the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Long-term incubations provide insight into the mechanisms of anaerobic oxidation of methane in methanogenic lake sediments
Hanni Vigderovich
Werner Eckert
Michal Elul
Maxim Rubin-Blum
Marcus Elvert
Orit Sivan
Anaerobic oxidation of methane (AOM) is among the main processes limiting the release of the greenhouse gas methane from natural environments. Geochemical profiles and experiments with fresh sediments from Lake Kinneret (Israel) indicate that iron-coupled AOM (Fe-AOM) sequesters 10 %–15 % of the methane produced in the methanogenic zone (>20 cm sediment depth). The oxidation of methane in this environment was shown to be mediated by a combination of mcr-gene-bearing archaea and pmoA-gene-bearing aerobic bacterial methanotrophs. Here, we used sediment slurry incubations under controlled conditions to elucidate the electron acceptors and microorganisms that are involved in the AOM process over the long term (∼ 18 months). We monitored the process with the addition of 13C-labeled methane and two stages of incubations: (i) enrichment of the microbial population involved in AOM and (ii) slurry dilution and manipulations, including the addition of several electron acceptors (metal oxides, nitrate, nitrite and humic substances) and inhibitors (2-bromoethanesulfonate, acetylene and sodium molybdate) of methanogenesis, methanotrophy and sulfate reduction and sulfur disproportionation. Carbon isotope measurements in the dissolved inorganic carbon pool suggest the persistence of AOM, consuming 3 %–8 % of the methane produced at a rate of 2.0 ± 0.4 nmol per gram of dry sediment per day. Lipid carbon isotopes and metagenomic analyses point towards methanogens as the sole microbes performing the AOM process by reverse methanogenesis. Humic substances and iron oxides, although not sulfate, manganese, nitrate or nitrite, are the likely electron acceptors used for this AOM. Our observations support the contrast between methane oxidation mechanisms in naturally anoxic lake sediments, with potentially co-existing aerobes and anaerobes, and long-term incubations, wherein anaerobes prevail.
- Article
(2406 KB) - Full-text XML
-
Supplement
(924 KB) - BibTeX
- EndNote
Methane (CH4) is an important greenhouse gas (Wuebbles and Hayhoe, 2002), which has both anthropogenic and natural sources, the latter of which account for about 50 % of the emission of this gas to the atmosphere (Saunois et al., 2020). Naturally occurring methane is mainly produced biogenically via the methanogenesis process, which is performed by methanogenic archaea. Traditionally acknowledged as the terminal process anchoring carbon remineralization (Froelich et al., 1979), methanogenesis occurs primarily via the reduction of carbon dioxide by hydrogen in marine sediments and via acetate fermentation in freshwater systems (Whiticar et al., 1986).
Methanotrophy, the aerobic oxidation of methane and the anaerobic oxidation of methane (AOM) by microbes, naturally controls the release of this gas to the atmosphere (Conrad, 2009; Reeburgh, 2007; Knittel and Boetius, 2009). In marine sediments, up to 90 % of the upward methane flux is consumed anaerobically by sulfate, and in established diffusive profiles, that methane consumption occurs within a distinct sulfate–methane transition zone (Valentine, 2002). While sulfate-dependent AOM, catalyzed by the archaeal anaerobic methanotrophs (ANMEs) 1–3, is widespread chiefly in marine sediments (Hoehler et al., 1994; Boetius et al., 2000; Orphan et al., 2001; Treude et al., 2005, 2014), methane oxidation in other environments can be coupled to other electron acceptors (e.g., Raghoebarsing et al., 2006; Ettwig et al., 2010; Sivan et al., 2011; Crowe et al., 2011; Norði and Thamdrup, 2014; Valenzuela et al., 2017).
In freshwater sediments, sulfate is often depleted, and methanogenesis may be responsible for most of the organic carbon remineralization, resulting in high concentrations of methane in shallow sediments (Sinke et al., 1992). Indeed, lakes and wetlands are responsible for 33 %–55 % of naturally emitted methane (Rosentreter et al., 2021). A large portion of this produced methane is oxidized by aerobic (type I, type II and type X) methanotrophic bacteria via oxygen. Aerobic methanotrophy is generally observed at the sediment–water interface (Damgaard et al., 1998) and/or in the water column thermocline (Bastviken, 2009). AOM, however, can also consume over 50 % of the produced methane (Segarra et al., 2015).
