the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
The effects of decomposing invasive jellyfish on biogeochemical fluxes and microbial dynamics in an ultra-oligotrophic sea
Tamar Guy-Haim
Maxim Rubin-Blum
Eyal Rahav
Natalia Belkin
Jacob Silverman
Guy Sisma-Ventura
Over the past several decades, jellyfish blooms have intensified spatially and temporally, affecting functions and services of ecosystems worldwide. At the demise of a bloom, an enormous amount of jellyfish biomass sinks to the seabed and decomposes. This process entails reciprocal microbial and biogeochemical changes, typically enriching the water column and seabed with large amounts of organic and inorganic nutrients. Jellyfish decomposition was hypothesized to be particularly important in nutrient-impoverished ecosystems, such as the Eastern Mediterranean Sea – one of the most oligotrophic marine regions in the world. Since the 1970s, this region has been experiencing the proliferation of a notorious invasive scyphozoan jellyfish, Rhopilema nomadica. In this study, we estimated the short-term decomposition effects of R. nomadica on nutrient dynamics at the sediment–water interface. Our results show that the degradation of R. nomadica has led to increased oxygen demand and acidification of overlying water as well as high rates of dissolved organic nitrogen and phosphate production. These conditions favored heterotrophic microbial activity and bacterial biomass accumulation, and triggered a shift towards heterotrophic biodegrading bacterial communities, whereas autotrophic picophytoplankton abundance was moderately affected or reduced. This shift may further decrease primary production in the water column of the Eastern Mediterranean Sea. Deoxygenation, acidification, nutrient enrichment, and microbial community shifts at the sediment–water interface may have a detrimental impact on macrobenthic communities. Based on these findings, we suggest that jelly-falls and their decay may facilitate an additional decline in ecosystem functions and services.
- Article
(5681 KB) - Full-text XML
- BibTeX
- EndNote
Marine jellyfish often form massive aggregations, known as jellyfish blooms, with profound implications to human health, recreation and tourism, fisheries, aquaculture, and coastal installations (Purcell, 2012; Purcell et al., 2007; Richardson et al., 2009). Over the past three decades, a substantial increase in the frequency and intensity of jellyfish blooms has been documented worldwide (Attrill et al., 2007; Brotz et al., 2012; Licandro et al., 2010; Lynam et al., 2006; Quiñones et al., 2015; Shiganova et al., 2001) and was attributed to the growth in shipping, aquaculture, and coastal protection (Duarte et al., 2013) or to natural global oscillations (Condon et al., 2013; Sanz-Martín et al., 2016). These blooms typically occur in “boom-and-bust” cycles, where individuals suddenly appear in large numbers and shortly after disappear (Condon et al., 2013; Hamner and Dawson, 2009; Schnedler-Meyer et al., 2018). This rapid collapse of jellyfish blooms en masse and their sinking to the seabed is a process commonly termed as “jelly-falls” (Lebrato and Jones, 2011; Lebrato et al., 2012; Sweetman and Chapman, 2011).
During the blooms, jellyfish propagate by assimilating organic compounds of their prey, thus acting as a nutrient sink of organic carbon (C), nitrogen (N), and phosphorus (P) (Lebrato and Jones, 2011; Lucas et al., 2011; Pitt et al., 2009). The death and sinking of jellyfish, followed by bacterial decomposition of their carcasses, lead to microbial community shifts (Kramar et al., 2019; Tinta et al., 2012; Titelman et al., 2006), resulting in oxygen depletion and acidification (Qu et al., 2015; Sweetman et al., 2016; West et al., 2008). On the seabed, jellyfish carcasses can be consumed by scavengers, thus acting as a rich carbon source that sustains benthic food webs (Hays et al., 2018; Sweetman et al., 2016, 2014). Both in the water column and on the sediment, jelly-falls undergo bacterial decomposition, directly affecting nutrient cycling (Qu et al., 2015; West et al., 2008), potentially altering plankton community composition (Xiao et al., 2019) and stimulating algal blooms (Møller and Riisgård, 2007). Changes in the sediment conditions may result in migration or mortality of infauna (Chelsky et al., 2016), which in turn affect indirectly nutrient cycling (Stief, 2013; Welsh, 2003). The contribution of jellyfish degradation to nutrient cycling was hypothesized to be particularly important in nutrient-depleted, oligotrophic ecosystems (Pitt et al., 2009), such as the ultra-oligotrophic Eastern Mediterranean Sea (EMS), where microbial production is mainly limited by organic carbon (Sisma-Ventura and Rahav, 2019) or nitrogen (Rahav et al., 2018b), or co-limited by nitrogen and phosphorus (Kress et al., 2005).
The most prominent jellyfish blooms in the Mediterranean Sea, particularly in its eastern basin, are caused by the scyphozoan Rhopilema nomadica (Edelist et al., 2020; Katsanevakis et al., 2014) (Fig. 1). R. nomadica was first recorded in Israel in 1977 as a Lessepsian invader, introduced via the Suez Canal (Galil et al., 1990). Since then, it has expanded its distribution westwards with more frequent blooming occurrences (Balistreri et al., 2017; Edelist et al., 2020; Yahia et al., 2013). This species is venomous and its nematocysts contain active toxins, inflicting painful stinging on humans, as well as other adverse health problems, negatively affecting coastal recreation and tourism (Galil, 2018; Ghermandi et al., 2015). During blooms, clogged intake pipes of power and desalination plants were reported in Israel (Angel et al., 2016; Galil, 2012). Reduced fishing harvests were also reported from Israel and Egypt, mostly due to net damage, loss of fishing days, and physical injury to the fishermen (Angel et al., 2016; Madkour et al., 2019; Nakar et al., 2011).
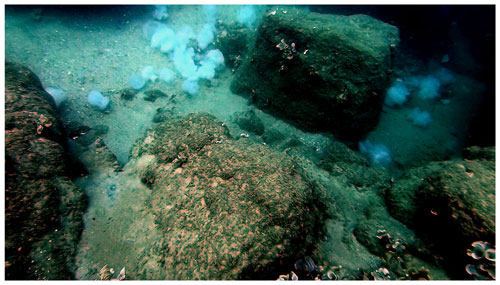
Figure 1Jelly-falls (carcasses) of approximately 30 Rhopilema nomadica on the Mediterranean coast of Caesarea, Israel. The image shows a depth of 8–9 m and was photographed on 27 July 2019 after the typical peak summer bloom (Photo: Zvika Fayer).
Although labeled as one of the worst invasive species in the Mediterranean Sea (Streftaris and Zenetos, 2006; Zenetos et al., 2010), the post-bloom decomposition dynamics of R. nomadica have never been investigated before. Here, we used incubation experiments at the sediment–water interface to estimate the short-term decomposition effects of the invasive jellyfish R. nomadica, on (1) organic and inorganic nutrient dynamics and derived benthic fluxes, (2) bacterial abundance and production, and (3) microbial community composition, in the nutrient-impoverished EMS. We hypothesize that decomposed R. nomadica will trigger a rapid release of limiting nutrients, leading to enhanced fluxes to the sediment and overlying water, a substantial increase in bacterial abundance and production, and a shift in the microbial community composition and functions.
2.1 Specimen collection and experimental setup
Three individuals of the scyphozoan jellyfish Rhopilema nomadica (Galil et al., 1990) of medium size (bell diameter 20–25 cm) were collected on 29 July 2019, at Tel Shikmona, Haifa, near the Israel Oceanographic and Limnological Research Institute, on the shore of the easternmost Mediterranean Sea (lat. 32∘49′32′′ N, long. 34∘57′26′′ E). The specimens were weighed and cut to pieces of 4–5 g to ensure representation of all body parts. Processed 25 g wet weight (approximately 1.25 g dry weight) of R. nomadica (including umbrellas, tentacles, and oral arms, following Qu et al., 2015) were placed each in three Perspex cylinders (9.45 cm internal diameter; 50 cm length) that were filled up to 10 cm height with coastal sediments (Fig. 2), that were collected 1 week prior to the experiment, allowing the re-establishment of natural sediment profiles. Three additional cylinders with sediments did not include jellyfish and functioned as controls. The setup was completed by topping off the cylinders with oxygen saturated Mediterranean coastal water (approximately 3.14 L) pumped from 1 m depth and pre-filtered to remove large-size zooplankton (67 µm). The cores were sealed with gas-tight sealing caps and placed in a lab with a relatively constant temperature of 27–28 ∘C, which is similar to the summer mean coastal water temperatures of the easternmost EMS (Raveh et al., 2015). The setup was acclimatized for 24 h to insure similar initial conditions in the chambers before jellyfish addition. Nutrient fluxes were measured using the whole-core incubation technique previously described by Denis et al. (2001). Although restricting this study for testing short-term responses, this method follows the best practices for measuring oxygen and nutrient fluxes and dynamics at the sediment–water interface (Glud, 2008; Hammond et al., 2004; Pratihary et al., 2014; Skoog and Arias-Esquivel, 2009). Pre-filtered coastal water was transferred to a reserve tank and stored under the same conditions as the incubated cores. The incubation cores were connected by tubing to the dedicated reserve tank, which replaced the water in the incubation chambers during each sampling. The cores were incubated under PAR = 100 µmol photons m−2 s−1 with a photoperiodicity of 14:10 (L:D).
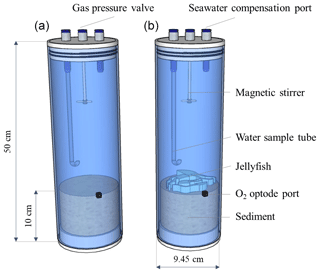
Figure 2Experimental setup. Incubation cylinders including jellyfish treatment (a, N=3) and controls (b, N=3).
Within each chamber, the overlying water was continuously mixed with a magnetic stirrer fixed 10 cm below the upper cap (75 rpm, Hammond et al., 2004) and was sampled at the following intervals: 0, 5, 10, 18, 26, 34, and 44 h, with dedicated sampling tubing. The reserve tank was sampled only at three intervals (0, 20, and 44 h). At each sampling, 200 mL water samples were transferred to acid-washed transparent Nalgene bottles (250 mL) and subsampled by filtering (Minisart® 0.45 µm) for the following chemical analyses: PO4, NO2+NO3 (NOx), Si(OH)4, NH4, TDP (DOP), and TN (DON). Nutrient samples were immediately frozen after collection for later analysis. Biological measurements were collected using unfiltered water for picophytoplankton (Synechococcus, Prochlorococcus, pico- and nanoeukaryotes), heterotrophic bacterial abundance, and bacterial production measurements. Oxygen consumption rates at the sediment–water interface were continuously monitored using oxygen sensor spots (FireSting, PyroScience, Germany) adapted for measuring oxygen in closed containers through a transparent window (plastic or glass). The sensor spots were fixed to the inner side of the window with silicone glue. Four optical fibers continuously measured the oxygen in the three jellyfish chambers and one of the control incubations. The system was calibrated with saturated de-ionized water (DIW). The pH was measured with a sensor (MultiLine WTW, Germany) calibrated with National Bureau of Standards (NBS) buffers.
