the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Plant genotype controls wetland soil microbial functioning in response to sea-level rise
Susanne Liebner
Svenja Reents
Stefanie Nolte
Kai Jensen
Fabian Horn
Peter Mueller
Climate change can strongly alter soil microbial functioning via plant–microbe interactions, often with important consequences for ecosystem carbon and nutrient cycling. Given the high degree of intraspecific trait variability in plants, it has been hypothesized that genetic shifts within plant species yield a large potential to control the response of plant–microbe interactions to climate change. Here we examined if sea-level rise and plant genotype interact to affect soil microbial communities in an experimental coastal wetland system, using two known genotypes of the dominant salt-marsh grass Elymus athericus characterized by differences in their sensitivity to flooding stress – i.e., a tolerant genotype from low-marsh environments and an intolerant genotype from high-marsh environments. Plants were exposed to a large range of flooding frequencies in a factorial mesocosm experiment, and soil microbial activity parameters (exo-enzyme activity and litter breakdown) and microbial community structure were assessed. Plant genotype mediated the effect of flooding on soil microbial community structure and determined the presence of flooding effects on exo-enzyme activities and belowground litter breakdown. Larger variability in microbial community structure, enzyme activities, and litter breakdown in soils planted with the intolerant plant genotype supported our general hypothesis that effects of climate change on soil microbial activity and community structure can depend on plant intraspecific genetic variation. In conclusion, our data suggest that adaptive genetic variation in plants could suppress or facilitate the effects of sea-level rise on soil microbial communities. If this finding applies more generally to coastal wetlands, it yields important implications for our understanding of ecosystem–climate feedbacks in the coastal zone.
- Article
(2584 KB) - Full-text XML
-
Supplement
(1039 KB) - BibTeX
- EndNote
Climate change strongly affects soil microbial decomposition, with important consequences for global carbon (C) and nutrient cycles (Davidson and Janssens, 2006; Dijkstra et al., 2010). Plant–microbe interactions in the rhizosphere are particularly susceptible to various climate change factors (Philippot et al., 2013; Pugnaire et al., 2019; Wieder, 2014). It is therefore not sufficient to only study the direct effects of abiotic climate change drivers on soil microbial communities and resulting changes in ecosystem functioning. Plant-mediated indirect effects of climate change on soil microbial communities also need to be examined (Bardgett et al., 2008; Van der Putten et al., 2013). Prior work on a wide range of ecosystems indicated that changes in plant productivity and community composition control soil microbial functioning in response to climate change, often with marked effects on ecosystem C as well as greenhouse-gas and nutrient dynamics (Fuchslueger et al., 2014; Mueller et al., 2020; Stagg et al., 2018; Ward et al., 2013).
Climate change is known to affect the intraspecific genetic structure within plant populations (Bustos-Korts et al., 2018; Crutsinger et al., 2006; Jump and Peñuelas, 2005), and it has been hypothesized that these intraspecific genetic shifts can translate into important changes in soil microbial functioning (Fischer et al., 2014; terHorst and Zee, 2016; Van Nuland et al., 2016; Ware et al., 2019). This hypothesis is based on studies demonstrating differences in soil microbial community structure or activity in soils of different plant genotypes (Madritch and Lindroth, 2011; Pérez-Izquierdo et al., 2019; Schweitzer et al., 2008; Seliskar et al., 2002; Zogg et al., 2018). Furthermore, genotype effects on soil C and nitrogen (N) stocks as well as N transformations have been observed to be variable across multiple common garden sites (Pregitzer et al., 2013). However, experimental evidence for the interaction effects of plant genotype and climate change factors on soil microbial C cycling is virtually absent.
Plant-mediated climate change effects on soil microbial functioning are expected to be particularly pronounced in wetlands, because here plants not only control the microbial substrate (i.e., electron donor) supply, but they also regulate the availability of electron acceptors by providing oxygen to an otherwise reducing rhizosphere (Kirwan and Megonigal, 2013; Wolf et al., 2007). At the same time, wetland soil microbial functioning plays a disproportionately large role in the global climate system (Freeman et al., 2001; Megonigal et al., 2003). In recent years, climate change research in tidal wetlands and other so-called blue carbon ecosystems has gained increasing attention by the scientific community (Kirwan et al., 2013, 2014; Spivak et al., 2019). These ecosystems are among the most effective long-term C sinks of the biosphere (Chmura et al., 2003; McLeod et al., 2011), but the impacts of accelerated rates of sea-level rise (SLR) destabilize tidal wetlands worldwide (Kirwan and Megonigal, 2013).
SLR affects the flooding frequency of tidal wetlands and represents the overriding climate change factor impacting tidal wetlands (Kirwan and Megonigal, 2013). Its effects on ecosystem functioning are largely plant-mediated and extremely variable, ranging from strong positive effects on soil C sequestration to ecosystem destabilization and ultimately loss (Rogers et al., 2019). SLR and the resulting flooding frequency alter plant primary production and microbial decomposition, the two primary factors controlling C sequestration in coastal marine ecosystems (Kirwan and Megonigal, 2013). Primary production often follows a unimodal (i.e., optimum) response to SLR, although interspecific variability is high (Kirwan and Guntenspergen, 2012; Morris et al., 2013). The microbial decomposition response to SLR is less understood. The prevailing notion is that decomposition rates are inversely related to flooding. However, recent studies demonstrated that the responses of decomposition and primary production to SLR are coupled (Janousek et al., 2017; Mueller et al., 2016; Stagg et al., 2017). For instance, Mueller et al. (2016) have demonstrated soil microbial activity is not directly affected by SLR and its control on soil oxygen availability but indirectly by the aboveground-biomass response to flooding frequency, which determines the input of both oxygen and labile substrates to soil microbial communities.
Considering the low plant community-level diversity of many wetland types, such as salt marshes and ombrotrophic peatlands (Wanner et al., 2014; Warner and Asada, 2006), and the strong plant control of microbial C cycling in wetland soils, it is possible that intraspecific variation and adaptive capacity function as important yet largely overlooked mediators of wetland–climate feedbacks. Here, we study the interaction effect of flooding frequency and plant genotype on soil microbial community structure and functioning, using the dominant tidal wetland grass Elymus athericus as a model species (Bockelmann and Neuhaus, 1999). Two genotypes of Elymus athericus, which differ in their adaptation to flooding frequency, have been identified: a flooding-sensitive genotype from the high marsh (to simplify hereafter referred to as intolerant genotype) and a less flooding-sensitive genotype from the low marsh (hereafter tolerant genotype) (Bockelmann et al., 2003; Reents et al., 2021). Given the overriding control of plant processes on microbial functioning in wetland soils, we hypothesized that flooding effects on microbial decomposition and microbial community structure are strong in soils with the intolerant plant genotype but absent or buffered in soils of the tolerant plant genotype (Fig. 1).
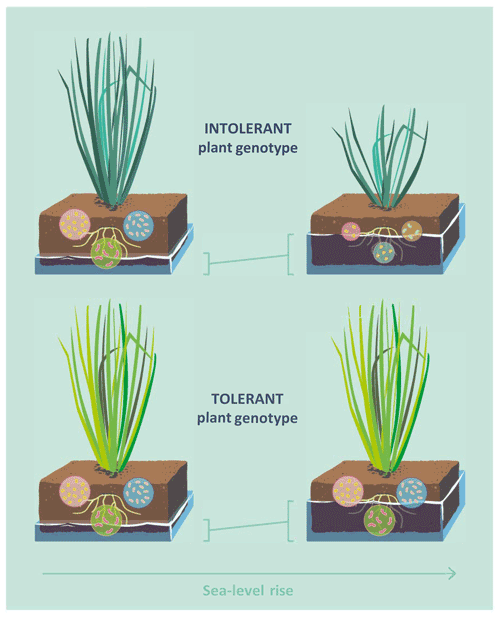
Figure 1Conceptual diagram illustrating the hypothesis that effects of sea-level rise on soil microbial functioning are mediated by intraspecific genetic variation in plants. Two genotypes of the dominant tidal wetland grass Elymus athericus have been identified, an intolerant plant genotype, found in high-marsh environments, and an tolerant plant genotype, found in low-marsh environments. The tolerant genotype shows no reduction of aboveground biomass even in response to extreme increases in flooding frequency (Reents et al., 2021). Given the overriding control of plant aboveground processes on microbial functioning in tidal wetland soils, we hypothesized that the tolerant genotype buffers the response of the soil microbial community to increasing flooding frequency.