Sulfate can be an electron acceptor of AOM in freshwater sediments, as was shown for example in Lake Cadagno (Schubert et al., 2011; Su et al., 2020). Alternative electron acceptors for AOM in natural freshwater environments and cultures include humic substances, nitrate, nitrite and metals (such as iron, manganese and chromium). Natural humic substances and their synthetic analogs were shown to function as terminal electron acceptors for AOM in soils, wetlands and cultures (Valenzuela et al., 2017, 2019; Bai et al., 2019; Zhang et al., 2019; Fan et al., 2020). Nitrate-dependent AOM has been demonstrated in a consortium of archaea and denitrifying bacteria from a canal (Raghoebarsing et al., 2006), in freshwater lake sediments (Norði and Thamdrup, 2014) and in a sewage enrichment culture of ANME-2d (Haroon et al., 2013; Arshad et al., 2015). Nitrite is exploited to oxidize methane by the aerobic bacteria Methylomirabilis (NC10), which split the nitrite into N2 and O2 and then use the produced oxygen to oxidize the methane (Ettwig et al., 2010). ANME-2d archaea were also suggested to be involved in Cr(VI)-coupled AOM, either alone or with a bacterial partner (Lu et al., 2016). Iron- and/or manganese-coupled AOM has also been suggested in lakes (Sivan et al., 2011; Crowe et al., 2011; Norði et al., 2013), sometimes by supporting sulfate-coupled AOM (Schubert et al., 2011; Su et al., 2020; Mostovaya et al., 2021). Iron-coupled AOM was also shown to occur in enriched, denitrifying cultures from sewage where it was performed by ANME-2 (Ettwig et al., 2016) and in a bioreactor with natural sediments (Cai et al., 2018).
The mechanism and role of iron-coupled AOM in lake sediments have been studied with a variety of tools in the sediments of Lake Kinneret. In situ porewater profiles and top-core experiments (Sivan et al., 2011), diagenetic models (Adler et al., 2011), and batch incubation experiments with fresh sediment slurries (Bar-Or et al., 2017) suggest that iron-coupled AOM (Fe-AOM) removes 10 %–15 % of the produced methane in the deeper part of the methanogenic zone (>20 cm below the water–sediment interface). Analysis of the microbial community structure suggested that both methanogenic archaea and methanotrophic bacteria are potentially involved in methane oxidation (Bar-Or et al., 2015). Analyses of stable isotopes in fatty acids, 16S rRNA gene amplicons and metagenomics showed that both reverse methanogenesis by archaea and bacterial type-I aerobic methanotrophy by Methylococcales play important roles in methane cycling (Bar-Or et al., 2017; Elul et al., 2021). Aerobic methanotrophy, which has also been observed in the hypolimnion and sediments of several other lakes that are considered anoxic (Beck et al., 2013; Oswald et al., 2016; Martinez-Cruz et al., 2017; Cabrol et al., 2020), may be driven by the presence of oxygen at nanomolar levels (Wang et al., 2018). Pure cultures of the ubiquitous aerobic methanotrophs Methylococcales have indeed been shown to survive under hypoxia conditions by oxidizing methane and with nitrate (Kits et al., 2015), by switching to iron reduction (Zheng et al., 2020), or even by exploiting their methanobactins to generate their own oxygen to fuel their methanotrophic activity (Dershwitz et al., 2021). The latter study also showed that the alphaproteobacterial methanotroph Methylocystis sp., strain SB2, can couple methane oxidation and iron reduction. However, whether these aerobic methanotrophic bacteria are able to oxidize methane under strictly anoxic conditions and which electron acceptors facilitate that activity are still not known.
In the current study, we used long-term anaerobic incubations to assess the dynamics of methane-oxidizing microbes under anoxic conditions and to quantify the respective availabilities of different electron acceptors for AOM. To that end, we diluted fresh methanogenic sediments from Lake Kinneret with original porewater from the same depth and amended the sediment with 13C-labeled methane. Our experiment design comprised two stages, the first of which included the enrichment of the microbial population involved in AOM, while the second involved an additional slurry dilution and several manipulations with different electron acceptors and inhibitors. We measured methane oxidation rates (based on 13C-DIC (dissolved inorganic carbon) enrichment), determined the characteristics of each electron acceptor (via its turnover) and evaluated changes in microbial diversity over various incubation periods (based on metagenomics and lipid biomarkers). The results from the long-term anaerobic incubations were compared to those of batch and semi-continuous bioreactor experiments.