Fluxes (mmol m−2 d−1) were determined by regressing the change in overlying water concentration (C) through time multiplied by the chamber height (Volume∕Area), following Eq. (1):
A correction for water replacement from the reserve tank was not applied, as the consequent error was less than 5 %.
2.2 Inorganic and organic nutrient analysis
Nutrient concentrations were determined using a three-channel segmented flow auto-analyzer system (AA-3 Seal Analytical) following Kress et al. (2014). The limit of detection (LOD), measured as 3 times the standard deviation of 10 measurements of the blank (low-nutrient seawater collected from the offshore EMS), was 8 nM for PO4, 50 nM for total dissolved phosphorus (TDP) and Si(OH)4, 80 nM for NO2+NO3 (NOx) 90 nM for NH4, and 0.74 µmol for total dissolved phosphorus (TDN). The accuracy of the analyses was determined using certified reference materials (CRMs): MOOS 3 (seawater certified reference material for nutrients) (PO4, NOx and Si(OH)4), VKI (Visual Kinematics, USA) 4.1 (NOx), and VKI 4.2 (PO4 and Si(OH)4). Results were accepted when measured CRMs were within ±5 % of the certified values.
TDN and TDP were measured following potassium persulfate digestion and ultraviolet (UV) photo-oxidation, using a digestion block system (Seal Analytical, UK). The reproducibility of the analyses was examined with VKI 4.2 and deep seawater reference (DSR) material. One of the TDP samples was lost (t=44 h). DON concentrations were determined by subtracting NOx and NH4 from TDN concentrations and DOP concentrations were determined by subtracting PO4 from TDP concentrations.
2.3 Pico-/nanophytoplankton and heterotrophic bacterial abundance
Samples (1.8 mL) were fixed with flow-cytometry-grade glutaraldehyde (0.02 % final concentration, G7651, Sigma-Aldrich, USA), frozen in liquid nitrogen, and stored at −80 ∘C until analysis within 2 weeks. Synechococcus and Prochlorococcus, autotrophic pico-/nanoeukaryotes (maximal size ∼ 70 µm), and heterotrophic bacterial abundances were determined using an Attune® Acoustic Focusing Flow Cytometer (Applied Biosystems, USA) as described in Bar-Zeev and Rahav (2015). Samples of Synechococcus, Prochlorococcus, and pico-/nanoeukaryotes were run at 100 µL min−1. Their taxonomic discrimination for based on the orange fluorescence of phycoerythrin (585 nm), the red fluorescence of chlorophyll a (630 nm), side scatter (SSC, a proxy of cell volume), and forward scatter (FSC, a proxy of cell size). Heterotrophic bacterial samples were run at 25 µL min−1 using a discrimination threshold of green fluorescence (520 nm) and FSC. Beads (0.93 µm, Polysciences) were run in parallel as a size standard. Blank samples of sterile seawater (0.2 µm) were also run and their reads were removed from the total bacterial counts.
2.4 Bacterial production (BP)
Bacterial production was estimated using the 3H-leucine incorporation method (Perkin Elmer, specific activity 123 Ci mmol−1) followed by microcentrifugation (Simon, 1990). Samples (1.7 mL) were incubated with 10 nmol leucine L−1 for 4–5 h under ambient temperature in the dark. Triplicate additions of trichloroacetic acid (TCA) were performed at each time point and served as controls. The incubations were terminated with 100 µL of concentrated (100 %) TCA. After adding 1 mL of scintillation cocktail (Ultima-Gold, PerkinElmer, USA) to each vial, the samples were counted using a TRI-CARB 2100 TR (Packard Biocience, USA) scintillation counter. A conversion factor of 3 kg C mol−1 per every mole leucine incorporated was used, assuming an isotopic dilution of 2.0 (Simon and Azam, 1989).
2.5 DNA extraction and sequencing
Approximately 300 mL of overlying seawater were collected with a sterile syringe and passed through 0.22 µm Sterivex filter. The membranes were removed from the cases, cut into pieces under sterile conditions, and transferred into the extraction tubes. A total of 250 mg from 0–1 and 1–2 cm sediment sections were transferred into the extraction tube. DNA was extracted from water and sediment using the DNeasy PowerSoil Kit (Qiagen, California, USA) using the manufacturer's protocol that included a FastPrep-24™ (MPBIO, Ohio, USA) bead-beating step (2×40 s at 5.5 m s−1, with a 5 min interval). The V4 region of the 16S rRNA gene was amplified using the modified primer pair 515F-806R (Apprill et al., 2015; Parada et al., 2016) in combination with CS1/CS2 tags (CS1_515Fc 5'-ACACTGACGACATGGTTCTACA GTGYCAGCMGCCGCGGTAA, CS2_806Rc 5'-TACGGTAGCAGAGACTTGGTCT GGACTACNVGGGTWTCTAAT), using the following polymerase chain reaction (PCR) amplification protocol: initial denaturation at 94 ∘C for 45 s, 30 cycles of denaturation (94 ∘C for 15 s), annealing (15 cycles at 50 ∘C and 15 cycles at 60 ∘C for 20 s), and extension (72 ∘C for 30 s). The 18S rRNA gene sequences were amplified using the 1391f-EukBr primer pair (Amaral-Zettler et al., 2009; Stoeck et al., 2010) in combination with CS1/CS2 tags (1391fc 5'-ACACTGACGACATGGTTCTACA GTACACACCGCCCGTC, EukBr 5'-TACGGTAGCAGAGACTTGGTCT TGATCCTTCTGCAGGTTCACCTAC), using the following PCR amplification protocol: initial denaturation at 94 ∘C for 45 s, 30 cycles of denaturation (94 ∘C for 15 s), annealing (60 ∘C for 20 s), and extension (72 ∘C for 30 s). Library preparation from the PCR products and sequencing of 2×250 bp Illumina MiSeq reads were performed at HyLabs (Israel).
2.6 Statistical and bioinformatic analyses
Demultiplexed paired-end reads were processed in Quantitative Insights Into Microbial Ecology (QIIME2) V2019.7 environment (Bolyen et al., 2018). Reads were truncated based on quality plots, checked for chimeras, merged, and grouped into amplicon/environmental sequence variants (A/ESVs) with DADA2 (Callahan et al., 2016), as implemented in QIIME2. After removing the low-quality sequences, a total of 361 335 (106 169 in 6 and 255 166 in 12 seawater and sediment samples, respectively) high-quality 16S rRNA gene amplicon reads with an average length of 260 bp, and a total of 658 251 (162 313 in 6 and 495 938 in 12 seawater and sediment samples, respectively) high-quality 18S rRNA gene amplicon reads with an average length of 207 bp, were generated. The 16S and 18S amplicons were classified with the naïve Bayes classifiers that were trained on the Silva 132 database, clustered at 99 % (515F/806R region for the 16S and full-length sequences for the 18S rRNA gene amplicons). Downstream statistical analyses, calculation of alpha diversity indices (Chao1, Shannon, and Simpson), and plotting were performed in R (Core Team, 2020), using packages phyloseq (McMurdie and Holmes, 2013), ampvis2 (Andersen et al., 2018) and ggplot2 (Wickham, 2016). Systematic changes across experimental conditions were estimated with DESeq2 (Love et al., 2014). The metabolic functions and pathways of the bacterial communities were predicted using Tax4Fun2 based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Wemheuer et al., 2018). Pearson correlations and similarity percentage (SIMPER) analysis (to assess the contribution of KEGG pathways to the dissimilarity between treatments and controls) were performed in R using packages Hmisc (Harrell, 2004) and vegan (Oksanen et al., 2010). Principal component analysis (PCA) of the metabolic functions was performed with PAST V4 (Hammer et al., 2001).
3.1 Dissolved oxygen and pH dynamics
Dissolved oxygen (DO) levels in the jellyfish treatments decreased from an initial average concentration of 261.5 ± 4.5 µmol L−1 to null within 40 h, at an average rate of 5.9 ± 0.1 µmol L−1 h−1, whereas the DO levels in the control chambers decreased slightly at an average rate of 0.7 ± 0.1 µmol L−1 h−1 (mean ± SD, Fig. 3a). The variability within the treatment replicates and within the controls was small and non-significant (treatment replicates: , p=0.98; controls: , p=0.59). The calculated average DO flux from the water column in the jellyfish treatment was −56.9 ± 1.0 mmol m−2 d−1 versus −6.7 ± 0.3 mmol m−2 d−1 in the controls (Table 1). In accordance with the decrease in DO, pH levels in the jellyfish treatments decreased from an initial average level of 8.10 ± 0.02 to 7.88 ± 0.01 and remained relatively stable (8.10–8.15) in the controls (Fig. 3b).
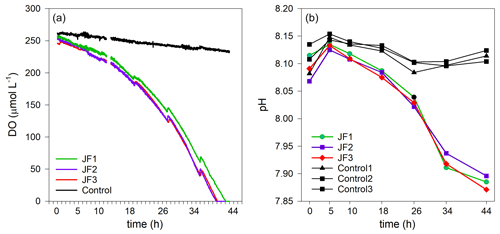
Figure 3The decomposition of the jellyfish R. nomadica leads to oxygen depletion and acidification in the seawater overlying the sediment. (a) Continuous dissolved oxygen (DO) record in the experimental cylinders enriched with carcasses of the jellyfish R. nomadica (JF1–JF3) and in the controls. (b) pH dynamics in the experimental cylinders, including jellyfish and in the controls. N=3. The temperature was kept relatively constant at 27–28 ∘C. The slight increases in DO concentrations throughout the incubation period indicate water compensation during discrete sampling events.
3.2 Nutrient dynamics
Nutrient levels significantly increased in the jellyfish-enriched chambers, whereas in the controls they remained stable and low (Fig. 4). These increases were non-linear and characterized by multiple phases, including in some cases changes of the flux direction. Therefore, nutrient flux rates were calculated in different linear phases over time.
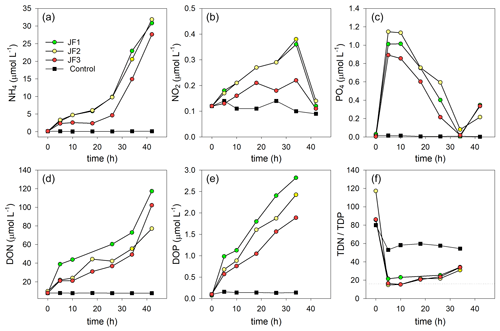
Figure 4Changes in the concentrations (µmol L−1) of organic and inorganic nutrients in the experimental cylinders enriched with carcasses of the jellyfish R. nomadica and in the controls: (a) ammonium, (b) nitrite, (c) orthophosphate, (d) DON, (e) DOP, and (f) TDN : TDP ratio. (N=3).
