the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
On giant shoulders: how a seamount affects the microbial community composition of seawater and sponges
Kathrin Busch
Ulrike Hanz
Furu Mienis
Benjamin Mueller
Andre Franke
Emyr Martyn Roberts
Hans Tore Rapp
Seamounts represent ideal systems to study the influence and interdependency of environmental gradients at a single geographic location. These topographic features represent a prominent habitat for various forms of life, including microbiota and macrobiota, spanning benthic as well as pelagic organisms. While it is known that seamounts are globally abundant structures, it still remains unclear how and to which extent the complexity of the sea floor is intertwined with the local oceanographic mosaic, biogeochemistry, and microbiology of a seamount ecosystem. Along these lines, the present study aimed to explore whether and to what extent seamounts can have an imprint on the microbial community composition of seawater and of sessile benthic invertebrates, sponges. For our high-resolution sampling approach of microbial diversity (16S rRNA gene amplicon sequencing) along with measurements of inorganic nutrients and other biogeochemical parameters, we focused on the Schulz Bank seamount ecosystem, a sponge ground ecosystem which is located on the Arctic Mid-Ocean Ridge. Seawater samples were collected at two sampling depths (mid-water, MW, and near-bed water, BW) from a total of 19 sampling sites. With a clustering approach we defined microbial microhabitats within the pelagic realm at Schulz Bank, which were mapped onto the seamount's topography and related to various environmental parameters (such as suspended particulate matter, SPM; dissolved inorganic carbon, DIC; silicate, ; phosphate, ; ammonia, ; nitrate, ; nitrite, ; depth; and dissolved oxygen, O2). The results of our study reveal a “seamount effect” (sensu stricto) on the microbial mid-water pelagic community at least 200 m above the sea floor. Further, we observed a strong spatial heterogeneity in the pelagic microbial landscape across the seamount, with planktonic microbial communities reflecting oscillatory and circulatory water movements, as well as processes of bentho-pelagic coupling. Depth, , , and O2 concentrations differed significantly between the determined pelagic microbial clusters close to the sea floor (BW), suggesting that these parameters were presumably linked to changes in microbial community structures. Secondly, we assessed the associated microbial community compositions of three sponge species along a depth gradient of the seamount. While sponge-associated microbial communities were found to be mainly species-specific, we also detected significant intra-specific differences between individuals, depending on the pelagic near-bed cluster they originated from. The variable microbial phyla (i.e. phyla which showed significant differences across varying depth, , , O2 concentrations, and different from local seawater communities) were distinct for every sponge species when considering average abundances per species. Variable microbial phyla included representatives of both those taxa traditionally counted for the variable community fraction and taxa counted traditionally for the core community fraction. Microbial co-occurrence patterns for the three examined sponge species Geodia hentscheli, Lissodendoryx complicata, and Schaudinnia rosea were distinct from each other. Over all, this study shows that topographic structures such as the Schulz Bank seamount can have an imprint (seamount effect sensu lato) on both the microbial community composition of seawater and sessile benthic invertebrates such as sponges by an interplay between the geology, physical oceanography, biogeochemistry, and microbiology of seamounts.
- Article
(6477 KB) - Full-text XML
-
Supplement
(910 KB) - BibTeX
- EndNote
Seamounts and mid-ocean ridges are prominent geologic features that add to the complexity of the sea floor. In the traditional sense, seamounts are defined as isolated submarine volcanic features with a minimum height of 1000 m from base to summit (Menard, 1964; Wessel et al., 2010). However, geologic features with a height of 50–100 m may also be considered seamounts (Staudigel et al., 2010; Smith and Cann, 1992; Wessel et al., 2010). There may be up to 100 000 to >25 million seamounts present in the oceans (IUCN, 2013), although the error rate associated with these estimations is high (IUCN, 2013). Despite the lack of accurate numbers, there is no doubt that with an estimated 10 million km2 coverage, the area occupied by these habitats is globally significant. Elevated topographic features in the open ocean are often hotspots of biological diversity and productivity (IUCN, 2013; Morato et al., 2010). It appears that interaction of the topography with the hydrography creates a combination of amplified tidal flow, increased current speed, and the formation of internal waves, which strongly enhances vertical mixing around seamounts (Lavelle and Mohn, 2010; Van Haren et al., 2017; Roberts et al., 2018). Consequential upwelling of nutrient-rich deep waters stimulates primary productivity in this layer of enhanced mixing (IUCN, 2013). In addition to vertical mixing processes, horizontal fluxes of organic matter may also be affected by the presence of seamounts, as they may promote enclosed or semi-enclosed oceanographic circulation patterns, like Taylor caps or columns (Chapman and Haidvogel, 1992; Roberts et al., 2018, and references therein), leading to a retention of organic and inorganic matter.
The above-mentioned processes make seamounts important habitats for pelagic as well as benthic species (Morato et al., 2010; Rogers, 2018) due to beneficial prevailing conditions. Particularly areas with strong water flows (evoked by interactions of currents and tides with elevated topography), in combination with a steep and irregular hard substrate, represent suitable habitats for benthic suspension feeders, which indeed densely populate most seamounts (Genin et al., 1986; IUCN, 2013). Sponges (Porifera; Grant, 1836) often dominate these suspension feeder communities and are increasingly recognized as key components of shallow and deep marine ecosystems (DeGoeij et al., 2017; Maldonado et al., 2016). Due to their high filtering capacity and association with diverse microbial communities, sponges are considered to substantially influence the carbon, nitrogen, and silicate cycling in marine systems (Taylor et al., 2007; Maldonado et al., 2012, 2019; De Goeij et al., 2013; Rix et al., 2016a) and to contribute to bentho-pelagic coupling by actively removing particulate organic matter (POM) from the water column (Pile et al., 1997; Reiswig, 1971; Ribes et al., 1999). In addition to their influence on particulate organic matter pools, many sponges have been identified to primarily feed on dissolved organic matter (DOM) (De Goeij et al., 2008; Mueller et al., 2014; Hoer et al., 2018; Gantt et al., 2019). Energy and nutrients stored in this DOM are then transferred into particulate detritus, which fuels benthic food webs (De Goeij et al., 2013; Rix et al., 2016b).
Intimate sponge–microbe associations have been observed throughout diverse habitats, reaching from coastal shallow sites in tropical and temperate regions to the deep sea and polar seas (Helber et al., 2019; Kennedy et al., 2014; Moitinho-Silva et al., 2014; Naim et al., 2014; Schmitt et al., 2012; Steinert et al., 2019; Thomas et al., 2016). According to their microbiome, sponges can be classified to either feature high microbial abundance (HMA) or low microbial abundance (LMA) (Hentschel et al., 2003; Moitinho-Silva et al., 2017; Weisz et al., 2008). The dichotomy between HMA and LMA sponges is considered a main driver of the microbial community structure associated with shallow-water sponges (Moitinho-Silva et al., 2017). In comparison to shallow waters, few studies have been conducted on the microbiology of deep-sea sponges (Borchert et al., 2017; Jackson et al., 2013; Kennedy et al., 2014; Reveillaud et al., 2014; Steinert et al., 2020; Tian et al., 2016). However, for example for deep-sea sponges of the genus Geodia (G. barretti, G. macandrewii, G. phlegraei, G. atlantica), similar microbial phyla have been observed as in HMA shallow-water sponges, such as Acidobacteria, Poribacteria, and Chloroflexi (Luter et al., 2017; Radax et al., 2012; Schöttner et al., 2013). In addition to the HMA–LMA dichotomy, an important factor in structuring the microbiomes of shallow-water sponges is host taxonomy, which is manifested in ubiquitous species-specific sponge microbiomes (Easson and Thacker, 2014; Steinert et al., 2017; Thomas et al., 2016). Systematic analyses of the influence of biogeochemical parameters (particularly dissolved inorganic substances) on sponge-associated microbial diversity and interactivity are still lacking, particularly in deep-sea sponges. Seamounts provide an ideal study system in this regard, as they offer the potential of examining steep environmental gradients over small spatial scales. Sponge ground ecosystems are areas harbouring high densities of structure-forming sponge individuals. The Arctic Schulz Bank seamount has been observed to host a rich and diverse community of sponges (Roberts et al., 2018; Meyer et al., 2019) and may be considered a sponge ground ecosystem harbouring a reservoir of yet unexamined microbial biodiversity.
The present study aimed to characterize the microbial community composition of seawater surrounding the Schulz Bank seamount ecosystem, located on the Arctic Mid-Ocean Ridge. Seawater samples were collected at two sampling depths from a total of 19 sampling sites and the corresponding microbiome data were mapped onto the topography of the Schulz Bank seamount ecosystem. Secondly, we assessed the associated microbial community compositions of three sponge species along a depth gradient of the seamount. Diversity metrics, as well as changes in the abundance of individual microbial taxa, were related with a set of biogeochemical parameters. This study explores whether topographic structures such as the Schulz Bank seamount can have an imprint on the microbial community composition both of seawater and of sessile benthic invertebrates, sponges.
