the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Microbial labilization and diversification of pyrogenic dissolved organic matter
Aleksandar I. Goranov
Andrew S. Wozniak
Kyle W. Bostick
Andrew R. Zimmerman
Siddhartha Mitra
Patrick G. Hatcher
With the increased occurrence of wildfires around the world, interest in the chemistry of pyrogenic organic matter (pyOM) and its fate in the environment has increased. Upon leaching from soils by rain events, significant amounts of dissolved pyOM (pyDOM) enter the aquatic environment and interact with microbial communities that are essential for cycling organic matter within the different biogeochemical cycles. To evaluate the biodegradability of pyDOM, aqueous extracts of laboratory-produced biochars were incubated with soil microbes, and the molecular changes to the composition of pyDOM were probed using ultrahigh-resolution mass spectrometry (Fourier transform–ion cyclotron resonance–mass spectrometry). Given that solar irradiation significantly affects the composition of pyDOM during terrestrial-to-marine export, the effects of photochemistry were also evaluated in the context of pyDOM biodegradability.
Ultrahigh-resolution mass spectrometry revealed that many different (both aromatic and aliphatic) compounds were biodegraded. New labile compounds were produced, 22 %–40 % of which were peptide-like. These results indicated that a portion of pyDOM has been labilized into microbial biomass during the incubations. Fluorescence excitation–emission matrix spectra revealed that some fraction of these new bio-produced molecules is associated with proteinaceous fluorophores. Two-dimensional 1H–1H total correlation nuclear magnetic resonance (NMR) spectroscopy identified a peptidoglycan-like backbone within the microbially produced compounds. These results are consistent with previous observations of peptidoglycans within the soil and ocean nitrogen cycles where remnants of biodegraded pyDOM are expected to be observed.
Interestingly, the exact nature of the bio-produced organic matter was found to vary drastically among samples indicating that the microbial consortium used may produce different exudates based on the composition of the initial pyDOM. Another potential explanation for the vast diversity of molecules is that microbes only consume low molecular-weight compounds, but they also produce reactive oxygen species (ROS), which initiate oxidative and recombination reactions that degrade high molecular-weight compounds and produce new molecules. Some of the bio-produced molecules (212–308 molecular formulas) were identified in estuarine and marine (surface and abyssal oceanic), and 81–192 of these formulas were of molecular composition attributed to carboxyl-rich alicyclic molecules (CRAM). These results indicate that some of the pyDOM biodegradation products have an oceanic fate and can be sequestered into the deep ocean. The observed microbially mediated diversification of pyDOM suggests that pyDOM contributes to the observed large complexity of natural organic matter observed in riverine and oceanic systems. More broadly, our research shows that pyDOM can be substrate for microbial growth and be incorporated into environmental food webs within the global carbon and nitrogen cycles.
- Article
(4397 KB) - Full-text XML
-
Supplement
(5736 KB) - BibTeX
- EndNote
Pyrogenic organic matter (pyOM), the carbonaceous solid residue that is left after biomass burning (e.g., wildfires, biochar production), has been gaining attention in recent years as an important active component of the global biogeochemical cycles. Compositionally, pyOM is mainly comprised of condensed aromatic compounds (ConAC) of various degrees of condensation and functionalization (Masiello, 2004; Schneider et al., 2010; Wagner et al., 2018; Wozniak et al., 2020). ConAC have been found in various environmental matrices such as soils and sediments (Schmidt and Noack, 2000; Skjemstad et al., 2002; Reisser et al., 2016) and atmospheric aerosols (Wozniak et al., 2008; Bao et al., 2017). In the terrestrial environmental matrices, particularly in soils and sediments, ConAC were originally thought to be highly stable (“recalcitrant”) due to their condensed character (Goldberg, 1985; Masiello and Druffel, 1998). However, more and more studies report the presence of pyrogenic molecules in different aquatic environments (Hockaday et al., 2006; Dittmar and Paeng, 2009; Roebuck et al., 2017; Wagner et al., 2017; Li et al., 2019). These studies support the proposition that pyOM can be solubilized upon rain events and be leached as pyrogenic dissolved organic matter (pyDOM) resulting in large annual riverine fluxes of pyDOM from global riverine systems to the open ocean (Dittmar et al., 2012; Jaffé et al., 2013; Wang et al., 2016; Marques et al., 2017; Jones et al., 2020). During export, pyDOM is likely altered by various processes resulting in its degradation and alteration of its physico-chemical characteristics (Masiello, 2004; Coppola et al., 2019; Wagner et al., 2019). Using laboratory-prepared biochars and conservative assumptions, Bostick et al. (2018) approximated that > 85 % of the leached pyDOM is degradable (e.g., mineralizable to CO2), which indicates that pyDOM is a very active component within the global carbon cycle, as previously suggested (Druffel, 2004; Lehmann, 2007; Riedel et al., 2016).
In sunlit aquatic environments, photodegradation is the most significant sink for the fraction of ConAC of pyDOM (Stubbins et al., 2012). The photochemistry of ConAC and pyDOM has been studied utilizing either laboratory-prepared pyDOM (Ward et al., 2014; Fu et al., 2016; Li et al., 2019; Bostick et al., 2020; Goranov et al., 2020; Wang et al., 2020) or natural organic matter rich in ConAC (Stubbins et al., 2010, 2012; Wagner and Jaffé, 2015). These studies have reported that ConAC are exceptionally photolabile and they degrade through a series of oxygenation, ring opening, and decarboxylation reactions leading to a pool of smaller aliphatic by-products. Additionally, pyDOM photochemistry has been associated with the production of high fluxes of reactive oxygen species (ROS), important transients involved in the photo-transformation and photodegradation of pyDOM (Fu et al., 2016; Li et al., 2019; Goranov et al., 2020; Wang et al., 2020). These studies have contributed to a better understanding of the biogeochemical cycling of pyDOM in the presence of sunlight in the environment. Microbial (biotic) pathways are another degradative pathway with high potential for altering and/or mineralizing pyDOM, but these pathways are far less understood.
Biotic reworking of organic molecules is a key mechanism for producing the diverse molecular composition of natural organic matter (Lechtenfeld et al., 2015; Hach et al., 2020). Due to the highly condensed character of pyOM, it is often regarded as bio-recalcitrant, though several studies have shown that a fraction of it (about 0.5 % to 10 %) is indeed biodegradable (Kuzyakov et al., 2009, 2014; Zimmerman, 2010; Zimmerman et al., 2011). PyOM is mainly comprised of ConAC (Bostick et al., 2018; Wozniak et al., 2020), which contributes to its low biodegradability (Zimmerman, 2010). By contrast, pyDOM is highly heterogeneous (Wozniak et al., 2020), and, in addition to ConAC, it contains numerous low molecular-weight (LMW) species (e.g., acetate, methanol, formate; Bostick et al., 2018; Goranov et al., 2020) as well as various pyrogenic aliphatic compounds and inorganic nutrients (Hockaday et al., 2007; Mukherjee and Zimmerman, 2013; Goranov et al., 2020; Wozniak et al., 2020). The high solubility of pyDOM is imparted by the greater abundance of polar functional groups, which would also allow for greater microbial accessibility. To date, there is no study that evaluates the molecular-scale biodegradability of pyDOM. It is unknown whether and how (e.g., mechanistic pathways, kinetic rates) the different compound groups of pyDOM are biodegraded and/or biotransformed.
In addition to the unexplored biodegradability of pyDOM, there are concerns that biochar leachates may be toxic due to the presence of condensed and ligninaceous aromatics. It has been shown that cellulose- and pinewood-derived biochar water extracts (i.e., pyDOM) inhibit the growth of cyanobacteria, while pyDOM of lignin-derived biochar has no inhibitory effects (Smith et al., 2016). The toxicity of pyDOM has been mainly attributed to polysubstituted phenols present in the cellulose- and pinewood-derived biochars. In natural systems, however, it is likely that other pyDOM components also play a role in controlling the biodegradability and toxicity of pyDOM. An important very recent finding is that pyOM and pyDOM contain organochlorine compounds (both aliphatic and aromatic; Wozniak et al., 2020), which may enhance the toxicity of pyDOM. Thus, biotic incubations of pyDOM are needed to reveal if microbial growth can be sustained in pyDOM and environments rich in ConAC.
To explore these questions, we incubated aqueous biochar leachates (i.e., pyDOM) with a soil-derived microbial consortium and evaluated the compositional changes to pyDOM using numerous analytical techniques. Laboratory-produced biochars can be considered model pyrogenic substances as they are similar to what is produced during natural wildfires (Santín et al., 2017) but have not experienced environmental aging which would have impacted their physico-chemical properties (Ascough et al., 2011). We have used oak wood because most riverine dissolved organic matter (DOM) is exported from forested catchments (Hedges et al., 1997). We used two pyrolysis temperatures (400 and 650 ∘C) representative of forest fire temperatures (Santín et al., 2015, 2016). As photochemistry has been shown to increase the bio-lability of various types of DOM (Kieber et al., 1989; Lindell et al., 1995; Wetzel et al., 1995; Benner and Biddanda, 1998; Moran and Covert, 2003; Qualls and Richardson, 2003; Obernosterer and Benner, 2004; Abboudi et al., 2008; Chen and Jaffé, 2014; Antony et al., 2018), we also incubated pyDOM that had been photo-irradiated. Previous studies showed that significant compositional changes occur to pyDOM during photo-irradiation, which certainly implies different biodegradability (Bostick et al., 2020; Goranov et al., 2020).
In a parallel study of the same incubations (Bostick et al., 2021), we quantified the dissolved organic carbon (DOC) loss, respired CO2 quantities and the changes to the bulk structural composition of pyDOM as determined by one-dimensional 1H nuclear magnetic resonance (NMR) spectroscopy. Additionally, in that study, benzenepolycarboxylic acid (BPCA) molecular markers were used to quantify the changes specific to the fraction of ConAC pyDOM. It was found that the pyDOM leachates derived from the biochar of the higher pyrolysis temperature (650 ∘C) were less biodegradable than the pyDOM leachates from the lower temperature (400 ∘C) biochar. As expected, photo-irradiation increased the bio-lability of pyDOM. Over the 96 d incubation, up to 48 % of the carbon was respired to CO2. The degradation followed first-order kinetics, with LMW compounds (e.g., acetate, formate, methanol) being preferentially degraded. To elucidate the molecular-level changes taking place during the bio-incubation of pyDOM and assess the various molecules that are being degraded or produced by soil biota, we employed ultrahigh-resolution mass spectrometry (Fourier transform–ion cyclotron resonance–mass spectrometry, FT-ICR-MS), two-dimensional NMR and fluorescence spectroscopy. The collective results from these two studies improve our understanding of the degradative pathways of pyDOM and ConAC in the environment and allow us to better interpret observations pertaining to terrestrial-to-marine transfers and global cycling of organic matter.
2.1 Preparation of pyDOM samples
Two biochars were prepared by heating laurel oak wood (Quercus hemisphaerica) under N2 atmosphere at 400 and 650 ∘C for 3 h. After grinding and sieving to particles of uniform size (0.25–2.00 mm), the biochars were leached in 18.1 mΩ MilliQ laboratory-grade water (5 g in 500 mL) over 50 h on a shaker table. The obtained pyDOM leachates, hereafter referred to as “Oak 400 Fresh” and “Oak 650 Fresh”, were filtered using 0.2 µm Millipore GSWP mixed cellulose ester filters. Physico-chemical characteristics of similarly produced solid biochars and their leachates were reported in several previous studies (Zimmerman, 2010; Mukherjee et al., 2011; Bostick et al., 2018; Wozniak et al., 2020). A fraction of each leachate was also subjected to photo-irradiation for 5 d in a custom-made solar simulator equipped with Q-Lab Corporation UV-A lamps (295–365 nm, λMAX=340 nm, 40 W) equivalent to natural photo-irradiation of 12 d. Photo-transformation rates, structural changes, photo-irradiation apparatus design and other relevant information have been published previously (Bostick et al., 2020; Goranov et al., 2020). Photo-irradiated pyDOM samples will be hereafter referred to as “Oak 400 Photo” and “Oak 650 Photo”. The four samples (Oak 400 Fresh, Oak 650 Fresh, Oak 400 Photo, Oak 650 Photo) were diluted to a uniform DOC concentration of 4.7 mgC L−1 prior to inoculation.
2.2 Microbial incubations of pyDOM
Microbial incubations were performed using a soil-derived microbial consortium as an inoculum. Soil from the Austin Cary Memorial Forest (Gainesville, FL) was chosen because this area is frequently subjected to prescribed burns (Johns, 2016), and its soil microbes likely interact with pyOM and pyDOM on a regular basis. Taxonomic details of the soil used have been published previously (Khodadad et al., 2011). The collected soil was treated to remove roots and detritus, and its water extract was centrifuged to obtain a pellet. The pellet was then dissolved in 10 mL MilliQ laboratory-grade water to obtain an inoculate, 100 µL of which was used to spike 50 mL of each pyDOM substrate. Additionally, microbial nutrients (KH2PO4 and (NH4)2SO4) were provided following Zimmerman (2010) to support a healthy growth medium. Samples were incubated in gas-sealed amber vials on a shaker table at 28±5 ∘C for 10 d in the dark. Using a double-needle assembly, CO2-free air (Airgas, Zero) was flushed through the samples on days 0, 2, 5 and 10, which oxygenated the samples and removed dissolved inorganic carbon for its measurement (reported by Bostick et al., 2021). A procedural blank and control samples were prepared in the exact same way but were poisoned with HgCl2 immediately following the mixing of the different components (pyDOM, inoculate, nutrients). Additionally, a solution of sucrose (0.5 g C12H22O11 in 40 mL MilliQ laboratory-grade water) was also incubated in the same manner to serve as a positive control. All incubated samples were poisoned with HgCl2 to terminate microbial activity before shipment to Old Dominion University (Norfolk, VA) for analysis. Prior to spectroscopic analysis (see Sect. 2.3 and 2.5 below) or spectrometric analysis (see Sect. 2.4 below), samples were filtered using acid-washed 0.1 µm Teflon (PTFE) syringe filters. Further details about sample preparation can be found in the parallel study (Bostick et al., 2021).
2.3 Analysis of chromophoric and fluorophoric dissolved organic matter
Chromophoric DOM (CDOM) measurements were performed on a Thermo Scientific Evolution 201 ultraviolet–visible (UV–Vis) spectrophotometer operated in a double-beam mode. A matched Starna quartz cuvette with MilliQ water was used as a reference during all spectral measurements. Spectra were recorded from 230–800 nm using a 1 nm step, 0.12 s integration time and 500 nm min−1 scan speed. In addition to the double-beam referencing, the average noise in the 700–800 nm spectral region was subtracted from the spectra to correct for any instrument baseline drifts, temperature fluctuations, as well as scattering and refractive effects (Green and Blough, 1994; Helms et al., 2008). After consecutive procedural-blank corrections, the spectra (kept in decadic units) were normalized to the cuvette path length (1.0 cm) and DOC content (in mgC L−1) to convert them to specific absorbance spectra (L mgC−1 cm−1; Weishaar et al., 2003). CDOM was quantified by integrating the spectra from 250–450 nm (Helms et al., 2008), and CDOM quantity is reported in L mgC−1 cm−1 nm units.