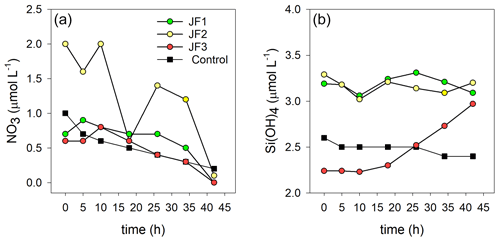
Ammonium was the dominant form of dissolved inorganic nitrogen in the experimental chambers. During the first 10 h from the onset of the experiment, NH4 levels increased at a rate of 0.39 ± 0.12 µmol L−1 h−1, after which (10–26 h) the rate of NH4 release slowed to 0.26 ± 0.10 µmol L−1 h−1 and then (26–44 h) sharply increased to 1.33 ± 0.31 µmol L−1 h−1 (Fig. 4a). NO2 levels steadily increased at a rate of µmol L−1 h−1 and decreased to background levels after 34 h (Fig. 4b). NO3 levels were generally higher in the jellyfish treatment than in the controls but did not present any significant trend over time (Appendix A, Fig. A1). One of the jellyfish treatments (JF2) showed higher (2-fold) concentrations of NO3 throughout the experiment, likely due to a different initial NO3 content derived from the mixture of jellyfish tissue, as some parts have shown to include higher concentrations of dissolved nitrogen (MacKenzie et al., 2017). Nevertheless, this has not affected the overall nutrient fluxes nor triggered different responses to the microbial communities (thus, the same direction and strength of responses were observed in all jellyfish addition treatments). Silicic acid concentrations remained overall stable throughout the experiment, and higher in two of the jellyfish-enriched chambers (Appendix A, Fig. A1).
Within the first 5 h following the jellyfish enrichment, orthophosphate levels increased by 2 orders of magnitude from 0.02 ± 0.01 to 1.02 ± 0.13 µmol L−1 (Fig. 4c). Throughout the rest of the experiment, PO4 was fully consumed and its levels decreased to the background levels within 34 h (0.04 µmol L−1), after which an increase was recorded (0.30 µmol L−1).
The majority of TDN and TDP released from the jellyfish was organic, where 84 % of the TDN was DON, (Fig. 4d), and 71 % of the TDP was DOP (Fig. 4e). Both organic nutrient levels significantly increased in the jellyfish enriched chambers, whereas their concentrations in the control chambers remained stable and low. During the incubation period, DON concentrations increased 12-fold in the jellyfish treatment compared to the controls (Fig. 4d) and DOP concentrations increased 18-fold (Fig. 4e). The ratio between TDN and TDP (TDN : TDP) decreased from an initial average value of to an average value of in the jellyfish treatments, whereas in the controls it decreased to (Fig. 4f).
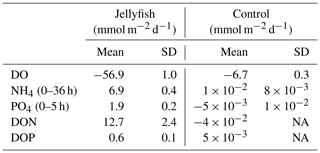
The rates of nutrient release (remineralization rates) standardized to jellyfish biomass are detailed in Table 1, and the calculated nutrient fluxes (mmol m−2 d−1) in the jellyfish enriched cylinders and in the controls are summarized in Table 2.
Table 2Calculated oxygen and nutrient fluxes in the seawater of jellyfish (R. nomadica)-enriched and control experimental cylinders. Positive flux represents water column enrichment (source); negative flux represents removal from the water column (sink). N=3. SD denotes standard deviation. NA – not available.
3.3 Autotrophic and heterotrophic abundance and bacterial production
Heterotrophic bacterial abundance increased linearly in the jellyfish treatments (R2=0.98, p<0.01) and reached cells mL−1 after 44 h, whereas the controls remained stable at a concentration of (Fig. 5a). Synechococcus abundance dropped in both jellyfish-enriched and control cylinders; however, after 44 h, the number of Synechococcus cells in the jellyfish treatment was 5-fold larger compared to the controls (Fig. 5b). Prochlorococcus cell numbers increased in both jellyfish-enriched and control cylinders, and after 44 h it was lower in the jellyfish treatment (Fig. 5c). Both cell numbers of pico- and nanoeukaryotes dropped throughout the experiment; nonetheless, they were higher in the jellyfish treatment than in the controls by 50 % (Fig. 5d–e).
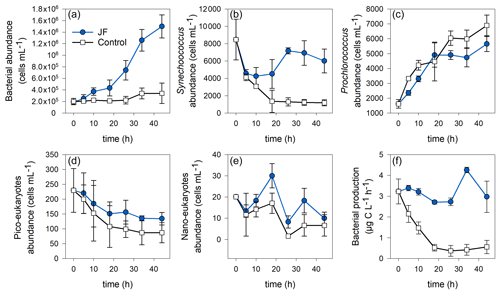
Figure 5Microbial abundance (cell mL−1) and production (µg C L−1 h−1) in the jellyfish R. nomadica-enriched (blue) and control (black) experimental cylinders over the experimental period: (a) total bacterial abundance, (b) Synechococcus, (c) Prochlorococcus, (d) picoeukaryotes, (e) nanoeukaryotes, and (f) bacterial production. N=3, the error bars denote standard deviation.
Bacterial production remained stable in the jellyfish treatments at a rate of 3.1 ± 0.3 µg C L−1 h−1 during the first 26 incubation hours, increased to 4.3 ± 0.1 µg C L−1 h−1, and decreased again after 34 h. In contrast, in the controls, the bacterial production decreased immediately from the onset of the experiment and reached a rate of 0.4 ± 0.2 µg C L−1 h−1 after 18 h that remained stable until the experiment ended (Fig. 5f).
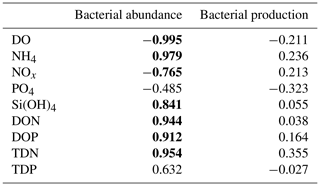
The temporal dynamics of DO and nutrient concentrations strongly correlated with total bacterial abundance but not with bacterial production (Appendix B, Table B1).
3.4 Microbial diversity
Bacterial alpha diversity (Fig. 6) was significantly lower in the jellyfish-enriched seawater than in the controls (p<0.05), but in the sediment samples there was no significant difference (p>0.05). The vast majority (93 %–97 %) of the 18S sequence variants in seawater (Appendix C, Fig. C2) belonged to Scyphozoa, hindering alpha diversity evaluation. In the sediment, no significant difference (p>0.05) in alpha diversity was observed between treatments (Appendix C, Fig. C1). These findings were confirmed with rarefaction curves (Appendix C, Figs. C3, C4).
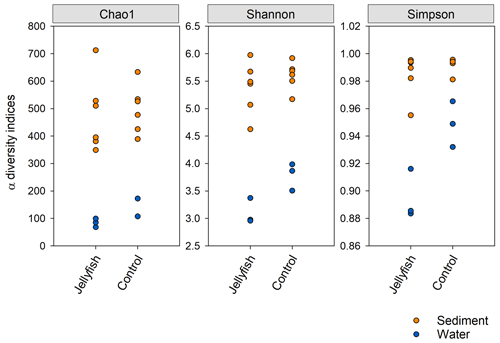
Figure 6Bacterial alpha diversity indices (Chao, Shannon, Simpson) in water and sediment samples from experimental cylinders enriched with carcasses of the jellyfish and in the controls (N=3).
The distribution of the 30 most abundant bacterial genera measured in seawater in the jellyfish-enriched and control chambers is presented in a heatmap (as inferred from read abundance estimates, Fig. 7). Lineages for which significant changes in abundance (p<0.05) between the treatment and control were detected by DESeq2 (Fig. 7, yellow star symbols: lineages more abundant in the controls, purple star symbols: lineages more abundant in the jellyfish treatment). Nine lineages were significantly more abundant in the jellyfish treatment, whereas 12 lineages were significantly more abundant in the controls. The relative abundance of the common marine bacteria, including the primary producers Synechococcus and the heterotrophic bacteria SAR11, HIMB11, and SAR86 (Dupont et al., 2012; Durham et al., 2014; Giovannoni, 2017), has all diminished following jellyfish additions. Mostly opportunistic lineages (Kordiimonadaceae, Pseudoalteromonadaceae, Saccharospirillaceae, and Nitrincolaceae) that use multiple carbon sources, including xenobiotics, were enriched in jellyfish-amended incubations and are often associated with oil discharge (Yakimov et al., 2007). Algicola (Pseudoalteromonadaceae) and Kordiimonas (Kordiimonadaceae) appear to be the most abundant degraders of the jellyfish biomass based on the marked change observed in the abundance of their relative amplicon sequence variants.
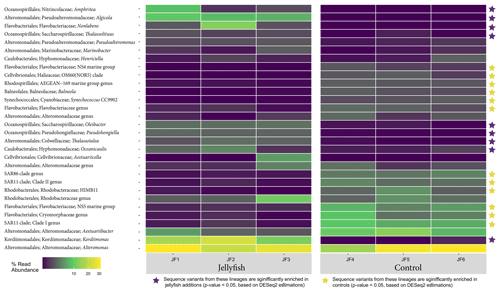
Figure 7Diversity of bacteria in the jellyfish-enriched and control experimental cylinder seawater. The 30 most abundant lineages are presented and organized by hierarchical clustering. Color scale denotes the relative abundance of reads (%). The star symbols on the right-side panel indicate lineages significantly more abundant in the jellyfish treatment (in purple) or the controls (in yellow) based on DESeq2 estimations.
Heatmap showing the distribution of the 30 most abundant genera in the sediment, measured in the 0–1 cm below-surface layer and in the 1–2 cm below-surface layer (inferred from 16S sequences), in the jellyfish-enriched and control chambers is presented in Fig. C1 (Appendix C). Among the 30 most abundant taxa, only Fusimonas and Algicola genera were significantly more abundant in the jellyfish treatments in the 0–1 cm layer; however, in the 1–2 cm layer, there was no significant difference between the treatments and controls.
The distribution of the 30 most abundant eukaryotic genera (inferred from the 18S rRNA amplicon read abundance) measured in seawater and sediment in the jellyfish-enriched and control chambers is presented in Fig. C2 (Appendix C). Both sediment layers showed no difference between treatment and controls, whereas in the seawater samples, four lineages of dinoflagellates, Ciliophora, and Labyrinthulomycetes were more abundant in the jellyfish than the controls.