2.1 Description of the Schulz Bank seamount
Schulz Bank is located on the Arctic Mid-Ocean Ridge (73.8∘ N, 7.5∘ E) between the Greenland and Norwegian seas (Fig. 1). It is exposed to three main water masses: (i) the Norwegian Deep Water (NwDW) that is present at the base and flanks of the seamount; (ii) the intermediate water mass (NwArIW), which is most likely Norwegian Arctic Intermediate Water and occurs at the summit and shallower areas; and (iii) the warmer surface water mass (NwAtW) which is Norwegian Atlantic Water. Notably near-bed water masses at Schulz Bank's summit have unusually low temperatures of around 0 to −1 ∘C (Roberts et al., 2018). Estimations of the seamount's basal dimensions state conservative values of 10 km × 4 km to 15 km × 6 km (Roberts et al., 2018), but it may however also be larger as Schulz Bank belongs to a ridge system. The summit of the seamount is located at around 600 m below the water surface, and the base depth is at more than 2500 m below the water surface. In a two-dimensional view, Schulz Bank has a broadly elliptical shape (Roberts et al., 2018). Bathymetry data presented in this study were derived from the Bathymetry Data Portal of the European Marine Observation and Data Network (EMODnet), and spatial analyses were performed in QGIS (version 3.4.4) as well as ArcGIS (version 10.6).
2.2 Sampling procedures
Three cruises were undertaken on board RV G. O. Sars (campaign names: GS2016109A, GS2017110, and GS2018108) during Northern Hemisphere summer in the years 2016–2018. Seawater samples were collected with a rosette water sampler equipped with 12×10 L Niskin bottles combined with a conductivity–temperature–depth (CTD) sensor system (SBE 9, Sea-Bird Electronics Inc., Washington, USA). In total, 19 CTD stations were covered, carried out along transects aligned with the seamounts' minor and major axes, and also with the 73.8∘ N line of latitude. At each of the 19 stations, seawater samples for microbial analyses and biogeochemical parameters were collected at two water depths during the CTD upcast (Fig. 3c and PANGAEA for metadata): (i) 400 m below the seawater surface (mid-water) and (ii) correspondingly at 10 m above the sea floor (near-bed water). Naturally, the near-bed depths varied along with seamount topography and ranged from 575 to 2966 m. Sponges were sampled between 580 and 2184 m water depth, along the CTD transects (and therefore in close spatial vicinity to the seawater samples), by a remotely operated vehicle (ROV Ægir 6000, University of Bergen). A total of 36 sponge individuals representing the most abundant species were randomly selected from a larger collection effort. This subset included 16 Geodia hentscheli (Cárdenas et al., 2010) (Demospongiae), 8 Lissodendoryx complicata (Hansen, 1885) (Demospongiae), and 12 Schaudinnia rosea (Fristedt, 1887) (Hexactinellida). The sponges were taxonomically identified by visual inspection on board the ship. In addition, whole specimens and additional sponge samples were fixed in 99 % EtOH for deposition in the collections of the University of Bergen.
2.3 Biogeochemical analyses and measurements of environmental parameters
The following nine environmental parameters were analysed: depth, suspended particulate matter (SPM), dissolved inorganic carbon (DIC), silicate (Si), phosphate (), ammonia (), nitrate (), nitrite (), and dissolved oxygen (O2). Depth and dissolved O2 data were recorded during in situ water column profiling. Depth (pressure) was recorded with the CTD sensor system mentioned above. O2 concentrations were derived from a dissolved oxygen sensor (SBE 43, Sea-Bird Electronics Inc., Washington, USA) that was attached to the rosette water sampler. For the analysis of suspended particulate matter (SPM), 2×10 L of water were filtered over pre-weighed combusted GF/F filters, which were rinsed with demineralized water to remove salts (47 mm Whatman™ GF/F filters pre-combusted at 450 ∘C, stored at −20 ∘C). Filters were freeze-dried and weighed before further analysis. For the analysis of inorganic nutrients (ammonia, ; phosphate, ; nitrate, ; nitrite, ; and silicate, Si), seawater samples were filtered over 0.2 µm filters. Water samples for , , and NOx analysis were stored at −20 ∘C and for Si analyses at 4 ∘C. Nutrients were measured with a QuAAtro gas segmented continuous-flow analyser (Seal Analytical, Norderstedt, Germany). Measurements were made simultaneously on four channels for (Murphy and Riley, 1962), (Helder and de Vries, 1979), and combined with (Grasshoff et al., 2009) and separately for Si (Strickland and Parsons, 1972). A freshly diluted mixed-nutrient standard containing Si, , and was added to each run. The cocktail served as a guide to monitor the performance of the standards. All measurements were calibrated with standards diluted in low-nutrient seawater (LNSW). For the analysis of dissolved inorganic carbon (DIC), seawater samples were transferred into a glass vial containing 15 µL HgCl2 (mercury chloride) and analysed on a TechniconTraacs800 auto-analyser (Technicon Instruments Corporation, Tarrytown, USA) following the methodology of Stoll et al. (2001). Analyses of variance (ANOVAs) were performed to test for statistical differences in the biogeochemical and physical parameters between the determined microbial near-bed water clusters (see below). As numbers of samples per mid-water cluster and per near-bed water cluster were unequal, we calculated Type III sums of squares for ANOVAs (unbalanced ANOVAs). We further calculated Spearman's rank correlations between depth and those biogeochemical parameters which turned out to differ significantly across the determined near-bed water clusters in the ANOVA analyses (see below).
2.4 Amplicon sequencing
Seawater samples were collected in triplicates from different Niskin bottles, yielding a total of 114 samples from all stations (i.e. 19 stations × 2 sampling depths × 3 biological replicates per station and depth = 114 seawater samples in total). A total of 2 L of seawater sample was filtered onto polyvinylidene fluoride (PVDF) filter membranes (Merck Millipore) with a pore size of 0.22 µm and a diameter of 47 mm and stored at −80 ∘C. For sponge collection, cubes of approximately 1 cm3 were cut from the mesohyl with a scalpel, rinsed (sterile seawater), flash-frozen in liquid nitrogen, and stored at −80 ∘C. DNA was extracted from half a seawater filter or ∼0.25 g of sponge tissue by using the DNeasy PowerSoil Kit (Qiagen, Venlo, the Netherlands). The quality of the DNA extraction was assessed based on the 260∕280 ratio using a NanoDrop spectrophotometer as well as by polymerase chain reaction with universal 16S primers and subsequent gel electrophoresis. The V3–V4 variable regions of the 16S rRNA gene were then amplified in a one-step polymerase chain reaction (PCR) using the primer pair 341F–806R (dual-barcoding approach; Kozich et al., 2013; primer sequences: 5'-CCTACGGGAGGCAGCAG-3' and 5'-GGACTACHVGGGTWTCTAAT-3'). After verification of the presence of PCR products by gel electrophoresis, normalization (SequalPrep normalization plate kit; Thermo Fisher Scientific, Waltham, USA) and equimolar pooling were performed. Sequencing was conducted on the MiSeq platform (MiSeq FGx; Illumina, San Diego, USA) with v3 chemistry. The settings for demultiplexing were 0 mismatches in the barcode sequences.
2.5 Bioinformatic analyses
For computation of microbial core-diversity metrics, sequences were processed within the QIIME2 environment (version 2018.11; Bolyen et al., 2019). Amplicon sequence variants (ASVs) were generated from forward reads (truncated to 270 nt) with the DADA2 algorithm (Callahan et al., 2016). Phylogenetic trees were calculated based on resulting ASVs with the FastTree2 plugin. Representative ASVs were classified using the Silva 132 99 % OTUs 16S database (Quast et al., 2013) with the help of a primer-specific trained naïve Bayes taxonomic classifier. Alpha and beta diversity indices (e.g. Faith's phylogenetic diversity and weighted UniFrac distances, respectively) were calculated within QIIME2. To evaluate sample separation in ordination space, non-metric multidimensional scaling (nMDS) was performed on weighted UniFrac distances for seawater and sponge-associated microbiomes separately.
A machine learning approach was used to define microbial microhabitats within the pelagic realm. Seawater microbiomes were clustered based on weighted UniFrac distances. The NbClust function was applied in R (version 3.0.2; R Development Core Team, 2008) to generate 30 indices to identify the best number of clusters based on the majority rule. A coordinate grid was set up as a basis for a georeferenced extrapolation of sampling points. In detail, we used the sampling coordinates and the determined cluster affiliation of our 114 in situ measured seawater samples as the input for our predictions of seawater microbial cluster affiliation in space. Clustering regions were set up with the help of the k-nearest-neighbour algorithm. The machine learning approach was fine-tuned in several ways: (i) the algorithm was trained in a way that in situ measured data points always belong to the cluster actually determined based on the sequencing data; (ii) a normalization was applied with the help of a distance-weighted function meaning that closer data points have a higher weight; (iii) the probability of class membership was calculated and plotted as indication of confidence. Permutational multivariate analyses of variance (PERMANOVAs) were performed with 999 permutations to determine whether microbiomes of selected clusters were statistically significantly different from each other. In detail, pairwise tests across the determined clusters were conducted for the following samples separately: mid-water samples, near-bed water samples, G. hentscheli, L. complicata, and S. rosea. A significance level of α=0.05 was applied for all statistical analyses in this study.
To evaluate co-occurrence patterns between microbial taxa across environmental gradients (i.e. determined near-bed water clusters), networks were constructed separately for every sponge species and seawater. Mean relative abundances of microbial phyla were calculated for all biological replicates of each sample type and for the corresponding near-bed water cluster. Microbial phyla, which showed significantly different enrichment between clusters, were determined and ranked using the linear discriminant analysis effect size (LEfSe) algorithm (Segata et al., 2011). A correlation matrix was established for those taxa that differed significantly between clusters, to assess co-occurrences. In particular, the direction and strength of correlations were characterized for any significant phylum with all other significant taxa (as well as the relations with depth).