2.1 Experimental design
The experiment was conducted from July to October 2017 (12 weeks) at the Institute of Plant Science and Microbiology (IPM), Universität Hamburg, Germany. We used platforms positioned at three elevations in a 12 m3 tidal tank to induce three flooding-frequency treatments capturing the full range of flooding frequencies of a typical NW European salt marsh: daily (two floods every day, simulating pioneer-zone conditions), weekly (two floods on one day per week, simulating low-marsh conditions), and monthly (two floods on one day every 2 weeks, simulating high-marsh conditions). Mesocosms (Ø = 15 cm; h=17 cm) were filled with soils collected from the upper 25 cm soil layer of the high-marsh platform of a salt marsh near Sönke-Nissen-Koog, Germany (DE, 54∘36′ N, 8∘49′ E). The collected soils had low organic matter contents of 3 %–4 %, low C:N ratios of 14–16, and a relatively high pH of 7.5–8.0, which are typical features of the minerogenic marshes of the European North Sea (Mueller et al., 2019). Soils were sieved using a 1 cm mesh to remove roots, rhizomes, and other coarse materials and homogenized before being transferred to the mesocosms. Mesocosms were planted with either tolerant or intolerant genotypes of the grass Elymus athericus. The design included a total of 48 planted mesocosms (24 per genotype, 16 per flooding treatment) resulting from n=8 intolerant versus n=8 tolerant genotypes per flooding treatment. We additionally added four unplanted mesocosms (n=4) to each flooding treatment to understand the direct (i.e., plant-independent) effect of flooding on soil microbial communities and thus gain more mechanistic insight into potential genotype effects.
Plants were collected in April 2015 from Elymus athericus stands on the island of Schiermonnikoog, the Netherlands, that have previously been demonstrated to be dominated by genetically distinct populations of Elymus, i.e., flooding-tolerant genotypes from the low marsh and intolerant genotypes from the high marsh (Bockelmann et al., 2003; Reents et al., 2021). In their natural environments, intolerant genotypes are grey blue in color and produce tall shoots in dense stands, whereas tolerant genotypes are light green, produce more ramets, and grow in a patchier distribution (Bockelmann et al., 2003). Recent common-garden experiments could demonstrate that some phenotypic differences between the genotypes are heritable. These include leaf color, shoot mass and length, and rhizome and root production (Mueller et al., 2021; Reents et al., 2021).
2.2 Soil sampling and processing
Soil sampling took place in October 2017 after 12 weeks of exposure to different flooding treatments and plant genotypes. Plant biomass and litter were removed prior to sampling. From each mesocosm, one soil sample was taken as a 5 cm diameter and 5 cm deep core using a volumetric steel ring. Subsamples of 20 g were homogenized and stored frozen until used for microbial enzyme assays and DNA extraction. The residual sample was passed through a 2.5 mm sieve, air-dried at 65 ∘C to constant weight, and used to determine dry mass and other soil properties.
2.3 Microbial exo-enzyme activity and belowground litter decomposition
Potential exo-enzyme activity (EEA) of ß-glucosidase, cellobiosidase, leucine-aminopeptidase, and chitinase was determined in fluorometric assays following Mueller et al. (2017). Briefly, 1:20 soil slurries were produced using 50 mmol/L bicarbonate buffer (pH = 8) (Sinsabaugh et al., 2003). Well-plate assays were conducted to measure potential EEA. Plates were incubated in the dark at 20 ∘C for 16 h and read on a multi-detection microplate reader (BioTek Synergy HT, Winooski, USA). The emission and excitation wavelengths were set at 460 and 365 nm, respectively. The four enzymes assayed are commonly used as proxies for microbial C- and N-acquisition activities that reflect the microbial C and N demand (Sinsabaugh et al., 2008, 2009).
We assessed the decomposition of standardized plant litter in the rhizosphere to evaluate if genotype effects on soil microbial exo-enzyme activity translate into altered organic matter turnover and thus into ecosystem functioning (Ochoa-Hueso et al., 2020). The decomposition rate constant (k) and stabilization factor (S) were assessed following the tea bag index (TBI) method (Keuskamp et al., 2013). The TBI is a standardized litter-decay assay using commercially available tea materials as standardized plant litter. The TBI has widely been applied to characterize and compare decomposition dynamics within and across ecosystems (Keuskamp et al., 2013; Mueller et al., 2018; Ochoa-Hueso et al., 2020). The advantages and limitations of the TBI and other standardized decomposition assays, such as cotton- and cellulose-strip assays, have been extensively discussed elsewhere (Clark, 1970; Mueller et al., 2018; Ochoa-Hueso et al., 2020; Risch et al., 2007). Each pot received two polypropylene tea bags (55 mm × 50 mm), one containing green tea (EAN: 8 714100 770542; Lipton, Unilever) and one containing rooibos (EAN: 8 722700 188438; Lipton, Unilever). Bags were deployed in 5 cm soil depth. The initial weight of the contents was determined by subtracting the mean content weight of five empty bags (green tea: 1.69 ± 0.005 g; rooibos tea: 1.79 ± 0.009 g). Bags were retrieved after an incubation period of 90 d, carefully separated from roots and soil, dried for 48 h at 70 ∘C, and weighed. The TBI parameters k and S were calculated following the tidal-wetland-adapted TBI protocol (Mueller et al., 2018).
2.4 Microbial community structure – Illumina sequencing
Soil DNA was extracted from n=3 randomly chosen mesocosms per treatment combination using the PowerSoil DNA extraction kit (Quiagen). From each mesocosm, two samples (technical replicates) were taken to assess within-mesocosm variability. DNA quality and yield were assessed using a fluorometer (Qubit 2.0, Thermo Fisher Scientific). PCR amplification of the prokaryotic 16S rRNA gene region was conducted using the barcoded primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Caporaso et al., 2010). The PCR protocol (PCR mix and cycling conditions) followed Meier et al. (2019). PCR products were purified using the Agencourt AMPure XP– PCR purification kit (Beckman Coulter, Inc.) and were pooled into a single sequencing library at equimolar concentrations (20 ng DNA per sample). Sequencing was conducted by Eurofins Scientific (Konstanz, Germany) using an Illumina HiSeq platform and Miseq v3 kits (2×300 bp). Sequence analysis and bioinformatics followed Holm et al. (2020). Briefly, the library was demultiplexed using Cutadapt (Martin, 2011), and samples were error-corrected using the DADA2 pipeline (Callahan et al., 2016). Paired-end reads were merged, and low-quality sequences and chimeras were removed. Amplicon sequence variants (ASVs) were assigned to the SILVA database (version 132) (Quast et al., 2013) applying vsearch (Rognes et al., 2016) as implemented in the QIIME2 framework (Bolyen et al., 2019). Taxonomic assignment of sequences was based on a 99 % similarity threshold. Raw sequencing data are available at the European Nucleotide Archive (ENA) under BioProject accession number PRJEB38150 and sample accession numbers ERS4541081–ERS4541134.
2.5 Statistical analyses
We used two-way ANOVA or two-way PERMANOVA to analyze the data of our two-factorial design (two genotypes and three flooding frequencies). Normal distribution of residuals was assessed visually prior to ANOVA testing. Due to the fully balanced study design, potential moderate deviations from homogeneity of variance between groups were considered unimportant for both ANOVA and PERMANOVA testing (Anderson, 2017; Box, 1954; McGuinness, 2002). Along with ANOVA tests, we used Cochran's C test with α=0.01 to test for single large variances (sensu McGuinness, 2002). When Cochran's test remained significant after log-transformation of data, nonparametric Kruskal–Wallis tests were conducted instead of ANOVA, which was only the case for a single parameter (decomposition rate, k). Beta dispersion tests conducted along with PERMANOVA indicated no significant heterogeneity of variances.
Two-way ANOVA was conducted to test for effects of flooding frequency, plant genotype, and their interaction on EEAs, k, and S. Data on EEAs, k, and S are presented both as absolute values and in relation to the mean of the unplanted mesocosms of each flooding treatment (i.e., percentage change versus unplanted conditions =ΔEEA, Δk, ΔS). This was done to explore potential differences in magnitude and direction of plant effects between genotypes. Two-way PERMANOVA, based on Bray–Curtis dissimilarities, was used to test for effects of flooding frequency and genotype on microbial community composition. Data of technical replicates were averaged for two-way PERMANOVA. Data were visualized using nonmetric multidimensional scaling (NMDS) displaying all technical replicates. In addition to these two-factorial tests, we conducted a paired t test to compare effect sizes of the flooding treatment on EEAs between genotypes and Pearson correlation and canonical correspondence analysis (CCA) to explore the relationships between soil microbial parameters (i.e., activity of ß-glucosidase, cellobiosidase, chitinase, and leucine-aminopeptidase, k, and S) and plant biomass parameters (i.e., aboveground, belowground, and total biomass; taken from Reents et al., 2021). Bonferroni correction was applied for multiple comparisons. The analysis of flooding effects on soil microbial parameters in the absence of plants can provide additional mechanistic insight but was not the primary objective of our study. To facilitate a clearer presentation of genotype and genotype–flooding interaction effects, flooding effects in the absence of plants were analyzed separately (i.e., not as part of our two-factorial design) using one-way ANOVA or one-way PERMANOVA.