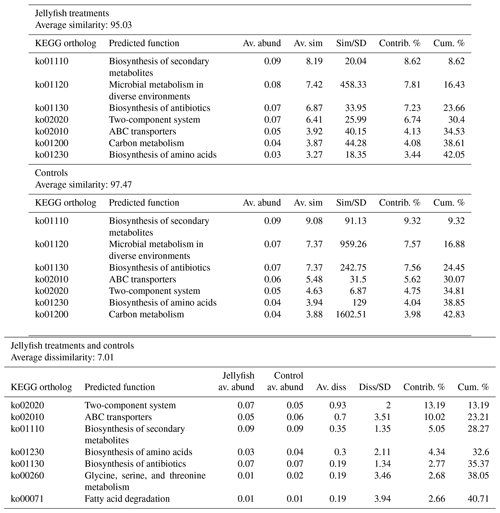
Predicted functions were classified as KEGG orthologs (KOs) resulting in the identification of 346 KOs across all samples, 160 of which were associated with prokaryotic functions. The principal component analysis (including 324 KOs across all samples, after removal of rare KOs that appear in only one of the replicates to avoid zero-inflated dimensionality) showed that jellyfish-treated and control samples significantly differed based on microbial predicted functions (Fig. 8). Photosynthesis (ko00195) and carbon fixation in photosynthetic organisms (ko00710) were enriched in controls, while catabolic functions, such as fatty acid degradation (ko00071), valine, leucine and isoleucine degradation (ko00362), and xenobiotic degradation pathways, benzoate degradation (ko00650) in particular, were enriched in jellyfish additions (Fig. 8). SIMPER analysis (Appendix C, Table C1) showed that the pathways mostly contributing to the difference between the jellyfish treatments and controls were signal transduction two-component system (ko02020) and ABC transporters (ko02010), contributing to 13 % and 10 % of the dissimilarity between the groups, respectively.
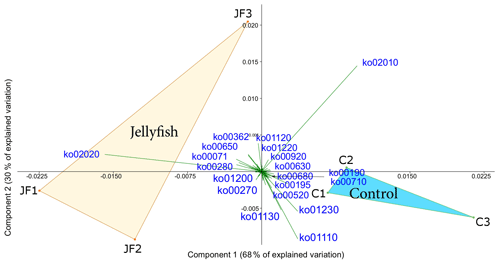
Figure 8Principle component analysis biplot of functional diversity based on taxonomy-based functional predictions using Tax4Fun2. JF indicates jellyfish samples; C indicates control samples. The vectors present KEGG pathways. The within-group similarity is 95 % between the jellyfish treatments (red) and 97 % between the controls (light blue). The following KEGG pathways are shown: ko00071 – fatty acid degradation; ko00190 – oxidative phosphorylation; ko00195 – photosynthesis; ko00270 – cysteine and methionine metabolism; ko00280 – valine, leucine, and isoleucine degradation; ko00362 – benzoate degradation; ko00520 – amino sugar and nucleotide sugar metabolism; ko00630 – glyoxylate and dicarboxylate metabolism; ko00650 – butanoate metabolism; ko00680 – methane metabolism; ko00710 – carbon fixation in photosynthetic organisms; ko00920 – sulfur metabolism; ko01110 – biosynthesis of secondary metabolites; ko01120 – microbial metabolism in diverse environments; ko01130 – biosynthesis of antibiotics; ko01200 – carbon metabolism; ko01220 – degradation of aromatic compounds; ko01230 – biosynthesis of amino acids; ko02010 – ABC transporters; ko02020 – two-component system.
4.1 The effects of R. nomadica decomposition on oxygen and nutrient fluxes
Jellyfish blooms trigger substantial changes in dissolved oxygen, inorganic carbon, and nutrient concentrations in the water column (Condon et al., 2011; Pitt et al., 2009). Post-bloom processes, by comparison, modify the oxygen, carbon, and nutrient fluxes in the benthic boundary layer and the sediment–water interface (Chelsky et al., 2015; Lebrato and Jones, 2011; Qu et al., 2015; West et al., 2008). Here, we found that the decomposition of the invasive jellyfish Rhopilema nomadica triggered deoxygenation of the seawater overlying the sediment to hypoxic and eventually anoxic levels, although the complete dissipation of oxygen is likely due to the experimental conditions. Similarly, increased sediment oxygen demand following jellyfish decomposition was measured by West et al. (2008) in Catostylus mosaicus and by Tinta et al. (2016) in the moon jellyfish (Aurelia aurita). Qu et al. (2015) that studied the effects of Cyanea nozakii decomposition in the Yellow Sea using incubations found that oxygen was depleted in both sediment and seawater. They hypothesized that the metabolism and propagation of heterotrophic bacteria led to enhanced oxygen consumption. Indeed, our experimental results support this hypothesis, as bacterial abundance was strongly correlated with oxygen levels, whereas the abundance of autotrophic cyanobacteria decreased as they were likely outcompeted by the heterotrophic bacteria (Sisma-Ventura and Rahav, 2019; Thingstad et al., 2005). Thus, jelly-falls can generate hypoxic areas on the seabed and overlying waters (Pitt et al., 2009), and affect the benthic infauna (Chelsky et al., 2016). Although the Eastern Mediterranean coastal waters are well oxygenated (Kress et al., 2014), the collapse of massive R. nomadica blooms could potentially create local hypoxic or even anoxic hotspots on the seabed, thereby affecting the surrounding biota (Feely et al., 2010).
In addition to deoxygenation, our experiment showed a significant reduction in pH to levels that are considered detrimental to various organisms, mainly calcifies (Kroeker et al., 2010; Zunino et al., 2017). Acidification as a result of jellyfish decomposition was also observed by Qu et al. (2015) that speculated that the release of amino acids and fatty acids from proteins and lipid metabolism of jellyfish tissue is the root cause for the observed decrease in pH. Nonetheless, hypoxia and acidification are biogeochemically coupled via the production of inorganic carbon in the process of respiration (Feely et al., 2010; Gobler and Baumann, 2016). In addition, the increase in NH4, as was measured in our experiment, increases total alkalinity and pH, whereas nitrate and silicate decrease pH, but they were comparably scarce. Based on oxygen-to-carbon conversion (1:1.3) and alkalinity change due to NH4 addition, it is estimated that the observed decrease in pH in our experiment can be solely attributed to inorganic carbon and carbonic acid production (due to bacterial respiration) and ammonium release. The combination of hypoxia and acidification may have synergistic additive negative effects on the benthic fauna (Gobler et al., 2014; Melzner et al., 2013). Furthermore, ammonium in high concentrations may have toxic effects on various marine organisms, from bacteria to fish (Brun et al., 2002; Eddy, 2005; Ferretti and Calesso, 2011; Müller et al., 2006).
The decomposition of dead R. nomadica tissue generated an immediate rapid release of organic and inorganic phosphate after which the inorganic phosphate (PO4) was completely consumed, while the efflux of organic and inorganic (mostly ammonium) nitrogenous compounds gradually increased throughout the experiment. Similar dynamics were observed in C. mosaicus by West et al. (2008) and Chelsky et al. (2015), and by Tinta et al. (2010) in Aurelia solida, where organic and inorganic phosphate peaked and completely abolished within 24 h, presumably due to bacterial uptake. The production of NOx in our experiment was evident only in the jellyfish treatment while oxygen levels were conducive, suggesting that nitrification plays an important role in nutrient dynamics following jellyfish decomposition, as was found in different jellyfish species (Hubot et al., 2020; Welsh et al., 2009). The non-linearity of nutrient fluxes that was evident in our experiment (especially in the multi-rate flux of NH4 and in the bi-directional flux of NO2 and PO4) as well as in other jellyfish degradation studies (Blanchet et al., 2015; Chelsky et al., 2016; Qu et al., 2015; Tinta et al., 2010), indicating a sequential nature of decomposition, likely due to microbial colonization and non-linear growth rates. The shift from nitrate production to nitrate consumption 36 h from the onset of the experiment likely reflects the shift from aerobic to anaerobic processes due to the low, hypoxic (and eventually anoxic) levels and may be regarded as an experimental artifact, although such changes were previously showed in surface sediments (Chelsky et al., 2016). The stoichiometric relationship between TDN and TDP decreased from 57:1 to 23:1 as a result of R. nomadica decomposition, as was also found by West et al. (2009) and Qu et al. (2015). This decrease can be explained by the elemental body composition of scyphozoan jellyfish, in general, is 2.48 N % DW (dry weight) and 0.22 P % DW and hence has an N:P ratio of 25:1 (Lucas et al., 2011).
The rates of nutrient release from R. nomadica decomposition found in this study were comparable to jellyfish-decomposition-driven rates found in former studies (e.g., Blanchet et al., 2015; Pitt et al., 2009; Qu et al., 2015; Tinta et al., 2012, 2016; Titelman et al., 2006; West et al., 2008). Ammonium release rate in R. nomadica (1.96 µmol g−1 WW d−1) was slightly higher than the rate measured by Tinta et al. (2012) in Rhizostoma pulmo (1.6 µmol g−1 WW d−1), another common Mediterranean scyphozoan. Reported densities of R. nomadica aggregations from the EMS are 1.6×105 km−2 on the Israeli coast (Lotan et al., 1992, 1994), 1×106 km−2 on the Lebanese coast (Lakkis and Zeidane, 1991), and 9×105 km−2 on the Egyptian Mediterranean coast (Madkour et al., 2019). The average wet weight of R. nomadica changes seasonally, 1340 ± 953 g ind−1 during summer and 2450 ± 1854 g ind−1 during winter (N=40, Guy-Haim, unpublished data), yielding approximately 1.3 kt km−2. We can therefore estimate that the collapse of R. nomadica bloom potentially releases ammonium and phosphate in concentrations of 2.5 and 0.8 kmol km−2, respectively.
Nutrient remineralization during jelly-fall decomposition, as was found in this study and others, can be inhibitory or toxic to some organisms (e.g., dissolved sulfides and ammonium in Chelsky et al., 2016), but on the other hand, they can stimulate primary production and induce algal blooms in the water column and on the sediment. Møller and Riisgård (2007) found that following blooms of A. aurita, peak concentrations of chlorophyll a were measured in a heavily eutrophic Danish fjord. Using mesocosm experiments, West et al. (2009) found that excretion of jellyfish C. mosaicus led to a 10-fold increase in diatom abundance. In the EMS, R. nomadica typically peaks in the summer months and collapses at the end of July (Edelist et al., 2020), whereas peak chlorophyll a concentrations in the water column are measured during wintertime (Ignatiades et al., 2009; Rahav et al., 2018a, 2015). This may result from the competitive exclusion of phytoplankton by heterotrophic bacteria (Sisma-Ventura and Rahav, 2019). Thus, fertilization of the water column due to nutrient release from R. nomadica decomposition may fail to trigger an algal bloom in the EMS. In contrast to the water column, maximum chlorophyll concentrations were measured in the sediment of the shallow Israeli coastal shelf during the late spring–summer (Hyams-Kaphzan et al., 2009; Tadir et al., 2017). This discrepancy was explained by the spring bloom of benthic primary producers. However, the results of this study could provide another plausible explanation for the high summer chlorophyll concentrations in the sediment, which may be the post-bloom nutrient boost to the benthic ecosystem.
