the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
The contribution of microbial communities in polymetallic nodules to the diversity of the deep-sea microbiome of the Peru Basin (4130–4198 m depth)
Massimiliano Molari
Felix Janssen
Tobias R. Vonnahme
Frank Wenzhöfer
Antje Boetius
Industrial-scale mining of deep-sea polymetallic nodules will remove nodules in large areas of the sea floor. The regrowth of the nodules by metal precipitation is estimated to take millions of years. Thus, for future mining impact studies, it is crucial to understand the role of nodules in shaping microbial diversity and function in deep-sea environments. Here we investigated microbial-community composition based on 16S rRNA gene sequences retrieved from sediments and nodules of the Peru Basin (4130–4198 m water depth). The nodule field of the Peru Basin showed a typical deep-sea microbiome, with dominance of the classes Gammaproteobacteria, Alphaproteobacteria, Deltaproteobacteria, and Acidimicrobiia. Nodules and sediments host distinct bacterial and archaeal communities, with nodules showing lower diversity and a higher proportion of sequences related to potential metal-cycling Bacteria (i.e. Magnetospiraceae, Hyphomicrobiaceae), bacterial and archaeal nitrifiers (i.e. AqS1, unclassified Nitrosomonadaceae, Nitrosopumilus, Nitrospina, Nitrospira), and bacterial sequences found in the oceanic crust, nodules, hydrothermal deposits, and sessile fauna. Sediment and nodule communities overall shared a low proportion of operational taxonomic units (OTUs; 21 % for Bacteria and 19 % for Archaea). Our results show that nodules represent a specific ecological niche (i.e. hard substrate, high metal concentrations, and sessile fauna), with a potentially relevant role in organic-carbon degradation. Differences in nodule community composition (e.g. Mn-cycling bacteria, nitrifiers) between the Clarion–Clipperton Fracture Zone (CCZ) and the Peru Basin suggest that changes in environmental setting (e.g. sedimentation rates) also play a significant role in structuring the nodule microbiome.
- Article
(2101 KB) - Full-text XML
-
Supplement
(482 KB) - BibTeX
- EndNote
Polymetallic nodules (or manganese nodules) occur in abyssal plains (4000–6000 m water depth) and consist primarily of manganese and iron as well as many other metals and rare earth elements (Crerar and Barnes, 1974; Kuhn et al., 2017). Nodules are potato- or cauliflower-shaped structures with typical diameters of 4–20 cm and are typically found at the sediment surface or occasionally buried in the uppermost 10 cm of the sediment horizon. The mechanisms of nodule formation are not completely elucidated. The current understanding is that they are formed via mineral precipitation from bottom waters (hydrogenetic growth) or pore waters (diagenetic growth) involving both abiotic and microbiological processes (Crerar and Barnes, 1974; Riemann, 1983; Halbach et al., 1988; Wang et al., 2009). The formation of nodules is a slow process that is estimated to range between thousands and millions of years per millimetre of growth (Kerr, 1984; Boltenkov, 2012).
Rising global demand for metals has renewed interests in commercial mining of deep-sea nodule deposits. Mining operations would remove nodules, disturb or erode the top decimetres of sediment, and create near-bottom sediment plumes that would resettle and cover the sea floor (Miller et al., 2018). Although the first nodules were discovered in the 1870s (Murray and Renard, 1891), only little is known about the biodiversity, biological processes, and ecological functions of the nodules and their surrounding sediments as a specific deep-see habitat. Major questions remain, for example as to spatial turnover on local and global scales, the role of the microbial community in and around nodules, and the role of nodules as substrate for endemic species. Hence, there is the need to thoroughly characterise baseline conditions as a requirement for any mining operations as these will require assessments of impacts associated with mining.
Extensive and dense nodule fields are found in different areas of the Pacific and Indian oceans. Nodule accumulations of economic interest have been found in four geographical locations: the Clarion–Clipperton Fracture Zone (CCZ) and the Penrhyn Basin in the north-central and south-central Pacific Ocean, respectively; the Peru Basin in the south-eastern Pacific; and in the centre of the northern Indian Ocean (Miller et al., 2018). To our knowledge the Peru Basin is the only region that does not have exploration activities and plans for mining so far. Previous work on the structure of microbial communities of nodule fields by 16S rRNA gene sequencing has focused on the CCZ and the south-central Pacific Ocean (Xu et al., 2007; Wu et al., 2013; Tully and Heidelberg, 2013; Blöthe et al., 2015; Shulse et al., 2017; Lindh et al., 2017). All studies showed that polymetallic nodules harbour microorganisms that are distinct from the surrounding sediments and overlying water. They indicate that nodule communities show a pronounced spatial variability, but these results are so far not conclusive. Similar microbial communities were observed in nodules collected at distances of 6000 and 30 km (Wu et al., 2013; Shulse et al., 2017), while Tully and Heidelberg (2013) found that nodule communities varied among sampling sites (<50 km). Besides, potential Mn oxidisers and reducers such as Alteromonas, Pseudoalteromonas, Shewanella, and Colwellia were proposed as a core of the nodule microbiome involved in the formation of nodules (Wu et al., 2013; Blöthe et al., 2015), but they were not found in all nodules sampled so far (Tully and Heidelberg, 2013; Shulse et al., 2017). The lack of knowledge on the diversity and composition of microbial assemblages of other nodule provinces makes it difficult to assess whether observed differences within the CCZ may reflect regional differences in environmental conditions (e.g. input of organic matter, bathymetry, topography, sediment type), in the abundance and morphology of nodules, or in the colonisation of the nodules by epifauna and protozoans.
In this study we investigated the diversity and composition of bacterial and archaeal communities associated with manganese nodule fields of the Peru Basin. The Peru Basin is located about 3000 km off the coast of Peru and covers about half of the size of the CCZ, which is 5000–9000 km away. The present-day organic carbon flux in this area is approximately 2 times higher than in the CCZ, resulting in higher content of organic carbon in the surface sediments (>1 % vs. 0.2 %–0.6 % in the CCZ) and a shallower oxic–sub-oxic front (10 cm vs. tens of metres of sediment depth in the CCZ; Müller et al., 1988; Haeckel et al., 2001; Volz et al., 2018). As a consequence of differences in environmental conditions (e.g. organic carbon flux, carbonate compensation depth, sediment type, topography, and near-bottom currents), the Peru Basin and the CCZ host manganese nodules with different geological features (Kuhn et al., 2017): (i) nodules from the Peru Basin are often larger, with a typical cauliflower shape, compared to those in the CCZ, which have a discoidal shape and a size of 2–8 cm (Kuhn et al., 2017); (ii) the average nodule abundance in the Peru Basin is lower (10 kg m−2) than in the CCZ (15 kg m−2; Kuhn et al., 2017); (iii) Mn nodules from the Peru Basin are thought to be mainly formed by sub-oxic diagenesis, whereas CCZ nodules apparently exhibit a mixture of diagenetic and hydrogenetic origin (von Stackelberg, 1997; Chester and Jickells, 2012); (iv) while Peru Basin and CCZ nodules consist of the same type of mineral (disordered phyllomanganates), they have a different metal content (Wegorzewski and Kuhn, 2014; Wegorzewski et al., 2015).
An increasing number of studies and policy discussions address the scientific basis of ecological monitoring in deep-sea mining, highlighting the need to identify appropriate indicators and standards for environmental impact assessments and ecological management. A key aspect is avoiding harmful effects to the marine environment, which will have to include loss of species and ecosystem functions. The primary aims of this study were to assess the structure and similarity of benthic microbial communities of nodules and sediments of the Peru Basin nodule province and to compare them with those of other global deep-sea sediments and nodules in the CCZ. The focus was on similarity comparisons in order to investigate endemism and potential functional taxa that could be lost due to the removal of manganese nodules by mining activities. To achieve this, the hypervariable 16S rRNA gene regions V3–V4 for Bacteria and V3–V5 for Archaea were amplified from DNA extracted from nodules and surrounding sediments and sequenced using the Illumina paired-end MiSeq platform. The hypotheses tested were that (i) nodules shape deep-sea microbial diversity and (ii) nodules host a specific microbial community compared to the surrounding sediments. The secondary aim of this study was to investigate the nodule features that may play a major role in shaping microbial-community composition and microbially mediated functions.
2.1 Sample collection
Sediment samples and polymetallic nodules were collected as a part of the MiningImpact project of the Joint Programming Initiative Healthy and Productive Seas and Oceans (JPI Oceans) on board the R/V Sonne (expedition SO242/2; 28 August–1 October 2015) in the Peru Basin around 7∘ S and 88.5∘ W. Samples were collected at three sites outside the sea floor area selected in 1989 for a long-term disturbance and recolonisation experiment (DISCOL; Thiel et al., 2001). For this reason they were called “Reference Sites”: Reference East, Reference West, and Reference South. Sediment samples were collected using a TV-guided multiple corer (TV MUC) at three stations per site (Table 1). The cores were sliced on board in a temperature-controlled room (set at in situ temperature), and aliquots of sediment were stored at −20 ∘C for DNA extraction. Manganese nodules were sampled using a TV MUC or a remotely operated vehicle (ROV; KIEL 6000, GEOMAR, Germany): one nodule at Reference West and four nodules at Reference South. The nodules were partly located at the surface or buried down to 3 cm below the sea floor (b.s.f.) and had diameters of a few centimetres. Nodules were gently rinsed with 0.22 µm filtered cold bottom seawater to remove adhering sediment, stored in sterile plastic bags at −20 ∘C, and crushed before DNA extraction in the home lab. From the nodules collected with the ROV, only the surface layer was scraped off using a sterile spoon and subsequently crushed and frozen (−20 ∘C). Sedimentary metadata (e.g. cell counts, pigments and organic carbon content, porewater profiles, and porosity) and a map of the study area are available in Vonnahme at al. (2020). Focusing entirely on sediments, that publication also includes a discussion of the variability of environmental settings and microbial communities.