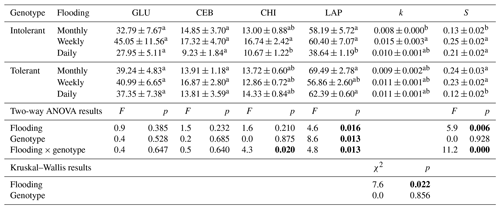
Table 1Exo-enzyme activities (nmol g DW−1 h−1) of ß-glucosidase (GLU), cellobiosidase (CEB), chitinase (CHI), and leucine-aminopeptidase (LAP) as well as the litter-breakdown parameters k (decomposition rate constant) and S (stabilization factor) in soils planted with intolerant and tolerant plant genotypes of Elymus athericus exposed to three different flooding frequencies (monthly, weekly, and daily). Values are means and SE (n=8). Values not connected by the same letter within one column are significantly different at p≤0.05 based on Tukey's HSD tests. Corresponding two-way ANOVA results are included below (p values highlighted in bold at p≤0.05). Nonparametric Kruskal–Wallis tests were conducted instead of ANOVA and Tukey's tests for k.
3.1 Soil microbial enzyme activity and litter decomposition
Enzyme activities were only affected by flooding frequency in soils planted with the intolerant genotype, whereas none of the four EEAs were affected in soils planted with the tolerant genotype (Table 1). In soils with the intolerant genotype, all four EEAs showed a unimodal response to flooding: they were always highest at the intermediate (i.e., weekly) flooding frequency and always lowest at the highest (i.e., daily) flooding frequency, whereas no consistent pattern was found in soils of the tolerant genotype (Table 1, Fig. 2). Overall, the effect size of flooding frequency (i.e., the difference between highest and lowest mean activity of the three flooding treatments) was 1.7–4.7 times greater in the intolerant vs. tolerant genotype (Fig. 3).
C-acquisition enzymes (ß-glucosidase and cellobiosidase, sensu Sinsabaugh et al., 2009) showed different responses than N-acquisition enzymes (leucine-aminopeptidase and chitinase, sensu Sinsabaugh et al., 2009). The activity of C-acquisition enzymes was not affected by flooding frequency, genotype, and their interaction (Fig. 2a, Table 1), whereas N-acquisition enzymes were significantly reduced by the highest flooding frequency (Fig. 2b, Table 1). The reduction of N-acquisition activities by increasing flooding frequency was only observed in the intolerant genotype, whereas activities remained unchanged throughout flooding treatments in the tolerant genotype (Fig. 2b).
Analyzing the EEA data in relation to the activity under unplanted conditions revealed contrasting plant effects between genotypes (Fig. 2). Specifically, at our highest flooding frequency, the activity change in relation to the unplanted condition was negative in the intolerant genotype but positive in the tolerant genotype (Fig. 2). This contrasting pattern in the direction of plant effects was generally found for all enzymes assayed, but it was significant in the N enzymes only (Fig. 2). The absolute values of enzyme activities under unplanted conditions are presented in the Supplement. None of the four enzymes assayed showed a significant response to changes in flooding frequency under unplanted conditions (Fig. S2, Table S1).
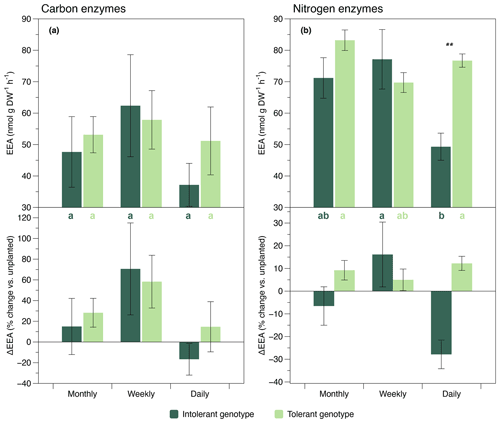
Figure 2Exo-enzyme activity (EEA) of C-acquisition enzymes (a, sum of ß-glucosidase and cellobiosidase) and N-acquisition enzymes (b, sum of leucine-aminopeptidase and chitinase) in soils planted with intolerant and tolerant plant genotypes of Elymus athericus exposed to three different flooding frequencies (monthly, weekly, and daily). Upper panels show absolute values, and lower panels show activities in relation to the unplanted control (i.e., percentage change vs. the mean value of n=4 unplanted mesocosms per flooding treatment). Values are means and SE (n=8). Asterisks denote significant genotypic differences within the same flooding treatment (; ). Bars not labeled by the same letter are significantly different at p≤0.05. All statistical results refer to the absolute enzyme data shown in the upper panels and are based on Tukey's HSD tests following two-way ANOVA.
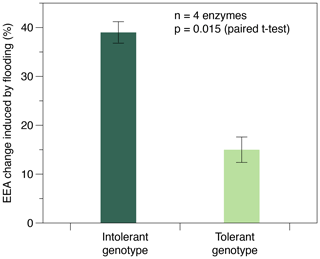
Figure 3Maximum change in exo-enzyme activity (EEA) induced by the flooding treatment in soils planted with flooding-intolerant vs. flooding-tolerant genotypes of Elymus athericus. EEA change (%) refers to the difference between max and min average EEA of the three flooding treatments determined for each of the n=4 exo-enzymes assayed (compare Table 1). Values are means and SE.
The initial belowground litter decomposition rate, k (sensu Keuskamp et al., 2013), was significantly affected by flooding frequency. However, based on pairwise comparisons, this effect was only significant in the intolerant plant genotype (Fig. 4a), reflecting the greater flooding sensitivity of the soil microbial community that has also been observed in EEAs. A strong interaction effect of flooding frequency and genotype was detected on the litter stabilization factor, S (Keuskamp et al., 2013) (Fig. 4b). At the highest flooding frequency, S was markedly lower in the rhizosphere of the tolerant versus intolerant genotype, whereas the reversed pattern was found at our lowest (i.e., monthly) flooding frequency (Fig. 4b).
Significant relationships between plant biomass parameters (taken from Reents et al., 2021), soil EEAs, and litter-breakdown parameters have been observed (Table 2). C enzymes were not significantly related to any plant biomass parameter, reflecting the missing plant genotype effect on microbial C-enzyme activities, whereas N-enzyme activities were significantly positively related to plant aboveground biomass (Table 2). Relationships between plant biomass parameters and litter-breakdown parameters (k, S) were only significant when controlling for direct (i.e., plant-independent) flooding effects (Table 2). Specifically, ΔS was significantly related to both above- and belowground biomass (Table 2). k and Δk were most strongly related to C-enzyme activities, whereas S was not significantly related to EEAs, and ΔS was significantly related to N-enzyme activities (Table S2).
Table 2Correlations between plant biomass parameters and soil microbial activity parameters using a Bonferroni correction for multiple comparisons (α=0.05/number of pairwise comparisons). Shown are Pearson correlation coefficients (r). Significant correlations are highlighted in bold (p≤α).
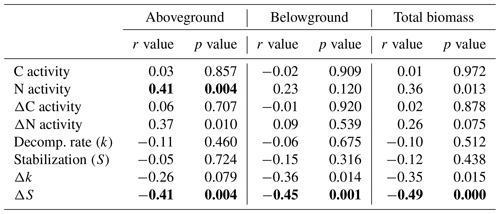
C activity: sum of C-acquisition enzyme activities (ß-glucosidase + cellobiosidase); N activity: sum of N-acquisition enzyme activities (aminopeptidase + chitinase); Decomp. rate (k): decomposition rate constant (sensu Keuskamp et al. 2013); stabilization (S): stabilization factor (sensu Keuskamp et al., 2013); Δ: activity values in relation to the unplanted control (i.e., percentage change of planted vs. unplanted mesocosms) reflecting plant effects independent of direct (i.e., non-plant-mediated) flooding effects.