4.2 Decomposition-induced shifts in bacterial community abundance, production, composition, and functionality
Heterotrophic bacteria are major consumers of dissolved organic matter (DOM) in marine ecosystems and can therefore benefit from jellyfish decomposition. Previous studies have demonstrated a significant increase in bacterial abundance triggered by jellyfish degradation (Blanchet et al., 2015; Condon et al., 2011; Dinasquet et al., 2012; Frost et al., 2012; Kramar et al., 2019; Tinta et al., 2016, 2010; Titelman et al., 2006; West et al., 2009). Our study found that the decomposition of R. nomadica induced an increase of 2 orders of magnitude in the heterotrophic bacteria abundance. Autotrophic cyanobacteria, on the other hand, decreased (Synechococcus), or increased to a lower level than the unamended control (Prochlorococcus), likely due to deoxygenation (Bagby and Chisholm, 2015) or outcompetition by heterotrophic bacteria (Sisma-Ventura and Rahav, 2019; Thingstad et al., 2005).
The fate of jellyfish DOM consumed by bacteria depends on bacterial growth efficiency – the ratio of bacterial production to substrate assimilation (i.e., the sum of bacterial production and respiration) (Condon et al., 2011). While some studies have found that the succession of bacterial production mirrored bacterial abundance and respiration (Blanchet et al., 2015; Titelman et al., 2006), in our study, bacterial production reduced in the controls, whereas under jellyfish enrichment remained at a steady, 8-fold-higher level. This decoupling between bacterial abundance and production may indicate a shift in the functional diversity and metabolic demands of the jellyfish-associated bacterial communities during the experiment. In the shallow coastal waters of the EMS, bacterial production levels peak in winter and summer (Raveh et al., 2015), coinciding with, and potentially contributed by, the seasonal aggregations of R. nomadica (Edelist et al., 2020).
A significant reduction in the microbial alpha diversity indices of seawater during jellyfish decomposition was observed in this as well as in former studies (Blanchet et al., 2015; Kramar et al., 2019; Tinta et al., 2012). The decline in diversity can be attributed to the specialization of surface-colonizing bacteria, having the competitive advantage for settling from the surrounding seawater (Kramar et al., 2019) and was thus less evident in the sediment samples. Additionally, changes in bacterial diversity may result from bacterial antagonism, i.e., the production of antagonistic compounds and sensitivity or resilience to them (Titelman et al., 2006). In this study, we found a significant increase in the relative abundance of the Alphaproteobacterium Kordiimonas and the Gammaproteobacteria Algicola in the seawater enriched with R. nomadica. Similarly, the predominance of Alphaproteobacterium and Gammaproteobacteria stimulated by jellyfish decomposition was found in different studies (Basso et al., 2019; Blanchet et al., 2015; Condon et al., 2011; Dinasquet et al., 2012; Kramar et al., 2019; Tinta et al., 2012; Titelman et al., 2006). Gammaproteobacteria are conspicuous particle colonizers (Bižić-Ionescu et al., 2015; Simon et al., 2002), capable of degrading high-molecular-weight organic compounds (Cottrell and Kirchman, 2000; Reichenbach, 1992; Woyke et al., 2009), e.g., hydrocarbons (Niepceron et al., 2013). Kramar et al. (2019) found that Alphaproteobacteria and Gammaproteobacteria dominated the body surface of Aurelia, especially during the senescent phase. Blanchet et al. (2015) found a succession of bacterial diversity during the degradation of Aurelia and concluded that Alphaproteobacteria and Gammaproteobacteria have a major role in the succession of jellyfish DOM degradation. The link between the bacterial diversity of living R. nomadica at different life phases and the diversity of bacteria associated with its decomposed DOM is yet to be investigated.
Both genetic and functional diversity analyses of bacterial communities demonstrated a shift under R. nomadica degradation. We found that the predicted functions that dominated the decomposed jellyfish communities were signal transduction (two-component system), catabolic functions, such as fatty acid degradation, valine, leucine and isoleucine degradation, xenobiotic degradation pathways, and benzoate degradation. In the control communities, predominating functions were photosynthesis and carbon fixation in photosynthetic organisms. This functional shift can be explained by the fact that autotrophic cyanobacteria may be outcompeted by biodegrading heterotrophic bacteria. Once the jellyfish bloom decomposes, populations of these intrinsic microbial biodegraders become dominant and active, exploiting the carbon and nutrients released from the jellyfish. The 16S rRNA amplicon data for predicting functional profiles are a powerful tool for assessing bacterial functional diversity; nonetheless, their accuracy and resolution are dependent on the representation of sampled organisms in the 16S rRNA and KEGG databases (Sun et al., 2020; Wemheuer et al., 2018). Likely, jellyfish degraders are underrepresented in these databases. Further research using omics (e.g., whole-genome sequencing) will elucidate the metabolic potential of microbial degraders of the jellyfish necromass.
Although not to the same extent as bacterial diversity, eukaryotic diversity had also shifted during the decomposition of R. nomadica to a more flagellate-dominated community. Marine ciliates and parasitic protists (Labyrinthulomycetes) were also more abundant in the jellyfish decomposed community. Flagellate bacterivory represents the primary mechanism for the reintroduction of jellyfish carbon into the planktonic food web (Condon et al., 2011; Gasol and Kirchman, 2018). The increase in ciliates can be attributed to a “bottom-up” effect, where with the increase in flagellates, the abundance of their predators (e.g., ciliates) also increases (Epstein et al., 1992). Since jellyfish consume ciliates (Kamiyama, 2018; Stoecker et al., 1987), the flagellate carbon could be assimilated and recycled by the jellyfish, creating a positive-feedback loop termed as the “jelly-loop” (Condon et al., 2011; Lebrato and Jones, 2011).
Our study examined, for the first time, the decomposition effects of the bloom-forming invasive jellyfish R. nomadica on the oxygen and nutrient fluxes and microbial communities at the sediment–water interface. The geographical distribution of this venomous species is continuously expanding, and its outbreaks are becoming more frequent, large, and prolonged, with numerous negative impacts on human health, marine infrastructure, tourism, and fisheries.
We found that jellyfish degradation had a significant influence on the fluxes of organic and inorganic nutrients at the sediment–water interface, transforming the microbial community composition and functions. The high rates of organic nitrogen and phosphate release favored heterotrophic-dominated metabolism, leading to a shift towards heterotrophic biodegrading bacterial communities. This shift may further decrease primary production under the ultra-oligotrophic regime of the Eastern Mediterranean Sea. On the seabed, hotspots of deoxygenated, acidified, and nutrient-rich sediment may alter microbial and macrobenthic communities.
Future investigations on the decomposition dynamics of R. nomadica should be conducted in larger experimental systems (i.e., mesocosms) or in situ, under more realistic conditions. The effects of environmental change drivers, such as warming, acidification, or anthropogenic pollution, should also be tested. Additionally, the consumption of jelly-falls by scavengers in the Eastern Mediterranean Sea should be explored. This and future studies will shed light on the variable effects of the reoccurring massive blooms on the ecosystem functions and services in this rapidly changing environment.
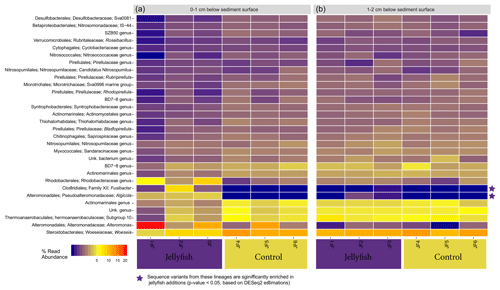
Figure C1Microbial diversity in sediment samples from the jellyfish-enriched and control experimental cylinders from 0–1 cm (a) and 1–2 cm (b) depth layers. The 30 most abundant lineages are presented and organized by hierarchical clustering. Color scale denotes the relative abundance of reads (%). The star symbols on the right-side panel indicate lineages significantly more abundant in the jellyfish treatment based on DESeq2 estimations.
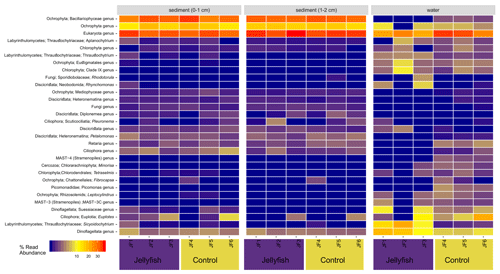
Figure C2Eukaryote diversity of seawater and sediment samples collected from jellyfish-enriched and control experimental cylinders. The 30 most abundant lineages are presented and organized by hierarchical clustering. Color scale denotes the relative abundance of reads (%).
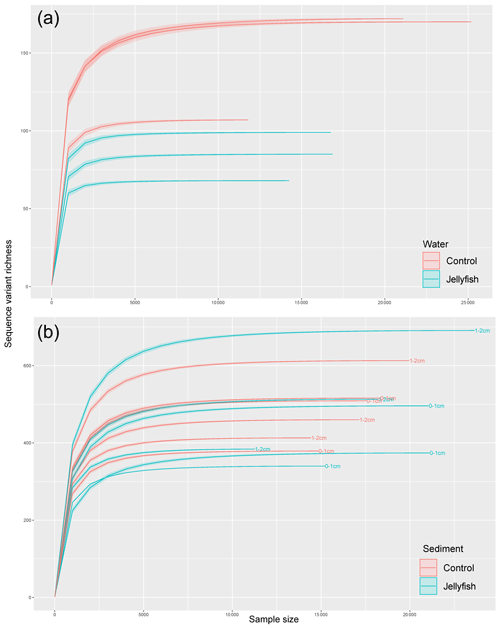
Figure C3Rarefaction curves of observed 16S rRNA sequence variants retrieved from the seawater (a) and sediment (b) samples.
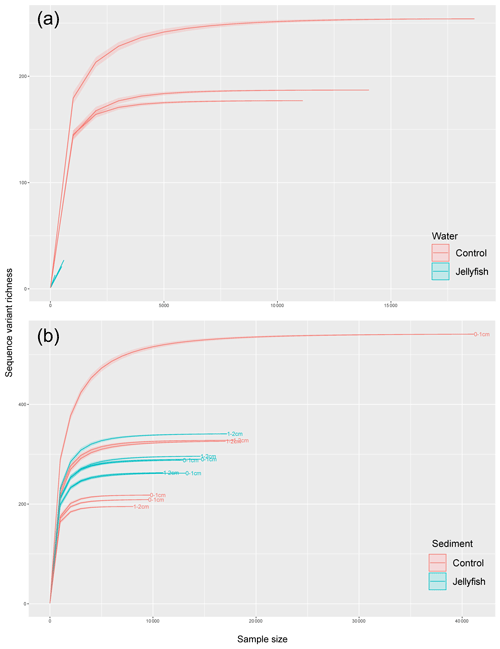
Figure C4Rarefaction curves of observed 18S rRNA sequence variants retrieved from the seawater (a) and sediment (b) samples.