2.2 DNA extraction and sequencing
The DNA was extracted from 1 g of wet sediment (0–1 cm layer) and from 1 g of wet nodule fragments using the FastDNA™ SPIN kit for soil (Q-BIOgene, Heidelberg, Germany) following the protocol provided by the manufacturer. An isopropanol precipitation was performed on the extracted DNA, and DNA samples were stored at −20 ∘C. To control for DNA contamination (negative control), DNA extraction was carried out on purified water after being in contact with a sterile scalpel and a plastic bag.
Amplicon sequencing was done at the CeBiTec laboratory (Center for Biotechnology, Bielefeld University) on an Illumina MiSeq machine. For the 16S rRNA gene amplicon library preparation we used the bacterial primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 785R (5′-GACTACHVGGGTATC TAATCC-3′) as well as the archaeal primers Arch349F (5′-GYGCASCAGKCGMGAAW-3′) and Arch915R (5′-GTGCTCCCCCGCCAATTCCT-3′; Wang and Qian, 2009; Klindworth et al., 2013), which amplify the 16S rRNA gene hypervariable region V3–V4 in Bacteria (400–425 bp fragment length) and the V3–V5 region in Archaea (510 bp fragment length). The amplicon library was sequenced with MiSeq v3 chemistry in a 2 bp×300 bp paired run with > 50 000 reads per sample, following the standard instructions of the 16S Metagenomic Sequencing Library Preparation protocol (Illumina, Inc., San Diego, CA, USA).
The quality cleaning of the sequences was performed with several software tools. CUTADAPT (Martin, 2011) was used for primer clipping. Subsequently the TRIMMOMATIC software (Bolger et al., 2014) was used to remove low-quality sequences starting with the following settings: SLIDINGWINDOW:4:10 MINLEN:300 (for Bacteria) and SLIDINGWINDOW:6:13 MINLEN:450 (for Archaea). In the case of Bacteria data, this step was performed before the merging of reverse and forward reads with a paired-end read merger (PEAR; J. Zhang et al., 2014). Low-quality archaeal sequences were removed after merging the reads in order to enhance the number of retained reads due to the increase in archaeal 16S rRNA gene fragment length. All sequences were quality-controlled with FastQC (Andrews, 2010). Where necessary, more sequences were removed with TRIMMOMATIC using larger sliding window scores until the FastQC quality control was passed (average quality score per sample: >34 for Bacteria and >22 for Archaea). Clustering of sequences into OTUs (operational taxonomic units) was done using the SWARM algorithm (Mahé et al., 2014). The taxonomic classification was based on the SILVA rRNA reference database (release 132) at a minimum alignment similarity of 0.9 and a last-common-ancestor consensus of 0.7 (Pruesse et al., 2012). The workflow and scripts applied in this study can be found in Hassenrück et al. (2016). Raw sequences with removed primer sequences were deposited at the European Nucleotide Archive (ENA) under accession number PRJEB30517 and PRJEB32680; the sequences were archived using the service of the German Federation for Biological Data (GFBio; Diepenbroek et al., 2014).
The total number of sequences obtained in this study is reported in Table S1 in the Supplement. Absolute singletons (SSOabs), i.e. OTUs consisting of sequences occurring only once in the full dataset (Gobet et al., 2014), were removed (Table S1). Similarly, contaminant sequences (as observed in the negative control) and unspecific sequences (i.e. bacterial sequences in the archaeal amplicon dataset as well as and archaeal, chloroplast, and mitochondrial sequences in the bacterial dataset) were removed from amplicon datasets before the analysis (Table S1). The dominant OTU sequences and OTU sequences highly abundant in the nodules were searched using the Basic Local Alignment Search Tool (BLASTn; GeneBank nucleotide database, 12 June 2019; Altschul et al., 1990) in order to identify the other habitats in which the most closely related (i.e. >99 %) sequences have been previously reported.
2.3 Data analysis
The first three Hill numbers, or the effective number of species, were used to describe alpha diversity: species richness (H0), the exponential of Shannon entropy (H1), and the inverse Simpson index (H2; Chao et al., 2014). Coverage-based and sample-size-based rarefaction (based on the actual number of sequences) and extrapolation (based on double the number of sequences) curves were calculated for the Hill numbers using the R package iNEXT (Hsieh et al., 2018). Calculation of the estimated richness (Chao1) and the identification of unique OTUs (present exclusively in one sample) were based on repeated (n=100) random subsampling of the amplicon datasets. Significant differences in alpha diversity indices between substrates (i.e. manganese nodules and sediments) were determined by analysis of variance (ANOVA) or by the non-parametric Kruskal–Wallis (KW) test when the ANOVA's assumptions were not satisfied.
Beta diversity in samples from different substrates and from different sites was quantified by calculating a Euclidean distance matrix based on OTU abundances that have underwent centred log-ratio (CLR) transformation (clr function in the R package compositions) and Jaccard dissimilarity based on a presence–absence OTU table. The latter was calculated with 100 sequence resamplings per sample on the smallest dataset (40 613 sequences for Bacteria and 1835 sequences for Archaea). Euclidean distance was used to produce non-metric multidimensional scaling (NMDS) plots. The Jaccard dissimilarity coefficient was used to perform hierarchical clustering (hclust function in the R package vegan using the complete linkage method), and the dissimilarity values for cluster nodes were used to calculate the number of shared OTUs between or within groups. The permutational multivariate analysis of variance (PERMANOVA; Anderson, 2001) was used to test difference in community structure and composition.
Differentially abundant OTUs and genera were detected using the R package ALDEx2 (Fernandes et al., 2014) at a significance threshold of 0.01 and 0.05 for Benjamini–Hochberg (BH)-adjusted parametric and non-parametric (KW) p values, respectively. We only discuss the taxa that were at least 2 times more abundant in nodules than in sediments, i.e. Log2(Nodule∕sediment) ≥1, and with a sequence contribution of total number of sequences of ≥1 % (for genera) or ≥0.1 % (for OTUs).
All statistical analyses were conducted in R using the core distribution with the additional packages vegan (Oksanen et al. 2015), compositions (Van den Boogaart et al., 2014), iNEXT (Hsieh et al., 2018), and ALDEx2 (Fernandes et al., 2014).
3.1 Microbial alpha diversity
Bacterial and archaeal communities in five nodules and nine sediment samples (Table 1) were investigated using specific sets of primers for Bacteria and Archaea on the same extracted pool of DNA per station. The number of bacterial sequences retrieved from DNA extracted from sediments and nodules was on average 5±5 and 25±14 times higher, respectively, than those obtained for Archaea (t test: p<0.001, df = 11, t=4.5).
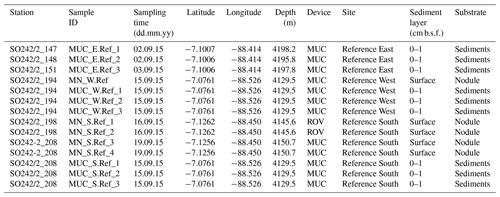
Table S1 shows the statistics of sequence abundance and proportion of singletons and cosmopolitan types. Sequence abundances of Bacteria were comparable between sediments and nodules. Cosmopolitan OTUs, i.e. those present in 80 % of the sediments and nodule samples, made up only 9 % of all taxa (77 % of all sequences), whereas rare OTUs occurring in only <20 % of all samples represented 50 % of the taxa (4 % of all sequences). Sediments and nodules contained only 4 % and 2 %, respectively, of endemic taxa, defined as those that were abundant in one substrate but rare in the other. Thus the contribution of unique OTUs to the total number of OTUs was lower in manganese nodules than in sediments samples (Table 2, Fig. 1a). Bacterial and archaeal diversity was investigated calculating the total number of OTUs (Hill number q=0; H0), the estimated richness (Chao1), and the unique OTUs (present exclusively at one station). For this analysis, the latter were calculated with sequence resampling to overcome differences in sequencing depth. Abundance-based coverage estimators – exponential Shannon (Hill number q=1; H1) and inverse Simpson (Hill number q=2; H2) – were also calculated. The rarefaction curve indicates that the richness (H0) of the less abundant and rare OTUs was somewhat underestimated both in nodules and in sediments (Fig. S1a–b in the Supplement). However, the bacterial and archaeal diversity was well described for the abundant OTUs (H1 and H2; Fig. S1a–b), with more than 90 % of the estimated diversity covered (Fig. S1c–d). In both sediments and nodules the alpha diversity indices were higher for Bacteria than for Archaea (t test: p<0.0001, df = 12, t = 8.0–16.0), while the contribution of unique OTUs to the total number of OTUs was comparable (Table 2). Bacterial communities in manganese nodules have lower Hill numbers and Chao1 indices compared to those associated with sediments (Table 2, Fig. 1a). Archaeal communities showed the same patterns for diversity indices and unique OTUs with the exception of the H2 index that did not show significant difference between nodules and sediments (Table 2, Fig. 1b).