3.1 Structure and composition of seawater microbial communities
A nMDS plot on weighted UniFrac distances separated the microbial communities of mid-water and near-bed water samples in ordination space with few exceptions (Fig. 2). Cluster analysis based on weighted UniFrac distances revealed two distinct clusters in the mid-water samples, of which one (MW1) was located precisely above the summit of Schulz Bank, while the other (MW2) covered the wider seamount region and vicinity (Fig. 3a). Four distinct microbiome clusters were detected in the near-bed water samples (BW1–4). In terms of similarity, cluster BW1 was most distinct from all other clusters while clusters BW2 and BW3 were most similar to each other (Fig. 3b). Moreover, BW1 cluster samples separated in ordination space in that they grouped with mid-water rather than near-bed water samples (i.e. consider the few black dots grouping together with the white dots in Fig. 2). Plotting the clusters on a spatial map revealed that near-bed water cluster BW1 was located near the summit of Schulz Bank seamount (average depth =575 m), while clusters BW2 and BW3 covered its flanks (average depth ± SE and 922±142 m, respectively), and cluster BW4 represented the vicinity close to the seamount (average depth ± SE m) (Fig. 3b). Statistical testing of the individual depth data points contributing to a given cluster revealed a significant difference in the depth parameter between the clusters (ANOVA, p=0.01, df1=3; df2=10).
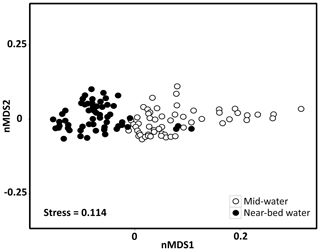
Figure 2Seawater microbial community composition of mid-water and near-bed water samples visualized by a non-metric multidimensional scaling plot on weighted UniFrac distances. Each marker is one microbial community, with colours indicating the sample sub-type (i.e. mid-water or near-bed water).
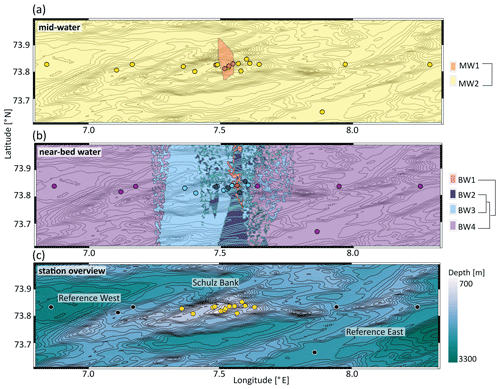
Figure 3Seawater microbial community structure across Schulz Bank. Contour lines in all three subplots represent the underlying topography. Colours in panels (a) and (b) represent clusters based on weighted UniFrac distances, where coloured dots indicate stations with in situ sampling and filled areas represent extrapolations based on machine learning. The further away predicted areas are from actual sample points, the higher the associated uncertainty of these predictions is. Panel (a) includes all mid-water samples derived during the CTD transects. Panel (b) includes all near-bed water samples. Here, the degree of cluster similarity can be deduced from the dendrogram to the right of the plot(s). Panel (c) provides an overview of the sampling area, showing the locations of all 19 CTD stations. Stations directly located on the Schulz Bank are coloured yellow, while reference stations (west and east of Schulz Bank) are indicated in black. Colouring in panel (c) was done according to depth.
Figure 3c shows the bathymetry highlighting the contour lines of Schulz Bank seamount and its vicinity (reference west and east) as well as the 19 sampling stations. As our samples originated from three different cruises, we also double-checked the extent of temporal variability in our dataset. We observe frequently that seawater samples of different cruises belong to the same cluster (Fig. S1a in the Supplement). From this we conclude that temporal variations are not the main driver of our observed similarities or dissimilarities in pelagic microbial community compositions.
Microbial richness was overall slightly lower in the mid-water samples (mean Faith's phylogenetic diversity ± standard error ) than in the near-bed water samples (54.4±0.9) (Fig. S2a). Near-bed water samples from the summit (BW1) displayed a slightly lower microbial richness than the other near-bed water samples (Fig. S2b). The mid-water samples collected above Schulz summit also showed a slightly lower microbial richness than the other mid-water samples. Pairwise comparisons (PERMANOVA) revealed that the seawater microbial community clusters within the mid-water and near-bed water samples were significantly different from each other in terms of their microbial community composition (Table S1). Furthermore, the pool of mid-water samples (MW1–MW2) was significantly different from the pool of near-bed water samples (BW1–BW4). Overall, the eight most dominant seawater microbial phyla, sorted in descending order of mean relative abundance, were Proteobacteria (54 % of total community), Bacteroidetes (17 %), Verrucomicrobia (7 %), Marinimicrobia (SAR406 clade) (6 %), Actinobacteria (5 %), Chloroflexi (4 %), Acidobacteria (2 %), and Planctomycetes (1 %).
3.2 Seawater biogeochemistry at Schulz Bank seamount
When comparing the biogeochemical parameters of the mid-water clusters, only dissolved O2 concentrations differed significantly (ANOVA, p=0.02, df1=1; df2=17) with slightly higher concentrations in MW1 (6.90±0.04 mL L−1) compared to MW2 (6.75±0.03 mL L−1) (Table S2). All other tested biogeochemical parameters SPM (2.22±1.39 mg L−1), DIC (2269.07±26.63 µmol L−1), (5.66±0.06 µmol L−1), (0.86±0.01 µmol L−1), (0.11±0.03 µmol L−1), (12.99±0.08 µmol L−1), and (0.02±0.01 µmol L−1) were not statistically different between MW1 and MW2. The values for mid-water samples are reported as average ± standard error.
Of the eight biogeochemical parameters tested, the following three differed significantly between the near-bed water clusters. These were (ANOVA, p=0.04, df1=3; df2=10), (ANOVA, p=0.03, df1=3; df2=10), and dissolved O2 (ANOVA, p=0.01, df1=3; df2=15). Nitrate (range =13.00–14.78 µmol L−1) and (range =6.00–10.65 µmol L−1) increased with depth, with the lowest concentrations at the summit (BW1), intermediate concentrations at the flanks (BW2, BW3), and highest concentrations in the seamount vicinity (BW4) (Fig. 4). Dissolved oxygen (range =6.48–6.99 mL L−1) showed the reverse pattern in that its concentration was highest at the summit (BW1), intermediate at the flanks (BW2, BW3), and lowest in the seamount vicinity sites (BW4). Spearman's rank correlations calculated between depth and the three other significant parameters indeed revealed significant correlations in all cases (: rho =0.77, p<0.01; : rho =0.85, p<0.01; dissolved O2: rho , p<0.01) (Fig. S3). The other biogeochemical parameters SPM (range =0.49–1.87 mg L−1), DIC (range =2248.00–2265.67 µmol L−1), (range =0.90–0.97 µmol L−1), (range =0.10–0.17 µmol L−1), and (range =0–0.05 µmol L−1) were not significantly different between the near-bed water clusters. At the summit, no pronounced differences in biogeochemical parameters were observed between the near-bed water (BW1) and mid-water samples (MW1) (Table S2).
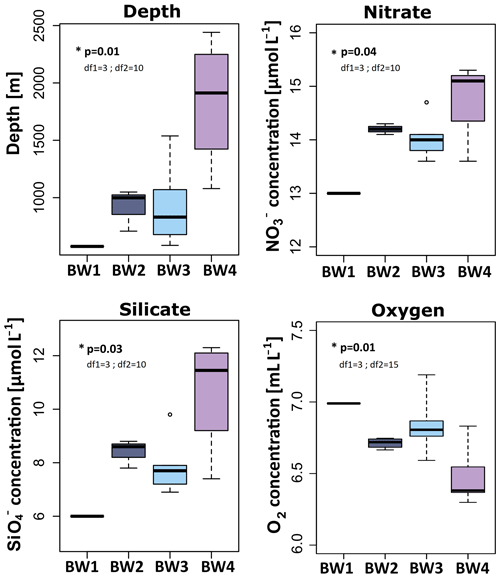
Figure 4Concentrations and measurements of significant (ANOVA, α=0.05) biogeochemical parameters for near-bed water samples, across the determined near-bed water clusters. The p values as well as degrees of freedom (df1 and df2) for these parameters are written into the respective graphs. Colouring is the same as for Figs. 2 and 3.
3.3 Structure and composition of sponge microbial communities
In order to analyse structure and microbial community composition of the sponges, we randomly selected at least four biological replicates per sponge species per BW cluster for statistical analysis. As sponges were sampled in the close vicinity but not exactly at the same locations as seawater samples, BW cluster affiliations of the sponge sampling locations were determined based on spatial extrapolations of the seawater data as described before (see Fig. S1b for exact sponge sampling locations). Overall, the three deep-sea sponge species S. rosea, G. hentscheli, and L. complicata showed host-species-specific microbiomes, as indicated by a clear separation of their microbial communities in ordination space (Fig. 5). Sub-structuring based on near-bed water clusters in the non-metric multidimensional scaling plot as well as pairwise comparisons (PERMANOVA) revealed that the sponge microbial communities within each species differed significantly depending on the near-bed water clusters from which they were collected (Table S1). The only exception was S. rosea, for which specimens from the flank (BW3) showed a microbial community composition that was intermediate between the summit (BW1) and the other flank cluster (BW2).