Fluorophoric DOM (FDOM) measurements were performed on a Shimadzu RF-6000 spectrofluorometer operated in 3D acquisition mode. Samples were analyzed without dilution as no sample yielded absorbance at 230 nm above 0.07 (Miller et al., 2010). Samples were excited from 230–500 nm (5 nm step) and emission was recorded over 250–650 nm (5 nm step) to obtain excitation–emission matrices (EEMs). Additionally, five replicate water Raman scans were acquired on MilliQ water in 2D emission mode by exciting the sample at 350 nm, and fluorescence intensity was monitored over 365–450 nm (0.5 nm steps). All measurements were done with 5 nm slit widths of the monochromators, with 600 nm min−1 scan speed and in high-sensitivity mode.
EEMs were processed in MATLAB using the drEEM toolbox (version 0.4.0.) using previously published routines (Murphy et al., 2010, 2013). Briefly, using the FDOMcorrrect function, the raw EEMs were adjusted for instrumental bias, blank-corrected using an EEM of the procedural blank and scaled to adjust for any inner-filter effects using the raw UV–Vis spectra (Kothawala et al., 2013). This function also normalized the EEMs to Raman units (RU) after the area of the water Raman peak (peak maximum at 397 nm) had been determined by the ramanintegrationrange function (Murphy, 2011) on the averaged water Raman spectrum. The EEMs were then processed using the smootheem function to remove first- and second-order Rayleigh signals and Raman scattering. EEMs are visualized and difference plots are generated using an in-house MATLAB script.
2.4 Fourier transform–ion cyclotron resonance–mass spectrometry (FT-ICR-MS)
Procedural-blank, control and incubated samples were loaded onto solid-phase extraction cartridges (Agilent Technologies Bond Elut Priority Pollutant (PPL), 100 mg styrene divinyl copolymer) as previously described (Dittmar et al., 2008). Cartridges were eluted with methanol (Fisher Scientific, Optima LC-MS grade) and infused into an Apollo II electrospray ionization (ESI) source interfaced with a Bruker Daltonics Apex Qe FT-ICR-MS operating at 10 T and housed in the College of Sciences Major Instrumentation Cluster (COSMIC) facility at Old Dominion University (Norfolk, VA). The instrument is externally calibrated daily with a polyethylene glycol standard, and a surrogate laboratory pyDOM standard was analyzed before and after the analytical sequence to verify for the lack of instrumental drift. Additionally, an instrumental blank of methanol was analyzed between samples to verify for the absence of sample carryover. Samples were analyzed in negative ionization mode. Source spray voltages were optimized for each sample to ensure consistent spray currents among all samples. For each sample, 300 transients with a 4MWord time domain were collected and co-added, and the resultant free induction decay was zero-filled and sine-bell apodized. After fast Fourier transformation, internal calibration of the resultant mass spectra was performed using naturally abundant fatty acids, dicarboxylic acids and compounds belonging to the CH2-homologous series as previously described (Sleighter et al., 2008). Then, using an in-house MATLAB script, salt, blank and isotopologue (13C, 37Cl) peaks were removed. Molecular formulas within ±1 ppm error were assigned to FT-ICR-MS spectral peaks () using the molecular formula calculator from the National High Magnetic Field Laboratory (Tallahassee, FL). Formula assignments were restricted to elemental composition of 12C5−∞, 1H1−∞, 14N0−5,16O0−30, 32S0−2, 31P0−2, and 35Cl0−4 and were refined using previously established rules (Stubbins et al., 2010). Any ambiguous peak assignments were refined by inclusion within homologous series (CH2, H2, COO, CH2O, O2, H2O, NH3, HCl) following Kujawinski and Behn (2006) and Koch et al. (2007). For all samples, at least 80 % of the mass spectral peaks were assigned, and they accounted for at least 93 % of the mass spectral magnitude.
Molecular composition was evaluated by plotting the molecular formulas on van Krevelen (vK) diagrams, scatterplots of the formulas' hydrogen-to-carbon () versus oxygen-to-carbon () ratios (Van Krevelen, 1950; Kim et al., 2003). Formulas were further categorized using the modified aromaticity index (AIMOD), a proxy for the aromatic character of molecules (Koch and Dittmar, 2006, 2016), and calculated as shown in Eq. (1).
Formulas were classified as follows: condensed aromatic compounds (ConAC, AIMOD ≥ 0.67, number of C-atoms ≥ 15), aromatic (0.67 < AIMOD≤0.50), olefinic alicyclic (0 < AIMOD<0.50) and aliphatic (AIMOD=0). Additionally, N-containing formulas falling in the ranges of 1.5 ≤ ≤ 2 and 0.1 ≤ ≤ 0.67 were classified as peptide-like. Statistical evaluation of means using one-way analysis of variance (ANOVA) was performed in MATLAB using the anova1 function. Post hoc Scheffé's assessments were performed using the multcompare function. Pearson correlations were performed using the corrcoef function in the same software. A confidence level of 95 % was used for all statistical assessments.
For the Kendrick mass defect (KMD) series analysis, Kendrick mass (KM) was first calculated using the molecular weight of each compound (i.e., calculated mass from its molecular formula) following Eq. (2). Then, the Kendrick nominal mass (KNM) was calculated as the integer (no decimals) of the KM. The KMD is the difference between KM and KNM, i.e., the decimals (Eq. 3). This analysis was performed for oxygen (O), carbonyl (CO) and carboxyl (COO) series (S).
where for O series, for CO series and for COO series.
2.5 Two-dimensional nuclear magnetic resonance (NMR) spectroscopy
One-dimensional 1H NMR spectra of the samples were published and evaluated in the parallel study (Bostick et al., 2021). For the study of this paper, a select sample was analyzed using two-dimensional 1H–1H total correlation spectroscopy (TOCSY) to further evaluate several functional groups of interest. Analyses were performed on a 400 MHz (9.4 Tesla) Bruker BioSpin AVANCE III spectrometer fitted with a double-resonance broadband z-gradient inverse (BBI) probe in the COSMIC facility. Samples were analyzed without pre-concentration and volumetrically diluted with deuterated water (D2O, Acros Organics, 100 % D) to obtain a 90 : 10 H2O : D2O solution. Further details of sample preparation and acquisition of 1D 1H spectra are published in the companion study (Bostick et al., 2021). To obtain ultraclean NMR spectra, NMR tubes were soaked with aqua regia, rinsed extensively with ultrapure water and individually tested as blanks to verify that no background peaks are present. While 1H spectra were originally processed using an exponential multiplication function (line broadening) of 5 Hz to obtain higher signal-to-noise for a more accurate and precise integration (Bostick et al., 2021), here they were re-processed using a multiplication function of 1.5 Hz to better observe the splitting (multiplicity) patterns of the peaks of interest. TOCSY spectra were acquired using the phase-sensitive gradient-enhanced mlevgpphw5 pulse program. It utilizes a 17-step Malcolm Levitt (MLEV-17) composite scheme (Bax and Davis, 1985) for magnetization transfer between any coupled nuclear spins and a W5-WATERGATE element for water suppression (Liu et al., 1998). Both short-range and long-range spin–spin couplings were observed using 30 and 100 ms mixing times, respectively. The data were then zero-filled to a 4096×1024 matrix and then fitted with a -shifted (SSB = 2) sine-squared window function. Linear prediction to 256 points was used in the F1 dimension. All spectra were internally calibrated to the sharp distinguishable methanol singlet at 3.34 ppm (Gottlieb et al., 1997), and then spectra were phased and baseline-corrected. T1-noise removal was performed by calculating the positive projection of rows with no resonances, and the summed projections were subtracted from all rows in the spectrum (Klevit, 1985). The same procedure was performed for all columns (i.e., the F2 dimension).
3.1 Molecular changes to pyDOM after microbial incubation
Ultrahigh-resolution mass spectrometric analysis of the bio-incubated and corresponding control pyDOM leachates revealed significant changes in molecular composition after the 10 d incubation (Fig. 1). The identified molecular formulas were classified into three groups using a presence–absence approach: bio-labile, bio-produced and bio-resistant (Stubbins et al., 2010; Sleighter et al., 2012). This approach identifies any common formulas among the two samples being compared (control and bio-incubated), as well as any formulas that are unique to each sample. It is important to note that the electrospray ionization (ESI) source is prone to biases, and the analytical window of FT-ICR-MS depends most critically on it. Thus, it may not identify compounds that are present if they are not ionizable (Stenson et al., 2002; Patriarca et al., 2020). Therefore, our observations are influenced by the limited analytical window, and it is essential that observations by FT-ICR-MS are always paired with supplementary quantitative techniques (optical analyses, NMR, etc.) in order to determine if the identified trends are real or an artifact of ESI charge competition (D'Andrilli et al., 2020).
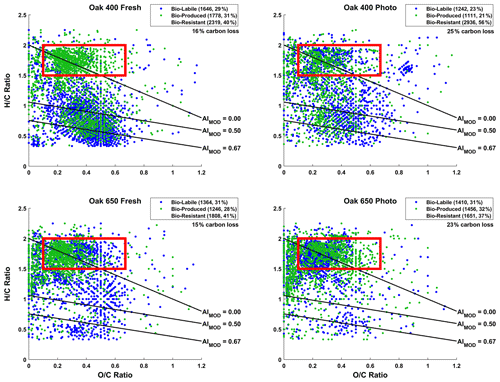
Figure 1Van Krevelen (vK) diagrams of 10 d microbially incubated pyDOM leachates. Formulas are classified as bio-labile (formulas only found in the control pyDOM leachates) and bio-produced (formulas only found in the bio-incubated samples). Formulas that are present in both the control and bio-incubated samples are operationally classified as bio-resistant and not shown for clarity. These three classes of molecules are separately plotted on vK diagrams and shown in Sect. S2 of the Supplement (Figs. S2–S4). The number of formulas in each of these pools is shown in the legends along with corresponding percentages (relative to total number of formulas in the two samples being compared). The carbon losses quantified by Bostick et al. (2021) are listed under the legends. The black lines indicate modified aromaticity index cutoffs (AIMOD; Koch and Dittmar, 2006, 2016), and the red box indicates the peptide region (valid only for N-containing formulas).
In all samples, nearly a third of the formulas (23 %–31 %) present in the control samples were not observed after the biotic incubations. This is somewhat proportional to the organic carbon losses observed over the 10 d incubation by Bostick et al. (2021). The organic carbon loss was also found to be equivalent to mineralized CO2 (± 4 %, Bostick et al., 2021) indicating that microbial respiration had occurred although CO2 mineralization can happen abiotically as well. Using the number of formulas lost as a proxy for bio-lability here, it appears that Oak 400 Fresh (1646 bio-labile formulas, 16 % carbon loss) is more bio-labile than Oak 650 Fresh (1364 bio-labile formulas, 15 % carbon loss). This was expected because of the richness of Oak 400 Fresh in smaller less aromatic compounds (Wozniak et al., 2020). Upon photo-irradiation, both Oak 400 Fresh and Oak 650 Fresh experience significant changes in their molecular composition as previously described in detail by Goranov et al. (2020). The photo-transformed pyDOM is much more aliphatic and richer in nitrogen and LMW compounds which render photo-irradiated pyDOM much more biologically labile (Goranov et al., 2020). Surprisingly, it was found that Oak 400 Fresh (1646 bio-labile formulas) is more bio-labile than its photo-irradiated counterpart (Oak 400 Photo, 1242 bio-labile formulas). However, this observation using molecular data does not agree with quantitative carbon loss results for the 10 d incubation (Oak 400 Fresh: 16 % carbon loss; Oak 400 Photo: 25 % carbon loss). The observed discrepancy is because LMW compounds contribute to a large fraction of the degraded carbon in the Oak 400 pyDOM systems and LMW species are not observed following the employed PPL sample preparation and FT-ICR-MS detection. A similar discrepancy is observed when comparing Oak 400 Photo (1242 bio-labile formulas, 25 % carbon loss) and Oak 650 Photo (1410 bio-labile formulas, 23 % carbon loss). In contrast, Oak 650 Fresh (1364 bio-labile formulas) was observed to be less bio-labile than Oak 650 Photo (1410 bio-labile formulas) via both FT-ICR-MS and the observed quantitative carbon losses (Oak 650 Fresh: 15 % carbon loss; Oak 650 Photo: 23 % carbon loss). LMW species are less abundant in the Oak 650 pyDOM systems resulting in consistent trends between the analyses. Biodegradability trends derived from FT-ICR-MS molecular data match those from the UV–Vis data from chromophoric pyDOM (Fig. S1 in the Supplement) revealing a similar inability of UV–Vis to detect LMW compounds which do not absorb UV–Vis light. In summary, we observe a degradation of a variety of different molecular classes as well as a production of many molecules that appear to be of high biological lability. However, we caution that there are observed discrepancies between carbon loss on the one hand and molecular and chromophoric data on the other for the Oak 400 pyDOM systems, an observation that highlights the need to clearly understand methodological analytical windows when interpreting molecular and spectroscopic data.
Interestingly for all leachates, the degraded (bio-labile) molecules were not from a specific area of the vK diagrams but rather represent a broad range of and ratios and compound types (see Fig. S2). This variety of compound characteristics among bio-labile molecules suggests that the degradation pathway may not be from microbial consumption alone. It would be unlikely for the soil microorganisms to utilize organic-matter compounds as food indiscriminately. Most interestingly, it is evident that large numbers of aromatic (AIMOD≥0.50) formulas are lost, in agreement with observed losses in CDOM (Fig. S1) and losses in aryl functional groups (measured by 1H NMR) reported in the parallel study (Bostick et al., 2021). ConAC were found to be resistant to biodegradation (Bostick et al., 2021) and therefore losses of ConAC observed via FT-ICR-MS are considered an artifact due the low ionizability of ConAC and competition processes in the ESI source (Stenson et al., 2002; Patriarca et al., 2020). However, the agreement between FT-ICR-MS and other quantitative data (UV–Vis, NMR, DOC) confirms the interpretation of bulk pyDOM degradation.
Approximately half of the formulas (37 %–56 %) in the original pyDOM leachates are classified as bio-resistant using the presence–absence approach (observed before and after biotic degradation). These formulas are located in all areas of the vK diagrams (Fig. S3) showing variable oxygenation and aromaticity. The relative peak magnitudes of these formulas did not change significantly following the incubations (R2 > 0.95, Fig. S9; Sleighter et al., 2012), also suggesting that a wide variety of pyDOM molecules are resistant to microbial degradation. Using the available molecular data, it is not possible to attribute the observed recalcitrance to any molecular property. Therefore, it is likely that some of these bio-resistant molecules are still bio-labile and would have degraded in due time if the incubations were sampled at later time points. Longer time series should be conducted in future studies to fully differentiate among bio-labile and bio-resistant pyDOM molecules.
The use of hydrogen-to-carbon ratio () versus molecular-weight (MW) plots has also been useful in interpreting ultrahigh-resolution mass spectrometry data (e.g., Gonsior et al., 2018; Powers et al., 2019; Valle et al., 2020). Such graphs are presented using the presence–absence approach in Figs. S5–S8 in Sect. S3. These graphics help evaluate how different types of compounds (aliphatic versus aromatic) change relative to their MW. For both Oak 400 leachates, it is clear that large aromatic molecules ( < 1.5, MW > 550 Da) are removed during the biotic degradation, and smaller (300 < MW < 550) aromatic compounds are produced. The consumption of large molecules indicates that microbes utilize extracellular enzymes to produce ROS, which degrade larger molecules into smaller substrates (Billen et al., 1990). The large aromatic molecules that are being degraded into smaller ones are mainly ligninaceous and not ConAC, in agreement with the insignificant changes in BPCA data published by Bostick et al. (2021). With regards to the aliphatic molecules ( > 1.5), it is clear that molecules of a wide range of MW are degraded and produced during the incubations suggesting that MW is not a critical factor in their biological fate. This is in apparent disagreement with the general knowledge that microbes preferentially consume LMW substrates (e.g., Søndergaard and Middelboe, 1995). Bostick et al. (2021) also concluded that LMW substances are preferentially degraded in the incubations of pyDOM. The observed production of higher MW aliphatics suggest that the microbial incubations were still very active at the point of sampling (10 d). This additionally suggests that future studies need to evaluate the molecular composition of biotically incubated pyDOM over a longer timescale.