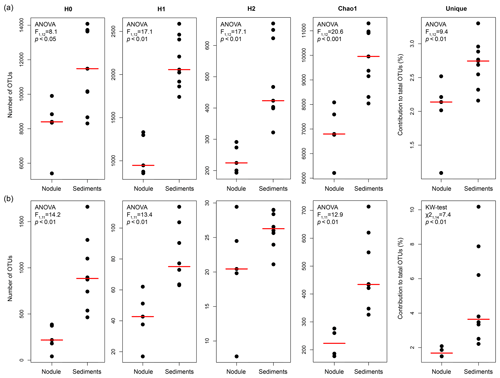
Figure 1Comparison of diversity indices and unique OTUs between manganese nodules and sediments for (a) bacterial and (b) archaeal communities. H0: number of OTUs (q=0); H1: exponential Shannon (q=1); H2: inverse Simpson (q=2); unique: OTUs present exclusively at each station (percentage relative to total OTUs of whole dataset). Chao1 and Unique OTUs were calculated with 100 sequence resamplings per sample for the smallest dataset (40 613 sequences for Bacteria and 1835 sequences for Archaea). The red line shows the median. F: statistic F ratio, with subscript numbers reporting the degrees of freedom between groups and within groups, respectively; p: probability level; KW test: Kruskal–Wallis test; χ2: chi-square test value, with subscript numbers reporting the degrees of freedom between groups and sample size, respectively.
Table 2Bacterial and archaeal diversity indices and unique OTUs for all nodules and sediment samples. Indices and unique OTUs were calculated without singletons.
H0: number of OTUs; H1: exponential Shannon; H2: inverse Simpson; unique: OTUs present exclusively at one station (percentage relative to total OTUs of whole dataset); NA: not available. a After the merging of forward and reverse reads. b After removal of unspecific and contaminant sequences (see Methods for details). c Calculated with 100 sequence resamplings per sample for the smallest dataset (40 613 sequences for Bacteria and 1835 sequences for Archaea; average data and standard deviation, SD, are given). d After quality trimming of merged forward and reverse reads. e Due to an extremely low number of sequences this sample was not included in analyses requiring sequence resampling.
3.2 Patterns in microbial-community composition
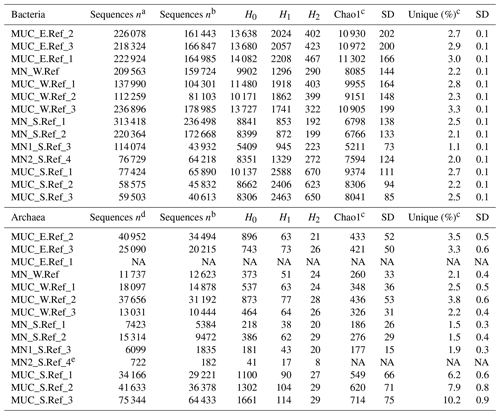
The changes in microbial-community structure at the OTU level (beta diversity) between substrates and samples were quantified by calculating Euclidean distances from CLR-transformed OTU abundance. Shared OTUs were estimated by calculating Jaccard dissimilarity from OTU presence or absence based on repeated random subsampling of the amplicon datasets. Microbial communities associated with manganese nodules differed significantly from those found in the sediments (Fig. 2, Table S2). Furthermore, significant differences were detected in sediment-microbial-community structure among the different sites (PERMANOVA; Bacteria: R2=0.384; p=0.003; ; Archaea: R2=0.480; p=0.013; ; Table S2) and between communities associated with nodules and sediment at the Reference South site (PERMANOVA; Bacteria: R2=0.341; p=0.023; ; Archaea: R2=0.601; p=0.029; ; Table S2), which was the only site where the number of samples allowed for the test. “Site” (defined by geographic location) and “substrate” (i.e. originating from sediments or nodules) explained a similar proportion of variation in bacterial-community structure (27 % and 23 %, respectively). “Substrate” had a more important role in shaping archaeal communities than “site” (explained variance: 35 % and 19 %, respectively; Table S2). The number of shared OTUs between nodules and sediments (Bacteria: 21 %; Archaea: 19 %) was lower than those shared within nodules (Bacteria: 30 %; Archaea: 30 %) and within sediments (Bacteria: 31 %; Archaea: 32 %; Fig. S2).
Figure 2Non-metric multidimensional scaling (NMDS) plot based on a Euclidean distance similarity matrix of (a) bacterial- and (b) archaeal-community structure at the OTU level. Sequence abundances of OTUs were CLR-transformed. Permutational multivariate analysis of variance (PERMANOVA) showed significant differences between nodule- and sediment-associated microbial communities (for details see Table S1 in the Supplement). Each sample (dot) is connected to the weighted averaged mean of the inter-group distances. Ellipses represent one standard deviation of the weighted averaged mean.
Bacterial communities in manganese nodules and sediments were dominated by the classes Gammaproteobacteria (26 %), Alphaproteobacteria (19 %), Deltaproteobacteria (9 %), Bacteroidia (5 %), Acidimicrobiia (4 %), Dehalococcoidia (4 %), Planctomycetacia (4 %), Nitrospinia (3 %), and Phycisphaerae (3 %), which accounted for more than 75 % of the total sequences (Fig. 3). All archaeal communities were dominated by Thaumarchaeota (Nitrosopumilales), which represented more than 95 % of all sequences. The remaining small proportion of sequences were taxonomically assigned to Woesearchaeia (Fig. S2b). Nodule and sediment samples showed similar compositions of most abundant bacterial genera (contribution to total number of sequence: ≥1 %; Fig. S2a). A total of 69 bacterial genera (9 % of all genera) were differentially abundant in the nodules and in the sediment, accounting for 36 % and 21 % of the total sequences retrieved from nodules and sediments, respectively (ALDEx2: ANOVA-adjusted p of <0.01 and KW-adjusted p of <0.05; Fig. 4 and Table S3). Of those only one unclassified genus within the family Sphingomonadaceae and the genus Filomicrobium were exclusively found in nodules and not in the sediment samples, and their contribution to the total number of sequences was less than 0.06 %. Genera that were more abundant in the nodules than in the sediments included unclassified Alphaproteobacteria (7 %), Nitrospina (4 %), unclassified SAR324 clade (marine group B; 3 %), unclassified Hyphomicrobiaceae (3 %), Pirellulaceae Pir4 lineage (2 %), unclassified Methyloligellaceae (1 %), unclassified Pirellulaceae (1 %), Acidobacteria, unclassified Subgroup 9 (1 %) and Subgroup 17 (1 %), Nitrosococcaceae AqS1 (1 %), Calditrichaceae JdFR-76 (1 %), and Cohaesibacter (1 %; Fig. 4 and Table S3). In the sediment we identified 21 genera that were more abundant than in the nodules, but all together they represented only 3 % of the total sequences recovered from sediments. A total of 128 OTUs were highly abundant in nodules (ALDEx2: ANOVA-adjusted p of <0.01 and KW-adjusted p of <0.05), which accounts for 24 % of the total sequences retrieved from nodules (Table 3a). The most closely related sequences (≥99 % similarity) were retrieved from the oceanic crust (30 %), from nodule fields (26 %), from hydrothermal or seep sediments and deposits (21 %), and from worldwide deep-sea sediments (16 %), and 7 % were found in association with invertebrates (Table 3b and Fig. 5).
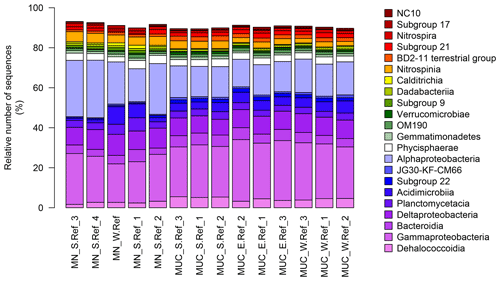
Figure 3Bacterial-community structure at the dominant class level (cut-off of ≥1 %). MN: manganese nodules; MUC: sediments.

Figure 4Taxa highly abundant in nodules (ALDEx2: glm-adjusted p of <0.01; KW-adjusted p of <0.05). The base 2 logarithm of the ratios between the sequence number of nodules (Nod) and sediments (Sed) centred by geometric mean as well as the average contribution of total number of sequences (%) retrieved in nodules and in sediments is shown. For details see Table S3.
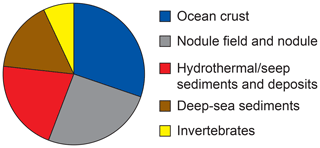
Figure 5Habitat coverage for the most closely related sequences (≥ 99 % similarity) to OTUs highly abundant in the nodules. For details see Table 3a–b.
Table 3(a) OTUs highly abundant in nodules (ALDEx2: glm-adjusted p of <0.01; KW-adjusted p of <0.05). Only OTUs of ≥ 0.1 % are reported. The base 2 logarithm of the ratios between the sequence number of nodules (Nod) and sediments (Sed) centred by geometric mean as well as the average contribution of total number of sequences (%) retrieved in nodules and in sediments is shown. (b) The most closely related sequences with identity (ID) >99%, as identified with BLASTn (GeneBank nucleotide database, 12 June 2019).