2.1 Study site
Lake Kinneret (Sea of Galilee) is a warm, monomictic, freshwater lake that is 21 km long and 13 km wide and located in northern Israel. Its maximum depth is ∼ 42 m at its center (station A, Fig. S1), and its average depth is 24 m. From March to December, the lake is thermally stratified, and from April to December, the hypolimnion is anoxic. Surface water temperatures range from 15 ∘C in the winter (January) to 32 ∘C in the summer (August), and the lake's bottom water temperatures remain in the range of 14–17 ∘C throughout the year. The sediment from the deep methanogenic zone used in this study (sediment samples taken from a sediment depth of ∼ 20 cm from the water–sediment interface at the lake's center) contains 50 % carbonates, 30 % clay and 7 % iron (Table S1). The dissolved organic carbon (DOC) concentration of the porewater increases with depth, ranging from ∼ 6 mg C L−1 at the sediment–water interface to 17 mg C L−1 at a depth of 25 cm (Adler et al., 2011). The concentrations of dissolved methane in the sediment porewater increase sharply with sediment depth, reaching a maximum of more than 2 mM at a depth of 15 cm, after which the amounts of dissolved methane gradually decrease with depth to 0.5 mM at a depth of 30 cm (Adler et al., 2011; Sivan et al., 2011; Bar-Or et al., 2015).
2.2 Experimental setup
2.2.1 General
In this study we compared three incubation strategies (Fig. 1a, b, c) in Lake Kinneret methanogenic sediments (sediment depths >20 cm), which were amended with original porewater from the same depth, 13C-labeled methane (0.05–2 mL; Table 1), different potential electron acceptors for AOM (nitrite, nitrate, iron and manganese oxides and humic substances) and activity inhibitors. We inhibited the mcr gene with 2-bromoethanesulfonate (BES), methanogenesis and methanotrophy with acetylene, and sulfate reduction and sulfur disproportionation with Na-molybdate (Nollet et al., 1997; Oremland and Capone, 1988; Lovley and Klug, 1983). Below we describe the three incubation strategies (Fig. 1).
- a.
Setup A. Long-term two-stage slurry incubations were performed with a 1:1 sediment-to-porewater ratio and high methane content for the first 3 months (first stage) to ensure the enrichment of the microorganisms involved in AOM. After 3 months, the slurry was diluted with porewater to a 1:3 ratio (second stage) and different reactants were added to the incubations, which were subsequently monitored for up to 18 months.
- b.
Setup B. Semi-continuous bioreactor experiments were performed in which sediments were collected up to 3 d before the experiment was set up (freshly sampled sediments). The sediment-to-porewater ratio was 1:4, and porewater was exchanged regularly.
- c.
Setup C. Batch incubation experiments were performed with freshly sampled sediments and porewater at a 1:5 ratio, respectively, and amended with hematite. This experimental setup was described in our previous studies (Bar-Or et al., 2017; Elul et al., 2021).
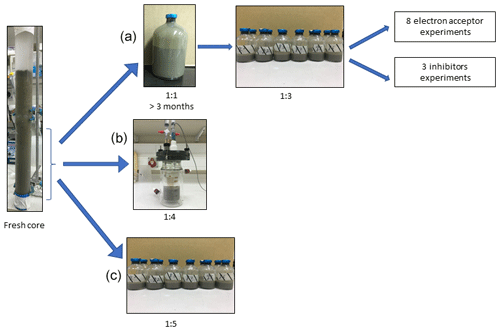
Figure 1Flow diagram of the experimental design. Three types of experiment were set up to investigate the methanogenic-zone sediments (deeper than 20 cm): (a) Two-stage slurry experiments, with 1:1 ratio of sediment-to-porewater incubations and then with diluted pre-incubated slurries and porewater (1:3 ratio of sediment to porewater). (b) Semi-continuous bioreactor experiment with freshly collected sediment. (c) Fresh-batch experiment – slurry experiment with freshly collected sediments (Bar-Or et al., 2017).