3.2 Composition of bio-produced organic matter
The bio-produced organic compounds can be evaluated in various ways to examine the processes that may have occurred during the incubations. Using a presence–absence approach (Sleighter et al., 2012), the bio-produced formulas of each sample are compared with those of the other samples (Table 1). No significant overlap was found (2–320 formulas in common, 0 %–12 %) among the molecules produced in the incubated pyDOM samples. Furthermore, no significant match was found between the bio-produced formulas of incubated pyDOM and those of incubated sucrose sample (63–94 formulas in common, 3 %, Table 1). These observations indicate that the products of the incubations were vastly different for each sample and likely depend on the starting substrate. An alternative explanation is that bio-produced formulas were further altered post-exudation by ROS to result in their molecular diversification.
Table 1Overlap of bio-produced molecular formulas among samples. The number of formulas corresponds to the formulas in common between the two samples being compared, and the percentage is relative to the total number of formulas in the two formula sets.

A significant fraction of the bio-produced organic matter was characterized as peptide-like (N-containing, 1.5 ≤ ≤ 2.0, 0.1 ≤ ≤ 0.67). This indicates that microbes convert a part of pyDOM into labile DOM (Moran et al., 2016; Vorobev et al., 2018), a process hereafter referred to as “microbial labilization”. Given that the pyDOM samples used in this study were poor in organic nitrogen, the microbes must have used the inorganic nitrogen (NH) that was provided as a nutrient and converted some or all of it into microbial biomass. Peptide-like formulas comprised 22 %–40 % of the bio-produced formulas (Table S2). The results of the comparative analyses (Table 1) imply that these proteinaceous formulas are of highly variable composition. Their molecular diversity is additionally evaluated using one-way analysis of variance (ANOVA) reported in Sect. S6. This statistical analysis revealed high molecular variability supporting the findings by the presence–absence comparisons presented earlier (Table 1). Collectively, these findings conclude that the microbial incubations of pyDOM created pools of new, very diverse molecules, a process hereafter referred to as “microbial diversification”. As FT-ICR-MS was performed with soft electrospray ionization with no fragmentation, the structure of the observed molecules is inferred from the elemental composition of the assigned molecular formulas. Another possibility for these N-containing molecules is that they were formed by coupling reactions among pyDOM molecules with the NH nutrient that was added to support microbial growth (e.g., via Michael addition reactions; McKee et al., 2014).
To confirm that the bio-produced formulas were associated with proteinaceous structures and are not just N-containing compounds that coincidentally plotted in the peptide region, spectrofluorometric analysis was performed to obtain excitation–emission matrices (EEMs) of the pyDOM samples before and after bio-incubation (Fig. 2). The data for Oak 650 Photo is not reported as the produced EEM spectra were of questionable quality, and as there were very limited amounts of the sample, analytical replication and quality assessment were not possible.
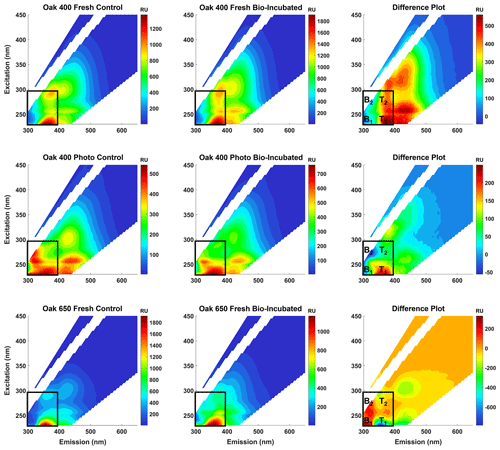
Figure 2Fluorescence excitation–emission matrices (EEMs) of control (left panels) and bio-incubated (middle panels) pyDOM samples. Difference spectra are shown in the right panels. The black box indicates the region where compounds of proteinaceous origin fluoresce (Coble, 1996; Coble et al., 2014), with tyrosine-like (B1 and B2) and tryptophan-like (T1 and T2) peaks labeled on the difference plots.
Proteinaceous organic matter has a highly characteristic fluorophoric signature due to the distinguishable signals of the aromatic amino acids tyrosine and tryptophan. The short Stokes' shifts of these fluorophores allow them to spectroscopically separate on the EEM plot allowing for the identification of related labile substances (Wünsch et al., 2019). Other amino acids, namely histidine and phenylalanine, are also fluorophoric but are not easily identified in EEM data of complex matrices. A practical approach to evaluate the change after the bio-incubation is to use difference plots (e.g., Hemmler et al., 2019). For all samples, strong proteinaceous signals evolve after biotic incubations indicating that molecules of autochthonous (microbial) origin are produced (Coble, 1996; Coble et al., 2014). This indicated that peptide-like molecules observed using FT-ICR-MS are not artificially produced by coupling reactions growth (e.g., via Michael addition reactions; McKee et al., 2014) or evolve due to less charge competition in the ESI source. Thus, the protein-like formulas are truly bio-produced, validating the findings of the presence–absence analysis.
There are subtle differences among the EEMs of all control and bio-incubated samples indicative of the high variability in fluorophoric content of these samples. This agrees with the observed variability in molecular composition described earlier. An interesting observation is that in the two Oak 400 pyDOM incubations, tyrosine-like fluorescence (peaks B1 and B2) decreases after biotic incubation, whereas tryptophan-like fluorescence (peaks T1 and T2) increases. In contrast, the tryptophan-like fluorophores are degraded and tyrosine-like ones are produced after biotic incubation of Oak 650 Fresh pyDOM. Proteinaceous compounds are highly bio-labile and aromatic compounds are susceptible to oxidation by ROS. Therefore, it is possible that tyrosine-like fluorophores (in Oak 400 pyDOM) and tryptophan-like fluorophores (in Oak 650 pyDOM) are still actively participating in biodegradation processes as the incubations were still active at the time of sampling (10 d). The loss of tyrosine-like fluorophores in the Oak 400 samples and loss of tryptophan-like fluorophores in the Oak 650 Fresh sample are indicative of different microbial physiology and exudates. The complexity of these EEM spectra and the compound-specific changes observed here indicate that proteomic and/or metabolomic analyses (e.g., Nalven et al., 2020) are necessary in future microbiological studies in order to fully understand the changes to the molecular composition of pyDOM during biotic incubations.
To determine if the bio-produced formulas are from true proteins or from compounds with residual proteinaceous fluorophores, FT-ICR-MS molecular formulas were evaluated in the context of possible combinations of amino acids that would be singly charged. Given that microbes exude large proteins (MW > 30 kDa) such as lignin peroxidases, manganese peroxidases and laccases (Higuchi, 2004), the peptide-like formulas observed by FT-ICR-MS (analytical window of 200–1000 Da) may have resulted from hydrolysis of the above-mentioned enzymes (or other proteinaceous exudates). If that is the case, the hydrolysates would likely have had a simple oligomeric composition. To test this, the bio-produced peptide-like formulas in each sample were compared to a library of 888 009 possible combinations of 20 amino acids (oligomeric sequences of two to seven residues). Only a small number of oligopeptides were identified (5–18 oligopeptides of 2–5 amino acids, Tables S2 and S3) which is counter to the proposed idea that hydrolysis of microbial exudates produced these newly observed peptide-like formulas. Therefore, the observed bio-produced formulas may represent compounds with residual proteinaceous fluorophores and are not true oligopeptides. The lack of identified oligopeptides also calls into question the idea that microbial processes were solely responsible for the high variability of the bio-produced organic matter observed after the microbial incubation of pyDOM.
To further elucidate the composition of these bio-produced N-containing substances, we re-evaluated the previously published 1H NMR data of these samples (Bostick et al., 2021) in greater detail. Additionally, the connectivity between previously observed functional groups was assessed using two-dimensional 1H–1H total correlation NMR spectroscopy (TOCSY) on a select sample. Figure 3 shows the TOCSY spectra of the bio-incubated Oak 650 Fresh sample.
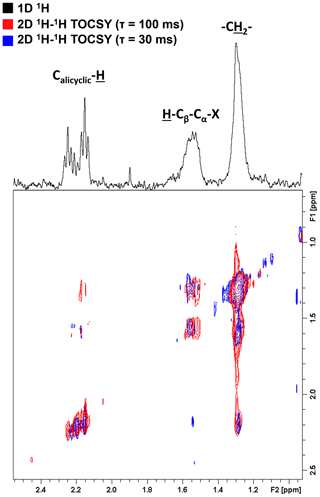
Figure 3Two-dimensional 1H–1H total correlation spectroscopy (TOCSY) NMR spectra of the bio-incubated Oak 650 Fresh sample. Short- and long-range couplings were allowed to evolve during mixing times (τ) of 30 (blue) and 100 ms (red), respectively. The 1D 1H spectrum is shown as a projection on top (black).
There are three groups of resonances representing an alicyclic structure, a β-hydrogen to a heteroatom and a methylene group that were found in all samples, even in the controls (although with small contributions relative to the total spectral signal). These resonances have not been previously observed in the 1H NMR spectra of these pyDOM samples (Bostick et al., 2018; Goranov et al., 2020) indicating that they represent by-products of the microbial incubations, likely microbial biomass. In the control samples, the compounds associated with these resonances must be from the soil inoculant that was added. The three resonances are observed to be in the same coupling network indicating that they are a part of the same or similar structures. Due to the very low concentration of these samples (3.5–4 mgC L−1), the NMR analysis did not allow for a high-resolution structural elucidation, but some distinct signatures were nonetheless observed. The deshielded aliphatic peaks at ppm have a complex multiplicity pattern, a characteristic feature of alicyclic structures. These are likely residual carbohydrate moieties which have lost most of their O-containing groups through various cleavage processes, and their backbone Calicyclic-H resonances have been shifted upfield. The peak at 1.55 ppm is from β-hydrogens to a heteroatom (H-Cβ-Cα-X, where X = O, N, S), and these are known to be associated with peptidoglycans (Spence et al., 2011). The TOCSY analysis was performed with two different mixing times (τ=30 and τ=100 ms) in order to evaluate short-range (2–3 bond) and long-range (4–6 bond) connectivities. Based on the couplings the observed resonances are vicinal to each other (three bonds away). This indicates that these functional groups are closely bound in the peptidoglycan substances they likely represent.
All of the assessments described above conclude that the observed biochemical processes in these pyDOM incubations are complex and difficult to unambiguously interpret. Based on our findings, we summarize that the bio-produced formulas (Fig. 1) can originate from three possible sources:
-
exoenzymes, which microbes use to extracellularly degrade larger molecules into smaller ones (Hyde and Wood, 1997; Higuchi, 2004);
-
peptidoglycans, which likely leached into solution after bacterial death and cell lysis (Yavitt and Fahey, 1984); and
-
other metabolites and exudates involved in the physiology of the different microbes in the consortium used (e.g., signaling compounds).
The significant degradation of pyDOM and production of these biological compounds indicates that microbes successfully converted the presumably carbon-rich recalcitrant pyrogenic molecules into more labile substances, a process we define as microbial labilization. However, the fact that the observed bio-produced labile molecules are not identifiable as simple oligopeptides and are present in significantly different composition among the four samples suggests that this molecular diversity may not be caused by predictable biotic reactions but by random radical-driven processes. Further evidence for the random radical-driven processes comes from the observed degradation of molecules across the whole vK space (Figs. 1 and S2), which is unusual because microbes preferentially consume smaller aliphatic species (Berggren et al., 2010a, b; Kirchman, 2018).
3.3 Radical oxygenation as a potential source of molecular diversity
Microbial physiology has been associated with the production of ROS, which have been shown to be important in the degradation of various types of organic compounds (e.g., Scully et al., 2003; McNally et al., 2005; Porcal et al., 2013; Trusiak et al., 2018; Xiao et al., 2020). Recent studies showed that radicals can degrade various types of ligninaceous molecules (Waggoner et al., 2015, 2017; Waggoner and Hatcher, 2017) suggesting that microbially induced radical reactions can target a variety of pyDOM molecules as well. While there were no ROS measurements made in this study, we have performed Kendrick mass defect (KMD) analysis of the FT-ICR-MS data (Kendrick, 1963; Hughey et al., 2001) to seek evidence for radical processes. The KMD analysis identifies formulas that differ by any repeating structural moiety (e.g., -CH2-). To identify potential products of radical attacks, we have evaluated the FT-ICR-MS data in the context of oxygenation, i.e., searched the mass lists for formulas differing by one oxygen atom (addition of hydroxyl group), carbonyl group (addition of aldehydes or ketones) and carboxyl group (Fig. 4).
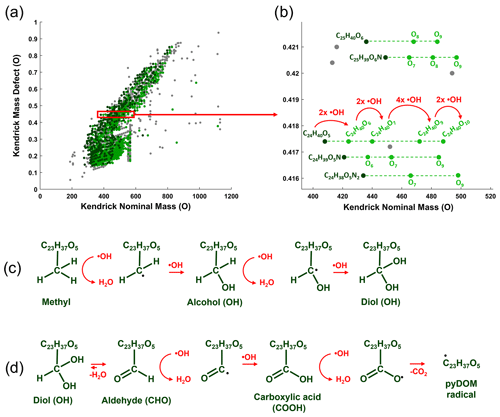
Figure 4Kendrick mass defect (KMD) analysis using oxygen (O) series of the bio-produced formulas of Oak 400 Fresh pyDOM. Panel (a) shows the whole KMD plot, while (b) shows an expanded region of it. Formulas not part of the O KMD series are colored in gray. Formulas in dark green are proposed substrates, and their oxygenation products are colored in light green. Only the molecular formulas for one of the series (KMD = 0.4174 Da) are labeled, while for the rest of the series, only the substrate formula and the number of oxygens in the oxygenation products are listed for clarity. The red arrows show the formation of the four oxygenation products of the C24H40O5 substrate after a sequential attack by hydroxyl radicals (•OH). Panel (c) shows possible chemical reactions that can cause an increase in the number of oxygens. Panel (d) shows further oxidative processes involving the formation of keto and carboxyl groups, processes which ultimately produce pyDOM radicals and CO2. The KMD plots for all samples are shown in Figs. S10–S12.