Table C1SIMPER analysis of main predicted functions based on KEGG
orthologs.
SIMPER analysis indicating the main
predicted functions characterizing the jellyfish and control communities
(N=3). Av. abund indicates average abundance, av. sim indicates average similarity,
sim/SD indicates similarity standard deviation, contrib. % indicates percent
contribution, and cum. % indicates cumulative contribution.
All data were deposited in an open-access data archiving and publication repository (Pangaea, a member of the ICSU World Data System) and are available at https://doi.org/10.1594/PANGAEA.915464 (Guy-Haim et al., 2020). All the 16S and 18S rRNA gene amplicon reads were submitted to NCBI Sequence Read Archive BioProject (PRJNA626084).
This work was conceived by all authors. TGH and GSV led the research and performed the experiments, MRB conducted the microbial diversity and bioinformatic analyses, ER and NB analyzed the microbial abundance and production, and JS contributed to the study conception. TGH wrote the manuscript with substantial contributions from all co-authors. All authors have read and approved the final submitted manuscript.
The authors declare that they have no conflict of interest.
We would like to thank Dina Kolker for helping with the nutrient analysis and Dar Golomb for preparing the incubation cylinder illustration. We would also like to thank David T. Welsh and the anonymous reviewer for their insightful comments that contributed to the improvement of the paper.
This research has been partially supported by the National Israeli monitoring program.
This paper was edited by Stefano Ciavatta and reviewed by David T. Welsh and one anonymous referee.
Amaral-Zettler, L. A., McCliment, E. A., Ducklow, H. W., and Huse, S. M.: A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes, PloS one, 4, e6372, https://doi.org/10.1371/journal.pone.0006372, 2009.
Andersen, K. S., Kirkegaard, R. H., Karst, S. M., and Albertsen, M.: ampvis2: an R package to analyse and visualise 16S rRNA amplicon data, bioRxiv 299537, https://doi.org/10.1101/299537, 2018.
Angel, D. L., Edelist, D., and Freeman, S.: Local perspectives on regional challenges: jellyfish proliferation and fish stock management along the Israeli Mediterranean coast, Reg. Environ. Change, 16, 315–323, 2016.
Apprill, A., McNally, S., Parsons, R., and Weber, L.: Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton, Aquat. Microbial Ecol., 75, 129–137, 2015.
Attrill, M. J., Wright, J., and Edwards, M.: Climate-related increases in jellyfish frequency suggest a more gelatinous future for the North Sea, Limnol. Oceanogr., 52, 480–485, 2007.
Bagby, S. C. and Chisholm, S. W.: Response of Prochlorococcus to varying CO2:O2 ratios, ISME J., 9, 2232–2245, 2015.
Balistreri, P., Spiga, A., Deidun, A., Gueroun, S. K., and Yahia, M. N. D.: Further spread of the venomous jellyfish Rhopilema nomadica Galil, Spannier & Ferguson, 1990 (Rhizostomeae, Rhizostomatidae) in the western Mediterranean, BioInvasions Records, 6, 19–24, 2017.
Bar-Zeev, E. and Rahav, E.: Microbial metabolism of transparent exopolymer particles during the summer months along a eutrophic estuary system, Front. Microbiol., 6, p. 403, 2015.
Basso, L., Rizzo, L., Marzano, M., Intranuovo, M., Fosso, B., Pesole, G., Piraino, S., and Stabili, L.: Jellyfish summer outbreaks as bacterial vectors and potential hazards for marine animals and humans health? The case of Rhizostoma pulmo (Scyphozoa, Cnidaria), Sci. Total Environ., 692, 305–318, 2019.
Bižić-Ionescu, M., Zeder, M., Ionescu, D., Orlić, S., Fuchs, B. M., Grossart, H. P., and Amann, R.: Comparison of bacterial communities on limnic versus coastal marine particles reveals profound differences in colonization, Environ. Microbiol., 17, 3500–3514, 2015.
Blanchet, M., Pringault, O., Bouvy, M., Catala, P., Oriol, L., Caparros, J., Ortega-Retuerta, E., Intertaglia, L., West, N., and Agis, M.: Changes in bacterial community metabolism and composition during the degradation of dissolved organic matter from the jellyfish Aurelia aurita in a Mediterranean coastal lagoon, Environ. Sci. Pollut. Res., 22, 13638–13653, 2015.
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C., Al-Ghalith, G. A., Alexander, H., Alm, E. J., Arumugam, M., and Asnicar, F.: QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science, PeerJ Preprints, 6, 2167–9843, 2018.
Brotz, L., Cheung, W. W., Kleisner, K., Pakhomov, E., and Pauly, D.: Increasing jellyfish populations: trends in large marine ecosystems, Jellyfish Blooms IV, Springer, 2012.
Brun, F. G., Hernández, I., Vergara, J. J., Peralta, G., and Pérez-Lloréns, J. L.: Assessing the toxicity of ammonium pulses to the survival and growth of Zostera noltii, Mar. Ecol. Prog. Ser., 225, 177–187, 2002.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P.: DADA2: high-resolution sample inference from Illumina amplicon data, Nature Methods, 13, 581–583, 2016.
Chelsky, A., Pitt, K. A., and Welsh, D. T.: Biogeochemical implications of decomposing jellyfish blooms in a changing climate, Estuarine, Coast. Shelf Sci., 154, 77–83, 2015.
Chelsky, A., Pitt, K. A., Ferguson, A. J., Bennett, W. W., Teasdale, P. R., and Welsh, D. T.: Decomposition of jellyfish carrion in situ: Short-term impacts on infauna, benthic nutrient fluxes and sediment redox conditions, Sci. Total Environ., 566, 929–937, 2016.
Condon, R. H., Steinberg, D. K., Del Giorgio, P. A., Bouvier, T. C., Bronk, D. A., Graham, W. M., and Ducklow, H. W.: Jellyfish blooms result in a major microbial respiratory sink of carbon in marine systems, P. Natl. Acad. Sci., 108, 10225–10230, 2011.
Condon, R. H., Duarte, C. M., Pitt, K. A., Robinson, K. L., Lucas, C. H., Sutherland, K. R., Mianzan, H. W., Bogeberg, M., Purcell, J. E., and Decker, M. B.: Recurrent jellyfish blooms are a consequence of global oscillations, P. Natl. Acad. Sci., 110, 1000–1005, 2013.
R Core Team: A language and environment for statistical computing version 4.0.2, R Foundation for Statistical Computing, Vienna, Austria, ISBN 3-900051-07-0, available at: http://www.R-project.org/ (last access: 15 June 2020), 2020.
Cottrell, M. T. and Kirchman, D. L.: Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low-and high-molecular-weight dissolved organic matter, Appl. Environ. Microbiol., 66, 1692–1697, 2000.
Denis, L., Grenz, C., Alliot, E., and Rodier, M.: Temporal variability in dissolved inorganic nitrogen fluxes at the sediment–water interface and related annual budget on a continental shelf (NW Mediterranean), Oceanologica Acta, 24, 85–97, 2001.
Dinasquet, J., Granhag, L. M., and Riemann, L.: Stimulated bacterioplankton growth and selection for certain bacterial taxa in the vicinity of the ctenophore Mnemiopsis leidyi, Front. Microbiol., 3, p. 302, 2012.
Duarte, C. M., Pitt, K. A., Lucas, C. H., Purcell, J. E., Uye, S.-i., Robinson, K., Brotz, L., Decker, M. B., Sutherland, K. R., and Malej, A.: Is global ocean sprawl a cause of jellyfish blooms?, Front. Ecol. Environ., 11, 91–97, 2013.
Dupont, C. L., Rusch, D. B., Yooseph, S., Lombardo, M.-J., Richter, R. A., Valas, R., Novotny, M., Yee-Greenbaum, J., Selengut, J. D., and Haft, D. H.: Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage, ISME J., 6, 1186–1199, 2012.
Durham, B. P., Grote, J., Whittaker, K. A., Bender, S. J., Luo, H., Grim, S. L., Brown, J. M., Casey, J. R., Dron, A., and Florez-Leiva, L.: Draft genome sequence of marine alphaproteobacterial strain HIMB11, the first cultivated representative of a unique lineage within the Roseobacter clade possessing an unusually small genome, Stand. Genomic Sci., 9, 632–645, 2014.
Eddy, F.: Ammonia in estuaries and effects on fish, J. Fish Biol., 67, 1495–1513, 2005.
Edelist, D., Guy-Haim, T., Kuplik, Z., Zuckerman, N., Nemoy, P., and Angel, D. L.: Phenological shift in swarming patterns of Rhopilema nomadica in the Eastern Mediterranean Sea, J. Plankton Res., 42, 211–219, 2020.
Epstein, S. S., Burkovsky, I. V., and Shiaris, M. P.: Ciliate grazing on bacteria, flagellates, and microalgae in a temperate zone sandy tidal flat: ingestion rates and food niche partitioning, Journal of experimental marine Biol. Ecol., 165, 103–123, 1992.
Feely, R. A., Alin, S. R., Newton, J., Sabine, C. L., Warner, M., Devol, A., Krembs, C., and Maloy, C.: The combined effects of ocean acidification, mixing, and respiration on pH and carbonate saturation in an urbanized estuary, Estuar. Coast. Shelf Sci., 88, 442–449, 2010.
Ferretti, J. A. and Calesso, D. F.: Toxicity of ammonia to surf clam (Spisula solidissima) larvae in saltwater and sediment elutriates, Mar. Environ. Res., 71, 189–194, 2011.
Frost, J. R., Jacoby, C. A., Frazer, T. K., and Zimmerman, A. R.: Pulse perturbations from bacterial decomposition of Chrysaora quinquecirrha (Scyphozoa: Pelagiidae), in: Jellyfish Blooms IV, ISBN 978-94-007-5315-0, Springer, Dordrecht, 2012.
Galil, B.: Poisonous and venomous: marine alien species in the Mediterranean Sea and human health, in: Invasive Species and Human Health, edited by: Mazza, G., 1–15, ISBN 9781786390981, CABI, Oxfordshire, UK, 2018.
Galil, B., Spanier, E., and Ferguson, W.: The Scyphomedusae of the Mediterranean coast of Israel, including two Lessepsian migrants new to the Mediterranean, Zoologische Mededelingen, 64, 95–105, 1990.
Galil, B. S.: Truth and consequences: the bioinvasion of the Mediterranean Sea, Integrative Zoology, 7, 299–311, 2012.
Gasol, J. M. and Kirchman, D. L.: Microbial ecology of the oceans, ISBN 9781119107187, USA John Wiley & Sons, Hoboken, 2018.
Ghermandi, A., Galil, B., Gowdy, J., and Nunes, P. A.: Jellyfish outbreak impacts on recreation in the Mediterranean Sea: welfare estimates from a socioeconomic pilot survey in Israel, Ecosystem Services, 11, 140–147, 2015.