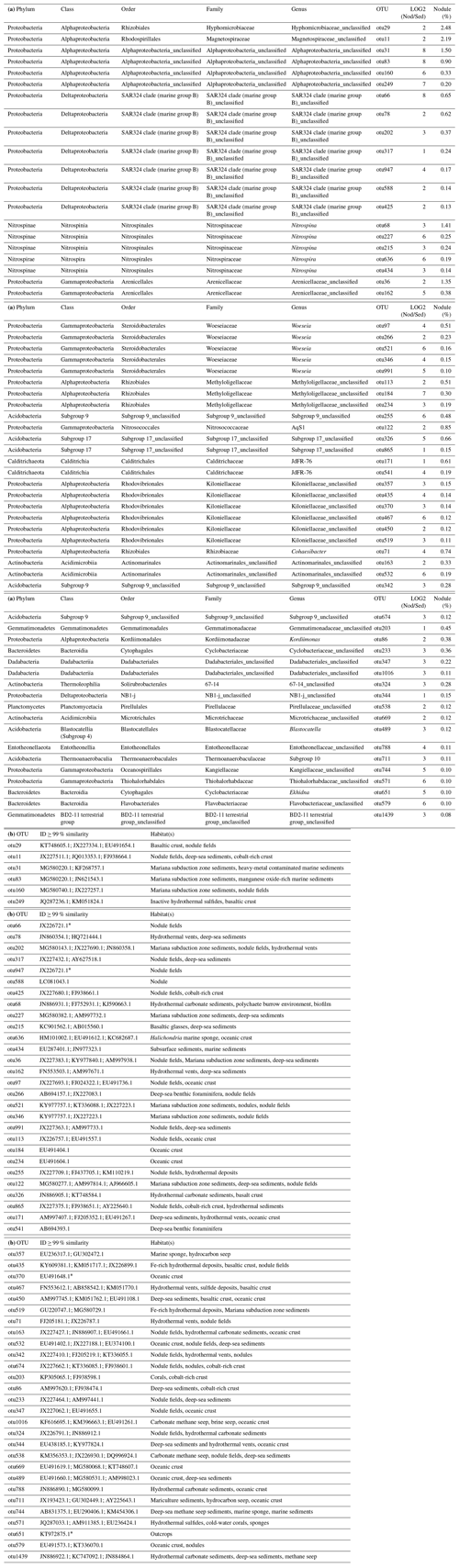
* ≥98 % similarity.
Industrial-scale mining of deep-sea polymetallic nodules may remove nodules and the active surface sea floor layer at a spatial scale ranging from ca. 50 000–75 000 km2 per claim to ca. 1×106 km2, including all current exploration licences (Miller et al., 2018). The regrowth of nodules will take millions of years; thus it is unknown whether the associated biota could recover at all (Simon-Lledo et al., 2019). The response of microbial communities to the loss of nodules and sea floor integrity is largely unknown. It may play an important role in the ecological state of the sea floor habitat due to the many functions Bacteria and Archaea hold in the food web, element recycling, and biotic interactions beyond representing the largest biomass in deep-sea sediments (Joergensen and Boetius, 2007). It is thus crucial to understand the role of nodules in shaping microbial diversity and in hosting microbes with important ecological functions. So far, only few studies have been carried out to investigate the microbiota of nodule fields, and most of them were focused on identifying microbes involved in metal cycling. Here, we investigated the similarity of microbial-community structures in sediments and nodules retrieved from the Peru Basin. The objectives of this study were (i) to compare the microbes of nodules fields with microbiota of deep-sea sediments in order to identify specific features of microbial diversity of nodule fields, (ii) to elucidate differences in diversity and in microbial-community structure between sediments and nodules, and (iii) to investigate potential microbially mediated functions and the major drivers in shaping microbial communities associated with the nodules.
4.1 Microbial diversity of nodule fields is distinct from other deep-sea areas
Benthic bacterial assemblages in sediments and nodules of the Peru Basin showed the typical dominance of the classes Gammaproteobacteria, Alphaproteobacteria, Deltaproteobacteria, and Acidimicrobiia as reported for deep-sea sediments worldwide (Bienhold et al., 2016; Fig. 3) and in the Pacific Nodule Province (Wang et al., 2010; Wu et al., 2013; Shulse et al., 2017; Lindh et al., 2017). However at higher taxonomic resolution we detected substantial differences in the microbial-community composition of other deep-sea regions. Sediments of the Peru Basin Bacteria classes were depleted in sequence abundances of Flavobacteria, Gemmatimonadetes, and Bacilli, whereas sequence abundances of the Chloroflexi (i.e. Dehalococcoidia) and Planctomycetes (i.e. Pirellulaceceae, Phycisphaeraceae) as well as the genus Nitrospina were higher compared to other deep-sea regions (Bienhold et al., 2016; Varliero et al., 2019). Dehalococcoidia and Planctomycetes were previously reported as important components of benthic microbial assemblages in the Pacific Ocean (Wang et al., 2010; Wu et al., 2013; Blöthe et al., 2015; Walsh et al., 2016; Lindh et al., 2017). Their contribution to the total community was found to increase in organic-matter-depleted subsurface sediments (Durbin and Teske, 2011; Walsh et al., 2016).
Dominant OTUs (>1 %) belonged to unclassified Actinomarinales, Gammaproteobacteria, Subgroup 21 (phylum Acidobacteria) and to the genus Woeseia (family Woeseiaceae). Members of Actinomarinales and Woeseiaceae are cosmopolitan types in deep-sea sediments (Bienhold et al., 2016). For Actinomarinales there are no cultured relatives, and the function of this group remains unknown. In the case of Woeseiaceae, one representative is in culture (Woeseia ocaeni). W. ocaeni is an obligate chemoorganoheterotroph (Du et al., 2016), suggesting a role in organic-carbon remineralisation for members of that family, as recently confirmed by analysis of deep-sea assembled genomes (Hoffmann et al., 2020). The most closely related sequences of Subgroup 21 have been reported in deep-sea sediments (Schauer et al., 2010) and across Pacific nodule fields (Wu et al., 2013) but also in association with deep-sea benthic giant foraminifera (xenophyophores) and in surrounding sediments (Hori et al., 2013). The Subgroup 21-like OTU was also one of the 10 most abundant OTUs retrieved from nodules (0.9 %). Xenophyophores have agglutinated tests and can grow to decimetre size, suggesting that members of Subgroup 21 may be colonists of biological and/or hard substrates.
Within the class Alphaproteobacteria the most abundant OTUs (>0.5 %) belonged to unclassified genera of the families Magnetospiraceae (order Rhodospirillales), Hyphomicrobiaceae (order Rhizobiales), and Kiloniellaceae (order Rhodovibrionales). Magnetospiraceae and Hyphomicrobiaceae are the most abundant families in nodules, with >2 % of OTUs. Closely related sequences have been reported previously across Pacific nodule provinces (Xu et al., 2007; Shulse et al., 2017). The family Magnetospiraceae includes microaerophilic heterotrophs, which are capable of magnetotaxis and iron reduction (i.e. genus Magnetospirillum; Matsunaga, 1991; Schüler and Frankel, 1999), and thus the members of this family could play a role in Fe(III) mobilisation, affecting its bioavailability. Hyphomicrobiaceae-like sequences found in this study are related to the genera Hyphomicrobium and Pedomicrobium (sequence identity: 97 %), which have been reported to be involved in manganese cycling (Tyler, 1970; Larsen et al., 1999; Stein et al., 2001). A potential contribution of these groups in metal cycling in manganese nodules is also suggested by the presence of the most closely related sequences in the oceanic crust (Santelli et al., 2008; Lee et al., 2015), which typically hosts epilithic and endolithic microbial communities of chemolithotrophic metal oxidisers (Staudigel et al., 2008). Similarly, Kiloniellaceae-related OTUs might be involved in metal cycling as closely related sequences were found in marine basalts (Mason et al., 2007; Santelli et al., 2008) and inside other manganese nodules (Blöthe et al., 2015). Most of the marine cultivates in the family Kiloniellaceae belong to the genus Kiloniella and have been isolated from marine macroalgae (Wiese et al., 2009), the guts of Pacific white shrimp (Wang et al., 2015), marine sponge (Yang et al., 2015), spider crab and clam (Gerpe et al., 2017), and from the surface water of a polynya in the Amundsen Sea in the West Antarctic Ice Sheet (Si et al., 2017). Furthermore, Kiloniellaceae-like sequences were found in sponges (Cleary et al., 2013), sea star larvae (Galac et al., 2016), and in the iron mats of seamounts (Scott et al., 2017). The presence of rich sessile and mobile metazoan communities associated with nodules offers various potential hosts for members of Kiloniellaceae. Kiloniella is a chemoheterotrophic aerobe, and the draft genome of an isolate from the gut of a Pacific white shrimp shows potential for denitrification and iron acquisition and metabolism (Wang et al., 2015). Thus, either as free-living or host-associated life, the potential contribution of Kiloniellaceae in metal cycling requires further investigation.
Archaea were also present in sediments of the Peru Basin, with Nitrosopumilaceae (phylum Thaumarchaeota) dominating the archaeal communities (Fig. S2b). Archaeal sequences comprised a lower portion of total sequences retrieved from sediments (6 %–45 %) and nodules (< 1 %–7 %) of the Peru Basin, and they were lower in nodules compared to sediments. Our data differed from what was reported by Shulse et al. (2017) for the CCZ, especially for nodules (ca. 20 %). We cannot rule out that the observed differences in microbial-community structure partly reflect the different sets of primers used in our study and by Shulse et al. (2017). As both primer sets amplified the same hypervariable region of the 16S rRNA gene (V4) we assume that biases are small enough to justify the comparison. The majority of members of Nitrosopumilaceae are believed to be capable of oxidation of ammonia to nitrite, the first step of nitrification (Offre et al., 2013). Archaeal ammonia oxidisers have a higher affinity for ammonia than bacterial ammonia oxidisers, and they are favoured in environments with low ammonia concentrations (Martens-Habbena et al., 2009). The Peru Basin has higher particulate organic-carbon fluxes as compared to the central Pacific Ocean (Haeckel et al., 2001; Mewes et al., 2014), which results in higher remineralisation rates and higher ammonia fluxes. These limit the thickness of oxygenated sediments to 10 cm in the Peru Basin, while they can reach up to 2–3 m depth in the CCZ (Haeckel et al., 2001; Mewes et al., 2014; Volz et al., 2018). Hence differences observed between CCZ and Peru Basin nodule fields in the contribution of archaeal sequences to microbial assemblages are likely due to ammonia availability, which is controlled by organic matter fluxes.