The sediments for the slurries conducted in the current work were collected during seven day-long sampling campaigns aboard the research vessel Lillian between 2017 and 2019 from the center of the lake (Station A, Fig. S1) using a gravity corer with a 50 cm Perspex core liner. The length of the sediment in each core was 35–45 cm. During each sampling campaign, 1–2 sediment cores were collected for the incubations and 10 cores were collected for the porewater extraction. Sediments from the deeper methanogenic zone (sediment depths >20 cm) for the experiments were diluted with porewater from the methanogenic zone of parallel cores sampled on the same day. The bottom part of the sediment cores (below 20 cm) was transferred, as a bulk, to a dedicated 5 L plastic container on board. The cores and the container were brought back to the lab, where the cores were kept at 4 ∘C, and the porewater was extracted on the same day of sampling. In the lab, sediments were collected from the container with 20 mL cutoff syringes and moved to 50 mL falcon tubes. The porewater was extracted by centrifugation at 9300 g for 15 min at 4 ∘C, syringe-filtered by 0.22 µM filters into 250 mL pre-autoclaved glass bottles, crimp-sealed with rubber stoppers and flushed for 30 min with N2. The extracted porewater was kept under anaerobic conditions at 4 ∘C until its use. The sediments for the incubations were subsampled from the liners, diluted no later than 3 d after their collection from the lake and treated further according to the experimental strategies described above (setup A or B).
2.2.2 Experiment type-A setup: long-term two-stage incubations (henceforth referred to as “two-stage” for simplicity)
Experiment A comprised 10 two-stage incubation experiments (experiment serial number (SN) 1–10; Table 1) with different treatments (electron acceptors/shuttling/inhibitors). In the first stage (pre-incubation slurry), the sediment core was sliced under continuous N2 flushing and sediments from depths >20 cm were collected into zipper bags. The sediment was homogenized by shaking the sediment in the bag, and between 80–100 g was transferred into 250 mL glass bottles under continuous N2 flushing. The sediments were diluted with the extracted porewater to create a 1:1 sediment-to-porewater slurry with a headspace of 70–90 mL (Fig. 1). The slurries were sealed with rubber stoppers and crimped caps and were flushed with N2 (99.999 %, Maxima, Israel) for 30 min. Methane (99.99 %, Maxima, Israel) was injected using a gastight syringe for a final content of 20 % in the headspace, where 10 % of the injected methane was 13C-labeled methane (99 %, Sigma-Aldrich). When significant AOM activity was observed based on the increase in δ13CDIC after approximately 3 months (Fig. S2), some of the incubations were further diluted during the second stage of the experiments. The remainder of the incubations continued to run with porewater exchange while the δ13CDIC values were monitored every 3 months.
All the experiments were set up similarly (see dates and detailed protocols in the Supplement): the pre-incubation bottle was opened, and subsamples (∼ 18 g each) were transferred with a syringe and a Tygon® tube under a laminar hood and continuous flushing of N2 gas into 60 mL glass bottles. The subsamples were then diluted with fresh anoxic porewater from the methanogenic zone (as described above) to achieve a 1:3 sediment-to-porewater ratio (Fig. 1) while leaving 24 mL of headspace in each bottle. The bottles were crimp-sealed, flushed with N2 gas for 5 min, shaken vigorously and flushed again (three times). Then 13C-labeled methane was added to all of the bottles as described in Table 1. The “killed” control slurries in each experiment were autoclaved twice and cooled, only after which they were amended with the appropriate treatments and 13C-labeled methane.
To the diluted (1:3) batch slurries, electron acceptors were added either as a powder (hematite – experiment no. 1, magnetite – experiment no. 2, clay and humic substances – experiment no. 7, MnO2 – experiment no. 3) or in dissolved form in double-distilled water (DDW) (KNO3 – experiment no. 4, NaNO2 – experiment no. 5). In addition, the potential involvement of sulfur cycling in the transfer of electrons was tested in experiment no. 2 via its inhibition with Na-molybdate (Lovley and Klug, 1983). The synthetic analog for humic substances, i.e., 9,10-anthraquinone-2,6-disulfonate (AQDS), was dissolved in DDW (detailed in the Supplement) and added to the bottles of experiment no. 6 until a final concentration of 5 mM was achieved in each bottle. Amorphous iron (Fe(OH)3) was prepared in the lab by dissolving FeCl3 in DDW that was then titrated with NaOH of 1.5 M up to pH 7 and injected into the bottles of experiment no. 2. The final concentration of each addition is detailed in Table 1. The 13C-labeled methane was injected into all of the experimental bottles at the beginning of each experiment (unless described otherwise) by using a gastight syringe from a stock bottle filled with 13C-labeled methane gas (which was replaced with saturated NaCl solution). Three different inhibitors were added to three different experiments: molybdate was added to experiment no. 1 (to one bottle of methane-only treatment, magnetite treatment and amorphous iron treatment) to detect the feasibility of an active sulfur cycle; BES was added to experiment no. 8 at the start of the experiment; and acetylene was added to experiment no. 9, wherein it was injected during the experiment into two bottles at different time points after 13C enrichment was observed in the DIC (Table 1).