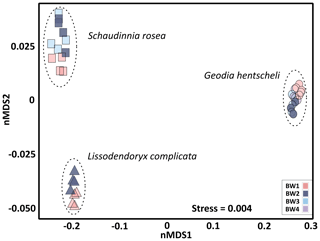
Figure 5Sponge microbial community composition visualized by a non-metric multidimensional scaling plot on weighted UniFrac distances. Each marker is one microbial community, with symbols representing the sample sub-type (i.e. sponge species) and colours indicating the near-bed water cluster present at the respective sponge sampling location.
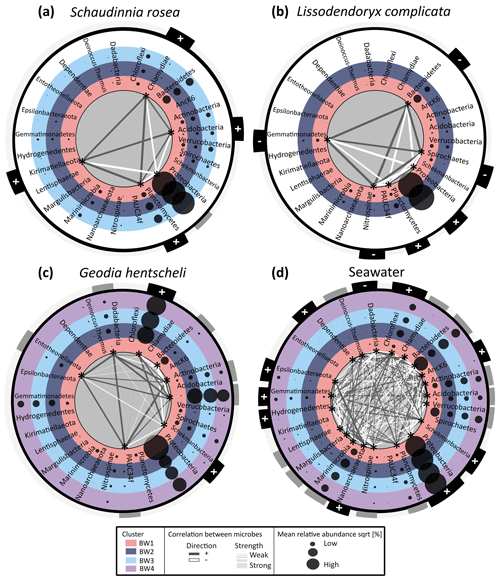
Figure 6Co-occurrence network and differential abundance of microbial phyla across the four determined near-bed water clusters. Panels (a)–(c) show sponge data, with panel (a) showing average Schaudinnia rosea data, panel (b) presenting average Lissodendoryx complicata data, and panel (c) illustrating Geodia hentscheli data. Panel (d) contains seawater data. Near-bed water clusters are represented by differently coloured rings. Each ring contains a list with microbial phyla which are alphabetically sorted. Average relative abundances of each of the respective phyla for the samples within a given cluster are indicated by bubble sizes. Those microbial phyla which are statistically significantly enriched or depleted across the four clusters (LEfSe analysis) are marked with an asterisk inside the inner most ring. Only for those taxa where the difference is significant are correlation strength (indicated by size of connecting lines) and direction (represented by colour of connecting lines: white is negative correlation; dark grey is positive correlation) with all other significant taxa plotted. For all microbial phyla relation with depth is indicated in the outer ring of each plot by + (meaning significant positive relation) or − (meaning significant negative relation).
The dominant microbial phylum in S. rosea and L. complicata was Proteobacteria (Fig. 6a and b), whereas G. hentscheli microbiomes were dominated by Chloroflexi, Acidobacteria, and Proteobacteria (Fig. 6c). Sponge microbiomes were more stable than seawater communities with fewer phyla exhibiting significant differences across the four near-bed water clusters or positively relating with depth (Fig. 6). For the hexactinellid S. rosea, the relative abundances of five bacterial phyla (Acidobacteria, Chlamydiae, Kirimatiellaota, Planctomyces, and Proteobacteria) were significantly different between individuals that were sampled from different near-bed water clusters. Out of these five phyla, Acidobacteria, Chlamydiae, Kirimatiellaota, and Planctomyces were positively related with depth while for Proteobacteria neither a positive nor negative relation with depth was discernable. Consequently, Proteobacteria showed a negative relation with the four other phyla in the network analysis. For the demosponge L. complicata, the relative abundances of Bacteroidetes, Gemmatimonadetes, Nitrospinae, Planctomyces, Proteobacteria, and Spirochaetes were significantly different between sponge individuals that were sampled from the different near-bed water clusters. For this sponge species, samples were only available from near-bed water clusters 1 and 2. Of the six phyla, Planctomyces and Proteobacteria were positively related with depth, while the other four were negatively related with depth, which is also reflected in the network analysis. For the demosponge G. hentscheli, the relative abundances of eight phyla (Acidobacteria, Actinobacteria, Bacteoidetes, Chloroflexi, Dadabacteria, Entotheonellota, PAUC34f, and Schekmanbacteria) were significantly different between sponge individuals sampled from the near-bed water clusters BW1–BW4. Of those, Chloroflexi and Schekmanbacteria were positively related with depth, while the others showed variable patterns over depth. The network analysis showed both positive and negative correlations between taxa for those increasing with depth as well as those displaying a variable response.
When analysing host-associated microbiomes, ambient seawater microbiomes are valuable references for comparison. In this study, a total of 21 microbial clades were identified in ambient seawater, whose relative abundances varied significantly between the four near-bed water clusters. A total of nine taxa showed a positive relation with depth, one (Dadabacteria) showed a negative relation with depth, and the remaining 11 taxa showed a variable response to depth. For seawater more phyla varied between near-bed water clusters than for the sponge samples. Overall, more microbial taxa showed significant positive relations with depth, , and and negative relations with O2 than vice versa. The microbial taxa showing a significant difference in relative abundance between the near-bed water clusters (as determined by LEfSe) were different between sponges and seawater and also between sponge species. Further, significant differences in relative abundances were observed for both abundant and less abundant sponge symbiont lineages.
Research records about seamount microbiology are sparse and comparably few studies have been conducted on deep-sea sponge microbiomes in general (Borchert et al., 2017; Jackson et al., 2013; Kennedy et al., 2014; Reveillaud et al., 2014). Our main aim was to assess whether and via which potential mechanisms a seamount can affect the community structure of pelagic and benthos (sponge)-associated microbial communities, using the Schulz Bank seamount as an exemplary field site. A total of 19 CTD sampling stations, each with two sampling depths, on and around Schulz Bank were analysed towards this goal and combined with sponge-associated microbial data gained during additional ROV dives.
4.1 A seamount imprint on seawater microbial communities
In this study we observed a pronounced similarity between the microbial community composition of the mid-water cluster located precisely above Schulz Bank's summit (MW1) and the microbial community composition of the near-bed water cluster at the summit (BW1). This is evident in Fig. 2, where few black dots representing the BW1 cluster group with mid-water samples rather than near-bed water samples. In addition, the microbial community in the mid-water cluster above Schulz Bank's summit (MW1) was distinct from the community in the mid-water cluster covering the wider seamount region and vicinity (MW2), despite similar prevailing biogeochemical conditions in both mid-water clusters (MW1 vs. MW2; an exception is the significant difference in O2 concentrations between both clusters). From these two observations we conclude that the presence of a seamount can have an imprint on the microbial community structure in the overlying water column (seamount effect sensu stricto). In particular we suspect that topography-induced vertical mixing processes occur at Schulz Bank seamount, which reallocate microbial communities within the water column and in turn influence the pelagic microbial diversity as far as at least 200 m above the seamount's summit. In support of these interpretations, oscillating currents relating to the barotropic and baroclinic (internal) tide have been reported previously at the summit of the Schulz Bank seamount (Roberts et al., 2018) and other seamounts (Van Haren et al., 2017). Although these mentioned studies suggest that respective oceanographic processes are particularly pronounced at the summit part of seamounts, we cannot exclude the following two aspects based on our own study: (i) since we only sampled one mid-water depth, vertical mixing of microbial communities could in theory extend far higher than 200 m above the seamount's summit. (ii) As the vertical distances between the mid-water and the near-bed water samples were larger at other parts of the seamount compared to the summit, vertical mixing of microbial communities may in fact not only be restricted to the summit of Schulz Bank.
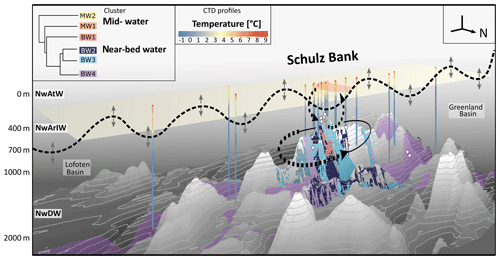
Figure 7Conceptual overview and vertical 3D section showing spatial distribution of microbial clusters and oceanographic patterns on the Schulz Bank seamount. Extrapolated seawater microbial clusters are indicated by coloured polygons: mid-water clusters are marked in orange (MW1) and yellow (MW2), while near-bed water clusters are marked in red (BW1), dark blue (BW2), light blue (BW3), and purple (BW4). The degree of cluster similarity can be deduced from the dendrogram in the left corner of the plot. Whole water column CTD profiles are indicated, showing the measured temperature values from surface to bottom at the respective sampling locations. Sponge sampling locations visible on this side of the seamount are indicated by white balls. Vertical positions of the major water masses Norwegian Atlantic Water (NwAtW), Norwegian Arctic Intermediate Water (NwArIW), and Norwegian Deep Water (NwDW) are indicated. To give a broad orientation in space, a north arrow is depicted, and the major geologic features (Lofoten Basin and Greenland Basin). For Schulz Bank, water flows, such as a potential Taylor column circulating around the seamount, mixing between summit and pelagic realm, as well as tidally driven internal motions (black horizontal line with bidirectional arrows), are indicated by dashed arrows and lines.