4.2 Microbial-community structure differs between sediments and nodules
Beta diversity of microbial-community structure in the Peru Basin sediments showed remarkable spatial OTU turnover already on a local scale (<60 km; Fig. S2), which is at the higher end of previous microbial beta diversity estimates for bathyal and abyssal sea floor assemblages (Jacob et al., 2013; Ruff et al., 2015; Bienhold et al., 2016; Walsh et al., 2016; Varliero et al., 2019). Here we focused specifically on the contribution of nodules to diversity, which could be a critical parameter in the ecological assessment of nodule removal. Analysis of community composition at OTU level shows that nodules and sediments host distinct bacterial and archaeal communities (Fig. 2), as previously reported for the CCZ as well (Wu et al., 2013; Tully and Heidelberg, 2013; Shulse et al., 2017; Lindh et al., 2017). Although the microbial communities in the sediment showed significant differences between sites, the low number of shared OTUs between sediments and nodules (<20 %) supports the presence of specific bacterial and archaeal communities associated with polymetallic nodule habitats. However, the proportion of truly endemic, unique nodule OTUs was also low (Fig. 1a, Table S1); nonetheless it is relevant to highlight that nodule removal would lead to a loss of specific types of microbes in a mined deep-sea region (Blöthe et al., 2015).
Microbial communities associated with nodules are significantly less diverse than those in the sediments, and the decrease in diversity was observed both in rare and abundant bacterial types (Figs. 1 and S1). This seems to be a common feature of polymetallic nodules (Wu et al., 2013; Tully and Heidelberg, 2013; G. Zhang et al., 2014; Shulse et al., 2017; Lindh et al., 2017). However, a recent meta-analysis of 16S rRNA gene diversity reports no significant differences in microbial biodiversity between nodules and sediments within the studied habitats in the CCZ (Church et al., 2019). Church and colleagues also pointed out that the findings are so far not conclusive due to the limited number of studies and differences in methods (e.g. PCR primers, sequencing approaches), which may also be a reason for the differences between the meta-analysis and the results of this study. Tully and Heidelberg (2013) suggested that lower microbial diversity in the nodules might be due to less availability of potential energy sources (e.g. organic matter) compared to sediments. Despite the fact that the sedimentation rate exceeds the growth rate of nodules, the nodules are typically exposed to bottom water and not covered by sediments (Peukert et al., 2018). Although, it is unknown whether physical mechanisms (e.g. current regime or seasonal events) or biological processes (e.g. grazing, active cleaning) are responsible for the lack of sediment accumulation on nodules, the decrease in microbial diversity with the decrease in organic matter availability is in accordance with the positive energy–diversity relationship reported for deep-sea sediments (Bienhold et al., 2012). However, the presence of foraminiferal assemblages (Gooday et al., 2015) and specific sessile metazoan communities (Vanreusel et al., 2016) on the surface of nodules may represent a potential source of transformed organic matter (e.g. dissolved organic matter) and catabolic products, which may represent a much more valuable energy source for microbes than refractory particulate organic matter sinking from the water column. Furthermore, higher microbial diversity in the sediments than in the nodules could be the result of the accumulation of allochthonous microbes, as suggested by the higher proportion of rare and unique OTUs in the sediments. Lastly, the nodules offer hard substrate and the presence of metals, which can select for specific Bacteria and Archaea. Similarly, hydrothermal deposits have typically lower bacterial diversity than deep-sea sediments despite chemical energy sources being highly available (Ruff et al., 2015; Wang et al., 2018). We propose that the decreased diversity of abundant OTUs in nodules, observed especially for Bacteria, suggests selection for colonists adapted to specific ecological niches associated with nodules (e.g. high metal concentration, hard substrate, presence of sessile fauna).
4.3 Potential functions of microbial communities associated with nodules
The presence of a large proportion of bacterial community with low abundance in the sediments but enriched in the nodules both at the level of genera (35 %) and OTUs (24 %; Fig. 4, Tables S3 and 3a) indicates niche specialisation. The most abundant OTUs (13 % of the bacterial community) in nodules include unclassified Hyphomicrobiaceae, Magnetospiraceae, Alphaproteobacteria, Arenicellaceae and SAR324, Nitrospina, AqS1, Methyloligellaceae, Subgroup 9, Subgroup 17, Kiloniellaceae, Cohaesibacter, and JdFR-76, of which the most closely related sequences have been retrieved from Pacific nodules (e.g. Wu et al., 2013; Blöthe et al., 2015), basaltic rocks (e.g. Mason et al., 2007; Santelli et al., 2008; Mason et al., 2009; Lee et al., 2015), sulfide and carbonate hydrothermal deposits (e.g. Sylvan et al., 2012; Kato et al., 2015), and giant foraminifera (Hori et al., 2013; Table 3b and Fig. 5). There are currently no cultivated representatives and metabolic information for these members of the Bacteria, and it is not known whether they have metal tolerance mechanisms or they are actively involved in metal cycling. The high abundance of potential metal-reducing Bacteria (i.e. Magnetospiraceae) and oxidisers (i.e. Hyphomicrobiaceae) and the presence of encrusting protozoans (Gooday et al., 2015), microbial eukaryotes (Shulse et al., 2017) and metazoans (Vanreusel et al., 2016) create specific ecological niches, which may be at least partially responsible for the observed selection of microbial taxa in nodules. Overall, these findings suggest that bacterial groups adapted to lithic or biological substrates preferentially colonise nodules, likely favoured by manganese and iron availability, formation of biofilms, and the presence of sessile fauna communities.
The reduction and dissolution of Mn oxides by dissolved organic matter (e.g. humic compounds) typically occur in photic or reducing aquatic environments (Sunda et al., 1983; Stone and Morgan, 1984; Stone, 1987; Sunda and Huntsman, 1994). However reductive dissolution of Mn oxides by dissolved organic substrates has also been observed in dark oxygenated seawater (Sunda et al., 1983; Sunda and Huntsman, 1994), suggesting that it could be a relevant abiotic process in manganese nodules. Indeed, this reaction yields manganese(II) and low-molecular-weight organic compounds (Sunda and Kieber, 1994), which may potentially favour Mn-oxidising Bacteria and microbial exploitation of refractory dissolved organic matter (DOM). Intense extracellular enzymatic activities have been reported for sea-floor-exposed basalts (Jacobson Meyers et al., 2014), raising the question of whether the closely related microbes associated with nodules might have comparable degradation rates. Furthermore, nodules host diversified communities of suspension feeders such as serpulid tubeworms, sponges, corals, and crinoids (Vanreusel et al., 2016), which filter microbes and particulate organic carbon (POC) from the bottom water and release DOM and catabolic metabolites (e.g. ammonia). Thus, nodules may act as hotspots of organic-carbon degradation. Although metabolic activity has never been quantified on nodules, and sequence abundances are lower, the increased abundance of nitrifiers in nodules compared to the sediments reported for the Pacific Nodule Province (Tully and Heidelberg, 2013; Shulse et al., 2017) and in this study could indicate high metabolic activity. Nitrifiers catalyse the oxidation of ammonia, a catabolic product of heterotrophic metabolism, to nitrite and eventually to nitrate. In the CCZ the nitrifier community was composed of archaeal ammonia-oxidising Nitrosopumilus, which represented a large portion of the microbial assemblages (up to 20 %) and a minor contribution of bacterial nitrite-oxidising Nitrospira (Tully and Heidelberg, 2013; Shulse et al., 2017). Peru Basin sediments and nodules showed more diversified nitrifier communities, which are enriched by ammonia-oxidising AqS1 (1 %) and unclassified Nitrosomonadaceae (1 %) and by nitrite-oxidising Nitrospina (4 %) and Nitrospira (1 %; Fig. 4, Tables S3 and 3a). Nitrospina are not commonly reported for deep-sea sediments, but they are the dominant nitrite oxidisers in the oceans (Luecker et al., 2013). They have recently been reported as symbionts of deep-sea glass sponges (Tian et al., 2016), which also commonly colonise FeMn nodules (Vanreusel et al., 2016). The Nitrospina-related OTUs detected in the nodules showed only low similarity with pelagic Nitrospina gracilis and Nitrospina-like sequences found in deep-sea glass sponges (sequence identity of 93 %) but were closely related with sequences recovered from marine basalts (Mason et al., 2007; Santelli et al., 2008; Mason et al., 2009), suggesting nodules as a native habitat.
The sediments of nodule fields in the Peru Basin host a specific microbial community of bacterial taxa reported for organic-carbon-poor environments (i.e. Chloroflexi, Planctomycetes) and potentially involved in metal cycling (i.e. Magnetospiraceae, Hyphomicrobiaceae). Nodule communities were distinct from sediments and showed a higher proportion of sequences from potential Mn-cycling Bacteria including bacterial taxa found in the oceanic crust, nodules, and hydrothermal deposits. Our results are in general agreement with previous studies in the CCZ, confirming that nodules provide a specific ecological niche. However remarkable differences in microbial-community composition (e.g. Mn-cycling bacteria, nitrifiers) between the CCZ and the Peru Basin also show that environmental settings (e.g. POC flux) and features of FeMn nodules (e.g. metal content, nodule attached fauna) may play a significant role in structuring the nodule microbiome. Due to limitations in the available datasets and methodological differences in the studies existing to date, findings are not yet conclusive and cannot be generalised. However, they indicate that microbial-community structure and function would be impacted by nodule removal. Future studies need to look at these impacts in more detail and should address regional differences to determine the spatial turnover and its environmental drivers as well as the consequences regarding endemic types.