Giovannoni, S. J.: SAR11 bacteria: the most abundant plankton in the oceans, Annu. Rev. Mar. Sci., 9, 231–255, 2017.
Glud, R. N.: Oxygen dynamics of marine sediments, Mar. Biol. Res., 4, 243–289, 2008.
Gobler, C. J. and Baumann, H.: Hypoxia and acidification in ocean ecosystems: coupled dynamics and effects on marine life, Biol. Lett., 12, 20150976, https://doi.org/10.1098/rsbl.2015.0976, 2016.
Gobler, C. J., DePasquale, E. L., Griffith, A. W., and Baumann, H.: Hypoxia and acidification have additive and synergistic negative effects on the growth, survival, and metamorphosis of early life stage bivalves, PloS one, 9, e83648, https://doi.org/10.1371/journal.pone.0083648, 2014.
Guy-Haim, T., Rubin-Blum, M., Rahav, E., Belkin, N., Silverman, J., and Sisma-Ventura, G.: Experiment on biogeochemical changes following Rhopilema nomadica decomposition, PANGAEA, https://doi.org/10.1594/PANGAEA.915464, 2020.
Hammer, Ø., Harper, D. A., and Ryan, P. D.: PAST: Paleontological statistics software package for education and data analysis, Palaeontologia Electronica, 4, 1–9, 2001.
Hammond, D. E., Cummins, K. M., McManus, J., Berelson, W. M., Smith, G., and Spagnoli, F.: Methods for measuring benthic nutrient flux on the California Margin: Comparing shipboard core incubations to in situ lander results, Limnol. Oceanogr.-Methods, 2, 146–159, 2004.
Hamner, W. M. and Dawson, M. N.: A review and synthesis on the systematics and evolution of jellyfish blooms: advantageous aggregations and adaptive assemblages, Hydrobiologia, 616, 161–191, 2009.
Harrell, F.: Hmisc: Harrell Miscellaneous library for R statistical software, R package, 2, 2–3, 2004.
Hays, G. C., Doyle, T. K., and Houghton, J. D.: A paradigm shift in the trophic importance of jellyfish?, Trends Ecol. Evol., 33, 874–884, 2018.
Hubot, N., Giering, S. L. C., Lucas, C., Robidart, J., and Fuessel, J.: Evidence of nitrification associated with jellyfish, Ocean Sciences Meeting Proceedings, available at: https://agu.confex.com/agu/osm20/meetingapp.cgi/Paper/639039 (last access: 18 February 2020), 2020.
Hyams-Kaphzan, O., Almogi-Labin, A., Benjamini, C., and Herut, B.: Natural oligotrophy vs. pollution-induced eutrophy on the SE Mediterranean shallow shelf (Israel): Environmental parameters and benthic foraminifera, Mar. Pollut. Bull., 58, 1888–1902, 2009.
Ignatiades, L., Gotsis-Skretas, O., Pagou, K., and Krasakopoulou, E.: Diversification of phytoplankton community structure and related parameters along a large-scale longitudinal east–west transect of the Mediterranean Sea, J. Plankton Res., 31, 411–428, 2009.
Kamiyama, T.: Planktonic ciliates as food for the scyphozoan Aurelia coerulea: feeding and growth responses of ephyra and metephyra stages, J. Oceanogr., 74, 53–63, 2018.
Katsanevakis, S., Wallentinus, I., Zenetos, A., Leppäkoski, E., Çinar, M. E., Oztürk, B., Grabowski, M., Golani, D., and Cardoso, A. C.: Impacts of invasive alien marine species on ecosystem services and biodiversity: a pan-European review, Aquatic Invasions, 9, 391–423, 2014.
Kramar, M. K., Tinta, T., Lučić, D., Malej, A., and Turk, V.: Bacteria associated with moon jellyfish during bloom and post-bloom periods in the Gulf of Trieste (northern Adriatic), PloS one, 14, e0198056, https://doi.org/10.1371/journal.pone.0198056, 2019.
Kress, N., Thingstad, T. F., Pitta, P., Psarra, S., Tanaka, T., Zohary, T., Groom, S., Herut, B., Mantoura, R. F. C., and Polychronaki, T.: Effect of P and N addition to oligotrophic Eastern Mediterranean waters influenced by near-shore waters: a microcosm experiment, Deep Sea Res. Pt. II, 52, 3054–3073, 2005.
Kress, N., Gertman, I., and Herut, B.: Temporal evolution of physical and chemical characteristics of the water column in the Easternmost Levantine basin (Eastern Mediterranean Sea) from 2002 to 2010, J. Mar. Syst., 135, 6–13, 2014.
Kroeker, K. J., Kordas, R. L., Crim, R. N., and Singh, G. G.: Meta-analysis reveals negative yet variable effects of ocean acidification on marine organisms, Ecol. Lett., 13, 1419–1434, 2010.
Lakkis, S. and Zeidane, R.: Jellyfish swarm along the Lebanese coast (Abstract) Lebanese Association for the Advancement of Science 11th Science Meeting American University of Beirut, 1991.
Lebrato, M. and Jones, D.: Jellyfish biomass in the biological pump: Expanding the oceanic carbon cycle, Biochem. Soc. J., 33, 35–39, 2011.
Lebrato, M., Pitt, K. A., Sweetman, A. K., Jones, D. O., Cartes, J. E., Oschlies, A., Condon, R. H., Molinero, J. C., Adler, L., and Gaillard, C.: Jelly-falls historic and recent observations: a review to drive future research directions, Hydrobiologia, 690, 227–245, 2012.
Licandro, P., Conway, D., Daly Yahia, M., Fernandez de Puelles, M. L., Gasparini, S., Hecq, J.-H., Tranter, P., and Kirby, R.: A blooming jellyfish in the northeast Atlantic and Mediterranean, Biol. Lett., 6, 688–691, 2010.
Lotan, A., Ben-Hillel, R., and Loya, Y.: Life cycle of Rhopilema nomadica: a new immigrant scyphomedusan in the Mediterranean, Mar. Biol., 112, 237–242, 1992.
Lotan, A., Fine, M., and Ben-Hillel, R.: Synchronization of the life cycle and dispersal pattern of the tropical invader scyphomedusan Rhopilema nomadica is temperature dependent, Mar. Ecol. Prog. Ser., 109, 59–59, 1994.
Love, M. I., Huber, W., and Anders, S.: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 15, p. 550, 2014.
Lucas, C. H., Pitt, K. A., Purcell, J. E., Lebrato, M., and Condon, R. H.: What's in a jellyfish? Proximate and elemental composition and biometric relationships for use in biogeochemical studies, Ecology, 92, 1704, https://doi.org/10.1890/11-0302.1, 2011.
Lynam, C. P., Gibbons, M. J., Axelsen, B. E., Sparks, C. A., Coetzee, J., Heywood, B. G., and Brierley, A. S.: Jellyfish overtake fish in a heavily fished ecosystem, Current Biol., 16, R492–R493, 2006.
MacKenzie, K. M., Trueman, C. N., Lucas, C. H., and Bortoluzzi, J.: The preparation of jellyfish for stable isotope analysis, Mar. Biol., 164, p. 219, 2017.
Madkour, F. F., Safwat, W., and Hanafy, M. H.: Record of Aggregation of Alien Tropical Schyphozoan Rhopilema nomadica Galil, 1990 in the Mediterranean Coast of Egypt, Int. Mar. Sci. J., 1, p. 1, 2019.
McMurdie, P. J. and Holmes, S.: phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data, PloS one, 8, e61217, https://doi.org/10.1371/journal.pone.0061217, 2013.
Melzner, F., Thomsen, J., Koeve, W., Oschlies, A., Gutowska, M. A., Bange, H. W., Hansen, H. P., and Körtzinger, A.: Future ocean acidification will be amplified by hypoxia in coastal habitats, Mar. Biol., 160, 1875–1888, 2013.
Møller, L. F. and Riisgård, H. U.: Impact of jellyfish and mussels on algal blooms caused by seasonal oxygen depletion and nutrient release from the sediment in a Danish fjord, J. Exp. Mar. Biol. Ecol., 351, 92–105, 2007.
Müller, T., Walter, B., Wirtz, A., and Burkovski, A.: Ammonium toxicity in bacteria, Current Microbiol., 52, 400–406, 2006.
Nakar, N., Disegni, D., and Angel, D.: Economic evaluation of jellyfish effects on the fishery sector – Case study from the eastern Mediterranean, in: Proceedings of the Thirteenth Annual BIOECON Conference, 10 May 2011, 11–13, 2011.
Niepceron, M., Martin-Laurent, F., Crampon, M., Portet-Koltalo, F., Akpa-Vinceslas, M., Legras, M., Bru, D., Bureau, F., and Bodilis, J.: GammaProteobacteria as a potential bioindicator of a multiple contamination by polycyclic aromatic hydrocarbons (PAHs) in agricultural soils, Environ. Pollut., 180, 199–205, 2013.
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., O'hara, R., Simpson, G. L., Solymos, P., Stevens, M. H. H., and Wagner, H.: Vegan: community ecology package, R package version 1.17-4, available at: http://CRAN.R-project.org/package=vegan (last access: March 2016), 2010.
Parada, A. E., Needham, D. M., and Fuhrman, J. A.: Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples, Environ. Microbiol., 18, 1403–1414, 2016.
Pitt, K. A., Welsh, D. T., and Condon, R. H.: Influence of jellyfish blooms on carbon, nitrogen and phosphorus cycling and plankton production, Hydrobiologia, 616, 133–149, 2009.
Pratihary, A. K., Naqvi, S. W. A., Narvenkar, G., Kurian, S., Naik, H., Naik, R., and Manjunatha, B. R.: Benthic mineralization and nutrient exchange over the inner continental shelf of western India, Biogeosciences, 11, 2771–2791, https://doi.org/10.5194/bg-11-2771-2014, 2014.
Purcell, J. E.: Jellyfish and ctenophore blooms coincide with human proliferations and environmental perturbations, Annu. Rev. Mar. Sci., 4, 209–235, 2012.
Purcell, J. E., Uye, S.-I., and Lo, W.-T.: Anthropogenic causes of jellyfish blooms and their direct consequences for humans: a review, Mar. Ecol. Prog. Ser., 350, 153–174, 2007.
Qu, C.-F., Song, J.-M., Li, N., Li, X.-G., Yuan, H.-M., Duan, L.-Q., and Ma, Q.-X.: Jellyfish (Cyanea nozakii) decomposition and its potential influence on marine environments studied via simulation experiments, Mar. Pollut. Bull., 97, 199–208, 2015.
Quiñones, J., Mianzan, H., Purca, S., Robinson, K. L., Adams, G. D., and Acha, E. M.: Climate-driven population size fluctuations of jellyfish (Chrysaora plocamia) off Peru, Mar. Biol., 162, 2339–2350, 2015.