In addition to tide-induced vertical hydrodynamic processes, horizontal flow patterns can also help to explain the presence of seamount-specific microbial communities. Roberts et al. (2018) calculated that a Taylor cap or Taylor column may be (temporarily) present at Schulz Bank. This oceanographic phenomenon describes an isolated anticyclonic flow circulation pattern over a seamount and hence may promote temporary spatial isolation of a seamount ecosystem from adjacent waters. Our conceptual schematic overview of hydrodynamics at Schulz Bank seamount is shown in Fig. 7 (dashed lines). In this figure, a digital elevation model of Schulz Bank seamount is depicted in combination with the overlying water column structure and oceanographic context. Temperature profiles derived from whole water column sensing by CTD casts are plotted. Based on these profiles the vertical distributions of the surface water (NwAtW), intermediate water (NwArIW), and Norwegian Deep Water (NwDW) were deduced in combination with the identified water masses as described in Roberts et al. (2018) (Fig. 7). In addition to oceanographic insights, Fig. 7 includes a 3D visualization of our determined microbiome clusters at and around Schulz Bank seamount. This figure recapitulates that similarities and dissimilarities in microbial signatures of seawater in this study (cluster dendrogram Fig. 7) were consistent with the oscillatory water movements (i.e. due to internal tide-induced mixing) and possible circulatory flows (Taylor column) as predicted by Roberts et al. (2018).
Microbial richness was overall slightly lower at the summit of the seamount (BW1) than at the deeper locations (i.e. BW2–BW4). A similar trend was observed for the mid-water samples, where microbial richness was slightly lower for the microbial community above Schulz's summit in comparison to samples in its vicinity (MW1 vs. MW2). On a macroscopic level, Morato et al. (2010) and others have described seamounts as hotspots of pelagic biodiversity; much less is however known about microbial diversity at seamounts. Our results of a lower microbial richness above the seamount summit might seem contradictory at first. However, Schulz Bank is a recognized sponge ground ecosystem, with a peak in sponge density and diversity at the seamount summit (Roberts et al., 2018; Meyer et al., 2019). Sponges are very efficient suspension feeders and are known for removing large amounts of particulate organic matter including prokaryotes and small eukaryotes from the water column (Leys et al., 2018). Benthic–pelagic coupling mediated by selective feeding of sponges on seawater microorganisms (McMurray et al., 2016; Van Oevelen et al., 2018) in combination with the discussed hydrodynamic patterns (vertical mixing) might explain the slightly reduced microbial richness of the water body residing directly above a sponge ground (BW1 and MW1). Sponge density and community composition changes along the topography of Schulz Bank seamount in that density is highest at the summit (Meyer et al., 2019; Roberts et al., 2018). This natural variation can further explain the observed differences in microbial community composition between the other near-bed water samples (BW2–4). For these samples, we observed distinct pelagic microbial communities at a finer resolution than can be explained by the pure water mass distribution (consider depth of intermediate (NwArIW) and deep water (NwDW) layers in Fig. 7). In particular the near-bed clusters BW2 and BW3 originated from a similar depth range and both were located at the seamount's flanks. Besides ecologically rooted explanations (i.e. variable presence of benthic organisms that influence biogeochemical cycles), hydrodynamic processes (i.e. local flow direction linked to small-scale topography and/or spatial orientation of the seamount's flanks) can also explain the observation of distinct microbial community compositions within the near-bed water. We hence conclude that besides patterns related to water masses, we observe a much higher spatial heterogeneity of pelagic microbial communities than previously recognized. We call this kind of imprint on the pelagic microbial community composition, which is probably based on the topography combined with bentho-pelagic coupling processes and hydrodynamics, a “seamount effect sensu lato”. These observations suggest that the presence of a seamount can have profound impacts on the distribution of microbial landscapes in the open ocean.
Seamounts are recognized as unique habitats in terms of ecosystem dynamics (Genin and Boehlert, 1985) and macroecology (Morato et al., 2010). The present study reveals that seamounts also have a unique microbial signature that extends hundreds of metres up into the overlying water column. In addition, we detected three distinct microbial clusters in seawater samples taken near the seabed directly above Schulz Bank which were distinctly different from those of seawater collected in the vicinity of the seamount (near-bed water clusters BW1–3 vs. BW4). , , and dissolved O2 concentrations differed significantly between the four near-bed water clusters. The observation that the seamount is intersecting with different biogeochemical properties and microbial communities is particularly interesting in regard to benthic organisms. Along these lines, the Schulz Bank seamount provides a platform for sponges and their associated microbial communities to respond to topography-enabled environmental gradients (also a seamount effect sensu lato).
4.2 A seamount imprint on sponge-associated microbial communities
The investigated sponge species S. rosea, G. hentscheli, and L. complicata were selected for this study as they represent key taxa of the sponge community at Schulz Bank. Microbiomes of these three species clustered clearly apart from each other in ordination space, indicating a dominant host species effect on the associated microbial community structure. S. rosea and L. complicata showed characteristic microbial signatures of LMA sponges (as defined in Moitinho-Silva et al., 2017) that are being dominated by Proteobacteria. On the contrary, G. hentscheli displayed a microbial signature characteristic of HMA sponges with dominant clades such as Chloroflexi and Acidobacteria, which is consistent with previous reports on sponges of the genus Geodia (Luter et al., 2017; Radax et al., 2012; Schöttner et al., 2013).
When analysing each sponge species separately, sponge specimen microbiomes differed significantly between each other and depended on the near-bed water clusters to which they belonged. This finding suggests that an environmental signature is also detectable in sponge-associated microbial communities (seamount signature sensu lato). This observation is striking, as sponge-associated microbial communities are considered to be highly stable associations (Cárdenas et al., 2014; Erwin et al., 2012, 2015; Pita et al., 2013; Steinert et al., 2016).
Previous studies have shown that abiotic factors (i.e. depth, geographical location) influence the microbial community structure in shallow-water sponges but stated that the core community is shaped by the intimate interaction with the sponge host (Lurgi et al., 2019). Interestingly, in our study the major microbial players in terms of abundance, such as Chloroflexi in G. hentscheli, show significant enrichment–depletion patterns across the four clusters. Traditionally (shallow water) sponge-associated microbes have been classified into core, variable, and species-specific communities (Schmitt et al., 2012). The present study reveals that for the three investigated deep-sea sponges at Schulz Bank seamount the variable community overlaps with the core community when considering high taxonomy ranks. In addition, network analyses showed both positive and negative correlations between taxa for those increasing with depth as well as those displaying a variable response. We suspect that primary responders to environmental parameters have cascading effects on microbial lineages that are not directly affected by water biogeochemistry.
In this study, silicate, oxygen, and nitrate concentrations, as well as depth, differed significantly between the four near-bed water clusters. While and were positively correlated with depth, O2 showed a negative relationship. An increase in nutrient concentrations with depth is consistent with our previous expectations (consider, e.g. Bristow et al., 2017) and can be explained by remineralization processes of sinking marine snow within the deep water layers. Decreasing O2 concentrations from the intermediate water (NwArIW) to the deep water (NwDW) are also consistent with our expectation based on physical oceanography, as water layers more recently oxygenated at the ocean surface at their site of formation typically carry more oxygen (Jeansson et al., 2017). In general, the absolute differences in the concentrations of all three significant environmental parameters were comparably small (especially and O2). However, as microbial communities were significantly different between the clusters, we posit that the observed biogeochemical differences, albeit small, should be considered drivers of sponge microbial community composition. In support of our hypothesis, the process of denitrification is for example known to be highly sensitive to nanomolar concentrations of O2 concentrations (Dalsgaard et al., 2013). In addition, a previous study on the sponge Xestospongia muta demonstrated that changing NOx concentrations over depth contribute to shaping the microbial community composition (Morrow et al., 2016). Furthermore, several studies have noted that depth is an important factor in structuring sponge-associated microbiomes (Indraningrat et al., 2019; Lesser et al., 2020; Steinert et al., 2016).
This is, to our knowledge, the first study that explores the impact of seawater biogeochemistry on deep-sea sponge microbiomes. We have used LEfSe analyses to identify sponge symbiont taxa whose relative abundance varies with depth, this being used as a proxy for selected biogeochemical parameters. We have further used co-occurrence networks to identify positively or negatively co-varying microbial clades. In the following we discuss one representative example of a sponge-associated (and seawater-associated) microbial phylum which was significantly enriched–depleted across the near-bed water clusters: Chloroflexi in G. hentscheli. Chloroflexi (among several other phyla) relative abundance differed significantly between the four near-bed water clusters for G. hentscheli and seawater, showing a positive relation with , , and depth and a negative relation with O2. Members of the phylum Chloroflexi have been attributed to a relevant role in the degradation of organic matter, particularly in the deep-ocean pelagic realm and within HMA sponges (Bayer et al., 2018; Landry et al., 2017). High degradation rates of organic matter are often related to low O2 and high nutrient concentrations, owing to biogeochemical feedbacks where nutrients enhance oxygen demand by increasing biological production and oxygen consumption during decomposition. Taken together, the differences in relative abundances of Chloroflexi in G. hentscheli and seawater could be driven by the , , and O2 concentrations in ambient seawater. While sponge microbiomes are generally considered to be highly stable in time and space, we provide the first evidence that small differences in water biogeochemistry may affect sponge microbiome composition. However, no uniform shifts in relative abundances of microbial taxa were observed for G. hentscheli, L. complicata, and S. rosea but rather an individual response of each host species related to biogeochemical parameters. One explanation is that biological interactions between the sponge host and its microbiome, or between the microbes themselves, might have masking effects.