The mathematics behind the KMD analysis (see Sect. 2.4) convert the mass of the molecular formula (also known as the IUPAC mass) to a “Kendrick” mass placing the formula on a scale that is specific for the selected structural moiety. In Fig. 4a, an example is shown with the KMD analysis for molecules differing by one oxygen (-O-). On the regular (IUPAC) mass scale, such formulas would differ by 15.994915 Da, but on the Kendrick “O” mass scale, they differ by 16 Da. The difference between the Kendrick mass (e.g., KM = 408.2876 Da) and the Kendrick nominal mass (e.g., KNM = 408 Da) is the Kendrick mass defect, KMD (i.e., KMD = 0.2876 Da). Formulas with the exact same KMD differ by one or more oxygens and lie on a KMD series. Visually these formulas would plot on horizontal lines on the KMD plot as indicated by the dashed lines in Fig. 4b. Taking the series of KMD = 0.4174 Da as an example, the KMD evaluation shows that there are five formulas in this particular KMD series that differ in the number of oxygens (C24H40O5–10). This implies that once C25H40O5 is produced, it acts as a substrate and the other four formulas (C24H40O6–10) are produced by oxygenation (likely in a sequential manner: C24H40O5 → C24H40O6 → C24H40O7 → C24H40O9 → C24H40O10). Such formulas differing in the number of oxygens can be formed via oxygenation by hydroxyl radical (•OH) attacks (Waggoner et al., 2015, 2017; Waggoner and Hatcher, 2017). This ROS can abstract a hydrogen from C-H bonds and the hydrogen is substituted with an OH group, resulting in the formation of alcohols (C-OH) as shown in Fig. 4c. This is the suggested pathway of how the oxygenation products shown in Fig. 4a and b have formed. Evidence for such reactions will be found on the KMD plots as the evolution of new molecules within the same KMD series but with a different number of oxygens. Further radical attacks would produce polyols (Fig. 4c). In the case of formation of geminal diols (two alcohol groups on the same carbon atom), they can rearrange to aldehydes or ketones via keto-enol tautomerism (Fig. 4d). Further radical attacks would produce carboxyl groups, which can also be radically cleaved, and pyDOM radicals would be formed. PyDOM radicals (as well as any other radical intermediate in this pathway) can be then further paired with hydrogen radicals (•H) from the solution, with other •OH radicals or with other radicalized pyDOM or proteinaceous species.
Using KMD analysis, formulas that could have been produced by oxygenation were identified and plotted individually (Fig. 5). It is assumed that the smallest molecule in each series is the substrate and any molecules with increasing number of oxygens are oxygenation products.
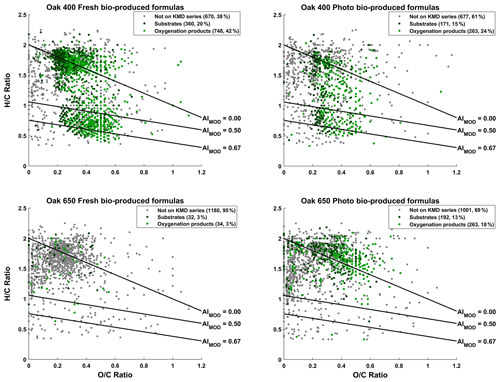
Figure 5Van Krevelen diagrams showing oxygenation products among the bio-produced formulas of the four incubated pyDOM samples. Formulas not part of any of the oxygenation KMD series (O, CO or COO) are colored in gray. Formulas in dark green are substrates with their oxygenation products colored in light green. The number of formulas in each of these pools is shown in the legends (along with corresponding percentages). The black lines indicate modified aromaticity index cutoffs (AIMOD; Koch and Dittmar, 2006, 2016).
KMD analysis revealed that about a third (34–748, 3 %–42 %) of the bio-produced formulas could be classified as products of oxygenation reactions, likely driven by ROS species such as the hydroxyl radical (•OH). This is in agreement with previously observed cross-linking of microbial compounds through oxidative processes (Sun et al., 2017). The majority of the bio-produced formulas, however, were not found to be products of oxidation as they did not lie on any of the evaluated KMD series (O, CO or COO). Therefore, the majority of the bio-produced formulas are likely formulas of exudates which were resistant to radical attacks or are formulas of compounds which have already been radically coupled with other compounds to result in unrecognizable molecules by the KMD analysis.
Additional evidence for intense radical processes in these systems is the evolution of bio-produced unsaturated aliphatic compounds (1 < < 2, < 2) on the vK diagrams (Figs. 1 and S4). ROS can attack aliphatic and aromatic compounds, open aromatic and alicyclic rings, cleave oxygen- or nitrogen-containing functionalities, and produce highly aliphatic molecules, as previously observed after photo-irradiation of pyDOM (Goranov et al., 2020), ConAC (Zeng et al., 2000a, b), and radical-based degradation of lignin (Waggoner et al., 2015, 2017; Waggoner and Hatcher, 2017; Khatami et al., 2019a, b). ROS can also attack any of the proteinaceous exudates and peptidoglycans, cleaving them from many of their functional groups and converting them into the observed unsaturated aliphatic compounds. These produced aliphatic compounds could also contribute to the newly produced N-containing peptide-like compounds observed by FT-ICR-MS if they are oxygenated by ROS post-formation. However, this seems unlikely as data from the supplementary fluorescence and NMR analyses support the formation of microbial biomass. The KMD analysis shown here strongly suggests the presence of intense radical processes as formulas with increasing numbers of oxygen atoms are known to be formed following radical oxygenation (Waggoner et al., 2015, 2017; Waggoner and Hatcher, 2017). However, it must be noted that this KMD analysis does not directly prove the existence of radical processes and the suggested radical processes are speculation based only on indirect observations. Future studies need to directly test for the presence of radical reactions by performing biotic incubations of pyDOM with radical quenchers as well as by quantifying radical fluxes in these microbiological systems.
While FT-ICR-MS peak magnitudes are a function of molecular ionizability, making it generally impossible to quantify the different bio-labile and bio-produced compounds of our study, the ultrasensitivity of this technique ensures the detection of all compounds that are within the FT-ICR-MS analytical window. Here, the number of molecular formulas can be used as a quantitative measure for molecular diversity (e.g., Gurganus et al., 2015). Previously published liquid-state 1H NMR data for the same samples provide a quantitative measure of functional group content (Bostick et al., 2021). Significant positive and negative correlations were observed between the numbers of bio-labile and bio-produced formulas and the percent NMR spectral signal accounted for by olefinic functionalities and methanol (Fig. 6 and Table S4). These correlations suggest that the diversity of biodegraded (bio-labile) and bio-produced molecules was related in some way with a process related to the availability of methanol (CH3OH) and olefinic functionalities (C=C) in pyDOM.
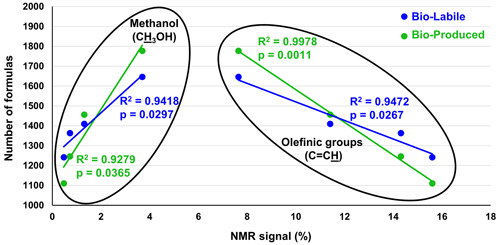
Figure 6Pearson correlation analysis between the number of bio-labile and bio-produced formulas detected by FT-ICR-MS and relative intensity (in %) of olefinic functionalities (C=C) and methanol (CH3OH) as measured by liquid-state 1H NMR and reported by Bostick et al. (2021). No significant correlations were found between other functional groups and the number of bio-produced or bio-labile formulas (data shown in Table S4).
Olefinic functionalities have been recently identified as important structural motifs in the composition of pyDOM and were observed to degrade in photochemical experiments likely due to their high reactivity with ROS species (Goranov et al., 2020). Although olefins are in low abundance in pyDOM (<10 %), it is likely that they act as important intermediates in the degradative pathways of pyDOM (Goranov et al., 2020). Olefinic bonds can be homolytically cleaved when attacked by radicals and effectively act as radical accelerators that further propagate radical-mediated organic-matter transformations. Thus, the abundance of olefins can further increase the abundance of radicals and contribute to the elevated molecular diversity resulting in the significant correlation shown in Fig. 6.
The other significant correlation between molecular diversity and NMR data is observed to be with methanol (CH3OH), a very sharp highly distinguishable singlet at δ=3.34 ppm in 1H NMR spectra (Gottlieb et al., 1997). As it is a common contaminant in NMR analysis, special precautions were taken to obtain ultraclean spectra (see Sect. 2.5). Methanol is a species that is naturally present in pyDOM (Bostick et al., 2018), and while it is generally considered to be toxic to microbes (Dyrda et al., 2019), there are methylotrophic bacteria and fungi (microbes of the families Methylococcaceae and Methylobacteriaceae) that can utilize it as a substrate (Chistoserdova et al., 2003; Kolb and Stacheter, 2013; Chistoserdova and Kalyuzhnaya, 2018). These species have been previously observed in the soil from the area where the microbial inoculum was extracted (Khodadad et al., 2011), suggesting that the degradation of methanol may be biotic. In fact, in these samples, methanol, along with the other two measured LMW substances, acetate and formate, was almost completely degraded over the 10 d incubation (Bostick et al., 2021).
The inverse relationship between the content of methanol and molecular diversity (Fig. 6) can be interpreted in several ways. Firstly, methanol could be exhibiting toxicity to the microbes that assimilate pyDOM, as has been observed previously (Dyrda et al., 2019). This, however, is unlikely for the pyDOM systems studied here because the sample with the highest amount of methanol (Oak 400 Photo, ∼ 3.7 % CH3OH) was the second-most bio-reactive (Bostick et al., 2021). Instead, the observed significant negative correlation may be explained by the fact that methanol is a known radical scavenger (Múčka et al., 2013). If, as we propose, the molecular diversity results from the activity of radical processes, an increasing concentration of methanol would quench ROS, thereby decreasing the radical activity and limiting the molecular diversity in these systems.
4.1 Multiple pathways for the alteration of pyDOM by microbes
Using a variety of analytical platforms in this and the parallel study (Bostick et al., 2021), significant quantitative and qualitative losses were observed when pyDOM was subjected to incubation with a microbial consortium collected from a site impacted by wildfires. Additionally, labile and diverse compounds were produced during these incubations. Due to the high complexity of pyDOM, the changes are not straightforward, and there are at least two degradation pathways at play: (1) degradation through microbial assimilation (consumption of pyDOM) and (2) degradation and/or transformation via radical-mediated reactions (e.g., oxygenation) by ROS produced from microbial exoenzymes. These two pathways are discussed in the context of degradation of pyDOM and the formation of new labile and diverse molecules.
4.1.1 Molecular degradation of pyDOM
A surprising observation in this study is that there was a uniform loss of pyDOM molecules from all regions of the vK diagrams. Microbes, it is generally presumed, preferentially assimilate small non-aromatic substances such as carbohydrates, proteins and LMW acids (Berggren et al., 2010a, b; Kirchman, 2018). Thus, the aromatic fraction of pyDOM, mainly the ConAC, are generally considered to be bio-resistant (Goldberg, 1985; Masiello, 2004). In addition to the condensed character of many of the molecules, there are significant numbers of potentially toxic organochlorine compounds, of both aliphatic and aromatic character, in pyDOM (Wozniak et al., 2020). Thus, the finding of major biological activity in these pyDOM systems and the significant amount of carbon, including aromatic carbon, that was mineralized is a very significant finding for the wildfire biogeochemistry community (Bostick et al., 2021).
Although pyDOM is highly heterogeneous (Wozniak et al., 2020), the observation of diverse molecular biodegradation is not unique to it. In a recent microbial degradation study of snow DOM, Antony et al. (2017) observed that both aromatic and aliphatic formulas were biodegraded. This is likely due to microbes evolving chemical mechanisms to thrive under extreme glacial conditions (Antony et al., 2016). Analogously, as there have been previous prescribed fires in the area from which the microbes for this study were extracted (Johns, 2016), it is also possible that our microbes had adapted to the presence of pyOM/pyDOM by developing mechanisms for assimilating the carbon in large, complex molecules, including ConAC (Judd et al., 2007).
While direct microbial assimilation of pyDOM compounds is certainly likely to have occurred, our molecular and spectroscopic findings suggest a second degradative pathway contributing to the extensive molecular alteration and to the significant loss of carbon that was quantified in the parallel study (Bostick et al., 2021). While some microbial exoenzymes operate via hydrolytic pathways (amylases, lipases, proteases, cellulases, β-galactosidases, etc.), many other enzymes operate through oxidative (electron-withdrawing) pathways. Examples of such enzymes are the various lignin-modifying enzymes in the peroxidase (lignin peroxidases, manganese peroxidases, etc.) and phenol oxidase (e.g., laccases) families (Higuchi, 2004). Thus, ROS are usually produced and involved in the microbial degradation of organic matter in the environment.
The bio-labile molecules in the studied pyDOM samples are of highly variable degrees of oxygenation, aromaticity and size. There were large MW compounds (MW > 550 Da) that were degraded indicating that microbial exoenzymes producing ROS would have been needed to reduce the size of these large substrates into smaller units that could pass through microbial cell membranes and be consumed by the biota (Sinsabaugh et al., 1997; Fuchs et al., 2011; Burns et al., 2013). The presence of enzymatic compounds is confirmed by observation of peptide-like compounds (FT-ICR-MS analysis) and proteinaceous fluorophores (spectrofluorometric analysis). An important finding is that a preferential degradation of ConAC of smaller MW was observed (Bostick et al., 2021). As small ConAC (i.e., oxygenated polycondensed aromatic hydrocarbons) are known to be toxic (e.g., Idowu et al., 2019), it is unlikely that they were directly consumed by the microbes. Oxygenated polycondensed aromatic hydrocarbons are highly susceptible to attacks by ROS, which is likely how they were degraded in these samples. Thus, we speculate that microbes are most likely not directly consuming the smaller ConAC, but, rather, smaller ConAC are degraded indirectly via ROS oxidation. Furthermore, ROS can oxygenate pyDOM with various functional groups (e.g., hydroxy, aldehyde/keto, carboxyl), and can also cleave functional groups (e.g., methoxy functionalities), open aromatic rings and completely mineralize compounds to inorganic carbon (CO, CO2, HCO and CO) as shown in Fig. 4. ROS have been previously shown to be very important in pyDOM photochemistry (Ward et al., 2014; Fu et al., 2016; Wang et al., 2020), and we speculate that they play an important role in the microbial degradation of pyDOM as well.
More indirect evidence for radical species involvement is provided by the peptidoglycan molecules produced during the pyDOM incubations. Peptidoglycan molecules are generally large (Vollmer et al., 2008) and would not be detected as singly charged ions using FT-ICR-MS (analytical window covering only 200–1000). The hydrolytic products of peptidoglycans (small oligopeptides) would be observed. Very few peptide sequences (5–18 oligopeptides of 2–5 residues) were identified among the bio-produced formulas, indicating that such hydrolysates did not exist in the samples at the time of measurement. However, if there were abundant radical reactions occurring in the system, as we suggest, it is very possible that these hydrolysates were altered into unrecognizable organic structures that would still be classified as peptide-like but would have different molecular composition than the predicted linear peptide sequences. It is also possible that instead of peptidoglycan hydrolysis followed by consecutive oxygenation, ROS directly cleaved the peptidoglycans into smaller non-linear substances of peptide-like molecular composition.
It must be noted that the results of our study were acquired using negative-mode ESI which is only effective for electronegative (carboxyl-rich, hydroxyl-rich) compounds (Stenson et al., 2002; Patriarca et al., 2020). Thus, the trends of degradation and labilization are skewed to fit this criterion and do not provide a complete overview of all molecules that are bio-labile or bio-produced. Future studies should employ positive-mode ESI (Ohno et al., 2016) and/or different ionization sources (such as atmospheric pressure photoionization, Hockaday et al., 2009) to better elucidate the molecular degradability of pyDOM.
4.1.2 Labilization and diversification of pyDOM
The production of labile unrecognizable biological substances during these incubations correlates well with previous findings showing the formation of thousands of new biological compounds during biotic incubations unrelated to microbial metabolic pathways (Lechtenfeld et al., 2015; Wienhausen et al., 2017; Patriarca et al., 2021). There was no significant overlap among the bio-produced formulas of the four pyDOM samples (2–320 formulas in common, 0 %–12 % overlap). Insignificant numbers of bio-produced formulas from pyDOM were also found in the bio-produced formulas of an incubation of sucrose with the same soil microbes (63–94 formulas, 3 % overlap). This indicates that microbes diversified the composition of the incubated pyDOM.