Rahav, E., Belkin, N., Paytan, A., and Herut, B.: Phytoplankton and Bacterial Response to Desert Dust Deposition in the Coastal Waters of the Southeastern Mediterranean Sea: A Four-Year In Situ Survey, Atmosphere, 9, p. 305, 2018a.
Rahav, E., Raveh, O., Hazan, O., Gordon, N., Kress, N., Silverman, J., and Herut, B.: Impact of nutrient enrichment on productivity of coastal water along the SE Mediterranean shore of Israel-A bioassay approach, Mar. Pollut. Bull., 127, 559–567, 2018b.
Raveh, O., David, N., Rilov, G., and Rahav, E.: The temporal dynamics of coastal phytoplankton and bacterioplankton in the Eastern Mediterranean Sea, PLoS One, 10, e0140690, https://doi.org/10.1371/journal.pone.0140690, 2015.
Reichenbach, H.: The order cytophagales, in: The prokaryotes, Springer, New York, NY, ISBN 978-1-4757-2193-5, 1992.
Richardson, A. J., Bakun, A., Hays, G. C., and Gibbons, M. J.: The jellyfish joyride: causes, consequences and management responses to a more gelatinous future, Trends Ecol. Evol., 24, 312–322, 2009.
Sanz-Martín, M., Pitt, K. A., Condon, R. H., Lucas, C. H., Novaes de Santana, C., and Duarte, C. M.: Flawed citation practices facilitate the unsubstantiated perception of a global trend toward increased jellyfish blooms, Global Ecol. Biogeogr., 25, 1039–1049, 2016.
Schnedler-Meyer, N. A., Kiørboe, T., and Mariani, P.: Boom and Bust: Life History, Environmental Noise, and the (un) Predictability of Jellyfish Blooms, Front. Mar. Sci., 5, p. 257, 2018.
Shiganova, T., Mirzoyan, Z., Studenikina, E., Volovik, S., Siokou-Frangou, I., Zervoudaki, S., Christou, E., Skirta, A., and Dumont, H.: Population development of the invader ctenophore Mnemiopsis leidyi, in the Black Sea and in other seas of the Mediterranean basin, Mar. Biol., 139, 431–445, 2001.
Simon, M.: Improved assessment of bacterial production: combined measurements of protein synthesis via leucine and cell multiplication via thymidine incorporation, Ergebnisse der Limnologie ERLIA, 6, 151–155, 1990.
Simon, M. and Azam, F.: Protein content and protein synthesis rates of planktonic marine bacteria, Marine ecology progress series, Oldendorf, 51, 201–213, 1989.
Simon, M., Grossart, H.-P., Schweitzer, B., and Ploug, H.: Microbial ecology of organic aggregates in aquatic ecosystems, Aquat. Microbial Ecol., 28, 175–211, 2002.
Sisma-Ventura, G. and Rahav, E.: DOP stimulates heterotrophic bacterial production in the oligotrophic southeastern Mediterranean coastal waters, Front. Microbiol., 10, 1913, https://doi.org/10.3389/fmicb.2019.01913, 2019.
Skoog, A. C. and Arias-Esquivel, V. A.: The effect of induced anoxia and reoxygenation on benthic fluxes of organic carbon, phosphate, iron, and manganese, Sci. Total Environ., 407, 6085–6092, 2009.
Stief, P.: Stimulation of microbial nitrogen cycling in aquatic ecosystems by benthic macrofauna: mechanisms and environmental implications, Biogeosciences, 10, 7829–7846, https://doi.org/10.5194/bg-10-7829-2013, 2013.
Stoeck, T., Bass, D., Nebel, M., Christen, R., Jones, M. D., Breiner, H. W., and Richards, T. A.: Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water, Molecular Ecol., 19, 21–31, 2010.
Stoecker, D. K., Michaels, A. E., and Davis, L. H.: Grazing by the jellyfish, Aurelia aurita, on microzooplankton, J. Plankton Res., 9, 901–915, 1987.
Streftaris, N. and Zenetos, A.: Alien marine species in the Mediterranean-the 100 `Worst Invasives' and their impact, Mediterranean Mar. Sci., 7, 87–118, 2006.
Sun, S., Jones, R. B., and Fodor, A. A.: Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories, Microbiome, 8, 1–9, 2020.
Sweetman, A. K. and Chapman, A.: First observations of jelly-falls at the seafloor in a deep-sea fjord, Deep Sea Res. Pt. I, 58, 1206–1211, 2011.
Sweetman, A. K., Smith, C. R., Dale, T., and Jones, D. O.: Rapid scavenging of jellyfish carcasses reveals the importance of gelatinous material to deep-sea food webs, Proc. Roy. Soc. B, 281, 20142210, https://doi.org/10.1098/rspb.2014.2210, 2014.
Sweetman, A. K., Chelsky, A., Pitt, K. A., Andrade, H., van Oevelen, D., and Renaud, P. E.: Jellyfish decomposition at the seafloor rapidly alters biogeochemical cycling and carbon flow through benthic food-webs, Limnol. Oceanogr., 61, 1449–1461, 2016.
Tadir, R., Benjamini, C., Almogi-Labin, A., and Hyams-Kaphzan, O.: Temporal trends in live foraminiferal assemblages near a pollution outfall on the Levant shelf, Mar. Pollut. Bull., 117, 50–60, 2017.
Thingstad, T. F., Krom, M., Mantoura, R., Flaten, G. F., Groom, S., Herut, B., Kress, N., Law, C., Pasternak, A., and Pitta, P.: Nature of phosphorus limitation in the ultraoligotrophic eastern Mediterranean, Science, 309, 1068–1071, 2005.
Tinta, T., Malej, A., Kos, M., and Turk, V.: Degradation of the Adriatic medusa Aurelia sp. by ambient bacteria, in: Jellyfish Blooms: New Problems and Solutions, ISBN 978-90-481-9540-4, Springer, Dordrecht, 2010.
Tinta, T., Kogovšek, T., Malej, A., and Turk, V.: Jellyfish modulate bacterial dynamic and community structure, PloS one, 7, e39274, https://doi.org/10.1371/journal.pone.0039274, 2012.
Tinta, T., Kogovšek, T., Turk, V., Shiganova, T. A., Mikaelyan, A. S., and Malej, A.: Microbial transformation of jellyfish organic matter affects the nitrogen cycle in the marine water column—A Black Sea case study, J. Exp. Mar. Biol. Ecol., 475, 19–30, 2016.
Titelman, J., Riemann, L., Sørnes, T. A., Nilsen, T., Griekspoor, P., and Båmstedt, U.: Turnover of dead jellyfish: stimulation and retardation of microbial activity, Mar. Ecol. Prog. Ser., 325, 43–58, 2006.
Welsh, D. T.: It's a dirty job but someone has to do it: the role of marine benthic macrofauna in organic matter turnover and nutrient recycling to the water column, Chem. Ecol., 19, 321–342, 2003.
Welsh, D. T., Dunn, R. J., and Meziane, T.: Oxygen and nutrient dynamics of the upside down jellyfish (Cassiopea sp.) and its influence on benthic nutrient exchanges and primary production, Hydrobiologia, 635, 351–362, 2009.
Wemheuer, F., Taylor, J. A., Daniel, R., Johnston, E., Meinicke, P., Thomas, T., and Wemheuer, B.: Tax4Fun2: a R-based tool for the rapid prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene marker gene sequences, BioRxiv 490037, 2018.
West, E. J., Welsh, D. T., and Pitt, K. A.: Influence of decomposing jellyfish on the sediment oxygen demand and nutrient dynamics, in: Jellyfish Blooms: Causes, Consequences, and Recent Advances, Springer, ISBN 978-1-4020-9748-5, 2008.
West, E. J., Pitt, K. A., Welsh, D. T., Koop, K., and Rissik, D.: Top-down and bottom-up influences of jellyfish on primary productivity and planktonic assemblages, Limnol. Oceanogr., 54, 2058–2071, 2009.
Wickham, H.: ggplot2: elegant graphics for data analysis, Springer, New York, ISBN 9783319242750, 2016.
Woyke, T., Xie, G., Copeland, A., Gonzalez, J. M., Han, C., Kiss, H., Saw, J. H., Senin, P., Yang, C., and Chatterji, S.: Assembling the marine metagenome, one cell at a time, PloS one, 4, e5299, https://doi.org/10.1371/journal.pone.0005299, 2009.
Xiao, W., Zeng, Y., Liu, X., Huang, X., Chiang, K.-P., Mi, T., Zhang, F., Li, C., Wei, H., and Yao, Q.: The impact of giant jellyfish Nemopilema nomurai blooms on plankton communities in a temperate marginal sea, Mar. Pollut. Bull., 149, 110507, https://doi.org/10.1016/j.marpolbul.2019.110507, 2019.
Yahia, M. N. D., Yahia, O. K.-D., Gueroun, S. K. M., Aissi, M., Deidun, A., Fuentes, V., and Piraino, S.: The invasive tropical scyphozoan Rhopilema nomadica Galil, 1990 reaches the Tunisian coast of the Mediterranean Sea, BioInvasions Records, 2, 319–323, 2013.
Yakimov, M. M., Timmis, K. N., and Golyshin, P. N.: Obligate oil-degrading marine bacteria, Current Opinion in Biotechnology, 18, 257–266, 2007.
Zenetos, A., Gofas, S., Verlaque, M., Çinar, M. E., Raso, J. G., Bianchi, C., Morri, C., Azzurro, E., Bilecenoglu, M., and Froglia, C.: Alien species in the Mediterranean Sea by 2010. A contribution to the application of European Union's Marine Strategy Framework Directive (MSFD). Part I. Spatial distribution, Mediterranean Mar. Sci., 11, 381–493, 2010.
Zunino, S., Canu, D. M., Bandelj, V., and Solidoro, C.: Effects of ocean acidification on benthic organisms in the Mediterranean Sea under realistic climatic scenarios: a meta-analysis, Reg. Stud. Mar. Sci., 10, 86–96, 2017.
- Abstract
- Introduction
- Methods and materials
- Results
- Discussion
- Conclusions
- Appendix A: Additional nutrient data
- Appendix B: Nutrient–bacteria correlations
- Appendix C: Bacterial and eukaryotic diversity in water and sediment samples
- Data availability
- Author contributions
- Competing interests
- Acknowledgements
- Financial support
- Review statement
- References
- Abstract
- Introduction
- Methods and materials
- Results
- Discussion
- Conclusions
- Appendix A: Additional nutrient data
- Appendix B: Nutrient–bacteria correlations
- Appendix C: Bacterial and eukaryotic diversity in water and sediment samples
- Data availability
- Author contributions
- Competing interests
- Acknowledgements
- Financial support
- Review statement
- References