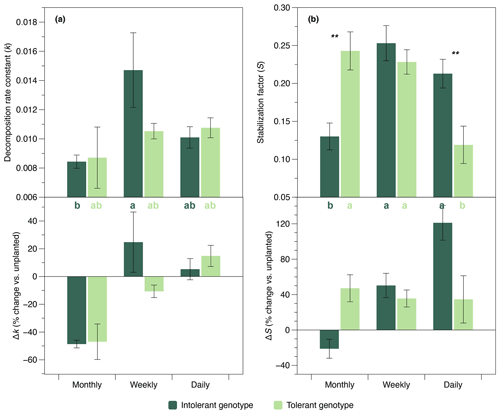
Figure 4Initial decomposition rate constant (k) and stabilization factor (S) (sensu Keuskamp et al., 2013) in soils planted with intolerant and tolerant plant genotypes of Elymus athericus exposed to three different flooding frequencies (monthly, weekly, and daily). Upper panels show absolute values, and lower panels show values in relation to the unplanted control (i.e., percentage change vs. the mean value of n=4 unplanted mesocosms per flooding treatment). Values are means and SE (n=8). Asterisks denote significant genotypic differences within the same flooding treatment (; ). Bars not labeled by the same letter are significantly different at p≤0.05. All statistical results refer to the absolute data shown in the upper panels and are based on Tukey's HSD tests following two-way ANOVA for S and Kruskal–Wallis tests for k.
3.2 Soil microbial community structure
Flooding frequency (two-way PERMANOVA, F=2.33, p≤0.001) and plant genotype (F=2.09, p≤0.001) significantly affected microbial community structure (Fig. 5). In accordance with the findings on EEAs, genotype effects were most pronounced at the highest (i.e., daily) flooding-frequency treatment (Fig. 5). By contrast, differences between genotypes were absent at the lowest, i.e., monthly flooding frequency, suggesting an interaction of genotype and flooding frequency on soil microbial community structure (Fig. 5), which was, however, not statistically significant based on two-way PERMANOVA (F=1.08; p≥0.1). Canonical correspondence analysis (Fig. S3) indicated that soil microbial community structure is significantly related to plant biomass parameters as well as to microbial C and N demands. Aboveground biomass exerted the strongest effect on community structure (Fig. S3). The overview of the most abundant prokaryotic taxa is shown in Fig. S4. However, owing to the artificial nature of the simulated tidal wetland system used in our study, it was not our objective to identify and discuss the specific microbial taxa affected by genotype or flooding treatments.
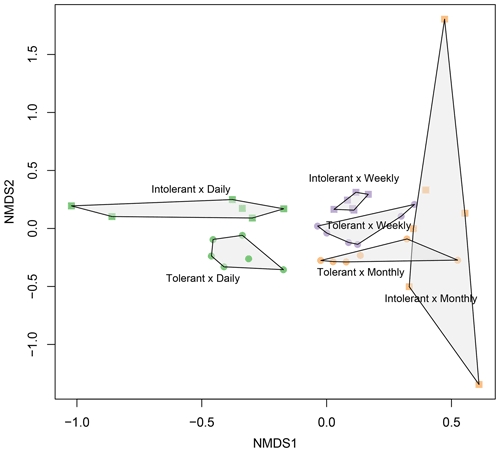
Figure 5NMDS plot showing prokaryotic (bacterial and archaeal) community composition in soils planted with intolerant and tolerant plant genotypes of Elymus athericus exposed to three different flooding frequencies (monthly, weekly, and daily). Plot shows all technical replicates (i.e., two samples from n=3 mesocosms; variability in technical replicates is illustrated in Fig. S1). For PERMANOVA analyses, data from technical replicates were averaged.
The present study provides experimental evidence of genotype–environment interaction effects on soil microbial enzyme activity (Figs. 2, 3) and belowground litter breakdown (Fig. 4), two key processes controlling ecosystem C and nutrient cycling. Specifically, plant genotype determined the presence or absence of flooding-frequency effects on microbial enzyme activities and litter breakdown. This result yields important implications for our understanding of soil–climate feedbacks in the coastal zone, because it shows that plant-genotype controls can mask or enhance the effects of SLR on soil microbial processes. Our data furthermore suggest genotype–SLR interaction effects on the soil microbial community structure (Fig. 5). This finding is in agreement with a recent observational study on genotype–environment interactions in terrestrial ecosystems, suggesting that climate-driven reduction of genetic variation in Populus angustifolia phenology affects soil fungi-to-bacteria ratios (Ware et al., 2019), and a laboratory experiment demonstrating interaction effects of drought and rapid evolution in Brassica rapa on soil microbial community structure (terHorst et al., 2014). Overall, larger variability in microbial enzyme activities (Fig. 2) and litter decomposition (Fig. 4a) in soils planted with the intolerant plant genotype support our central hypothesis that effects of climate change on soil microbial activity depend on plant intraspecific genetic variation. The results are less clear for microbial community structure (Fig. 5).
4.1 Genotype aboveground-biomass response controls flooding effects on soil microbial functioning
While the majority of studies on genotype–environment interactions are concerned with plant responses to temperature or latitudinal climate gradients in terrestrial ecosystems (Bauerle et al., 2007; Curasi et al., 2019; Taylor et al., 2019; Walker et al., 2019; Ware et al., 2019), the present work is focused on SLR, the overriding climate change factor in coastal ecosystems, such as tidal wetlands (Kirwan and Megonigal, 2013). The effects of SLR on soil microbial activity can be tightly controlled by the plant response to changes in flooding frequency, as demonstrated by recent studies showing strong positive correlations between aboveground biomass and soil litter decomposition (Janousek et al., 2017), cellulose decomposition (i.e., tensile strength loss; Jones et al., 2018), or recalcitrant soil organic matter decomposition (Mueller et al., 2016). The importance of plant processes in controlling soil microbial functioning in response to changing flooding frequency is reflected in the findings of the present study: in the absence of plants, flooding frequency affected neither soil microbial enzyme activities nor the soil microbial community structure (Figs. S2 and S5). In the presence of plants, however, flooding frequency and genotypic variation in plant biomass exerted significant effects on soil microbial activity and community structure. Most notably, microbial enzyme activities only responded to changes in flooding frequency when aboveground biomass responded. Aboveground and belowground biomass across flooding treatments was unchanged in the tolerant genotype, whereas the intolerant genotype showed a strong reduction of aboveground biomass at our highest flooding treatment (Reents et al., 2021). Consequently, only the flooding-sensitive intolerant genotype showed changes in soil microbial activity, whereas the tolerant genotype was able to maintain microbial enzyme activities at a constant level across the flooding gradient (Table 1; Fig. 2).
In support of the notion that the soil microbial activity response to increasing flooding frequency follows the response of plant aboveground processes, we found a significant relationship between aboveground biomass and microbial N-acquisition activity (aminopeptidase + chitinase activity, sensu Sinsabaugh et al., 2008, 2009) across all flooding treatments (r=0.41; p≤0.01, Table 2) and to an even larger degree within the daily flooding treatment (r=0.63; p=0.01), where effects on aboveground biomass and N-acquisition activity existed (Table 2 and Fig. 2; Reents et al., 2021). Soil enzyme activity is tightly controlled by the balance of nutrient supply and demand (Sinsabaugh et al., 2008, 2012). It is therefore possible that the maintenance of N-rich aboveground plant biomass increased the soil microbial N demand and thus stimulated the mineralization of N from soil organic matter, a mechanism that has been discussed in the context of rhizosphere priming effects (Kuzyakov, 2002).
4.2 Genotype–environment interactions control belowground litter breakdown
To evaluate if genotype effects on soil microbial communities translate into altered organic matter turnover and thus ecosystem functioning, we assessed the decomposition of standardized plant litter in the rhizosphere. The parameters S and k describe the initial transformation process of biomass to soil organic matter, which is a key component of many tidal wetland resilience models that have highlighted the critical role of the organic contribution to wetland elevation gain (Schile et al., 2014; Swanson et al., 2014). Although actual rates of S and k cannot be inferred from TBI assays using standardized litter, the approach has proven to be a powerful tool for characterizing the potential of the soil environment to transform and stabilize organic matter inputs (Keuskamp et al., 2013; Mueller et al., 2018; Ochoa-Hueso et al., 2020). Effect sizes of the flooding treatment on S and k observed here are similar in range to those reported from field sites (Tang et al., 2020), and genotype effect sizes were surprisingly large. Specifically, differences in S between genotypes within flooding treatments corresponded to ca. 20 % of the total range of S reported for tidal wetlands worldwide (Mueller et al., 2018). This result illustrates that the effects of plant genotype and genotype–SLR interactions on the C balance of tidal wetlands are not restricted to shifts in plant performance and primary production (Reents et al., 2021) but also concern soil C turnover.