The observed diversity of bio-produced formulas can be explained by a scenario wherein the microbes secreted molecules whose identities differed depending on the growth medium and/or food source, yielding high variability among bio-produced formulas after the incubation of pyDOM. Additionally, it is possible that different microbial species (different bacteria, fungi, archaea, etc.) have proliferated in response to the sample-specific pyDOM composition, yielding different microbial populations growing during each different incubation, sequentially producing different bio-produced compounds (Fitch et al., 2018). Another possible explanation for the observed diversification is the presence of ROS-driven oxygenation processes. ROS are known to be key species in the consumption of organic matter by microbes. Microbes can only consume small molecules that could pass through their cell membranes (Sinsabaugh et al., 1997; Fuchs et al., 2011; Burns et al., 2013). Thus, to utilize large molecules as food, microbes produce exoenzymes which generate ROS extracellularly (Hyde and Wood, 1997; Higuchi, 2004). These ROS then degrade the large molecules into smaller ones that are utilized as food. Though not directly proven to exist in this study, many of the observed trends in FT-ICR-MS, NMR and fluorescence data suggest the presence of radicals which diversify the composition of the bio-produced formulas.
Our finding of extreme molecular diversity contrasts with previous observations in a study by Lechtenfeld et al. (2015) evaluating the molecular composition of microbially produced DOM. In their study, marine microbes were supplied with two different substrates (glucose and glutamic acid; and a mixture of oligosaccharides and oligopeptides), and a significant overlap (67 %–69 %) in the bio-produced organic matter was observed. The difference in observations between the work presented in this paper and by Lechtenfeld et al. (2015) is likely caused by a large difference in the composition of the pyDOM substrates relative to those in the Lechtenfeld et al. (2015) study. While the four pyDOM samples used here are highly different to one another (Goranov et al., 2020; Wozniak et al., 2020), the substrates by Lechtenfeld et al. (2015) were of much higher similarity (glucose, glutamic acid, oligosaccharides and oligopeptides). Another possible reason is that due to different physiology the soil microbes used here may be producing more diverse biomass than the marine microbes used by Lechtenfeld et al. (2015). It is likely that aquatic microbes have a very different degradation strategy than soil microbes. As soils are far less rich in labile molecules, it is possible that soil microbes have evolved to produce much higher fluxes of ROS to degrade the more recalcitrant soil organic matter into consumable substrates, which can also explain the larger dissimilarity in bio-produced organic molecules after the incubations of pyDOM.
An important observation using the versus MW plots (Fig. S5) was that the bio-produced compounds after incubation of pyDOM were of various MW. Thus, it is likely that the microbial biomass produced during the incubation is radically coupled with ambient pyDOM molecules or their biochemical remnants. Radical coupling has recently been proposed as an important process in marine DOM cycling (Hach et al., 2020). In that study, when isotopically 13C-labeled organisms were incubated with oceanic surface waters, microbially produced compounds were quickly coupled to the ambient marine DOM molecules. This “recombination” process occurred within hours of the production of microbial exudates, followed by the observation of a highly diversified DOM pool. This process is likely driven by radical coupling reactions, and such pathways have also been observed in incubations in the presence of sunlight (Sun et al., 2017). Another possible explanation is that chemically reactive species, such as quinones, reacted with microbially produced compounds or the NH nutrient via nucleophile-driven reactions (such as the Michael addition; McKee et al., 2014) to produce highly diverse pools of molecules after each incubation.
Our results are also compared to previous work by Waggoner et al. (2017), where a ligninaceous sample was treated with three different ROS: hydroxyl radical (•OH), singlet oxygen (1O2) and superoxide (O). Each different radical degraded a specific pool of ligninaceous compounds, which showed that different ROS can degrade a variety of types of organic matter. However, there was a significant overlap observed between the three pools of molecules that were degraded (Waggoner et al., 2017) indicating that degradation pathways solely based on ROS attacks are still ordered. Thus, because ROS on their own do not produce completely diversified molecular pools, the combination of the two possible pathways (consumption and ROS degradation) must have occurred to produce the great variability in the bio-produced microbial biomass observed in our study.
Collectively, our results indicate that pyDOM can be both directly consumed by biota and secondarily degraded by ROS-driven processes. These pathways could not be explored at a mechanistic level in the current study, and we suggest that future studies focus on employing more specialized analytical techniques (e.g., genetic sequencing, ROS quantification) for deconvoluting the complexity of the biodegradation of pyDOM.
4.2 Implications for the cycling of pyDOM in the environment
The present study provides a detailed evaluation of the compounds that microbes degrade and produce in samples mimicking pyDOM in hydrologically dynamic systems such as riverine and groundwater systems. This work brings new knowledge regarding the properties and degradability of pyDOM and challenges the conventional idea that pyDOM is stable towards biotic degradation. Several studies have already shown that pyrogenic substances have soluble DOM components (Hockaday et al., 2007; Mukherjee and Zimmerman, 2013; Wagner et al., 2017; Bostick et al., 2018) and that more soluble components are produced with environmental aging (Abiven et al., 2011; Ascough et al., 2011; Roebuck et al., 2017; Quan et al., 2020). The experiments presented here, in parallel with Bostick et al. (2021), show that a large portion of pyDOM can be remineralized (biodegraded to CO2) without priming (Guenet et al., 2010; Bianchi, 2011), which indicates that pyrogenic molecules may be far less resistant to biodegradation than previously presumed.
The involvement of pyDOM within the global carbon cycle is complex and, for many biogeochemical processes, poorly understood. There is a growing body of literature showing that significant amounts of ConAC are solubilized and exported to the global ocean (Dittmar et al., 2012; Jaffé et al., 2013; Wang et al., 2016; Marques et al., 2017; Jones et al., 2020) suggesting constant global leaching of pyDOM in riverine systems from pyOM in soils. The global riverine flux of pyDOM is estimated using the recently reported global flux of ConAC (18 Tg C yr−1) and scaled using a factor of 7.5 as proposed by Bostick et al. (2018) to be 135 Tg C yr−1, a value that is much lower than the estimated annual pyDOM production and seepage flux of 1440 TgC yr−1 (Bostick et al., 2018). In addition to the implied 91 % loss of carbon during riverine export, a recent study also reported that the stable carbon isotopic signature (δ13C) of oceanic ConAC is not terrestrial but rather marine-like (Wagner et al., 2019). This suggests that either all riverine-exported ConAC are being mineralized before they reach the global ocean or ConAC are chemically altered significantly to change their δ13C isotopic signature (Jones et al., 2020). Microbial and photochemical processes have been found to transform DOM with characteristic terrestrial DOM composition (compounds with lower and higher ratios) into compounds having characteristics of marine-derived DOM (compounds with higher , lower ratios; Rossel et al., 2013). Thus, pyDOM may simply be losing its diagnostic molecular and isotopic terrestrial-like fingerprints during riverine export due to a variety of degradative post-production processes, and the observed molecular transformations in our study are likely one of them.
The cycling of organic matter in the environment has always been an enigma, and there has been a long-standing effort to explain the fate of terrestrial DOM (including pyDOM) in the global ocean (Hedges et al., 1997). In a previous paper evaluating the photochemical transformation of pyDOM (Goranov et al., 2020), we suggested that biotic consumption of photodegradation products of pyDOM (“small aliphatic compounds”) could result in the formation of marine-like DOM. This hypothesis was tested by comparing our incubation products (the bio-produced formulas) to FT-ICR-MS formulas of an estuarine transect of the Elizabeth River, VA (Sleighter and Hatcher, 2008) and another 10 oceanic DOM samples (reported in Sect. S5). A significant number of common formulas was observed in these comparisons (193–308 common formulas, 8 %–18 % overlap) confirming the hypothesis that bio-incubation of pyDOM can produce marine-like DOM. The observed common formulas were not condensed and 81–192 of them (42 %–70 % overlap) were of molecular composition attributed to carboxyl-rich alicyclic molecules (CRAM) per the definition of Hertkorn et al. (2006). These results indicate that biotic incubations of pyDOM (regardless of photo-irradiation) can contribute to some of the molecules observed in oceanic environments. The fact that some of these molecules were observed in both surface and abyssal oceanic DOM indicate that some pyDOM biodegradation products may be sequestered into the deep ocean as refractory DOM.
The observed bio-produced labile formulas in our study do not appear to be commonly observed in other environmental samples. This is likely because these labile molecules are part of the fast-cycling, labile DOM pool per Hansell's model (Hansell and Carlson, 2015) and are quickly depleted in the natural environment. This parallels the findings of a recently published study (Hach et al., 2020) observing that microbially produced molecules are extremely labile and are, within hours, broken down and recombined with ambient DOM molecules. The closed laboratory systems in our study may have enabled the observation of these highly labile molecules, whereas in the natural environment they would have been quickly transformed, diluted, or mineralized to inorganic carbon resulting in their removal from analytical detection. The richness in nitrogen and peptide-like character of the bio-produced molecules we observe suggest greater potential lability, and it is likely that the by-products of biotic degradation of pyDOM are readily incorporated into microbial food webs. This is consistent with the idea that terrestrial DOM is either mineralized to CO2 or incorporated into food webs (Berggren et al., 2010a; Ward et al., 2013; Fasching et al., 2014). It is also consistent with the suggestion that the majority of organic nitrogen in the oceans is derived from microbial peptidoglycans (McCarthy et al., 1997, 1998; Simpson et al., 2011) and with observations of nitrogen from peptidoglycans in soil and sedimentary porewater systems (Schulten and Schnitzer, 1998; Hu et al., 2018, 2020).
The production of highly variable and diverse bio-produced molecules is likely a contributing factor to the large complexity of organic matter in the environment (Hertkorn et al., 2007; Hawkes et al., 2018). Our observed bio-produced molecules likely contribute to the highly variable microbial exometabolomes observed previously (Antón et al., 2013; Watrous et al., 2013; Romano et al., 2014) and stimulate further questions about pyDOM's function and fate within the global carbon and nitrogen cycles. In this study, we have used soil microbes, as the corresponding degradation by-products can be observed in both soil and groundwater and partially in the upstream sections of rivers. Therefore, it would be critical to perform further studies with different microbial consortia (riverine, estuarine, marine, etc.) to fully understand the biological degradation of pyDOM in different environments. Additionally, the observed evidence for two possible degradative pathways (consumption and ROS degradation) indicates that these pyDOM incubations are extremely complex systems. Future microbiological studies must aim to investigate these pathways further by designing radical quenching experiments (to test for the presence/absence of radical oxygenation pathways) as well as to employ bio-analytical techniques (e.g., genetic sequencing; Nalven et al., 2020) for assessing what microbes are responsible for the labilization and diversification of pyDOM.
This study probing the molecular changes occurring after biotic degradation of pyDOM revealed that soil microbes can effectively recycle and transform a significant portion of pyDOM molecules into labile microbial biomass. After the 10 d incubations, it appears that a wide range of pyDOM molecules, both aromatic and aliphatic, were degraded and/or transformed, forming a highly diverse pool of compounds, including N-containing compounds with proteinaceous signatures and a peptidoglycan-like backbone. These observations are consistent with the previous identification of nitrogen from peptidoglycans in soils and oceans. These bio-produced compounds were highly specific for each pyDOM sample which was concluded by observing very few common bio-produced molecular formulas among incubated samples. The observed molecular labilization and diversification have implications for the biogeochemistry of pyDOM as we show that microbial reworking of pyDOM can contribute to the large complexity and variability of natural organic matter. This study reveals that (1) pyDOM can be a medium for microbial growth and (2) previously considered recalcitrant pyrogenic molecules can be broken down and the carbon and nitrogen therein can be incorporated into microbial food webs. This study suggests that pyDOM is a much more active component in the global carbon and nitrogen cycles and that some non-condensed pyDOM degradation products have an oceanic fate. Therefore, future studies need to further evaluate the biodegradability of pyDOM with microbial consortia of different environments as well as in the context of wetted soils, groundwater processes, cycling within the riverine and marine water columns, and other aspects of the global carbon and nitrogen cycles.
Research data associated with this article can be accessed at https://doi.org/10.17632/kjkhy3tfys.2 (Goranov et al., 2022).
The supplement related to this article is available online at: https://doi.org/10.5194/bg-19-1491-2022-supplement.
AIG, ASW, KWB, ARZ, SM and PGH conceptualized the study and designed the experiments; KWB carried out the preparation of pyDOM, photo-irradiations, microbial incubations and quantitative analyses (DOC, BPCA markers); AIG performed the extractions and spectrometric/spectroscopic analyses; AIG and KWB analyzed the data; AIG generated the original draft; AIG, ASW, KWB, ARZ, SM and PGH reviewed and edited the manuscript. Funding was acquired by ASW, ARZ, SM and PGH.
The contact author has declared that neither they nor their co-authors have any competing interests.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
We thank Rachel L. Sleighter for providing the estuarine and marine FT-ICR-MS data. We also thank Isaiah Ruhl, Deepti Varma and Ravi Garimella for assistance during instrumental analyses at the COSMIC facility (Old Dominion University) and Dow Van Arnam (University of Florida) for design and construction of the biochar pyrolyzer and solar simulator. Lastly, we would like to express our gratitude to the editor and referees for their time and feedback in improving this paper.
This research has been supported by the National Science Foundation (grant numbers EAR-1451452 and EAR-1451367) and the Old Dominion University (Frank Batten Endowment Fund to Patrick G. Hatcher and Dominion Fellowship to Aleksandar I. Goranov).
This paper was edited by Jianming Xu and reviewed by two anonymous referees.
Abboudi, M., Jeffrey, W. H., Ghiglione, J. F., Pujo-Pay, M., Oriol, L., Sempéré, R., Charrière, B., and Joux, F.: Effects of photochemical transformations of dissolved organic matter on bacterial metabolism and diversity in three contrasting coastal sites in the Northwestern Mediterranean Sea during summer, Microb. Ecol., 55, 344–357, https://doi.org/10.1007/s00248-007-9280-8, 2008.
Abiven, S., Hengartner, P., Schneider, M. P. W., Singh, N., and Schmidt, M. W. I.: Pyrogenic carbon soluble fraction is larger and more aromatic in aged charcoal than in fresh charcoal, Soil Biol. Biochem., 43, 1615–1617, https://doi.org/10.1016/j.soilbio.2011.03.027, 2011.
Antón, J., Lucio, M., Peña, A., Cifuentes, A., Brito-Echeverría, J., Moritz, F., Tziotis, D., López, C., Urdiain, M., Schmitt-Kopplin, P., and Rosselló-Móra, R.: High metabolomic microdiversity within co-occurring isolates of the extremely halophilic bacterium Salinibacter ruber, PLoS ONE, 8, 1–14, https://doi.org/10.1371/journal.pone.0064701, 2013.
Antony, R., Sanyal, A., Kapse, N., Dhakephalkar, P. K., Thamban, M., and Nair, S.: Microbial communities associated with Antarctic snow pack and their biogeochemical implications, Microbiol. Res., 192, 192–202, https://doi.org/10.1016/j.micres.2016.07.004, 2016.