All live treatments were set up in duplicate or triplicate, depending on the target amount of the pre-incubated slurry for each experiment, and the results are presented as the average with an error bar. In two experiments, only one killed control bottle was set up, and the remainder of the slurry was prioritized for other treatments because the killed controls repeatedly had shown no activity in several previous experiments. The humic substrate experiment used a natural (humic) substance that was extracted from a lake near Fairbanks, Alaska, where iron reduction was observed in the methanogenic zone. One experiment was set up without any additional electron acceptor to assess the rate of methanogenesis in the two-stage slurries. Porewater was sampled anaerobically for δ13CDIC and dissolved Fe(II) measurements in duplicate (2 mL), and methane was measured from the headspace. Variations in the δ13CDIC values between the experiments resulted from different amounts of 13C–labeled methane injected at the start of each experiment (geochemical measurements detailed in the analytical methods section below).
2.2.3 Experiment type-B setup: semi-continuous bioreactor
Semi-continuous bioreactors were used to monitor the redox state regularly at close-to-natural in situ conditions for 15 months in freshly collected sediments. Two 0.5 L semi-continuous bioreactors (Fig. 1) (Lenz, Wertheim, Germany) were set up with freshly sampled sediments from the methanogenic zone (25–40 cm) and extracted porewater from the same depth from Station A on Lake Kinneret immediately after their collection. Both reactors were filled, headspace-free, with a slurry at a 1:4 sediment-to-porewater ratio. One bioreactor was amended with 10 mM of hematite, while the second, which was a control, was not amended. To dissolve 13C-labeled methane in the porewater, 15 mL of porewater was replaced with 15 mL of methane gas (13 mL of 12CH4 and 2 mL of 13CH4) to produce a methane-only headspace for 24 h, during which time the reactors were shaken repeatedly. After 24 h, the gas was replaced with anoxic porewater, thus eliminating the headspace, which resulted in lower methane concentrations (0.2 mM) than in either the two-stage incubations or the fresh-batch experiment (∼ 2 mM). The redox potential was monitored continuously using a platinum/glass electrode (Metrohm, Herisau, Switzerland) to verify anoxic conditions and to determine the redox state throughout the incubation period. The bioreactors were subsampled weekly to bi-weekly, and the sample volume (5–10 mL) was replaced immediately by preconditioned anoxic (flushed with N2 gas for 15 min) porewater from the methanogenic zone. As outlined below, samples were analyzed for dissolved Fe(II), methane and δ13CDIC. Additional subsamples for metagenome and lipid analyses were taken at the beginning of the experiment and on days 151 and 382, respectively.
2.2.4 Experiment type-C setup: fresh-batch experiment
Sediments for this experiment were collected in August 2013 at Station A using a protocol similar to that used to collect the sediments for the pre-incubations. Sediments from depths greater than 26 cm were diluted under anaerobic conditions with porewater from the same depth to obtain a ratio of sediment to porewater of 1:5. The resulting slurry was then divided between 60 mL glass bottles (40 mL slurry in each bottle). The sampling and experimental setup are described in detail in our earlier study (Bar-Or et al., 2017). Here we present our results of the δ13CDIC, metagenome and lipid analyses of two treatments: natural (with only 13C-labeled methane) and hematite. The experiment ran for 15 months.