Although significant correlations between microbial activity and plant-biomass parameters were found, these are insufficient to clearly identify functional-trait differences between genotypes that control soil microbial functioning. Plants can control soil microbial activity and ultimately the decomposition of different soil organic matter pools via at least three non-exclusive mechanisms: (1) supplying oxygen to an otherwise anoxic soil system via root oxygen loss (Wolf et al., 2007), (2) competing with microbial communities for nutrients (Kuzyakov and Xu, 2013), and (3) supplying labile microbial substrates via rhizodeposition (Jones et al., 2004; Kuzyakov, 2002). Root oxygen loss (mechanism 1) is only relevant in oxygen-deficient soils, like those found in coastal marshes. This suggests that it might be the most important mechanism, but strong genotype effects on belowground litter decomposition were also present in our well-aerated monthly-flooding treatment (Fig. 4b). Therefore, root oxygen loss is unlikely to represent the primary and sole driver of the observed genotype effects. Differences in nutrient demand between genotypes (mechanism 2) are supported by the clear differences in aboveground biomass production (Reents et al., 2021) and soil microbial N-acquisition activities (Fig. 2). However, these differences in biomass production and microbial N-acquisition were restricted to our highest flooding frequency and cannot explain the changes in belowground litter decomposition we observed under lower flooding frequencies. We therefore hypothesize that genotypic differences in root exudation patterns (mechanism 3) could have played an important role in the studied system. Root exudates are a key component of the plant control on soil decomposition processes in terrestrial soils, and their quantity and quality are not necessarily related to plant biomass parameters (Henneron et al., 2020; Jones et al., 2004; Koelbener et al., 2010). Furthermore, differences in root-exudation patterns between genotypes are known to alter microbial community structures in terrestrial ecosystems (Micallef et al., 2009). For wetlands, however, the current understanding of root-exudate effects on soil decomposition dynamics is insufficient to explore this hypothesis more thoroughly without additional research (Dinter et al., 2019; Mueller et al., 2016). Taken together, our findings highlight the need for further investigations into rhizosphere-trait variability, plant–soil interactions, and the mechanisms of rhizosphere priming effects in wetland ecosystems.
4.3 Methodological considerations
We previously demonstrated realistic plant-productivity responses to variations in flooding frequency simulated by the tidal-tank facility at Hamburg University (Reents et al., 2021). Therefore, we argue that the present investigation on plant–soil interactions can also provide relevant mechanistic insight into flooding effects on tidal wetland functioning. However, owing to the artificial nature of the simulated tidal wetland system, absolute effect sizes reported here need to be considered with caution. For the same reason, we refrain from providing a detailed interpretation of changes in single microbial taxa. One important caveat in this context is the restriction of our study to a single soil type. Because plant–microbe interactions in the rhizosphere can reflect provenance (e.g., Di Lonardo et al., 2018), future investigations will need to assess the generality of our findings using different combinations of plant genotype and soil type, including the native home soils from the locations at which the plants are sampled. We furthermore recommend repeating this experiment in situ, e.g., in the form of reciprocal transplantations, in order to improve the quantitative understanding of plant-genotype-mediated SLR effects on soil microbial functioning.
Larger variability in microbial enzyme activities and litter decomposition in soils planted with the intolerant plant genotype support our general hypothesis that effects of changing abiotic conditions on soil microbial functioning depend on plant intraspecific genetic variation. Our findings suggest that intraspecific variation in wetland plants could represent an important factor determining the response of soil microbial communities and soil C turnover to climate change. If our findings apply more generally to coastal wetland ecosystems, they could yield important implications for experimental climate change research and models of soil C accumulation, because they show that plant-genotype controls can mask or enhance the effects of changing abiotic conditions on soil microbial processes. Future research will need to put more emphasis on the intraspecific variability in plant functional traits as well as climate-change-driven intraspecific genetic shifts in wetland plant communities.
All data presented in this paper are available upon reasonable request. Raw sequencing data are available at the European Nucleotide Archive (ENA) under BioProject accession number PRJEB38150 and sample accession numbers ERS4541081–ERS4541134.
The supplement related to this article is available online at: https://doi.org/10.5194/bg-18-6133-2021-supplement.
HT, SR, SN, KJ, and PM designed and set up the experiment. HT conducted enzyme and decomposition assays and analyzed the resulting data. SL, PM, and FH planned the molecular microbial work. PM conducted the molecular microbial lab work. FH carried out the bioinformatics and analyzed the molecular data. HT and PM wrote the original draft with input from all co-authors.
The authors declare that they have no conflict of interest.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
We thank Chris Smit and his colleagues from Groningen University for the provision of plants; Max Beiße, Marion Klötzl, and Maren Winnacker at Hamburg University for assisting in the experimental phase; and Anke Saborowski at GFZ for her assistance with lab work.
Hao Tang received financial support from the China Scholarship Council (grant no. CSC201606910043). Peter Mueller was supported by the DAAD (German academic exchange service) PRIME fellowship program funded through the German Federal Ministry of Education and Research (BMBF).
This paper was edited by Michael Bahn and reviewed by two anonymous referees.
Anderson, M. J.: Permutational Multivariate Analysis of Variance (PERMANOVA), Wiley StatsRef Stat. Ref. Online, 1–15, https://doi.org/10.1002/9781118445112.stat07841, 2017.
Bardgett, R. D., Freeman, C., and Ostle, N. J.: Microbial contributions to climate change through carbon cycle feedbacks, ISME J., 2, 805–814, https://doi.org/10.1038/ismej.2008.58, 2008.
Bauerle, W. L., Bowden, J. D., and Wang, G. G.: The influence of temperature on within-canopy acclimation and variation in leaf photosynthesis: Spatial acclimation to microclimate gradients among climatically divergent Acer rubrum L. genotypes, J. Exp. Bot., 58, 3285–3298, https://doi.org/10.1093/jxb/erm177, 2007.
Bockelmann, A. C. and Neuhaus, R.: Competitive exclusion of Elymus athericus from a high-stress habitat in a European salt marsh, J. Ecol., 87, 503–513, https://doi.org/10.1046/j.1365-2745.1999.00368.x, 1999.
Bockelmann, A. C., Reusch, B. H., Bijlsma, R., and Bakker, J. P.: Habitat differentiation vs. isolation-by-distance: The genetic population structure of Elymus athericus in European salt marshes, Mol. Ecol., 12, 505–515, https://doi.org/10.1046/j.1365-294X.2003.01706.x, 2003.
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., Alexander, H., Alm, E. J., Arumugam, M., Asnicar, F., Bai, Y., Bisanz, J. E., Bittinger, K., Brejnrod, A., Brislawn, C. J., Brown, C. T., Callahan, B. J., Caraballo-Rodríguez, A. M., Chase, J., Cope, E. K., Da Silva, R., Diener, C., Dorrestein, P. C., Douglas, G. M., Durall, D. M., Duvallet, C., Edwardson, C. F., Ernst, M., Estaki, M., Fouquier, J., Gauglitz, J. M., Gibbons, S. M., Gibson, D. L., Gonzalez, A., Gorlick, K., Guo, J., Hillmann, B., Holmes, S., Holste, H., Huttenhower, C., Huttley, G. A., Janssen, S., Jarmusch, A. K., Jiang, L., Kaehler, B. D., Kang, K. Bin, Keefe, C. R., Keim, P., Kelley, S. T., Knights, D., Koester, I., Kosciolek, T., Kreps, J., Langille, M. G. I., Lee, J., Ley, R., Liu, Y. X., Loftfield, E., Lozupone, C., Maher, M., Marotz, C., Martin, B. D., McDonald, D., McIver, L. J., Melnik, A. V., Metcalf, J. L., Morgan, S. C., Morton, J. T., Naimey, A. T., Navas-Molina, J. A., Nothias, L. F., Orchanian, S. B., Pearson, T., Peoples, S. L., Petras, D., Preuss, M. L., Pruesse, E., Rasmussen, L. B., Rivers, A., Robeson, M. S., Rosenthal, P., Segata, N., Shaffer, M., Shiffer, A., Sinha, R., Song, S. J., Spear, J. R., Swafford, A. D., Thompson, L. R., Torres, P. J., Trinh, P., Tripathi, A., Turnbaugh, P. J., Ul-Hasan, S., van der Hooft, J. J. J., Vargas, F., Vázquez-Baeza, Y., Vogtmann, E., von Hippel, M., Walters, W., Wan, Y. H., Wang, M. X., Warren, J., Weber, K. C., Williamson, C. H. D., Willis, A. D., Xu, Z. Z., Zaneveld, J. R., Zhang, Y. L., Zhu, Q. Y. Knight, R., Carporaso, J. G.: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 37, 852–857, https://doi.org/10.1038/s41587-019-0209-9, 2019.
