecular paleontology of the earliest danian at Geulhemmerberg (the Netherlands), Geol. Mijnbouw, 75, 255–267, 1996.
Zhu, F., Massana, R., Not, F., Marie, D., and Vaulot, D.: Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene, FEMS Microbiol. Ecol., 52, 79–92, https://doi.org/10.1016/j.femsec.2004.10.006, 2005.