2.3 Analytical methods
2.3.1 Geochemical measurements
Measurements of δ13CDIC were performed on a DELTA V Advantage Thermo Scientific isotope-ratio mass spectrometer (IRMS). Results are reported referred to the Vienna Pee Dee Belemnite (VPDB) standard. For these measurements, about 0.3 mL of filtered (0.22 µm) porewater was injected into a 12 mL glass vial with a He atmosphere and 10 µL of 85 % H3PO4 to acidify all the DIC species to CO2 (g). The headspace autosampler (CTC Analytics, type PC PAL) sampled the gas from the vials and measured the δ13CDIC of the sample on the GasBench interface with a precision of ± 0.1 ‰. DIC was measured on the IRMS using the peak height and a precision of 0.05 mM. Dissolved Fe(II) concentrations were determined using the ferrozine method (Stookey, 1970) by a Hanon i2 visible spectrophotometer at a 562 nm wavelength with a detection limit of 1 µmol L−1. A 100 µL headspace sample was taken for methane measurements with a gastight syringe and was analyzed by a gas chromatograph (FOCUS GC, Thermo Fisher) equipped with a flame ionization detector (FID) and a packed column (ShinCarbon ST) with a helium carrier gas (UHP) and a detection limit of 1 nmol of methane. Bottles to which acetylene was added were also measured by the GC with the same column and carrier gas for ethylene to determine the acetylene turnover with the N cycle.
2.3.2 Lipid analysis
A sub-set of samples (Table 3) was investigated for the assimilation of 13C-labeled methane into polar lipid-derived fatty acids (PLFAs) and intact ether lipid-derived hydrocarbons. A total lipid extract (TLE) was obtained from 0.4 to 1.6 g of the freeze-dried sediment or incubated sediment slurry using a modified Bligh and Dyer protocol (Sturt et al., 2004). Before extraction, 1 µg each of 1,2-diheneicosanoyl-sn-glycero-3-phosphocholine and 2-methyloctadecanoic acid was added as internal standards. PLFAs in the TLE were converted to fatty acid methyl esters (FAMEs) using saponification with KOH MeOH and derivatization with BF3 MeOH (Elvert et al., 2003). Intact archaeal ether lipids in the TLE were separated from the apolar archaeal lipid compounds using preparative liquid chromatography (Meador et al., 2014) followed by ether cleavage with BBr3 in dichloromethane-forming hydrocarbons (Lin et al., 2010). Both FAMEs and ether-cleaved hydrocarbons were analyzed by a GC–mass spectrometer (GC-MS; Thermo Finnigan TRACE GC coupled to a TRACE MS) for identification and by a GC-IRMS (Thermo Scientific TRACE GC coupled via a GC IsoLink interface to a DELTA V Plus) to determine δ13C values by using the column and temperature program settings described by Aepfler et al. (2019). The δ13C values are reported with an analytical precision better than 1 ‰ as determined by long-term measurements of an n-alkane standard with known isotopic composition of each compound. Reported fatty acid isotope data (Table 3) are corrected for the introduction of the methyl group during derivatization by mass balance calculation that is similar to Eq. (1) (see below) using the measured δ13C value of each FAME and the known isotopic composition of methanol as input parameters.
2.3.3 Metagenomic analysis
For the metagenomic analyses, total genomic DNA was extracted from the semi-aerobic bioreactor with hematite addition (duplicate samples), pre-incubation slurries (13CH4-only control, 13CH4 + hematite) and their respective initial slurries (t0) by using the DNeasy PowerLyzer PowerSoil Kit (Qiagen). Genomic DNA was eluted using 50 µL of elution buffer and stored at −20 ∘C. Metagenomics libraries were prepared at the sequencing core facility at the University of Illinois Chicago using the Nextera XT DNA library preparation kit (Illumina, USA). Between 19 and 40 million 2×150 bp paired-end reads per library were sequenced using Illumina NextSeq 500. Metagenomes were co-assembled from the concatenated reads of all of the metagenomic libraries with SPAdes v3.12 (Bankevich et al., 2012; Nurk et al., 2013) after decontamination, quality value (QV=10) and adapter trimming with the BBDuk tool from the BBMap suite (Brian Bushnell, http://sourceforge.net/projects/bbmap/, last access: 1 June 2020). Downstream analyses, including reading coverage estimates, automatic binning with MaxBin (Wu et al., 2014) and MetaBAT 2 (Kang et al., 2019) bin refining with the DAS Tool (Sieber et al., 2018), were performed within the SqueezeMeta framework (Tamames and Puente-Sánchez, 2019). GTDB-Tk was used to classify the metagenome-assembled genomes (MAGs) based on the Genome Taxonomy Database release 95 (Parks et al., 2021). The principal component analysis biplot was constructed with PAST v4.03 (Hammer et al., 2001).