Antony, R., Willoughby, A. S., Grannas, A. M., Catanzano, V., Sleighter, R. L., Thamban, M., Hatcher, P. G., and Nair, S.: Molecular insights on dissolved organic matter transformation by supraglacial microbial communities, Environ. Sci. Technol., 51, 4328–4337, https://doi.org/10.1021/acs.est.6b05780, 2017.
Antony, R., Willoughby, A. S., Grannas, A. M., Catanzano, V., Sleighter, R. L., Thamban, M., and Hatcher, P. G.: Photo-biochemical transformation of dissolved organic matter on the surface of the coastal East Antarctic ice sheet, Biogeochemistry, 141, 229–247, https://doi.org/10.1007/s10533-018-0516-0, 2018.
Ascough, P. L., Bird, M. I., Francis, S. M., Thornton, B., Midwood, A. J., Scott, A. C., and Apperley, D.: Variability in oxidative degradation of charcoal: Influence of production conditions and environmental exposure, Geochim. Cosmochim. Ac., 75, 2361–2378, https://doi.org/10.1016/j.gca.2011.02.002, 2011.
Bao, H., Niggemann, J., Luo, L., Dittmar, T., and Kao, S.-J.: Aerosols as a source of dissolved black carbon to the ocean, Nat. Commun., 8, 1–7, https://doi.org/10.1038/s41467-017-00437-3, 2017.
Bax, A. and Davis, D. G.: MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy, J. Magn. Reson., 65, 355–360, https://doi.org/10.1016/0022-2364(85)90018-6, 1985.
Benner, R. and Biddanda, B.: Photochemical transformations of surface and deep marine dissolved organic matter: Effects on bacterial growth, Limnol. Oceanogr., 43, 1373–1378, https://doi.org/10.4319/lo.1998.43.6.1373, 1998.
Berggren, M., Laudon, H., Haei, M., Ström, L., and Jansson, M.: Efficient aquatic bacterial metabolism of dissolved low-molecular-weight compounds from terrestrial sources, ISME J., 4, 408–416, https://doi.org/10.1038/ismej.2009.120, 2010a.
Berggren, M., Ström, L., Laudon, H., Karlsson, J., Jonsson, A., Giesler, R., Bergström, A.-K., and Jansson, M.: Lake secondary production fueled by rapid transfer of low molecular weight organic carbon from terrestrial sources to aquatic consumers, Ecol. Lett., 13, 870–880, https://doi.org/10.1111/j.1461-0248.2010.01483.x, 2010b.
Bianchi, T. S.: The role of terrestrially derived organic carbon in the coastal ocean: A changing paradigm and the priming effect, P. Natl. Acad. Sci. USA, 108, 19473–19481, https://doi.org/10.1073/pnas.1017982108, 2011.
Billen, G., Servais, P., and Becquevort, S.: Dynamics of bacterioplankton in oligotrophic and eutrophic aquatic environments: Bottom-up or top-down control?, Hydrobiologia, 207, 37–42, https://doi.org/10.1007/BF00041438, 1990.
Bostick, K. W., Zimmerman, A. R., Wozniak, A. S., Mitra, S., and Hatcher, P. G.: Production and composition of pyrogenic dissolved organic matter from a logical series of laboratory-generated chars, Front. Earth Sci., 6, 1–14, https://doi.org/10.3389/feart.2018.00043, 2018.
Bostick, K. W., Zimmerman, A. R., Goranov, A. I., Mitra, S., Hatcher, P. G., and Wozniak, A. S.: Photolability of pyrogenic dissolved organic matter from a thermal series of laboratory-prepared chars, Sci. Total Environ., 724, 1–9, https://doi.org/10.1016/j.scitotenv.2020.138198, 2020.
Bostick, K. W., Zimmerman, A. R., Goranov, A. I., Mitra, S., Hatcher, P. G., and Wozniak, A. S.: Biolability of fresh and photodegraded pyrogenic dissolved organic matter from laboratory-prepared chars, J. Geophys. Res.-Biogeo., 126, 1–17, https://doi.org/10.1029/2020JG005981, 2021.
Burns, R. G., DeForest, J. L., Marxsen, J., Sinsabaugh, R. L., Stromberger, M. E., Wallenstein, M. D., Weintraub, M. N., and Zoppini, A.: Soil enzymes in a changing environment: Current knowledge and future directions, Soil Biol. Biochem., 58, 216–234, https://doi.org/10.1016/j.soilbio.2012.11.009, 2013.
Chen, M. and Jaffé, R.: Photo- and bio-reactivity patterns of dissolved organic matter from biomass and soil leachates and surface waters in a subtropical wetland, Water Res., 61, 181–190, https://doi.org/10.1016/j.watres.2014.03.075, 2014.
Chistoserdova, L. and Kalyuzhnaya, M. G.: Current trends in methylotrophy, Trends Microbiol., 26, 703–714, https://doi.org/10.1016/j.tim.2018.01.011, 2018.
Chistoserdova, L., Chen, S.-W., Lapidus, A., and Lidstrom, M. E.: Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view, J. Bacteriol., 185, 2980–2987, https://doi.org/10.1128/JB.185.10.2980-2987.2003, 2003.
Coble, P. G.: Characterization of marine and terrestrial DOM in seawater using excitation-emission matrix spectroscopy, Mar. Chem., 51, 325–346, https://doi.org/10.1016/0304-4203(95)00062-3, 1996.
Coble, P. G., Lead, J., Baker, A., Reynolds, D. M., and Spencer, R. G. M. (Eds.): Aquatic Organic Matter Fluorescence, Cambridge University Press, New York, NY, https://doi.org/10.1017/CBO9781139045452, 2014.
Coppola, A. I., Seidel, M., Ward, N. D., Viviroli, D., Nascimento, G. S., Haghipour, N., Revels, B. N., Abiven, S., Jones, M. W., Richey, J. E., Eglinton, T. I., Dittmar, T., and Schmidt, M. W. I.: Marked isotopic variability within and between the Amazon River and marine dissolved black carbon pools, Nat. Commun., 10, 1–8, https://doi.org/10.1038/s41467-019-11543-9, 2019.
D'Andrilli, J., Fischer, S. J., and Rosario-Ortiz, F. L.: Advancing critical applications of high resolution mass spectrometry for DOM assessments: Re-engaging with mass spectral principles, limitations, and data analysis, Environ. Sci. Technol., 54, 11654–11656, https://doi.org/10.1021/acs.est.0c04557, 2020.
Dittmar, T. and Paeng, J.: A heat-induced molecular signature in marine dissolved organic matter, Nat. Geosci., 2, 175–179, https://doi.org/10.1038/ngeo440, 2009.
Dittmar, T., Koch, B., Hertkorn, N., and Kattner, G.: A simple and efficient method for the solid-phase extraction of dissolved organic matter (SPE-DOM) from seawater, Limnol. Oceanogr.-Meth., 6, 230–235, https://doi.org/10.4319/lom.2008.6.230, 2008.
Dittmar, T., de Rezende, C. E., Manecki, M., Niggemann, J., Coelho Ovalle, A. R., Stubbins, A., and Bernardes, M. C.: Continuous flux of dissolved black carbon from a vanished tropical forest biome, Nat. Geosci., 5, 618–622, https://doi.org/10.1038/ngeo1541, 2012.
Druffel, E.: Comments on the importance of black carbon in the global carbon cycle, Mar. Chem., 92, 197–200, https://doi.org/10.1016/j.marchem.2004.06.026, 2004.
Dyrda, G., Boniewska-Bernacka, E., Man, D., Barchiewicz, K., and Słota, R.: The effect of organic solvents on selected microorganisms and model liposome membrane, Mol. Biol. Rep., 46, 3225–3232, https://doi.org/10.1007/s11033-019-04782-y, 2019.
Fasching, C., Behounek, B., Singer, G. A., and Battin, T. J.: Microbial degradation of terrigenous dissolved organic matter and potential consequences for carbon cycling in brown-water streams, Sci. Rep.-UK, 4, 1–7, https://doi.org/10.1038/srep04981, 2014.
Fitch, A., Orland, C., Willer, D., Emilson, E. J. S., and Tanentzap, A. J.: Feasting on terrestrial organic matter: Dining in a dark lake changes microbial decomposition, Glob. Change Biol., 24, 5110–5122, https://doi.org/10.1111/gcb.14391, 2018.
Fu, H. Y., Liu, H. T., Mao, J. D., Chu, W. Y., Li, Q. L., Alvarez, P. J. J., Qu, X. L., and Zhu, D. Q.: Photochemistry of dissolved black carbon released from biochar: Reactive oxygen species generation and phototransformation, Environ. Sci. Technol., 50, 1218–1226, https://doi.org/10.1021/acs.est.5b04314, 2016.
Fuchs, G., Boll, M., and Heider, J.: Microbial degradation of aromatic compounds – from one strategy to four, Nat. Rev. Microbiol., 9, 803–816, https://doi.org/10.1038/nrmicro2652, 2011.
Goldberg, E. D.: Black carbon in the environment: Properties and distribution, J. Wiley, New York, NY, ISBN-13 978-0471819790, 1985.
Gonsior, M., Hertkorn, N., Hinman, N., Dvorski, S. E., Harir, M., Cooper, W. J., and Schmitt-Kopplin, P.: Yellowstone hot springs are organic chemodiversity hot spots, Sci. Rep.-UK, 8, 1–13, https://doi.org/10.1038/s41598-018-32593-x, 2018.
Goranov, A. I., Wozniak, A. S., Bostick, K. W., Zimmerman, A. R., Mitra, S., and Hatcher, P. G.: Photochemistry after fire: Structural transformations of pyrogenic dissolved organic matter elucidated by advanced analytical techniques, Geochim. Cosmochim. Ac., 290, 271–292, https://doi.org/10.1016/j.gca.2020.08.030, 2020.
Goranov, A., Wozniak, A., Bostick, K., Zimmerman, A., Mitra, S., and Hatcher, P.: Microbial labilization and diversification of pyrogenic dissolved organic matter, Mendeley Data [data set], https://doi.org/10.17632/kjkhy3tfys.2, 2022.
Gottlieb, H. E., Kotlyar, V., and Nudelman, A.: NMR chemical shifts of common laboratory solvents as trace impurities, J. Org. Chem., 62, 7512–7515, https://doi.org/10.1021/Jo971176v, 1997.
Green, S. A. and Blough, N. V.: Optical absorption and fluorescence properties of chromophoric dissolved organic matter in natural waters, Limnol. Oceanogr., 39, 1903–1916, https://doi.org/10.4319/lo.1994.39.8.1903, 1994.
Guenet, B., Danger, M., Abbadie, L., and Lacroix, G.: Priming effect: Bridging the gap between terrestrial and aquatic ecology, Ecology, 91, 2850–2861, https://doi.org/10.1890/09-1968.1, 2010.
Gurganus, S. C., Wozniak, A. S., and Hatcher, P. G.: Molecular characteristics of the water soluble organic matter in size-fractionated aerosols collected over the North Atlantic Ocean, Mar. Chem., 170, 37–48, https://doi.org/10.1016/j.marchem.2015.01.007, 2015.
Hach, P. F., Marchant, H. K., Krupke, A., Riedel, T., Meier, D. V., Lavik, G., Holtappels, M., Dittmar, T., and Kuypers, M. M. M.: Rapid microbial diversification of dissolved organic matter in oceanic surface waters leads to carbon sequestration, Sci. Rep.-UK, 10, 1–10, https://doi.org/10.1038/s41598-020-69930-y, 2020.
Hansell, D. A. and Carlson, C. A. (Eds.): Biogeochemistry of marine dissolved organic matter, 2nd Edn., Academic Press, Amsterdam, 712 pp., https://doi.org/10.1016/C2012-0-02714-7, 2015.
Hawkes, J. A., Patriarca, C., Sjöberg, P. J. R., Tranvik, L. J., and Bergquist, J.: Extreme isomeric complexity of dissolved organic matter found across aquatic environments, Limnol. Oceanogr. Lett., 3, 21–30, https://doi.org/10.1002/lol2.10064, 2018.
Hedges, J. I., Keil, R. G., and Benner, R.: What happens to terrestrial organic matter in the ocean?, Org. Geochem., 27, 195–212, https://doi.org/10.1016/S0146-6380(97)00066-1, 1997.
Helms, J. R., Stubbins, A., Ritchie, J. D., Minor, E. C., Kieber, D. J., and Mopper, K.: Absorption spectral slopes and slope ratios as indicators of molecular weight, source, and photobleaching of chromophoric dissolved organic matter, Limnol. Oceanogr., 53, 955–969, https://doi.org/10.4319/lo.2008.53.3.0955, 2008.
Hemmler, D., Gonsior, M., Powers, L. C., Marshall, J. W., Rychlik, M., Taylor, A. J., and Schmitt-Kopplin, P.: Simulated sunlight selectively modifies Maillard reaction products in a wide array of chemical reactions, Chemistry, 25, 13208–13217, https://doi.org/10.1002/chem.201902804, 2019.
Hertkorn, N., Benner, R., Frommberger, M., Schmitt-Kopplin, P., Witt, M., Kaiser, K., Kettrup, A., and Hedges, J. I.: Characterization of a major refractory component of marine dissolved organic matter, Geochim. Cosmochim. Ac., 70, 2990–3010, https://doi.org/10.1016/j.gca.2006.03.021, 2006.
Hertkorn, N., Ruecker, C., Meringer, M., Gugisch, R., Frommberger, M., Perdue, E., Witt, M., and Schmitt-Kopplin, P.: High-precision frequency measurements: Indispensable tools at the core of the molecular-level analysis of complex systems, Anal. Bioanal. Chem., 389, 1311–1327, https://doi.org/10.1007/s00216-007-1577-4, 2007.
Higuchi, T.: Microbial degradation of lignin: Role of lignin peroxidase, manganese peroxidase, and laccase, P. Jpn. Acad. B-Phys., 80, 204–214, https://doi.org/10.2183/pjab.80.204, 2004.
Hockaday, W. C., Grannas, A. M., Kim, S., and Hatcher, P. G.: Direct molecular evidence for the degradation and mobility of black carbon in soils from ultrahigh-resolution mass spectral analysis of dissolved organic matter from a fire-impacted forest soil, Org. Geochem., 37, 501–510, https://doi.org/10.1016/j.orggeochem.2005.11.003, 2006.
Hockaday, W. C., Grannas, A. M., Kim, S., and Hatcher, P. G.: The transformation and mobility of charcoal in a fire-impacted watershed, Geochim. Cosmochim. Ac., 71, 3432–3445, https://doi.org/10.1016/j.gca.2007.02.023, 2007.
Hockaday, W. C., Purcell, J. M., Marshall, A. G., Baldock, J. A., and Hatcher, P. G.: Electrospray and photoionization mass spectrometry for the characterization of organic matter in natural waters: A qualitative assessment, Limnol. Oceanogr.-Meth., 7, 81–95, https://doi.org/10.4319/lom.2009.7.81, 2009.
Hu, Y., Zheng, Q., Zhang, S., Noll, L., and Wanek, W.: Significant release and microbial utilization of amino sugars and D-amino acid enantiomers from microbial cell wall decomposition in soils, Soil Biol. Biochem., 123, 115–125, https://doi.org/10.1016/j.soilbio.2018.04.024, 2018.
Hu, Y., Zheng, Q., Noll, L., Zhang, S., and Wanek, W.: Direct measurement of the in situ decomposition of microbial-derived soil organic matter, Soil Biol. Biochem., 141, 1–10, https://doi.org/10.1016/j.soilbio.2019.107660, 2020.