Furthermore, our results suggest that the removal of nodules and potentially also the blanketing of nodules with plume sediments resuspended during the mining operations may affect the cycling of metal and other elements. Future work is needed to characterise metabolic activities on and in nodules and to understand factors and processes controlling nodule colonisation. Specifically, restoration experiments should take place to test whether artificial substrates favour the recovery of microbial and fauna communities and their related ecological functions.
Raw sequences with removed primer sequences were deposited at the European Nucleotide Archive (ENA) under accession numbers PRJEB30517 (https://www.ebi.ac.uk/ena/data/view/PRJEB30517; Centre for Polar and Marine Research, 2020) and PRJEB32680 (https://www.ebi.ac.uk/ena/data/view/PRJEB32680; Max Planck Institute for Marine Microbiology, 2020).
The supplement related to this article is available online at: https://doi.org/10.5194/bg-17-3203-2020-supplement.
AB, FJ, FW, and MM conceived the study. AB, FJ, FW, and TRV performed sampling activities. MM compiled and analysed the data. MM wrote the paper with contributions from all authors.
The authors declare that they have no conflict of interest.
The authors want to thank the captain and crew of the SO242/1 and 2 expedition as well as Martina Alisch, Jana Bäger, Jakob Barz, Fabian Schramm, Rafael Stiens, and Wiebke Stiens (HGF-MPG Joint Research Group for Deep-Sea Ecology and Technology) for technical support. The authors are also grateful to Halina Tegetmeyer for support with Illumina sequencing (CeBiTec laboratory; HGF-MPG Joint Research Group for Deep-Sea Ecology and Technology). Lastly, the authors would like to thank Denise Akob, Beth N. Orcutt, Tim D'Angelo, and an anonymous reviewer, whose comments and suggestions greatly helped in improving the quality of this paper.
This work was funded by the German Ministry of Research and Education (BMBF) (grant no. 03F0707A-G) as part of the MiningImpact project of the Joint Programming Initiative of Healthy and Productive Seas and Oceans (JPIOceans). Further financial support was provided by the Helmholtz Association (Alfred Wegener Institute Helmholtz Center for Polar and Marine Research, Bremerhaven) and the Max Planck Society (MPG) as well as from the ERC Advanced Investigator Grant ABYSS (grant no. 294757) to Antje Boetius for technology and sequencing. The research has also received funding from the European Union Seventh Framework Programme (FP7/2007–2013) under the MIDAS project (grant agreement no. 603418).
The article processing charges for this open-access
publication were covered by the Max Planck Society.
This paper was edited by Denise Akob and reviewed by Beth Orcutt and one anonymous referee.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J.: Basic local alignment search tool, J. Mol. Biol., 215, 403–410, 1990.
Anderson, M. J.: A new method for non-parametric multivariate analysis of variance, Austral Ecol., 26, 32–46, 2001.
Andrews, S.: FastQC: a quality control tool for high throughput sequence data, available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (last access: 15 April 2019), 2010.
Bienhold, C., Boetius, A., and Ramette, A.: The energy – diversity relationship of complex bacterial communities in Arctic deep-sea sediments, ISME J., 6, 724–732, https://doi.org/10.1038/ismej.2011.140, 2012.
Bienhold, C., Zinger, L., Boetius, A., and Ramette, A.: Diversity and Biogeography of Bathyal and Abyssal Seafloor Bacteria, PLoS One, 11, 1–20, https://doi.org/10.1371/journal.pone.0148016, 2016.
Blöthe, M., Wegorzewski, A., Müller, C., Simon, F., Kuhn, T., and Schippers, A.: Manganese-Cycling Microbial Communities Inside Deep-Sea Manganese Nodules, Environ. Sci. Technol., 49, 7692–7700, https://doi.org/10.1021/es504930v, 2015.
Bolger, A. M., Lohse, M., and Usadel, B.: Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 30, 2114–2120, https://doi.org/10.1093/bioinformatics/btu170, 2014.
Boltenkov, B. S.: Mechanisms of formation of deep-sea ferromanganese nodules: Mathematical modeling and experimental results, Geochem. Int., 50, 125–132, https://doi.org/10.1134/S0016702911120044, 2012.
Centre for Polar and Marine Research: Effects of simulated deep-sea mining impacts on microbial communities and functions in the DISCOL experimental area, ENA, available at: https://www.ebi.ac.uk/ena/data/view/PRJEB30517, last access: 19 June 2020.
Chao, A., Gotelli, N. J., Hsieh, T. C., Sander, E. L., Ma, K. H., Colwell, R., and Ellison, A. M.: Rarefaction and extrapolation with Hill numbers: A framework for sampling and estimation in species diversity studies, Ecol. Monogr., 84, 45–67, https://doi.org/10.1890/13-0133.1, 2014.
Chester, R. and Jickells, T.: Marine Geochemistry, 3rd Edn., Wiley-Blackwell, Oxford, 2012.
Church, M. J., Wear, E. K., Orcutt, B. N., Young, C. R., and Smith, J. M.: Taxonomic diversity of Bacteria and Archaea in the Clarion-Clipperton Zone of the North Pacific Ocean, Annex V in Deep CCZ Biodiversity Synthesis Workshop, 1–4 October 2019, Friday Harbor, Washington, USA, 2019.
Cleary, D. F. R., Becking, L. E., Voogd, N. J. De, Pires, A. C. C., Ana, R. M. P., Egas, C., and Gomes, N. C. M.: Habitat- and host-related variation in sponge bacterial symbiont communities in Indonesian waters, FEMS Microbiol. Ecol., 85, 465–482, https://doi.org/10.1111/1574-6941.12135, 2013.
Crerar, D. A. and Barnes, H. L.: Deposition of deep-sea manganese nodules, Geochim. Cosmochim. Ac., 38, 279–300, https://doi.org/10.1016/0016-7037(74)90111-2, 1974.
Diepenbroek, M., Glöckner, F. O., Grobe, P., Güntsch, A., Huber, R., König-Ries, B., Kostadinov, I., Nieschulze, J., Seeger, B., Tolksdorf, R., and Triebel, D.: Towards an Integrated Biodiversity and Ecological Research Data Management and Archiving Platform?: The German Federation for the Curation of Biological Data (GFBio), Inform. 2014 – Big Data Komplexität meistern, GI-Edition Lect. Notes Informatics – Proc., in: Informatik 2014, edited by: Plödereder, E., Grunske, L., Schneider, E. and Ull, D., Gesellschaft für Informatik e.V., Bonn, 1711–1724, 2014.
Du, Z., Wang, Z., Zhao, J.-X., and Chen, G.: Woeseia oceani gen. nov., sp. nov., a novel chemoheterotrophic member of the order Chromatiales, and proposal of Woeseiaceae fam. nov, Int. J. Syst. Evol. Micr., 66, 107–112, https://doi.org/10.1099/ijsem.0.000683, 2016.
Durbin, A. M. and Teske, A.: Microbial diversity and stratification of South Pacific abyssal marine sediments, Environ. Microbiol., 13, 3219–3234, https://doi.org/10.1111/j.1462-2920.2011.02544.x, 2011.
Fernandes, A. D., Reid, J. N. S., Macklaim, J. M., McMurrough, T. A., Edgell, D. R., and Gloor, G. B.: Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis, Microbiome, 2, 15, https://doi.org/10.1186/2049-2618-2-15, 2014.
Galac, M. R., Bosch, I., and Janies, D. A.: Bacterial communities of oceanic sea star (Asteroidea: Echinodermata) larvae, Mar. Biol., 163, 162, https://doi.org/10.1007/s00227-016-2938-3, 2016.
Gerpe, D., Buján, N., Diéguez, A. L., Lasa, A., and Romalde, J. L.: Kiloniella majae sp. nov., isolated from spider crab (Maja brachydactyla) and pullet carpet shell clam (Venerupis pullastra)?, Syst. Appl. Microbiol., 40, 274–279, https://doi.org/10.1016/j.syapm.2017.05.002, 2017.
Gobet, A., Boetius, A., and Ramette, A.: Ecological coherence of diversity patterns derived from classical fingerprinting and Next Generation Sequencing techniques, Environ. Microbiol., 16, 2672–2681, https://doi.org/10.1111/1462-2920.12308, 2014.
Gooday, A. J., Goineau, A., and Voltski, I.: Abyssal foraminifera attached to polymetallic nodules from the eastern Clarion Clipperton Fracture Zone: a preliminary description and comparison with North Atlantic dropstone assemblages, Mar. Biodivers., 45, 391–412, https://doi.org/10.1007/s12526-014-0301-9, 2015.
Haeckel, M., Konig, I., Reich, V., Weber, M. E., and Suess, E.: Pore water profiles and numerical modelling of biogeochemical processes in Peru Basin deep-sea sediments, Deep-Sea Res Pt. II, 48, 3713–3736, doi.org/10.1016/S0967-0645(01)00064-9, 2001.
Halbach, P., Friedrich, G., and von Stackelberg, U.: The manganese nodule belt of the Pacific Ocean, Enke, Stuttgart, p. 254, 1988.
Hassenrück, C., Quast, C., Rapp, J., and Buttigieg, P.: Amplicon, GitHub repository, available at: https://github.com/chassenr/NGS/tree/master/AMPLICON (last access: 15 April 2019), 2016.
Hoffmann, K., Bienhold, C., Buttigieg, P. L., Knittel, K., Laso-Pérez, R., Rapp, J. Z., Boetius, A., and Offre, P.: Diversity and metabolism of Woeseiales bacteria, global members of deep-sea sediment communities, ISME J., 14, 1042–1056, https://doi.org/10.1038/s41396-020-0588-4, 2020.
Hori, S., Tsuchiya, M., Nishi, S., Arai, W., and Takami, H.: Active Bacterial Flora Surrounding Foraminifera (Xenophyophorea) Living on the Deep-Sea Floor, Biosci. Biotech. Bioch., 77, 381–384, https://doi.org/10.1271/bbb.120663, 2013.