Box, G. E. P.: Some Theorems on Quadratic Forms Applied in the Study of Analysis of Variance Problems, I. Effect of Inequality of Variance in the One-Way Classification, Ann. Math. Stat., 25, 290–302, https://doi.org/10.1214/aoms/1177728786, 1954.
Bustos-Korts, D., Romagosa, I., Borràs-Gelonch, G., Casas, A. M., Slafer, G. A., and van Eeuwijk, F.: Genotype by Environment Interaction and Adaptation, in: Encyclopedia of Sustainability Science and Technology, edited by: Meyers, R. A., Springer New York, New York, NY, 1–44, 2018.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P.: DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 13, 581–583, https://doi.org/10.1038/nmeth.3869, 2016.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., Fierer, N., Peña, A. G., Goodrich, J. K., Gordon, J. I., Huttley, G. A., Kelley, S. T., Knights, D., Koenig, J. E., Ley, R. E., Lozupone, C. A., Mcdonald, D., Muegge, B. D., Pirrung, M., Reeder, J., Sevinsky, J. R., Turnbaugh, P. J., Walters, W. A., Widmann, J., Yatsunenko, T., Zaneveld, J. and Knight, R.: correspondence QIIME allows analysis of high- throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing, Nat. Publ. Gr., 7, 335–336, https://doi.org/10.1038/nmeth0510-335, 2010.
Chmura, G. L., Anisfeld, S. C., Cahoon, D. R., and Lynch, J. C.: Global carbon sequestration in tidal, saline wetland soils, Global Biogeochem. Cycles, 17, 1–11, https://doi.org/10.1029/2002gb001917, 2003.
Clark, F. E.: Decomposition of organic materials in grassland soil, Tech. Rep. (US Int. Biol. Progr. Grassl. Biome), no 61, 1970.
Crutsinger, G. M., Collins, M. D., Fordyce, J. A., Gompert, Z., Nice, C. C., and Sanders, N. J.: Plant genotypic diversity predicts community structure and governs an ecosystem process, Science, 313, 966–968, https://doi.org/10.1126/science.1128326, 2006.
Curasi, S. R., Parker, T. C., Rocha, A. V., Moody, M. L., Tang, J., and Fetcher, N.: Differential responses of ecotypes to climate in a ubiquitous Arctic sedge: implications for future ecosystem C cycling, New Phytol., 223, 180–192, https://doi.org/10.1111/nph.15790, 2019.
Davidson, E. A. and Janssens, I. A.: Temperature sensitivity of soil carbon decomposition and feedbacks to climate change, Nature, 440, 165–173, https://doi.org/10.1038/nature04514, 2006.
Dijkstra, F. A., Blumenthal, D., Morgan, J. A., Pendall, E., Carrillo, Y. and Follett, R. F.: Contrasting effects of elevated CO2 and warming on nitrogen cycling in a semiarid grassland, New Phytol., 187, 426–437, https://doi.org/10.1111/j.1469-8137.2010.03293.x, 2010.
Dinter, T., Geihser, S., Gube, M., Daniel, R., and Kuzyakov, Y.: Impact of sea level change on coastal soil organic matter, priming effects and prokaryotic community assembly, FEMS Microbiol. Ecol., 95, fiz129, https://doi.org/10.1093/femsec/fiz129, 2019.
Fischer, D. G., Chapman, S. K., Classen, A. T., Gehring, C. A., Grady, K. C., Schweitzer, J. A., and Whitham, T. G.: Plant genetic effects on soils under climate change, Plant Soil, 379, 1–19, https://doi.org/10.1007/s11104-013-1972-x, 2014.
Freeman, C., Ostle, N., and Kang, H.: An enzymic “latch” on a global carbon store: A shortage of oxygen locks up carbon in peatlands by restraining a single enzymes, Nature, 409, 149, https://doi.org/10.1038/35051650, 2001.
Fuchslueger, L., Bahn, M., Fritz, K., Hasibeder, R., and Richter, A.: Experimental drought reduces the transfer of recently fixed plant carbon to soil microbes and alters the bacterial community composition in a mountain meadow, New Phytol., 201, 916–927, https://doi.org/10.1111/nph.12569, 2014.
Henneron, L., Cros, C., Picon-Cochard, C., Rahimian, V., and Fontaine, S.: Plant economic strategies of grassland species control soil carbon dynamics through rhizodeposition, J. Ecol., 108, 528–545, https://doi.org/10.1111/1365-2745.13276, 2020.
Holm, S., Walz, J., Horn, F., Yang, S., Grigoriev, M. N., Wagner, D., Knoblauch, C., and Liebner, S.: Methanogenic response to long-term permafrost thaw is determined by paleoenvironment, FEMS Microbiol. Ecol., 96, 1–13, https://doi.org/10.1093/femsec/fiaa021, 2020.
Huang, G., Rymer, P. D., Duan, H., Smith, R. A., and Tissue, D. T.: Elevated temperature is more effective than elevated [CO2] in exposing genotypic variation in Telopea speciosissima growth plasticity: Implications for woody plant populations under climate change, Glob. Chang. Biol., 21, 3800–3813, https://doi.org/10.1111/gcb.12990, 2015.
Janousek, C. N., Buffington, K. J., Guntenspergen, G. R., Thorne, K. M., Dugger, B. D., and Takekawa, J. Y.: Inundation, Vegetation, and Sediment Effects on Litter Decomposition in Pacific Coast Tidal Marshes, Ecosystems, 20, 1296–1310, https://doi.org/10.1007/s10021-017-0111-6, 2017.
Jones, D. L., Hodge, A., and Kuzyakov, Y.: Plant and mycorrhizal regulation of rhizodeposition, New Phytol., 163, 459–480, https://doi.org/10.1111/j.1469-8137.2004.01130.x, 2004.
Jones, S. F., Stagg, C. L., Krauss, K. W., and Hester, M. W.: Flooding Alters Plant-Mediated Carbon Cycling Independently of Elevated Atmospheric CO2 Concentrations, J. Geophys. Res.-Biogeo., 123, 1976–1987, https://doi.org/10.1029/2017JG004369, 2018.
Jump, A. S. and Peñuelas, J.: Running to stand still: Adaptation and the response of plants to rapid climate change, Ecol. Lett., 8, 1010–1020, https://doi.org/10.1111/j.1461-0248.2005.00796.x, 2005.
Keuskamp, J. A., Dingemans, B. J. J., Lehtinen, T., Sarneel, J. M., and Hefting, M. M.: Tea Bag Index: A novel approach to collect uniform decomposition data across ecosystems, Methods Ecol. Evol., 4, 1070–1075, https://doi.org/10.1111/2041-210X.12097, 2013.
Kirwan, M. L. and Guntenspergen, G. R.: Feedbacks between inundation, root production, and shoot growth in a rapidly submerging brackish marsh, J. Ecol., 100, 764–770, https://doi.org/10.1111/j.1365-2745.2012.01957.x, 2012.
Kirwan, M. L. and Megonigal, J. P.: Tidal wetland stability in the face of human impacts and sea-level rise, Nature, 504, 53–60, https://doi.org/10.1038/nature12856, 2013.
Kirwan, M. L., Langley, J. A., Guntenspergen, G. R., and Megonigal, J. P.: The impact of sea-level rise on organic matter decay rates in Chesapeake Bay brackish tidal marshes, Biogeosciences, 10, 1869–1876, https://doi.org/10.5194/bg-10-1869-2013, 2013.
Kirwan, M. L., Guntenspergen, G. R., and Langley, J. A.: Temperature sensitivity of organic-matter decay in tidal marshes, Biogeosciences, 11, 4801–4808, https://doi.org/10.5194/bg-11-4801-2014, 2014.
Koelbener, A., Ström, L., Edwards, P. J., and Olde Venterink, H.: Plant species from mesotrophic wetlands cause relatively high methane emissions from peat soil, Plant Soil, 326, 147–158, https://doi.org/10.1007/s11104-009-9989-x, 2010.