Hughey, C. A., Hendrickson, C. L., Rodgers, R. P., Marshall, A. G., and Qian, K.: Kendrick mass defect spectrum: a compact visual analysis for ultrahigh-resolution broadband mass spectra, Anal. Chem., 73, 4676–4681, https://doi.org/10.1021/ac010560w, 2001.
Hyde, S. M. and Wood, P. M.: A mechanism for production of hydroxyl radicals by the brown-rot fungus Coniophora Puteana: Fe(III) reduction by cellobiose dehydrogenase and Fe(II) oxidation at a distance from the hyphae, Microbiology, 143, 259–266, https://doi.org/10.1099/00221287-143-1-259, 1997.
Idowu, O., Semple, K. T., Ramadass, K., O'Connor, W., Hansbro, P., and Thavamani, P.: Beyond the obvious: Environmental health implications of polar polycyclic aromatic hydrocarbons, Environ. Int., 123, 543–557, https://doi.org/10.1016/j.envint.2018.12.051, 2019.
Jaffé, R., Ding, Y., Niggemann, J., Vähätalo, A. V., Stubbins, A., Spencer, R. G. M., Campbell, J., and Dittmar, T.: Global charcoal mobilization from soils via dissolution and riverine transport to the oceans, Science, 340, 345–347, https://doi.org/10.1126/science.1231476, 2013.
Johns, G.: Austin Cary Forest Prescribed Burn (33/8S/21E), School of Forest Resources and Conservation, UF/IFAS, Prescribed Burn Prescription 721, 1–5, 2016.
Jones, M. W., Coppola, A. I., Santín, C., Dittmar, T., Jaffé, R., Doerr, S. H., and Quine, T. A.: Fires prime terrestrial organic carbon for riverine export to the global oceans, Nat. Commun., 11, 1–8, https://doi.org/10.1038/s41467-020-16576-z, 2020.
Judd, K. E., Crump, B. C., and Kling, G. W.: Bacterial responses in activity and community composition to photo-oxidation of dissolved organic matter from soil and surface waters, Aquat. Sci., 69, 96–107, https://doi.org/10.1007/s00027-006-0908-4, 2007.
Kendrick, E.: A mass scale based on CH2 = 14.0000 for high resolution mass spectrometry of organic compounds, Anal. Chem., 35, 2146–2154, https://doi.org/10.1021/ac60206a048, 1963.
Khatami, S., Deng, Y., Tien, M., and Hatcher, P. G.: Formation of water-soluble organic matter through fungal degradation of lignin, Org. Geochem., 135, 64–70, https://doi.org/10.1016/j.orggeochem.2019.06.004, 2019a.
Khatami, S., Deng, Y., Tien, M., and Hatcher, P. G.: Lignin contribution to aliphatic constituents of humic acids through fungal degradation, J. Environ. Qual., 48, 1565–1570, https://doi.org/10.2134/jeq2019.01.0034, 2019b.
Khodadad, C. L. M., Zimmerman, A. R., Green, S. J., Uthandi, S., and Foster, J. S.: Taxa-specific changes in soil microbial community composition induced by pyrogenic carbon amendments, Soil Biol. Biochem., 43, 385–392, https://doi.org/10.1016/j.soilbio.2010.11.005, 2011.
Kieber, D. J., McDaniel, J., and Mopper, K.: Photochemical source of biological substrates in sea water: Implications for carbon cycling, Nature, 341, 637–639, https://doi.org/10.1038/341637a0, 1989.
Kim, S., Kramer, R. W., and Hatcher, P. G.: Graphical method for analysis of ultrahigh-resolution broadband mass spectra of natural organic matter, the van Krevelen diagram, Anal. Chem., 75, 5336–5344, https://doi.org/10.1021/ac034415p, 2003.
Kirchman, D. L. (Ed.): Processes in microbial ecology, 2nd Edn., Oxford University Press, 318 pp., https://doi.org/10.1093/oso/9780198789406.001.0001, 2018.
Klevit, R. E.: Improving two-dimensional NMR spectra by t1 ridge subtraction, J. Magn. Reson., 62, 551–555, https://doi.org/10.1016/0022-2364(85)90227-6, 1985.
Koch, B. P. and Dittmar, T.: From mass to structure: An aromaticity index for high-resolution mass data of natural organic matter, Rapid Commun. Mass Sp., 20, 926–932, https://doi.org/10.1002/rcm.2386, 2006.
Koch, B. P. and Dittmar, T.: From mass to structure: An aromaticity index for high-resolution mass data of natural organic matter (Erratum), Rapid Commun. Mass Sp., 30, p. 250, https://doi.org/10.1002/rcm.7433, 2016.
Koch, B. P., Dittmar, T., Witt, M., and Kattner, G.: Fundamentals of molecular formula assignment to ultrahigh resolution mass data of natural organic matter, Anal. Chem., 79, 1758–1763, https://doi.org/10.1021/ac061949s, 2007.
Kolb, S. and Stacheter, A.: Prerequisites for amplicon pyrosequencing of microbial methanol utilizers in the environment, Front. Microbiol., 4, 1–12, https://doi.org/10.3389/fmicb.2013.00268, 2013.
Kothawala, D. N., Murphy, K. R., Stedmon, C. A., Weyhenmeyer, G. A., and Tranvik, L. J.: Inner filter correction of dissolved organic matter fluorescence, Limnol. Oceanogr.-Meth., 11, 616–630, https://doi.org/10.4319/lom.2013.11.616, 2013.
Kujawinski, E. B. and Behn, M. D.: Automated analysis of electrospray ionization Fourier transform ion cyclotron resonance mass spectra of natural organic matter, Anal. Chem., 78, 4363–4373, https://doi.org/10.1021/ac0600306, 2006.
Kuzyakov, Y., Subbotina, I., Chen, H., Bogomolova, I., and Xu, X.: Black carbon decomposition and incorporation into soil microbial biomass estimated by 14C labeling, Soil Biol. Biochem., 41, 210–219, https://doi.org/10.1016/j.soilbio.2008.10.016, 2009.
Kuzyakov, Y., Bogomolova, I., and Glaser, B.: Biochar stability in soil: Decomposition during eight years and transformation as assessed by compound-specific 14C analysis, Soil Biol. Biochem., 70, 229–236, https://doi.org/10.1016/j.soilbio.2013.12.021, 2014.
Lechtenfeld, O. J., Hertkorn, N., Shen, Y., Witt, M., and Benner, R.: Marine sequestration of carbon in bacterial metabolites, Nat. Commun., 6, 1–8, https://doi.org/10.1038/ncomms7711, 2015.
Lehmann, J.: A handful of carbon, Nature, 447, 143–144, https://doi.org/10.1038/447143a, 2007.
Li, M., Bao, F., Zhang, Y., Sheng, H., Chen, C., and Zhao, J.: Photochemical aging of soot in the aqueous phase: Release of dissolved black carbon and the formation of 1O2, Environ. Sci. Technol., 53, 12311–12319, https://doi.org/10.1021/acs.est.9b02773, 2019.
Lindell, M. J., Granéli, W., and Tranvik, L. J.: Enhanced bacterial growth in response to photochemical transformation of dissolved organic matter, Limnol. Oceanogr., 40, 195–199, https://doi.org/10.4319/lo.1995.40.1.0195, 1995.
Liu, M., Mao, X.-A., Ye, C., Huang, H., Nicholson, J. K., and Lindon, J. C.: Improved WATERGATE pulse sequences for solvent suppression in NMR spectroscopy, J. Magn. Reson., 132, 125–129, https://doi.org/10.1006/jmre.1998.1405, 1998.
Marques, J. S. J., Dittmar, T., Niggemann, J., Almeida, M. G., Gomez-Saez, G. V., and Rezende, C. E.: Dissolved black carbon in the headwaters-to-ocean continuum of Paraíba Do Sul River, Brazil, Front. Earth Sci., 5, 1–12, https://doi.org/10.3389/feart.2017.00011, 2017.
Masiello, C. A.: New directions in black carbon organic geochemistry, Mar. Chem., 92, 201–213, https://doi.org/10.1016/j.marchem.2004.06.043, 2004.
Masiello, C. A. and Druffel, E. R. M.: Black carbon in deep-sea sediments, Science, 280, 1911–1913, https://doi.org/10.1126/science.280.5371.1911, 1998.
McCarthy, M., Pratum, T., Hedges, J., and Benner, R.: Chemical composition of dissolved organic nitrogen in the ocean, Nature, 390, 150–154, https://doi.org/10.1038/36535, 1997.
McCarthy, M. D., Hedges, J. I., and Benner, R.: Major bacterial contribution to marine dissolved organic nitrogen, Science, 281, 231–234, https://doi.org/10.1126/science.281.5374.231, 1998.
McKee, G. A., Kobiela, M. E., and Hatcher, P. G.: Effect of Michael adduction on peptide preservation in natural waters, Environ. Sci.-Proc. Imp., 16, 2087–2097, https://doi.org/10.1039/C4EM00075G, 2014.
McNally, A. M., Moody, E. C., and McNeill, K.: Kinetics and mechanism of the sensitized photodegradation of lignin model compounds, Photochem. Photobio. S., 4, 268–274, https://doi.org/10.1039/b416956e, 2005.
Miller, M. P., Simone, B. E., McKnight, D. M., Cory, R. M., Williams, M. W., and Boyer, E. W.: New light on a dark subject: Comment, Aquat. Sci., 72, 269–275, https://doi.org/10.1007/s00027-010-0130-2, 2010.
Moran, M. A. and Covert, J. S.: Photochemically mediated linkages between dissolved organic matter and bacterioplankton, in: Aquatic Ecosystems, edited by: Findlay, S. E. G. and Sinsabaugh, R. L., Academic Press, Burlington, 243–262, https://doi.org/10.1016/B978-012256371-3/50011-1, 2003.
Moran, M. A., Kujawinski, E. B., Stubbins, A., Fatland, R., Aluwihare, L. I., Buchan, A., Crump, B. C., Dorrestein, P. C., Dyhrman, S. T., Hess, N. J., Howe, B., Longnecker, K., Medeiros, P. M., Niggemann, J., Obernosterer, I., Repeta, D. J., and Waldbauer, J. R.: Deciphering ocean carbon in a changing world, P. Natl. Acad. Sci. USA, 113, 3143–3151, https://doi.org/10.1073/pnas.1514645113, 2016.
Múčka, V., Bláha, P., Čuba, V., and Červenák, J.: Influence of various scavengers of •OH radicals on the radiation sensitivity of yeast and bacteria, Int. J. Radiat. Biol., 89, 1045–1052, https://doi.org/10.3109/09553002.2013.817702, 2013.
Mukherjee, A. and Zimmerman, A. R.: Organic carbon and nutrient release from a range of laboratory-produced biochars and biochar-soil mixtures, Geoderma, 193–194, 122–130, https://doi.org/10.1016/j.geoderma.2012.10.002, 2013.
Mukherjee, A., Zimmerman, A. R., and Harris, W.: Surface chemistry variations among a series of laboratory-produced biochars, Geoderma, 163, 247–255, https://doi.org/10.1016/j.geoderma.2011.04.021, 2011.
Murphy, K. R.: A note on determining the extent of the water Raman peak in fluorescence spectroscopy, Appl. Spectrosc., 65, 233–236, https://doi.org/10.1366/10-06136, 2011.
Murphy, K. R., Butler, K. D., Spencer, R. G. M., Stedmon, C. A., Boehme, J. R., and Aiken, G. R.: Measurement of dissolved organic matter fluorescence in aquatic environments: An interlaboratory comparison, Environ. Sci. Technol., 44, 9405–9412, https://doi.org/10.1021/es102362t, 2010.
Murphy, K. R., Stedmon, C. A., Graeber, D., and Bro, R.: Fluorescence spectroscopy and multi-way techniques, PARAFAC, Anal. Meth., 5, 6541–6882, https://doi.org/10.1039/c3ay41160e, 2013.
Nalven, S. G., Ward, C. P., Payet, J. P., Cory, R. M., Kling, G. W., Sharpton, T. J., Sullivan, C. M., and Crump, B. C.: Experimental metatranscriptomics reveals the costs and benefits of dissolved organic matter photo-alteration for freshwater microbes, Environ. Microbiol., 22, 3505–3521, https://doi.org/10.1111/1462-2920.15121, 2020.
Obernosterer, I. and Benner, R.: Competition between biological and photochemical processes in the mineralization of dissolved organic carbon, Limnol. Oceanogr., 49, 117–124, https://doi.org/10.4319/lo.2004.49.1.0117, 2004.
Ohno, T., Sleighter, R. L., and Hatcher, P. G.: Comparative study of organic matter chemical characterization using negative and positive mode electrospray ionization ultrahigh-resolution mass spectrometry, Anal. Bioanal. Chem., 408, 2497–2504, https://doi.org/10.1007/s00216-016-9346-x, 2016.
Patriarca, C., Balderrama, A., Može, M., Sjöberg, P. J. R., Bergquist, J., Tranvik, L. J., and Hawkes, J. A.: Investigating the ionization of dissolved organic matter by electrospray ionization, Anal. Chem., 92, 14210–14218, https://doi.org/10.1021/acs.analchem.0c03438, 2020.
Patriarca, C., Sedano-Núñez, V. T., Garcia, S. L., Bergquist, J., Bertilsson, S., Sjöberg, P. J. R., Tranvik, L. J., and Hawkes, J. A.: Character and environmental lability of cyanobacteria-derived dissolved organic matter, Limnol. Oceanogr., 66, 496–509, https://doi.org/10.1002/lno.11619, 2021.
Porcal, P., Dillon, P. J., and Molot, L. A.: Photochemical production and decomposition of particulate organic carbon in a freshwater stream, Aquat. Sci., 75, 469–482, https://doi.org/10.1007/s00027-013-0293-8, 2013.
Powers, L. C., Hertkorn, N., McDonald, N., Schmitt-Kopplin, P., Del Vecchio, R., Blough, N. V., and Gonsior, M.: Sargassum sp. act as a large regional surce of marine dissolved organic carbon and polyphenols, Global Biogeochem. Cy., 33, 1423–1439, https://doi.org/10.1029/2019GB006225, 2019.
Qualls, R. G. and Richardson, C. J.: Factors controlling concentration, export, and decomposition of dissolved organic nutrients in the Everglades of Florida, Biogeochemistry, 62, 197–229, https://doi.org/10.1023/A:1021150503664, 2003.
Quan, G., Fan, Q., Zimmerman, A. R., Sun, J., Cui, L., Wang, H., Gao, B., and Yan, J.: Effects of laboratory biotic aging on the characteristics of biochar and its water-soluble organic products, J. Hazard. Mater., 382, 1–9, https://doi.org/10.1016/j.jhazmat.2019.121071, 2020.
Reisser, M., Purves, R. S., Schmidt, M. W. I., and Abiven, S.: Pyrogenic carbon in soils: A literature-based inventory and a global estimation of its content in soil organic carbon and stocks, Front. Earth Sci., 4, 1–14, https://doi.org/10.3389/feart.2016.00080, 2016.
Riedel, T., Zark, M., Vähätalo, A. V., Niggemann, J., Spencer, R. G. M., Hernes, P. J., and Dittmar, T.: Molecular signatures of biogeochemical transformations in dissolved organic matter from ten world rivers, Front. Earth Sci., 4, 1–16, https://doi.org/10.3389/feart.2016.00085, 2016.