We provide insights into the variability of pelagic and benthic (sponge-associated) microbiomes at the Arctic Schulz Bank seamount based on the microbiome analyses of 114 seawater and 36 sponge samples. Interestingly, a seamount signature is detected within the microbial community composition of samples originating as far as 200 m above the seamount summit. Further, our correlation-based results suggest that the biogeochemistry of seawater which varies over depth (, , and O2 concentrations) has a detectable but variable influence on the composition of sponge-associated microbiomes. This study provides new perspectives on the influence of seamounts on the microbial diversity in their vicinity. We conclude that the geology, physical oceanography, biogeochemistry, and microbiology of seamounts and similar structures are even more closely linked than currently appreciated.
Sample metadata and biogeochemical data were deposited in the PANGAEA database: https://doi.org/10.1594/PANGAEA.911304 (Busch et al., 2020). Raw sequences were archived in the NCBI Sequence Read Archive under BioProject ID PRJNA600711.
The supplement related to this article is available online at: https://doi.org/10.5194/bg-17-3471-2020-supplement.
KB, FM, BM, and UHe designed the study. KB, UHa, FM, EMR, and HTR participated in sampling. UHa, FM, and KB focused on biogeochemical parameters. HTR conducted the sponge taxonomic analysis. UHe and KB were responsible for the microbial pipeline. AF was involved in sequencing. KB performed the data analysis (bioinformatics and visualizations). KB and UHe wrote the manuscript. BM, FM, EMR, UHa, and HTR reviewed and edited the manuscript.
The authors declare that they have no conflict of interest.
This document reflects only the authors' view and the Executive Agency for Small and Medium-sized Enterprises (EASME) is not responsible for any use that may be made of the information it contains.
This work is dedicated to Hans Tore Rapp, sponge taxonomist, deep-sea explorer, colleague, and friend. Samples included in this study were collected in compliance with the Nagoya Protocol. We thank the crews and scientific parties of RV G. O. Sars cruises GS2016109A, GS2017110, and GS2018108 for great technical support while at sea. We are grateful for sponge sampling by Stig Vågenes and his ROV Ægir 6000 team (UiB), Christine Rooks (UiB) for sampling assistance on board the ship during GS2016109A, and Jasper de Goeij (UvA) for interesting discussions. We further acknowledge Ina Clefsen, Andrea Hethke, Ilona Urbach, and Tonio Hauptmann (Kiel, Germany) for excellent laboratory support with the amplicon pipeline. Corinna Bang (IKMB, Kiel) provided valuable support with the sample sequencing and revised the manuscript before submission. We also thank the two anonymous reviewers for their comments, which helped to improve this paper.
This research has been supported by the European Union's Horizon 2020 Research
and Innovation Programme under grant agreement no. 679849 (the SponGES project).
The article processing charges for this open-access
publication were covered by a Research
Centre of the Helmholtz Association.
This paper was edited by Tina Treude and reviewed by two anonymous referees.
Bayer, K., Jahn, M. T., Slaby, B. M., Moitinho-Silva, L., and Hentschel, U.: Marine sponges as chloroflexi hot spots: genomic insights and high-resolution visualization of an abundant and diverse symbiotic clade, mSystems, 3, 1–19, https://doi.org/10.1128/msystems.00150-18, 2018.
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., Alexander, H., Alm, E. J., Arumugam, M., Asnicar, F., Bai, Y., Bisanz, J. E., Bittinger, K., Brejnrod, A., Brislawn, C. J., Brown, C. T., Callahan, B. J., Caraballo-Rodríguez, A. M., Chase, J., Cope, E. K., Da Silva, R., Diener, C., Dorrestein, P. C., Douglas, G. M., Durall, D. M., Duvallet, C., Edwardson, C. F., Ernst, M., Estaki, M., Fouquier, J., Gauglitz, J. M., Gibbons, S. M., Gibson, D. L., Gonzalez, A., Gorlick, K., Guo, J., Hillmann, B., Holmes, S., Holste, H., Huttenhower, C., Huttley, G. A., Janssen, S., Jarmusch, A. K., Jiang, L., Kaehler, B. D., Kang, K. B., Keefe, C. R., Keim, P., Kelley, S. T., Knights, D., Koester, I., Kosciolek, T., Kreps, J., Langille, M. G. I., Lee, J., Ley, R., Liu, Y.-X., Loftfield, E., Lozupone, C., Maher, M., Marotz, C., Martin, B. D., McDonald, D., McIver, L. J., Melnik, A. V., Metcalf, J. L., Morgan, S. C., Morton, J. T., Naimey, A. T., Navas-Molina, J. A., Nothias, L. F., Orchanian, S. B., Pearson, T., Peoples, S. L., Petras, D., Preuss, M. L., Pruesse, E., Rasmussen, L. B., Rivers, A., Robeson II, M. S., Rosenthal, P., Segata, N., Shaffer, M., Shiffer, A., Sinha, R., Song, S. J., Spear, J. R., Swafford, A. D., Thompson, L. R., Torres, P. J., Trinh, P., Tripathi, A., Turnbaugh, P. J., Ul-Hasan, S., vanderHooft, J. J. J., Vargas, F., Vázquez-Baeza, Y., Vogtmann, E., von Hippel, M., Walters, W., Wan, Y., Wang, M., Warren, J., Weber, K. C., Williamson, C. H. D., Willis, A. D., Xu, Z. Z., Zaneveld, J. R., Zhang, Y., Zhu, Q., Knight, R., and Caporaso, J. G.: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 37, 852–857, https://doi.org/10.1038/s41587-019-0209-9, 2019.
Borchert, E., Selvin, J., Kiran, S. G., Jackson, S. A., O'Gara, F., and Dobson, A. D. W.: A novel cold active esterase from a deep sea sponge Stelletta normani metagenomic library, Front. Mar. Sci., 4, 1–13, https://doi.org/10.3389/fmars.2017.00287, 2017.
Bristow, L. A., Mohr, W., Ahmerkamp, S., and Kuypers, M. M. M.: Nutrients that limit growth in the ocean, Curr. Biol., 27, 474–478, https://doi.org/10.1016/j.cub.2017.03.030, 2017.
Busch, K., Hanz, U., Mienis, F., Müller, B., Franke, A., Roberts, E. M., Rapp, H. T., and Hentschel, U.: Microbial diversity and biogeochemical parameters at Schulz Bank seamount (Arctic Mid-Ocean Ridge) measured in summer 2016, 2017 and 2018, PANGAEA, https://doi.org/10.1594/PANGAEA.911304, 2020.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P.: DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 13, 581–583, https://doi.org/10.1038/nmeth.3869, 2016.
Cárdenas, C. A., Bell, J. J., Davy, S. K., Hoggard, M., and Taylor, M. W.: Influence of environmental variation on symbiotic bacterial communities of two temperate sponges, FEMS Microbiol. Ecol., 88, 516–527, https://doi.org/10.1111/1574-6941.12317, 2014.
Cárdenas, P., Rapp, H. T., Schander, C., and Tendal, O. S.: Molecular Taxonomy and Phylogeny of the Geodiidae Gray, 1867 (Porifera, Demospongiae, Astrophorida) – combining Phylogenetic and Linnaean Classification, Zool. Scr., 39, 89–106, 2010.
Chapman, D. C. and Haidvogel, D. B.: Formation of taylor caps over a tall isolated seamount in a stratified ocean, Geophys. Astro. Fluid, 64, 31–65, https://doi.org/10.1080/03091929208228084, 1992.
Dalsgaard, T., De Brabandere, L., and Hall, P. O. J.: Denitrification in the water column of the central Baltic Sea, Geochim. Cosmochim. Ac., 106, 247–260, https://doi.org/10.1016/j.gca.2012.12.038, 2013.
De Goeij, J. M., Van Den Berg, H., Van Oostveen, M. M., Epping, E. H. G., and Van Duyl, F. C.: Major bulk dissolved organic carbon (DOC) removal by encrusting coral reef cavity sponges, Mar. Ecol. Prog. Ser., 357, 139–151, https://doi.org/10.3354/meps07403, 2008.
De Goeij, J. M., Van Oevelen, D., Vermeij, M. J. A., Osinga, R., Middelburg, J. J., De Goeij, A. F. P. M., and Admiraal, W.: Surviving in a marine desert: The sponge loop retains resources within coral reefs, Science, 342, 108–110, https://doi.org/10.1126/science.1241981, 2013.
DeGoeij, J. M., Lesser, M. P., and Pawlik, J. R.: Nutrient Fluxes and Ecological Functions of Coral Reef Sponges in a Changing Ocean, in: Climate Change, Ocean Acidification and Sponges: Impacts Across Multiple Levels of Organization, edited by: Carballo, J. L. and Bell, J. J., Springer International Publishing, Cham, Switzerland, 373–410, 2017.
Easson, C. G. and Thacker, R. W.: Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges, Front. Microbiol., 5, 1–11, https://doi.org/10.3389/fmicb.2014.00532, 2014.
Erwin, P. M., Pita, L., López-Legentil, S., and Turon, X.: Stability of sponge-associated bacteria over large seasonal shifts in temperature and irradiance, Appl. Environ. Microbiol., 78, 7358–7368, https://doi.org/10.1128/AEM.02035-12, 2012.
Erwin, P. M., Coma, R., López-Sendino, P., Serrano, E., and Ribes, M.: Stable symbionts across the HMA-LMA dichotomy: Low seasonal and interannual variation in sponge-associated bacteria from taxonomically diverse hosts, FEMS Microbiol. Ecol., 91, 1–11, https://doi.org/10.1093/femsec/fiv115, 2015.
Fristedt, K.: Sponges from the Atlantic and Arctic Oceans and the Behring Sea. Vega-Expeditionens Vetenskap, Iakttagelser (Nordenskiöld), plates 22-31, 4, 401–471, 1887.