Kuzyakov, Y.: Review: Factors affecting rhizosphere priming effects, J. Plant Nutr. Soil Sci., 165, 382–396, 2002.
Kuzyakov, Y. and Xu, X.: Competition between roots and microorganisms for nitrogen: Mechanisms and ecological relevance, New Phytol., 198, 656–669, https://doi.org/10.1111/nph.12235, 2013.
Di Lonardo, D. P., Manrubia, M., De Boer, W., Zweers, H., Veen, G. F., and Van der Wal, A.: Relationship between home-field advantage of litter decomposition and priming of soil organic matter, Soil Biol. Biochem., 126, 49–56, https://doi.org/10.1016/j.soilbio.2018.07.025, 2018.
Madritch, M. D. and Lindroth, R. L.: Soil microbial communities adapt to genetic variation in leaf litter inputs, Oikos, 120, 1696–1704, https://doi.org/10.1111/j.1600-0706.2011.19195.x, 2011.
Martin, M.: Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet.journal, 17, 10, https://doi.org/10.14806/ej.17.1.200, 2011.
McGuinness, K. A.: Of rowing boats, ocean liners and tests of the ANOVA homogeneity of variance assumption, Austral Ecol., 27, 681–688, https://doi.org/10.1046/j.1442-9993.2002.01233.x, 2002.
McLeod, E., Chmura, G. L., Bouillon, S., Salm, R., Björk, M., Duarte, C. M., Lovelock, C. E., Schlesinger, W. H., and Silliman, B. R.: A blueprint for blue carbon: Toward an improved understanding of the role of vegetated coastal habitats in sequestering CO2, Front. Ecol. Environ., 9, 552–560, https://doi.org/10.1890/110004, 2011.
Megonigal, J. P., Hines, M. E., and Visscher, P. T.: Anaerobic Metabolism: Linkages to Trace Gases and Aerobic Processes, Treatise on Geochemistry, 8, 317–424, https://doi.org/10.1016/B0-08-043751-6/08132-9, 2003.
Meier, L. A., Krauze, P., Prater, I., Horn, F., Schaefer, C. E. G. R., Scholten, T., Wagner, D., Mueller, C. W., and Kühn, P.: Pedogenic and microbial interrelation in initial soils under semiarid climate on James Ross Island, Antarctic Peninsula region, Biogeosciences, 16, 2481–2499, https://doi.org/10.5194/bg-16-2481-2019, 2019.
Micallef, S. A., Shiaris, M. P., and Colón-Carmona, A.: Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates, J. Exp. Bot., 60, 1729–1742, https://doi.org/10.1093/jxb/erp053, 2009.
Morris, J. T., Sundberg, K., and Hopkinson, C. S.: Salt marsh primary production and its responses to relative sea level and nutrientsin estuaries at plum island, Massachusetts, and North Inlet, South Carolina, USA, Oceanography, 26, 78–84, https://doi.org/10.5670/oceanog.2013.48, 2013.
Mueller, P., Jensen, K., and Megonigal, J. P.: Plants mediate soil organic matter decomposition in response to sea level rise, Glob. Chang. Biol., 22, 404–414, https://doi.org/10.1111/gcb.13082, 2016.
Mueller, P., Granse, D., Nolte, S., Do, H. T., Weingartner, M., Hoth, S., and Jensen, K.: Top-down control of carbon sequestration: Grazing affects microbial structure and function in salt marsh soils: Grazing, Ecol. Appl., 27, 1435–1450, https://doi.org/10.1002/eap.1534, 2017.
Mueller, P., Schile-Beers, L. M., Mozdzer, T. J., Chmura, G. L., Dinter, T., Kuzyakov, Y., de Groot, A. V., Esselink, P., Smit, C., D'Alpaos, A., Ibáñez, C., Lazarus, M., Neumeier, U., Johnson, B. J., Baldwin, A. H., Yarwood, S. A., Montemayor, D. I., Yang, Z., Wu, J., Jensen, K., and Nolte, S.: Global-change effects on early-stage decomposition processes in tidal wetlands – implications from a global survey using standardized litter, Biogeosciences, 15, 3189–3202, https://doi.org/10.5194/bg-15-3189-2018, 2018.
Mueller, P., Ladiges, N., Jack, A., Schmiedl, G., Kutzbach, L., Jensen, K. and Nolte, S.: Assessing the long-term carbon-sequestration potential of the semi-natural salt marshes in the European Wadden Sea, Ecosphere, 10, e02556, https://doi.org/10.1002/ecs2.2556, 2019.
Mueller, P., Mozdzer, T. J., Langley, J. A., Aoki, L. R., Noyce, G. L., and Megonigal, J. P.: Plant species determine tidal wetland methane response to sea level rise, Nat. Commun., 11, 1–9, https://doi.org/10.1038/s41467-020-18763-4, 2020.
Mueller, P., Do, H. T., Smit, C., Reisdorff, C., Jensen, K., and Nolte, S.: With a little help from my friends: physiological integration facilitates invasion of wetland grass Elymus athericus into flooded soils, Oikos, 130, 431–439, https://doi.org/10.1111/oik.07863, 2021.
Van Nuland, M. E., Wooliver, R. C., Pfennigwerth, A. A., Read, Q. D., Ware, I. M., Mueller, L., Fordyce, J. A., Schweitzer, J. A., and Bailey, J. K.: Plant–soil feedbacks: connecting ecosystem ecology and evolution, Funct. Ecol., 30, 1032–1042, https://doi.org/10.1111/1365-2435.12690, 2016.
Ochoa-Hueso, R., Borer, E. T., Seabloom, E. W., Hobbie, S. E., Risch, A. C., Collins, S. L., Alberti, J., Bahamonde, H. A., Brown, C. S., Caldeira, M. C., Daleo, P., Dickman, C. R., Ebeling, A., Eisenhauer, N., Esch, E. H., Eskelinen, A., Fernández, V., Güsewell, S., Gutierrez-Larruga, B., Hofmockel, K., Laungani, R., Lind, E., López, A., McCulley, R. L., Moore, J. L., Peri, P. L., Power, S. A., Price, J. N., Prober, S. M., Roscher, C., Sarneel, J. M., Schütz, M., Siebert, J., Standish, R. J., Velasco Ayuso, S., Virtanen, R., Wardle, G. M., Wiehl, G., Yahdjian, L., and Zamin, T.: Microbial processing of plant remains is co-limited by multiple nutrients in global grasslands, Glob. Chang. Biol., 1–11, https://doi.org/10.1111/gcb.15146, 2020.
Pérez-Izquierdo, L., Zabal-Aguirre, M., González-Martínez, S. C., Buée, M., Verdú, M., Rincón, A., and Goberna, M.: Plant intraspecific variation modulates nutrient cycling through its below ground rhizospheric microbiome, J. Ecol., 107, 1594–1605, https://doi.org/10.1111/1365-2745.13202, 2019.
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and Van Der Putten, W. H.: Going back to the roots: The microbial ecology of the rhizosphere, Nat. Rev. Microbiol., 11, 789–799, https://doi.org/10.1038/nrmicro3109, 2013.
Pregitzer, C. C., Bailey, J. K., and Schweitzer, J. A.: Genetic by environment interactions affect plant-soil linkages, Ecol. Evol., 3, 2322–2333, https://doi.org/10.1002/ece3.618, 2013.
Pugnaire, F. I., Morillo, J. A., Peñuelas, J., Reich, P. B., Bardgett, R. D., Gaxiola, A., Wardle, D. A., and van der Putten, W. H.: Climate change effects on plant-soil feedbacks and consequences for biodiversity and functioning of terrestrial ecosystems, Sci. Adv., 5, eaaz1834, https://doi.org/10.1126/sciadv.aaz1834, 2019.
Van der Putten, W. H., Bardgett, R. D., Bever, J. D., Bezemer, T. M., Casper, B. B., Fukami, T., Kardol, P., Klironomos, J. N., Kulmatiski, A., Schweitzer, J. A., Suding, K. N., Van de Voorde, T. F. J., and Wardle, D. A.: Plant-soil feedbacks: The past, the present and future challenges, J. Ecol., 101, 265–276, https://doi.org/10.1111/1365-2745.12054, 2013.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glöckner, F. O.: The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools, Nucleic Acids Res., 41, 590–596, https://doi.org/10.1093/nar/gks1219, 2013.
Reents, S., Mueller, P., Tang, H., Jensen, K., and Nolte, S.: Plant genotype determines biomass response to flooding frequency in tidal wetlands, Biogeosciences, 18, 403–411, https://doi.org/10.5194/bg-18-403-2021, 2021.