Roebuck, J. A., Podgorksi, D. C., Wagner, S., and Jaffé, R.: Photodissolution of charcoal and fire-impacted soil as a potential source of dissolved black carbon in aquatic environments, Org. Geochem., 112, 16–21, https://doi.org/10.1016/j.orggeochem.2017.06.018, 2017.
Romano, S., Dittmar, T., Bondarev, V., Weber, R. J. M., Viant, M. R., and Schulz-Vogt, H. N.: Exo-metabolome of Pseudovibrio sp. FO-BEG1 analyzed by ultra-high resolution mass spectrometry and the effect of phosphate limitation, PLoS ONE, 9, 1–11, https://doi.org/10.1371/journal.pone.0096038, 2014.
Rossel, P. E., Vähätalo, A. V., Witt, M., and Dittmar, T.: Molecular composition of dissolved organic matter from a wetland plant (Juncus effusus) after photochemical and microbial decomposition (1.25 yr): Common features with deep sea dissolved organic matter, Org. Geochem., 60, 62–71, https://doi.org/10.1016/j.orggeochem.2013.04.013, 2013.
Santín, C., Doerr, S. H., Preston, C. M., and Gonzalez-Rodriguez, G.: Pyrogenic organic matter production from wildfires: A missing sink in the global carbon cycle, Glob. Change Biol., 21, 1621–1633, https://doi.org/10.1111/gcb.12800, 2015.
Santín, C., Doerr, S. H., Merino, A., Bryant, R., and Loader, N. J.: Forest floor chemical transformations in a boreal forest fire and their correlations with temperature and heating duration, Geoderma, 264, 71–80, https://doi.org/10.1016/j.geoderma.2015.09.021, 2016.
Santín, C., Doerr, S. H., Merino, A., Bucheli, T. D., Bryant, R., Ascough, P., Gao, X., and Masiello, C. A.: Carbon sequestration potential and physicochemical properties differ between wildfire charcoals and slow-pyrolysis biochars, Sci. Rep.-UK, 7, 1–11, https://doi.org/10.1038/s41598-017-10455-2, 2017.
Schmidt, M. W. I. and Noack, A. G.: Black carbon in soils and sediments: Analysis, distribution, implications, and current challenges, Global Biogeochem. Cy., 14, 777–793, https://doi.org/10.1029/1999GB001208, 2000.
Schneider, M. P. W., Hilf, M., Vogt, U. F., and Schmidt, M. W. I.: The benzene polycarboxylic acid (BPCA) pattern of wood pyrolyzed between 200 ∘C and 1000 ∘C, Org. Geochem., 41, 1082–1088, https://doi.org/10.1016/j.orggeochem.2010.07.001, 2010.
Schulten, H. R. and Schnitzer, M.: The chemistry of soil organic nitrogen: A review, Biol. Fert. Soils, 26, 1–15, https://doi.org/10.1007/s003740050335, 1998.
Scully, N. M., Cooper, W. J., and Tranvik, L. J.: Photochemical effects on microbial activity in natural waters: The interaction of reactive oxygen species and dissolved organic matter, FEMS Microbiol. Ecol., 46, 353–357, https://doi.org/10.1016/s0168-6496(03)00198-3, 2003.
Simpson, A. J., McNally, D. J., and Simpson, M. J.: NMR spectroscopy in environmental research: From molecular interactions to global processes, Prog. Nucl. Mag. Res. Sp., 58, 97–175, https://doi.org/10.1016/j.pnmrs.2010.09.001, 2011.
Sinsabaugh, R. L., Findlay, S., Franchini, P., and Fischer, D.: Enzymatic analysis of riverine bacterioplankton production, Limnol. Oceanogr., 42, 29–38, https://doi.org/10.4319/lo.1997.42.1.0029, 1997.
Skjemstad, J., Reicosky, D. C., Wilts, A., and McGowan, J.: Charcoal carbon in U.S. agricultural soils, Soil Sci. Soc. Am. J., 66, 1249–1255, https://doi.org/10.2136/sssaj2002.1249, 2002.
Sleighter, R. L. and Hatcher, P. G.: Molecular characterization of dissolved organic matter (DOM) along a river to ocean transect of the lower Chesapeake Bay by ultrahigh resolution electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry, Mar. Chem., 110, 140–152, https://doi.org/10.1016/j.marchem.2008.04.008, 2008.
Sleighter, R. L., McKee, G. A., Liu, Z., and Hatcher, P. G.: Naturally present fatty acids as internal calibrants for Fourier transform mass spectra of dissolved organic matter, Limnol. Oceanogr.-Meth., 6, 246–253, https://doi.org/10.4319/lom.2008.6.246, 2008.
Sleighter, R. L., Chen, H., Wozniak, A. S., Willoughby, A. S., Caricasole, P., and Hatcher, P. G.: Establishing a measure of reproducibility of ultrahigh-resolution mass spectra for complex mixtures of natural organic matter, Anal. Chem., 84, 9184–9191, https://doi.org/10.1021/ac3018026, 2012.
Smith, C. R., Hatcher, P. G., Kumar, S., and Lee, J. W.: Investigation into the sources of biochar water-soluble organic compounds and their potential toxicity on aquatic microorganisms, ACS Sustain. Chem. Eng., 4, 2550–2558, https://doi.org/10.1021/acssuschemeng.5b01687, 2016.
Søndergaard, M. and Middelboe, M.: A cross-system analysis of labile dissolved organic carbon, Mar. Ecol.-Prog. Ser., 118, 283–294, https://doi.org/10.3354/meps118283, 1995.
Spence, A., Simpson, A. J., McNally, D. J., Moran, B. W., McCaul, M. V., Hart, K., Paull, B., and Kelleher, B. P.: The degradation characteristics of microbial biomass in soil, Geochim. Cosmochim. Ac., 75, 2571–2581, https://doi.org/10.1016/j.gca.2011.03.012, 2011.
Stenson, A. C., William, M., Marshall, A. G., and Cooper, W. T.: Ionization and fragmentation of humic substances in electrospray ionization Fourier transform-ion cyclotron resonance mass spectrometry, Anal. Chem., 74, 4397–4409, https://doi.org/10.1021/ac020019f, 2002.
Stubbins, A., Spencer, R. G. M., Chen, H. M., Hatcher, P. G., Mopper, K., Hernes, P. J., Mwamba, V. L., Mangangu, A. M., Wabakanghanzi, J. N., and Six, J.: Illuminated darkness: Molecular signatures of Congo River dissolved organic matter and its photochemical alteration as revealed by ultrahigh precision mass spectrometry, Limnol. Oceanogr., 55, 1467–1477, https://doi.org/10.4319/lo.2010.55.4.1467, 2010.
Stubbins, A., Niggemann, J., and Dittmar, T.: Photo-lability of deep ocean dissolved black carbon, Biogeosciences, 9, 1661–1670, https://doi.org/10.5194/bg-9-1661-2012, 2012.
Sun, L., Xu, C., Zhang, S., Lin, P., Schwehr, K. A., Quigg, A., Chiu, M.-H., Chin, W.-C., and Santschi, P. H.: Light-induced aggregation of microbial exopolymeric substances, Chemosphere, 181, 675–681, https://doi.org/10.1016/j.chemosphere.2017.04.099, 2017.
Trusiak, A., Treibergs, L., Kling, G., and Cory, R.: The controls of iron and oxygen on hydroxyl radical (•OH) production in soils, Soil Systems, 3, 1–23, https://doi.org/10.3390/soilsystems3010001, 2018.
Valle, J., Harir, M., Gonsior, M., Enrich-Prast, A., Schmitt-Kopplin, P., Bastviken, D., and Hertkorn, N.: Molecular differences between water column and sediment pore water SPE-DOM in ten Swedish boreal lakes, Water Res., 170, 1–11, https://doi.org/10.1016/j.watres.2019.115320, 2020.
Van Krevelen, D. W.: Graphical-statistical method for the study of structure and reaction processes of coal, Fuel Process. Technol., 29, 269–228, 1950.
Vollmer, W., Blanot, D., and De Pedro, M. A.: Peptidoglycan structure and architecture, FEMS Microbiol. Rev., 32, 149–167, https://doi.org/10.1111/j.1574-6976.2007.00094.x, 2008.
Vorobev, A., Sharma, S., Yu, M., Lee, J., Washington, B. J., Whitman, W. B., Ballantyne, F. t., Medeiros, P. M., and Moran, M. A.: Identifying labile DOM components in a coastal ocean through depleted bacterial transcripts and chemical signals, Environ. Microbiol., 20, 3012–3030, https://doi.org/10.1111/1462-2920.14344, 2018.
Waggoner, D. C. and Hatcher, P. G.: Hydroxyl radical alteration of HPLC fractionated lignin: Formation of new compounds from terrestrial organic matter, Org. Geochem., 113, 315–325, https://doi.org/10.1016/j.orggeochem.2017.07.011, 2017.
Waggoner, D. C., Chen, H., Willoughby, A. S., and Hatcher, P. G.: Formation of black carbon-like and alicyclic aliphatic compounds by hydroxyl radical initiated degradation of lignin, Org. Geochem., 82, 69–76, https://doi.org/10.1016/j.orggeochem.2015.02.007, 2015.
Waggoner, D. C., Wozniak, A. S., Cory, R. M., and Hatcher, P. G.: The role of reactive oxygen species in the degradation of lignin derived dissolved organic matter, Geochim. Cosmochim. Ac., 208, 171–184, https://doi.org/10.1016/j.gca.2017.03.036, 2017.
Wagner, S. and Jaffé, R.: Effect of photodegradation on molecular size distribution and quality of dissolved black carbon, Org. Geochem., 86, 1–4, https://doi.org/10.1016/j.orggeochem.2015.05.005, 2015.
Wagner, S., Ding, Y., and Jaffé, R.: A new perspective on the apparent solubility of dissolved black carbon, Front. Earth Sci., 5, 1–16, https://doi.org/10.3389/feart.2017.00075, 2017.
Wagner, S., Jaffé, R., and Stubbins, A.: Dissolved black carbon in aquatic ecosystems, Limnol. Oceanogr. Lett., 3, 168–185, https://doi.org/10.1002/lol2.10076, 2018.
Wagner, S., Brandes, J., Spencer, R. G. M., Ma, K., Rosengard, S. Z., Moura, J. M. S., and Stubbins, A.: Isotopic composition of oceanic dissolved black carbon reveals non-riverine source, Nat. Commun., 10, 1–8, https://doi.org/10.1038/s41467-019-13111-7, 2019.
Wang, H., Zhou, H., Ma, J., Nie, J., Yan, S., and Song, W.: Triplet photochemistry of dissolved black carbon and its effects on the photochemical formation of reactive oxygen species, Environ. Sci. Technol., 54, 4903–4911, https://doi.org/10.1021/acs.est.0c00061, 2020.
Wang, X., Xu, C., Druffel, E. M., Xue, Y., and Qi, Y.: Two black carbon pools transported by the Changjiang and Huanghe Rivers in China, Global Biogeochem. Cy., 30, 1778–1790, https://doi.org/10.1002/2016GB005509, 2016.
Ward, C. P., Sleighter, R. L., Hatcher, P. G., and Cory, R. M.: Insights into the complete and partial photooxidation of black carbon in surface waters, Environ. Sci.-Proc. Imp., 16, 721–731, https://doi.org/10.1039/C3EM00597F, 2014.
Ward, N. D., Keil, R. G., Medeiros, P. M., Brito, D. C., Cunha, A. C., Dittmar, T., Yager, P. L., Krusche, A. V., and Richey, J. E.: Degradation of terrestrially derived macromolecules in the Amazon River, Nat. Geosci., 6, 530–533, https://doi.org/10.1038/ngeo1817, 2013.
Watrous, J., Roach, P., Heath, B., Alexandrov, T., Laskin, J., and Dorrestein, P. C.: Metabolic profiling directly from the Petri dish using nanospray desorption electrospray ionization imaging mass spectrometry, Anal. Chem., 85, 10385–10391, https://doi.org/10.1021/ac4023154, 2013.
Weishaar, J. L., Aiken, G. R., Bergamaschi, B. A., Fram, M. S., Fujii, R., and Mopper, K.: Evaluation of specific ultraviolet absorbance as an indicator of the chemical composition and reactivity of dissolved organic carbon, Environ. Sci. Technol., 37, 4702–4708, https://doi.org/10.1021/es030360x, 2003.
Wetzel, R. G., Hatcher, P. G., and Bianchi, T. S.: Natural photolysis by ultraviolet irradiance of recalcitrant dissolved organic matter to simple substrates for rapidbacterial metabolism, Limnol. Oceanogr., 40, 1369–1380, https://doi.org/10.4319/lo.1995.40.8.1369, 1995.
Wienhausen, G., Noriega-Ortega, B. E., Niggemann, J., Dittmar, T., and Simon, M.: The exometabolome of two model strains of the Roseobacter group: A marketplace of microbial metabolites, Front. Microbiol., 8, 1–15, https://doi.org/10.3389/fmicb.2017.01985, 2017.
Wozniak, A. S., Bauer, J. E., Sleighter, R. L., Dickhut, R. M., and Hatcher, P. G.: Technical Note: Molecular characterization of aerosol-derived water soluble organic carbon using ultrahigh resolution electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry, Atmos. Chem. Phys., 8, 5099–5111, https://doi.org/10.5194/acp-8-5099-2008, 2008.
Wozniak, A. S., Goranov, A. I., Mitra, S., Bostick, K. W., Zimmerman, A. R., Schlesinger, D. R., Myneni, S., and Hatcher, P. G.: Molecular heterogeneity in pyrogenic dissolved organic matter from a thermal series of oak and grass chars, Org. Geochem., 148, 1–18, https://doi.org/10.1016/j.orggeochem.2020.104065, 2020.
Wünsch, U. J., Bro, R., Stedmon, C. A., Wenig, P., and Murphy, K. R.: Emerging patterns in the global distribution of dissolved organic matter fluorescence, Anal. Meth., 11, 888–893, https://doi.org/10.1039/C8AY02422G, 2019.
Xiao, Y., Carena, L., Näsi, M.-T., and Vähätalo, A. V.: Superoxide-driven autocatalytic dark production of hydroxyl radicals in the presence of complexes of natural dissolved organic matter and iron, Water Res., 177, 1–8, https://doi.org/10.1016/j.watres.2020.115782, 2020.
Yavitt, J. B. and Fahey, T. J.: An experimental analysis of solution chemistry in a lodgepole pine forest foor, Oikos, 43, 222–234, https://doi.org/10.2307/3544772, 1984.
Zeng, Y., Hong, P. K. A., and Wavrek, D. A.: Chemical-biological treatment of pyrene, Water Res., 34, 1157–1172, https://doi.org/10.1016/S0043-1354(99)00270-5, 2000a.
Zeng, Y., Hong, P. K. A., and Wavrek, D. A.: Integrated chemical-biological treatment of benzo[a]pyrene, Environ. Sci. Technol., 34, 854–862, https://doi.org/10.1021/es990817w, 2000b.
Zimmerman, A. R.: Abiotic and microbial oxidation of laboratory-produced black carbon (biochar), Environ. Sci. Technol., 44, 1295–1301, https://doi.org/10.1021/es903140c, 2010.
Zimmerman, A. R., Gao, B., and Ahn, M.-Y.: Positive and negative carbon mineralization priming effects among a variety of biochar-amended soils, Soil Biol. Biochem., 43, 1169–1179, https://doi.org/10.1016/j.soilbio.2011.02.005, 2011.