Gantt, S. E., McMurray, S. E., Stubler, A. D., Finelli, C. M., Pawlik, J. R., and Erwin, P. M.: Testing the relationship between microbiome composition and flux of carbon and nutrients in Caribbean coral reef sponges, Microbiome, 7, 1–13, https://doi.org/10.1186/s40168-019-0739-x, 2019.
Genin, A. and Boehlert, G. W.: Dynamics of temperature and chlorophyll structures above a seamount: an oceanic experiment, J. Mar. Res., 43, 907–924, https://doi.org/10.1357/002224085788453868, 1985.
Genin, A., Dayton, P. K., Lonsdale, P. F., and Spiess, F. N.: Corals on seamount peaks provide evidence of current acceleration over deep-sea topography, Nature, 322, 59–61, https://doi.org/10.1038/322059a0, 1986.
Grant, R. E.: Animal Kingdom, in: The Cyclopaedia of Anatomy and Physiology, edited by: Todd, R. B., Sherwood, Gilbert and Piper, London, UK, 107–118, 1836.
Grasshoff, K., Kremling, K., and Ehrhardt, M.: Methods of Seawater Analysis, 3rd edn., WILEY-VCH Verlag GmbH, Weinheim, Germany, 2009.
Hansen, G. A.: Spongidae. The Norwegian North-Atlantic Expedition 1876–1878, plates I–VII, Zoology, 13, 1–26, 1885.
Helber, S. B., Steinert, G., Wu, Y. C., Rohde, S., Hentschel, U., Muhando, C. A., and Schupp, P. J.: Sponges from Zanzibar host diverse prokaryotic communities with potential for natural product synthesis, FEMS Microbiol. Ecol., 95, fiz026, https://doi.org/10.1093/femsec/fiz026, 2019.
Helder, W. and de Vries, R. T. P.: An automatic phenol-hypochlorite method for the determination of ammonia in sea- and brackish waters, Neth. J. Sea Res., 13, 154–160, 1979.
Hentschel, U., Fieseler, L., Wehrl, M., Gernert, C., Steinert, M., Hacker, J., and Horn, M.: Microbial diversity of marine sponges, in: Sponges (Porifera), edited by: Mueller, W. E. G., Springer, Berlin, Heidelberg, Germany, 59–88, 2003.
Hoer, D. R., Gibson, P. J., Tommerdahl, J. P., Lindquist, N. L., and Martens, C. S.: Consumption of dissolved organic carbon by Caribbean reef sponges, Limnol. Oceanogr., 63, 337–351, https://doi.org/10.1002/lno.10634, 2018.
Indraningrat, A. A. G., Micheller, S., Runderkamp, M., Sauerland, I., Becking, L. E., Smidt, H., and Sipkema, D.: Cultivation of sponge-associated bacteria from Agelas sventres and Xestospongia muta collected from different depths, Mar. Drugs, 17, 1–17, https://doi.org/10.3390/md17100578, 2019.
IUCN: Seamounts Project: An Ecosystem Approach to Management of Seamounts in the Southern Indian Ocean, Int. Union Conserv. Nat. Nat. Resour., Gland, Switzerland, 1, 1–60, 2013.
Jackson, S. A., Flemer, B., McCann, A., Kennedy, J., Morrissey, J. P., O'Gara, F., and Dobson, A. D. W.: Archaea appear to dominate the microbiome of Inflatella pellicula deep sea sponges, PLoS One, 8, 1–8, https://doi.org/10.1371/journal.pone.0084438, 2013.
Jeansson, E., Olsen, A., and Jutterström, S.: Arctic Intermediate Water in the Nordic Seas, 1991–2009, Deep-Sea Res. Pt. I, 128, 82–97, https://doi.org/10.1016/j.dsr.2017.08.013, 2017.
Kennedy, J., Flemer, B., Jackson, S. A., Morrissey, J. P., O'Gara, F., and Dobson, A. D. W.: Evidence of a putative deep sea specific microbiome in marine sponges, PLoS One, 9, 1–13, https://doi.org/10.1371/journal.pone.0091092, 2014.
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D.: Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform, Appl. Environ. Microbiol., 79, 5112–5120, https://doi.org/10.1128/AEM.01043-13, 2013.
Landry, Z., Swa, B. K., Herndl, G. J., Stepanauskas, R., and Giovannoni, S. J.: SAR202 genomes from the dark ocean predict pathways for the oxidation of recalcitrant dissolved organic matter, MBio, 8, 1–19, https://doi.org/10.1128/mBio.00413-17, 2017.
Lavelle, J. W. and Mohn, C.: Motion, commotion, and biophysical connections at deep ocean seamounts, Oceanography, 23, 90–103, https://doi.org/10.5670/oceanog.2010.64, 2010.
Lesser, M. P., Mueller, B., Pankey, M. S., Macartney, K. J., Slattery, M., and de Goeij, J. M.: Depth-dependent detritus production in the sponge, Halisarca caerulea, Limnol. Oceanogr., 65, 1200–1216, 2020.
Leys, S. P., Kahn, A. S., Fang, J. K. H., Kutti, T., and Bannister, R. J.: Phagocytosis of microbial symbionts balances the carbon and nitrogen budget for the deep-water boreal sponge Geodia barretti, Limnol. Oceanogr., 63, 187–202, https://doi.org/10.1002/lno.10623, 2018.
Lurgi, M., Thomas, T., Wemheuer, B., Webster, N. S., and Montoya, J. M.: Modularity and predicted functions of the global sponge-microbiome network, Nat. Commun., 10, 1–12, https://doi.org/10.1038/s41467-019-08925-4, 2019.
Luter, H. M., Bannister, R. J., Whalan, S., Kutti, T., Pineda, M.-C., and Webster, N. S.: Microbiome analysis of a disease affecting the deep-sea sponge Geodia barretti, FEMS Microbiol. Ecol., 93, 1–6, https://doi.org/10.1093/femsec/fix074, 2017.
Maldonado, M., Ribes, M., and van Duyl, F. C.: Nutrient fluxes through sponges. Biology, budgets, and ecological implications, Adv. Mar. Biol., 62, 113–182, https://doi.org/10.1016/B978-0-12-394283-8.00003-5, 2012.
Maldonado, M., Aguilar, R., Bannister, R. J., Bell, J. J., Conway, K. W., Dayton, P. K., Díaz, C., Gutt, J., Kelly, M., Kenchington, E. L. R., Leys, S. P., Pomponi, S. A., Rapp, H. T., Rützler, K., Tendal, O. S., Vacelet, J., and Young, C. M.: Sponge grounds as key marine habitats: a synthetic review of types, structure, functional roles, and conservation concerns, in: Marine animal forests, edited by: Rossi, S., Bramanti, L., Gori, A., and Orejas, C., Springer International Publishing Switzerland, Cham, Switzerland, 2016.
Maldonado, M., López-Acosta, M., Sitjà, C., García-Puig, M., Galobart, C., Ercilla, G., and Leynaert, A.: Sponge skeletons as an important sink of silicon in the global oceans, Nat. Geosci., 12, 815–822, https://doi.org/10.1038/s41561-019-0430-7, 2019.
McMurray, S. E., Johnson, Z. I., Hunt, D. E., Pawlik, J. R., and Finelli, C. M.: Selective feeding by the giant barrel sponge enhances foraging efficiency, Limnol. Oceanogr., 61, 1271–1286, https://doi.org/10.1002/lno.10287, 2016.
Menard, H. W.: Marine Geology of the Pacific, McGraw-Hill, New York, USA, 271 pp., 1964.
Meyer, H. K., Roberts, E. M., Rapp, H. T., and Davies, A. J.: Spatial patterns of arctic sponge ground fauna and demersal fish are detectable in autonomous underwater vehicle (AUV) imagery, Deep-Sea Res. Pt. I, 153, 103137, https://doi.org/10.1016/j.dsr.2019.103137, 2019.
Moitinho-Silva, L., Bayer, K., Cannistraci, C. V., Giles, E. C., Ryu, T., Seridi, L., Ravasi, T., and Hentschel, U.: Specificity and transcriptional activity of microbiota associated with low and high microbial abundance sponges from the Red Sea, Mol. Ecol., 23, 1348–1363, https://doi.org/10.1111/mec.12365, 2014.
Moitinho-Silva, L., Steinert, G., Nielsen, S., Hardoim, C. C. P., Wu, Y. C., McCormack, G. P., López-Legentil, S., Marchant, R., Webster, N., Thomas, T., and Hentschel, U.: Predicting the HMA-LMA status in marine sponges by machine learning, Front. Microbiol., 8, 1–14, https://doi.org/10.3389/fmicb.2017.00752, 2017.
Morato, T., Hoyle, S. D., Allain, V., and Nicol, S. J.: Seamounts are hotspots of pelagic biodiversity in the open ocean, P. Natl. Acad. Sci. USA, 107, 9707–9711, https://doi.org/10.1073/pnas.0910290107, 2010.
Morrow, K. M., Fiore, C. L., and Lesser, M. P.: Environmental drivers of microbial community shifts in the giant barrel sponge, Xestospongia muta, over a shallow to mesophotic depth gradient, Environ. Microbiol., 18, 2025–2038, https://doi.org/10.1111/1462-2920.13226, 2016.