Hsieh, T. C., Ma, K. H., and Chao, A.: iNEXT: iNterpolation and EXTrapolation for species diversity, R package version 2.0.20, available at: http://chao.stat.nthu.edu.tw/wordpress/software_download (last access: 19 June 2020), 2018.
Jacob, M., Soltwedel, T., Boetius, A., and Ramette, A.: Biogeography of Deep-Sea Benthic Bacteria at Regional Scale (LTER HAUSGARTEN, Fram Strait, Arctic), PLoS One, 8, e72779, https://doi.org/10.1371/journal.pone.0072779, 2013.
Jacobson Meyers, M. E., Sylvan, J. B., and Edwards, K. J.: Extracellular enzyme activity and microbial diversity measured on seafloor exposed basalts from Loihi seamount indicate the importance of basalts to global biogeochemical cycling, Appl. Environ. Microb., 80, 4854–4864, https://doi.org/10.1128/AEM.01038-14, 2014.
Jørgensen, B. B. and Boetius, A.: Feast and famine–microbial life in the deep-sea bed, Nat. Rev. Microbiol., 5, 770–781, https://doi.org/10.1038/nrmicro1745, 2007.
Kato, S., Ikehata, K., Shibuya, T., Urabe, T., Ohkuma, M., and Yamagishi, A.: Potential for biogeochemical cycling of sulfur, iron and carbon within massive sulfide deposits below the seafloor, Environ. Microbiol., 17, 1817–1835, https://doi.org/10.1111/1462-2920.12648, 2015.
Kerr, R. A.: Manganese Nodules Grow by Rain from Above, Science, 223, 576–577, https://doi.org/10.1126/science.223.4636.576, 1984.
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., and Glöckner, F. O.: Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies, Nucleic Acids Res., 41, e1–e1, https://doi.org/10.1093/nar/gks808, 2013.
Kuhn, T., Wegorzewski, A., Rühlemann, C., and Vink, A.: Composition, Formation, and Occurrence of Polymetallic Nodules, in: Deep-Sea Mining: Resource Potential, Technical and Environmental Considerations, edited by: Sharma, R., 23–63, Springer International Publishing, Cham., 2017.
Larsen, E. I., Sly, L. I., and McEwan, A. G.: Manganese (II) adsorption and oxidation by whole cells and a membrane fraction of Pedomicrobium sp. ACM 3067, Arch. Microbiol., 171, 257–264, 1999.
Lee, M. D., Walworth, N. G., Sylvan, J. B., Edwards, K. J., and Orcutt, B. N.: Microbial Communities on Seafloor Basalts at Dorado Outcrop Reflect Level of Alteration and Highlight Global Lithic Clades, Front. Microbiol., 6, 1470, https://doi.org/10.3389/fmicb.2015.01470, 2015.
Lindh, M. V., Maillot, B. M., Shulse, C. N., Gooday, A. J., Amon, D. J., Smith, C. R., and Church, M. J.: From the surface to the deep-sea: Bacterial distributions across polymetallic nodule fields in the clarion-clipperton zone of the Pacific Ocean, Front. Microbiol., 8, 1–12, https://doi.org/10.3389/fmicb.2017.01696, 2017.
Luecker, S., Nowka, B., Rattei, T., Spieck, E., and Daims, H.: The Genome of Nitrospina gracilis Illuminates the Metabolism and Evolution of the Major Marine Nitrite Oxidizer, Front. Microbiol., 4, 27, https://doi.org/10.3389/fmicb.2013.00027, 2013.
Mahé, F., Rognes, T., Quince, C., de Vargas, C., and Dunthorn, M.: Swarm: robust and fast clustering method for amplicon-based studies, PeerJ, 2, e593, https://doi.org/10.7717/peerj.593, 2014.
Martens-Habbena, W., Berube, P., Urakawa, H., De la Torre, J., and Stahl, D.: Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria, Nature, 461, 976–979, https://doi.org/10.1038/nature08465, 2009.
Martin, M.: Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet.journal, 17, 1, https://doi.org/10.14806/ej.17.1.200, 2011.
Mason, O. U., Stingl, U., Wilhelm, L. J., Moeseneder, M. M., Di Meo-Savoie, C. A., Fisk, M. R., and Giovannoni, S. J.: The phylogeny of endolithic microbes associated with marine basalts, Environ. Microbiol., 9, 2539–2550, https://doi.org/10.1111/j.1462-2920.2007.01372.x, 2007.
Mason, O. U., Di Meo-Savoie, C. A., Van Nostrand, J. D., Zhou, J., Fisk, M. R., and Giovannoni, S. J.: Prokaryotic diversity, distribution, and insights into their role in biogeochemical cycling in marine basalts, ISME J., 3, 231–242, https://doi.org/10.1038/ismej.2008.92, 2009.
Matsunaga, T.: Applications of bacterial magnets Mognet, Trends Biotechnol., 9, 91–95, 1991.
Max Planck Institute for Marine Microbiology: Bacterial and archaeal community structure associated to manganese nodules at Peru Basin, ENA, available at: https://www.ebi.ac.uk/ena/data/view/PRJEB32680, last access: 19 June 2020.
Mewes, K., Mogollón, J. M., Picard, A., Rühlemann, C., Kuhn, T., Nöthen, K., and Kasten, S.: Impact of depositional and biogeochemical processes on small scale variations in nodule abundance in the Clarion-Clipperton Fracture Zone, Deep-Sea Res. Pt. I, 91, 125–141, https://doi.org/10.1016/j.dsr.2014.06.001, 2014.
Miller, K. A., Thompson, K. F., Johnston, P., and Santillo, D.: An Overview of Seabed Mining Including the Current State of Development, Environmental Impacts, and Knowledge Gaps, Front. Mar. Sci., 4, 418, https://doi.org/10.3389/fmars.2017.00418, 2018.
Müller, J., Hartmann, M. P., and Suess, E.: The chemical environment of pelagic sediments, in: The manganese nodule belt of the Pacific ocean: geological, environment, nodule formation, and mining aspects, edited by: Halbach, P., Friedrich, G., von Stackelberg, U., Enke, Stuttgart, 70–90, 1988.
Murray, J. and Renard, A. F.: Deep-sea deposits (based on the specimens collected during the voyage of HMS Challenger in the years 1872 to 1876). Report on the scientific results of the voyage of H.M.S. Challenger during the years 1873–76, John Menzies and Co., Endinburgh, UK, hdl:10013/epic.45942.d002, 1891.
Offre, P., Spang, A., and Schleper, C.: Archaea in biogeochemical cycles, Annu. Rev. Microbiol., 67, 437–457, https://doi.org/10.1146/annurev-micro-092412-155614, 2013.
Oksanen J., Blanchet, J. F., Friendly, M., Kindt, R., Legendre P., McGlinn, D., Minchin, P. R., O'Hara, R. B., Simpson, G. L., Solymos, P., Stevens, M. H. H., Szoecs, E., and Wagner, H.: vegan: Community Ecology Package, R package version 2.5-6, available at: https://cran.rstudio.com/web/packages/vegan/index.html (last access: 19 June 2020), 2015.
Peukert, A., Schoening, T., Alevizos, E., Köser, K., Kwasnitschka, T., and Greinert, J.: Understanding Mn-nodule distribution and evaluation of related deep-sea mining impacts using AUV-based hydroacoustic and optical data, Biogeosciences, 15, 2525–2549, https://doi.org/10.5194/bg-15-2525-2018, 2018.
Pruesse, E., Peplies, J., and Glöckner, F. O.: SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes, Bioinformatics, 28, 1823–1829, https://doi.org/10.1093/bioinformatics/bts252, 2012.
Riemann, F.: Biological aspects of deep-sea manganese nodule formation, Oceanol. Acta, 6, 303–311, 1983.
Ruff, S. E., Biddle, J. F., Teske, A. P., Knittel, K., Boetius, A., and Ramette, A.: Global dispersion and local diversification of the methane seep microbiome, P. Natl. Acad. Sci. USA, 112, 4015–4020, https://doi.org/10.1073/pnas.1421865112, 2015.
Santelli, C. M., Orcutt, B. N., Banning, E., Bach, W., Moyer, C. L., Sogin, M. L., Staudigel, H., and Edwards, K. J.: Abundance and diversity of microbial life in ocean crust, Nature, 453, 5–9, https://doi.org/10.1038/nature06899, 2008.
Schauer, R., Bienhold, C., Ramette, A., and Harder, J.: Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean, ISME J., 4, 159–170, https://doi.org/10.1038/ismej.2009.106, 2010.
Schüler, D. and Frankel, R.: Bacterial magnetosomes: Microbiology, biomineralization and biotechnological applications, Appl. Microbiol. Biot., 52, 464–473, https://doi.org/10.1007/s002530051547, 1999.
Scott, J. J., Glazer, B. T., and Emerson, D.: Bringing microbial diversity into focus: high-resolution analysis of iron mats from the Lō'ihi Seamount, Environ. Microbiol., 19, 301–316, https://doi.org/10.1111/1462-2920.13607, 2017.
Shulse, C. N., Maillot, B., Smith, C. R., and Church, M. J.: Polymetallic nodules, sediments, and deep waters in the equatorial North Pacific exhibit highly diverse and distinct bacterial, archaeal, and microeukaryotic communities, Microbiologyopen, 6, 1–16, https://doi.org/10.1002/mbo3.428, 2017.