Risch, A. C., Jurgensen, M. F., and Frank, D. A.: Effects of grazing and soil micro-climate on decomposition rates in a spatio-temporally heterogeneous grassland, Plant Soil, 298, 191–201, https://doi.org/10.1007/s11104-007-9354-x, 2007.
Rogers, K., Kelleway, J. J., Saintilan, N., Megonigal, J. P., Adams, J. B., Holmquist, J. R., Lu, M., Schile-Beers, L., Zawadzki, A., Mazumder, D. and Woodroffe, C. D.: Wetland carbon storage controlled by millennial-scale variation in relative sea-level rise, Nature, 567, 91–95, https://doi.org/10.1038/s41586-019-0951-7, 2019.
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F.: VSEARCH: A versatile open source tool for metagenomics, PeerJ, 10, 1–22, https://doi.org/10.7717/peerj.2584, 2016.
Schile, L. M., Callaway, J. C., Morris, J. T., Stralberg, D., Thomas Parker, V., and Kelly, M.: Modeling tidal marsh distribution with sea-level rise: Evaluating the role of vegetation, sediment, and upland habitat in marsh resiliency, PLoS One, 9, e88760, https://doi.org/10.1371/journal.pone.0088760, 2014.
Schweitzer, J. A., Bailey, J. K., Fischer, D. G., LeRoy, C. J., Lonsdorf, E. V., Whitham, T. G., and Hart, S. C.: Plant-soil-microorganism interactions: Heritable relationship between plant genotype and associated soil microorganisms, Ecology, 89, 773–781, https://doi.org/10.1890/07-0337.1, 2008.
Seliskar, D. M., Gallagher, J. L., Burdick, D. M., and Mutz, L. A.: The regulation of ecosystem functions by ecotypic variation in the dominant plant: A Spartina alterniflora salt-marsh case study, J. Ecol., 90, 1–11, https://doi.org/10.1046/j.0022-0477.2001.00632.x, 2002.
Sinsabaugh, R. L., Saiya-Cork, K., Long, T., Osgood, M. P., Neher, D. A., Zak, D. R., and Norby, R. J.: Soil microbial activity in a Liquidambar plantation unresponsive to CO2-driven increases in primary production, Appl. Soil Ecol., 24, 263–271, https://doi.org/10.1016/S0929-1393(03)00002-7, 2003.
Sinsabaugh, R. L., Lauber, C. L., Weintraub, M. N., Ahmed, B., Allison, S. D., Crenshaw, C., Contosta, A. R., Cusack, D., Frey, S., Gallo, M. E., Gartner, T. B., Hobbie, S. E., Holland, K., Keeler, B. L., Powers, J. S., Stursova, M., Takacs-Vesbach, C., Waldrop, M. P., Wallenstein, M. D., Zak, D. R., and Zeglin, L. H.: Stoichiometry of soil enzyme activity at global scale, Ecol. Lett., 11, 1252–1264, https://doi.org/10.1111/j.1461-0248.2008.01245.x, 2008.
Sinsabaugh, R. L., Hill, B. H., and Follstad Shah, J. J.: Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment, Nature, 462, 795–798, https://doi.org/10.1038/nature08632, 2009.
Sinsabaugh, R. L., Follstad Shah, J. J., Hill, B. H., and Elonen, C. M.: Ecoenzymatic stoichiometry of stream sediments with comparison to terrestrial soils, Biogeochemistry, 111, 455–467, https://doi.org/10.1007/s10533-011-9676-x, 2012.
Spivak, A. C., Sanderman, J., Bowen, J. L., Canuel, E. A., and Hopkinson, C. S.: Global-change controls on soil-carbon accumulation and loss in coastal vegetated ecosystems, Nat. Geosci., 12, 685–692, https://doi.org/10.1038/s41561-019-0435-2, 2019.
Stagg, C. L., Schoolmaster, D. R., Krauss, K. W., Cormier, N., and Conner, W. H.: Causal mechanisms of soil organic matter decomposition: deconstructing salinity and flooding impacts in coastal wetlands, Ecology, 98, 2003–2018, https://doi.org/10.1002/ecy.1890, 2017.
Stagg, C. L., Baustian, M. M., Perry, C. L., Carruthers, T. J. B., Hall, C. T., and Zanne, A.: Direct and indirect controls on organic matter decomposition in four coastal wetland communities along a landscape salinity gradient, J. Ecol., 106, 655–670, https://doi.org/10.1111/1365-2745.12901, 2018.
Swanson, K. M., Drexler, J. Z., Schoellhamer, D. H., Thorne, K. M., Casazza, M. L., Overton, C. T., Callaway, J. C., and Takekawa, J. Y.: Wetland Accretion Rate Model of Ecosystem Resilience (WARMER) and Its Application to Habitat Sustainability for Endangered Species in the San Francisco Estuary, Estuar. Coasts, 37, 476–492, https://doi.org/10.1007/s12237-013-9694-0, 2014.
Tang, H., Nolte, S., Jensen, K., Yang, Z., Wu, J., and Mueller, P.: Grazing mediates soil microbial activity and litter decomposition in salt marshes, Sci. Total Environ., 720, 137559, https://doi.org/10.1016/j.scitotenv.2020.137559, 2020.
Taylor, M. A., Cooper, M. D., and Schmitt, J.: Phenological and fitness responses to climate warming depend upon genotype and competitive neighbourhood in Arabidopsis thaliana, Funct. Ecol., 33, 308–322, https://doi.org/10.1111/1365-2435.13262, 2019.
terHorst, C. P. and Zee, P. C.: Eco-evolutionary dynamics in plant–soil feedbacks, Funct. Ecol., 30, 1062–1072, https://doi.org/10.1111/1365-2435.12671, 2016.
terHorst, C. P., Lennon, J. T., and Lau, J. A.: The relative importance of rapid evolution for plant-microbe interactions depends on ecological context, Proc. R. Soc. B Biol. Sci., 281, 20140028, https://doi.org/10.1098/rspb.2014.0028, 2014.
Walker, T. W. N., Weckwerth, W., Bragazza, L., Fragner, L., Forde, B. G., Ostle, N. J., Signarbieux, C., Sun, X., Ward, S. E., and Bardgett, R. D.: Plastic and genetic responses of a common sedge to warming have contrasting effects on carbon cycle processes, Ecol. Lett., 22, 159–169, https://doi.org/10.1111/ele.13178, 2019.
Wanner, A., Suchrow, S., Kiehl, K., Meyer, W., Pohlmann, N., Stock, M., and Jensen, K.: Scale matters: Impact of management regime on plant species richness and vegetation type diversity in Wadden Sea salt marshes, Agric. Ecosyst. Environ., 182, 69–79, https://doi.org/10.1016/j.agee.2013.08.014, 2014.
Ward, S. E., Ostle, N. J., Oakley, S., Quirk, H., Henrys, P. A., and Bardgett, R. D.: Warming effects on greenhouse gas fluxes in peatlands are modulated by vegetation composition, Ecol. Lett., 16, 1285–1293, https://doi.org/10.1111/ele.12167, 2013.
Ware, I. M., Van Nuland, M. E., Schweitzer, J. A., Yang, Z., Schadt, C. W., Sidak-Loftis, L. C., Stone, N. E., Busch, J. D., Wagner, D. M., and Bailey, J. K.: Climate-driven reduction of genetic variation in plant phenology alters soil communities and nutrient pools, Glob. Chang. Biol., 25, 1514–1528, https://doi.org/10.1111/gcb.14553, 2019.
Warner, B. G. and Asada, T.: Biological diversity of peatlands in Canada, Aquat. Sci., 68, 240–253, https://doi.org/10.1007/s00027-006-0853-2, 2006.
Wieder, W.: Soil carbon: Microbes, roots and global carbon, Nat. Clim. Chang., 4, 1052–1053, https://doi.org/10.1038/nclimate2454, 2014.
Wolf, A. A., Drake, B. G., Erickson, J. E., and Megonigal, J. P.: An oxygen-mediated positive feedback between elevated carbon dioxide and soil organic matter decomposition in a simulated anaerobic wetland, Glob. Chang. Biol., 13, 2036–2044, https://doi.org/10.1111/j.1365-2486.2007.01407.x, 2007.
Zogg, G. P., Travis, S. E., and Brazeau, D. A.: Strong associations between plant genotypes and bacterial communities in a natural salt marsh, Ecol. Evol., 8, 4721–4730, https://doi.org/10.1002/ece3.4105, 2018.