Mueller, B., De Goeij, J. M., Vermeij, M. J. A., Mulders, Y., Van Der Ent, E., Ribes, M., and Van Duyl, F. C.: Natural diet of coral-excavating sponges consists mainly of dissolved organic carbon (DOC), PLoS One, 9, 1–7, https://doi.org/10.1371/journal.pone.0090152, 2014.
Murphy, J. and Riley, J. P.: A modified single solution method for the determination of phosphate in natural waters, Anal. Chim. Ac., 27, 31–36, 1962.
Naim, M. A., Morillo, J. A., Sørensen, S. J., Waleed, A. A. S., Smidt, H., and Sipkema, D.: Host-specific microbial communities in three sympatric North Sea sponges, FEMS Microbiol. Ecol., 90, 390–403, https://doi.org/10.1111/1574-6941.12400, 2014.
Pile, A. J., Patterson, M. R., Savarese, M., Chernykh, V. I., and Fialkov, V. A.: Trophic effects of sponge feeding within Lake Baikal's littoral zone. Sponge abundance, diet, feeding efficiency, and carbon flux, Limnol. Oceanogr., 42, 178–184, 1997.
Pita, L., Turon, X., López-Legentil, S., and Erwin, P. M.: Host rules: Spatial stability of bacterial communities associated with marine sponges (Ircinia spp.) in the western Mediterranean sea, FEMS Microbiol. Ecol., 86, 268–276, https://doi.org/10.1111/1574-6941.12159, 2013.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glöckner, F. O.: The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools, Nucleic Acids Res., 41, 590–596, https://doi.org/10.1093/nar/gks1219, 2013.
Radax, R., Rattei, T., Lanzen, A., Bayer, C., Rapp, H. T., Urich, T., and Schleper, C.: Metatranscriptomics of the marine sponge Geodia barretti: Tackling phylogeny and function of its microbial community, Environ. Microbiol., 14, 1308–1324, https://doi.org/10.1111/j.1462-2920.2012.02714.x, 2012.
R Development Core Team: R: A language and environment for statistical computing, available at: http://www.r-project.org (last access: 10 October 2013), 2008.
Reiswig, H. M.: Particle feeding in natural populations of three marine demosponges, Biol. Bull., 141, 568–591, 1971.
Reveillaud, J., Maignien, L., Eren, M. A., Huber, J. A., Apprill, A., Sogin, M. L., and Vanreusel, A.: Host-specificity among abundant and rare taxa in the sponge microbiome, ISME J., 8, 1198–1209, https://doi.org/10.1038/ismej.2013.227, 2014.
Ribes, M., Coma, R., and Gili, J.: Natural diet and grazing rate of the temperate sponge Dysidea avara (Demospongiae, Dendroceratida) throughout an annual cycle, Mar. Ecol. Prog. Ser., 176, 179–190, 1999.
Rix, L., De Goeij, J. M., Mueller, C. E., Struck, U., Middelburg, J. J., Van Duyl, F. C., Al-Horani, F. A., Wild, C., Naumann, M. S., and Van Oevelen, D.: Coral mucus fuels the sponge loop in warm-and cold-water coral reef ecosystems, Sci. Rep., 6, 1–11, https://doi.org/10.1038/srep18715, 2016a.
Rix, L., Goeij, J. M. De, Oevelen, D. Van, Struck, U., Al-horani, F. A., Wild, C., and Naumann, M. S.: Differential recycling of coral and algal dissolved organic matter via the sponge loop, Funct. Ecol., 31, 1–12, https://doi.org/10.1111/1365-2435.12758, 2016b.
Roberts, E. M., Mienis, F., Rapp, H. T., Hanz, U., Meyer, H. K., and Davies, A. J.: Oceanographic setting and short-timescale environmental variability at an Arctic seamount sponge ground, Deep Sea Res. Pt. I, 98–113, https://doi.org/10.1016/j.dsr.2018.06.007, 2018.
Rogers, A. D.: The Biology of Seamounts: 25 Years on, Adv. Mar. Biol., 79, 137–224, https://doi.org/10.1016/bs.amb.2018.06.001, 2018.
Schmitt, S., Tsai, P., Bell, J., Fromont, J., Ilan, M., Lindquist, N., Perez, T., Rodrigo, A., Schupp, P. J., Vacelet, J., Webster, N., Hentschel, U., and Taylor, M. W.: Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges, ISME J., 6, 564–576, https://doi.org/10.1038/ismej.2011.116, 2012.
Schöttner, S., Hoffmann, F., Cárdenas, P., Rapp, H. T., Boetius, A., and Ramette, A.: Relationships between host phylogeny, host type and bacterial community diversity in cold-water coral reef sponges, PLoS One, 8, 1–11, https://doi.org/10.1371/journal.pone.0055505, 2013.
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., and Huttenhower, C.: Metagenomic biomarker discovery and explanation, Genome Biol., 12, R60, https://doi.org/10.1186/gb-2011-12-6-r60, 2011.
Smith, D. K. and Cann, J. R.: The role of seamount volcanism in crustal construction at the Mid-Atlantic Ridge (24∘–30∘ N), J. Geophys. Res.-Sol. Ea., 97, 1645–1658, 1992.
Staudigel, H., Koppers, A. A. P., William Lavelle, J., Pitcher, T. J., and Shank, T. M.: Defining the word “seamount”, Oceanography, 23, 20–21, 2010.
Steinert, G., Taylor, M. W., Deines, P., Simister, R. L., De Voogd, N. J., Hoggard, M., and Schupp, P. J.: In four shallow and mesophotic tropical reef sponges from Guam the microbial community largely depends on host identity, PeerJ, 2016, 1–25, https://doi.org/10.7717/peerj.1936, 2016.
Steinert, G., Rohde, S., Janussen, D., Blaurock, C., and Schupp, P. J.: Host-specific assembly of sponge-associated prokaryotes at high taxonomic ranks, Sci. Rep., 7, 1–9, https://doi.org/10.1038/s41598-017-02656-6, 2017.
Steinert, G., Wemheuer, B., Janussen, D., Erpenbeck, D., Daniel, R., Simon, M., Brinkhoff, T., and Schupp, P. J.: Prokaryotic diversity and community patterns in antarctic continental shelf sponges, Front. Mar. Sci., 6, 1–15, https://doi.org/10.3389/fmars.2019.00297, 2019.
Steinert, G., Busch, K., Bayer, K., Kodami, S., Arbizu, P. M., Kelly, M., Mills, S., Erpenbeck, D., Dohrmann, M., Wörheide, G., Hentschel, U., and Schupp, P. J.: Compositional and quantitative insights into bacterial and archaeal communities of South Pacific deep-sea sponges (Demospongiae and Hexactinellida), Front. Microbiol., 11, 716, https://doi.org/10.3389/fmicb.2020.00716, 2020.
Stoll, M. H. C., Bakker, K., Nobbe, G. H., and Haese, R. R.: Continuous-flow analysis of dissolved inorganic carbon content in seawater, Anal. Chem., 73, 4111–4116, https://doi.org/10.1021/ac010303r, 2001.
Strickland, J. and Parsons, T.: A practical handbook of seawater analysis, Fish. Res. Board Canada, 167, 65–70, 1972.
Taylor, M. W., Radax, R., Steger, D., and Wagner, M.: Sponge-Associated Microorganisms: Evolution, Ecology, and Biotechnological Potential, Microbiol. Mol. Biol. Rev., 71, 295–347, https://doi.org/10.1128/MMBR.00040-06, 2007.
Thomas, T., Moitinho-Silva, L., Lurgi, M., Björk, J. R., Easson, C., Astudillo-García, C., Olson, J. B., Erwin, P. M., López-Legentil, S., Luter, H., Chaves-Fonnegra, A., Costa, R., Schupp, P. J., Steindler, L., Erpenbeck, D., Gilbert, J., Knight, R., Ackermann, G., Victor Lopez, J., Taylor, M. W., Thacker, R. W., Montoya, J. M., Hentschel, U., and Webster, N. S.: Diversity, structure and convergent evolution of the global sponge microbiome, Nat. Commun., 7, 1–12, https://doi.org/10.1038/ncomms11870, 2016.
Tian, R. M., Sun, J., Cai, L., Zhang, W. P., Zhou, G. W., Qiu, J. W., and Qian, P. Y.: The deep-sea glass sponge Lophophysema eversa harbours potential symbionts responsible for the nutrient conversions of carbon, nitrogen and sulfur, Environ. Microbiol., 18, 2481–2494, https://doi.org/10.1111/1462-2920.13161, 2016.
Van Haren, H., Hanz, U., de Stigter, H., Mienis, F., and Duineveld, G.: Internal wave turbulence at a biologically rich Mid-Atlantic seamount, PLoS One, 12, 1–16, https://doi.org/10.1371/journal.pone.0189720, 2017.
Van Oevelen, D., Mueller, C. E., Lundälv, T., Van Duyl, F. C., De Goeij, J. M., and Middelburg, J. J.: Niche overlap between a cold-water coral and an associated sponge for isotopicallyenriched particulate food sources, PLoS One, 13, 1–16, https://doi.org/10.1371/journal.pone.0194659, 2018.
Weisz, J. B., Lindquist, N., and Martens, C. S.: Do associated microbial abundances impact marine demosponge pumping rates and tissue densities?, Oecologia, 155, 367–376, https://doi.org/10.1007/s00442-007-0910-0, 2008.
Wessel, P., Sandwell, D. T., and Kim, S.-S.: Seamount Census, Oceanography, 23, 24–33, 2010.