Si, O., Yang, H., Hwang, C. Y., Kim, S., Choi, S., Kim, J., Jung, M., Kim, S., Roh, S. W., and Rhee, S.: Kiloniella antarctica sp. nov., isolated from a polynya of Amundsen Sea in Western Antarctic Sea, Int. J. Syst. Evol. Micr., 67, 2397–2402, https://doi.org/10.1099/ijsem.0.001968, 2017.
Simon-Lledo, E., Bett, B. J., Huvenne, V. A. I., Köser, K., Schoening, T., Greinert, J., and Jones, D. O. B.: Biological effects 26 years after simulated deep-sea mining, Sci. Rep.-UK, 9, 8040, https://doi.org/10.1038/s41598-019-44492-w, 2019.
Staudigel, H., Furnes, H., McLoughlin, N., Banerjee, N. R., Connell, L. B., and Templeton, A.: 3.5 billion years of glass bioalteration: Volcanic rocks as a basis for microbial life?, Earth-Sci. Rev., 89, 156–176, https://doi.org/10.1016/j.earscirev.2008.04.005, 2008.
Stein, L. Y., La Duc, M. T., Grundl, T. J., and Nealson, K. H.: Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediments, Environ. Microbiol., 3, 10–18, 2001.
Stone, A. T.: Microbial metabolites and the reductive dissolution of manganese oxides: oxalate and pyruvate, Geochim. Cosmochim. Ac., 51, 919–925, 1987.
Stone, A. T. and Morgan, J. J.: Reduction and dissolution of Manganese(III) and manganese(IV) oxides by organics: 2. Survey of the reactivity of organics, Environ. Sci. Technol., 18, 617–624, 1984.
Sunda, W. G. and Huntsman, S. A.: Photoreduction of manganise oxides in seawater, Mar. Chem., 46, 133–152, 1994.
Sunda, W. G. and Kieber, D. J.: Oxidation of humic substrates by manganese oxides yields low-molecular-weight organic substrates, Nature, 367, 62–64, 1994.
Sunda, W. G., Huntsman, S. A., and Harvey, G. R.: Photoreduction of manganese oxides in seawater and its geochemical and biological implications, Nature, 301, 234–236, 1983.
Sylvan, J. B., Toner, B. M., and Edwards, K. J.: Life and Death of Deep-Sea Vents: Bacterial Diversity and Ecosystem Succession on Inactive Hydrothermal Sulfides, MBio, 3, e00279-11, https://doi.org/10.1128/mBio.00279-11, 2012.
Thiel, H., Schriever, G., Ahnert, A., Bluhm, H., Borowski, C., and Vopel, K.: The large-scale environmental impact experiment DISCOL – Reflection and foresight, Deep-Sea Res. Pt. II, 48, 3869–3882, https://doi.org/10.1016/S0967-0645(01)00071-6, 2001.
Tian, R.-M., Sun, J., Cai, L., Zhang, W.-P., Zhou, G., Qui, J.-W., and Qian, P.-Y.: The deep-sea glass sponge Lophophysema eversa harbours potential symbionts responsible for the nutrient conversions of carbon, nitrogen and sulfur, Environ. Microbiol., 18, 2481–2494, https://doi.org/10.1111/1462-2920.12911, 2016.
Tully, B. J. and Heidelberg, J. F.: Microbial communities associated with ferromanganese nodules and the surrounding sediments, Front. Microbiol., 4, 1–10, https://doi.org/10.3389/fmicb.2013.00161, 2013.
Tyler, P. A.: Hyphomicrobia and the oxidation of manganesse in aquatic ecosystems Mg/, Antonie von Leeuwenhoek, 36, 567–578, 1970.
Van den Boogaart, K. G., Tolosana, R., and Bren, M.: compositions: Compositional Data Analysis, R package version 1.40-5, available at: http://www.stat.boogaart.de/compositions/ (last access: 19 June 2020), 2014.
Vanreusel, A., Hilario, A., Ribeiro, P. A., Menot, L., and Arbizu, P. M.: Threatened by mining, polymetallic nodules are required to preserve abyssal epifauna, Sci. Rep.-UK, 6, 26808, https://doi.org/10.1038/srep26808, 2016.
Varliero, G., Bienhold, C., Schmid, F., Boetius, A., and Molari, M.: Microbial Diversity and Connectivity in Deep-Sea Sediments of the South Atlantic Polar Front, Front. Microbiol., 10, 1–18, https://doi.org/10.3389/fmicb.2019.00665, 2019.
Volz, J. B., Mogollón, J. M., Geibert, W., Arbizu, P. M., Koschinsky, A., and Kasten, S.: Natural spatial variability of depositional conditions, biogeochemical processes and element fluxes in sediments of the eastern Clarion-Clipperton Zone, Pacific Ocean, Deep-Sea Res. Pt., 140, 159–172, https://doi.org/10.1016/j.dsr.2018.08.006, 2018.
Vonnahme, T. R., Molari, M., Janssen, F., Wenzhöfer, F., Haeckel, M., Titschack, J., and Boetius, A.: Effects of a deep-sea mining experiment on seafloor microbial communities and functions after 26 years, Science Advances, in press, 2020.
Von Stackelberg, U.: Growth history of manganese nodules and crusts of the Peru Basin, Geol. Soc. Lond. Spec. Publ., 119, 153–176, https://doi.org/10.1144/GSL.SP.1997.119.01.11, 1997.
Walsh, E. A., Kirkpatrick, J. B., Rutherford, S. D., Smith, D. C., Sogin, M., and Hondt, S. D.: Bacterial diversity and community composition from seasurface to subseafloor, ISME J., 10, 979–989, https://doi.org/10.1038/ismej.2015.175, 2016.
Wang, C., Liao, L., Xu, H., Xu, X., Wu, M., and Zhu, L.: Bacterial Diversity in the Sediment from Polymetallic Nodule Fields of the Clarion-Clipperton Fracture Zone, J. Microbiol., 48, 573–585, https://doi.org/10.1007/s12275-010-0151-5, 2010.
Wang, L., Li, X., Lai, Q., and Shao, Z.: Kiloniella litopenaei sp. nov., isolated from the gut microflora of Pacific white shrimp, Litopenaeus vannamei, Antonie Van Leeuwenhoek, 108, 1293–1299, https://doi.org/10.1007/s10482-015-0581-5, 2015.
Wang, L., Yu, M., Liu, Y., Liu, J., Wu, Y., Li, L., Liu, J., Wang, M., and Zhang, X.-H.: Comparative analyses of the bacterial community of hydrothermal deposits and seafloor sediments across Okinawa Trough, J. Mar. Syst., 180, 162–172, https://doi.org/10.1016/j.jmarsys.2016.11.012, 2018.
Wang, X. H., Gan, L., and Müller, W. E. G.: Contribution of biomineralization during growth of polymetallic nodules and ferromanganese crusts from the Pacific Ocean, Front. Mater. Sci. China, 3, 109–123, https://doi.org/10.1007/s11706-009-0033-0, 2009.
Wang, Y. and Qian, P.-Y.: Conservative Fragments in Bacterial 16S rRNA Genes and Primer Design for 16S Ribosomal DNA Amplicons in Metagenomic Studies, PLoS One, 4, e7401, https://doi.org/10.1371/journal.pone.0007401, 2009.
Wegorzewski, A. V. and Kuhn, T.: The influence of suboxic diagenesis on the formation of manganese nodules in the Clarion Clipperton nodule belt of the Pacific Ocean, Mar. Geol., 357, 123–138, https://doi.org/10.1016/j.margeo.2014.07.004, 2014.
Wegorzewski, A. V., Kuhn, T., Dohrmann, R., Wirth, R., and Grangeon, S.: Mineralogical characterization of individual growth structures of Mn-nodules with different Ni+Cu content from the central Pacific Ocean, Am. Mineral., 100, 2497–2508, https://doi.org/10.2138/am-2015-5122, 2015.
Wiese, J., Thiel, V., Ga, A., Schmaljohann, R., and Imhoff, J. F.: alphaproteobacterium from the marine macroalga Laminaria saccharina, Int. J. Syst. Evol. Micr., 59, 350–356, https://doi.org/10.1099/ijs.0.001651-0, 2009.
Wu, Y. H., Liao, L., Wang, C. S., Ma, W. L., Meng, F. X., Wu, M., and Xu, X. W.: A comparison of microbial communities in deep-sea polymetallic nodules and the surrounding sediments in the Pacific Ocean, Deep-Sea Res. Pt. I, 79, 40–49, https://doi.org/10.1016/j.dsr.2013.05.004, 2013.
Xu, M., Wang, F., Meng, J., and Xiao, X.: Construction and preliminary analysis of a metagenomic library from a deep-sea sediment of east Pacific Nodule Province, FEMS Microbiol. Ecol., 62, 233–241, https://doi.org/10.1111/j.1574-6941.2007.00377.x, 2007.
Yang, S., Seo, H., Lee, J., Kim, S., and Kwon, K. K.: Kiloniella spongiae sp. nov., isolated from a marine sponge and emended description of the genus Kiloniella Wiese et al. 2009 and Kiloniella laminariae, Int. J. Syst. Evol. Micr., 65, 230–234, https://doi.org/10.1099/ijs.0.069773-0, 2015.
Zhang, G., He, J., Liu, F., and Zhang, L.: Iron-Manganese Nodules Harbor Lower Bacterial Diversity and Greater Proportions of Proteobacteria Compared to Bulk Soils in Four Locations Spanning from North to South China, Geomicrobiol. J., 31, 562–577, https://doi.org/10.1080/01490451.2013.854428, 2014.
Zhang, J., Kobert, K., Flouri, T., and Stamatakis, A.: PEAR: a fast and accurate Illumina Paired-End reAd mergeR, Bioinformatics, 30, 614–620, https://doi.org/10.1093/bioinformatics/btt593, 2014.