2.3.4 Rate calculations
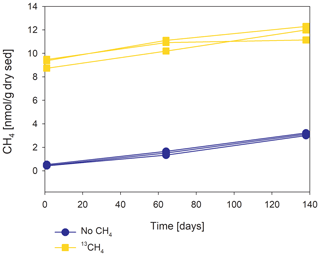
Methanogenesis rates were calculated from temporal changes in methane concentration in a representative pre-incubated slurry experiment (Fig. 2). The amount of methane oxidized was calculated by a simple mass balance calculation according to Eqs. (1) and (2):
The final DIC pool comprises two end-members, the initial DIC pool and the oxidized 13C CH4. The term x denotes the fraction of oxidized 13C CH4, while 1−x denotes the fraction of the initial DIC pool out of the final DIC pool. F13CH4 is the fraction of the total CH4 that is 13C at t0 (i denotes initial); FDI13Ci is the fraction of the total DIC that is 13C at t0; and FDI13Cf is the fraction of the total DIC that is 13C at tfinal. [CH4]ox is the amount (concentration in porewater) of the methane oxidized throughout the full incubation period, and [DIC]f is the DIC concentration at tfinal. It was assumed that the isotopic composition of the labeled CH4 did not change significantly throughout the incubation period.
In 10 sets of slurry incubation experiments, we followed the progress of the methane oxidation process in Lake Kinneret methanogenic sediments in type-A two-stage long-term incubations. This is done by monitoring the changes in δ13CDIC values and by running metagenomic and specific isotope lipid analyses. We also followed methane oxidation in a semi-continuous bioreactor system (type B) with freshly collected sediments with or without the addition of hematite (Fig. 3). The results were compared to those of fresh-batch slurry incubations (type C) from the same methanogenic zone, presented by Bar-Or et al. (2017) and Elul et al. (2021).
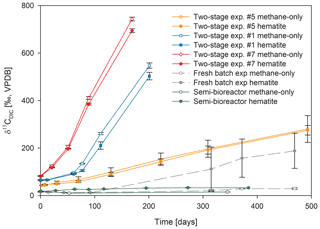
Figure 3Comparison of δ13CDIC values among the three types of experiments with 13C-labeled methane addition: setup A, three two-stage slurry experiments (at the second stage of the 1:3 ratio of sediment to porewater); setup B, the semi-continuous bioreactor experiment; and setup C, the slurry batch experiment with freshly collected sediments (Bar-Or et al., 2017). In each experiment, two treatments are shown, with hematite (filled symbol) and without hematite (empty symbols). The error bars represent the average deviation of the mean of duplicate/triplicate bottles.
3.1 Geochemical trends in the long-term two-stage experiments
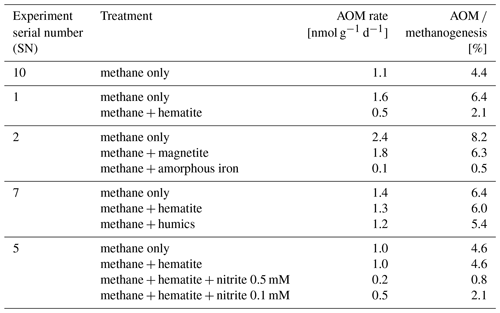
In the second-stage (1:3 ratio of sediment to porewater) long-term batch slurry experiments (type A) from the methanogenic zone, methanogenesis occurred with net methanogenesis rates of ∼ 25 nmol g−1 d−1 (grams are of dry weight here and throughout the rest of the paper; Fig. 2, Table S2), which are similar to those of fresh incubation experiments (Bar-Or et al., 2017). At the same time there was a conversion of 13C methane to 13C DIC in all the non-killed slurries amended with 13C methane, indicating AOM (Figs. 3 and 4). The δ13CDIC values of the “methane-only” control slurries reached values as high as 743 ‰. The average AOM rate in the methane-only controls was 2.0 ± 0.4 nmol g−1 d−1 (Table 2). AOM was also observed in these geochemical experiments with the addition of electron acceptors, and the potential of several electron acceptors to perform and stimulate the AOM process is detailed below.
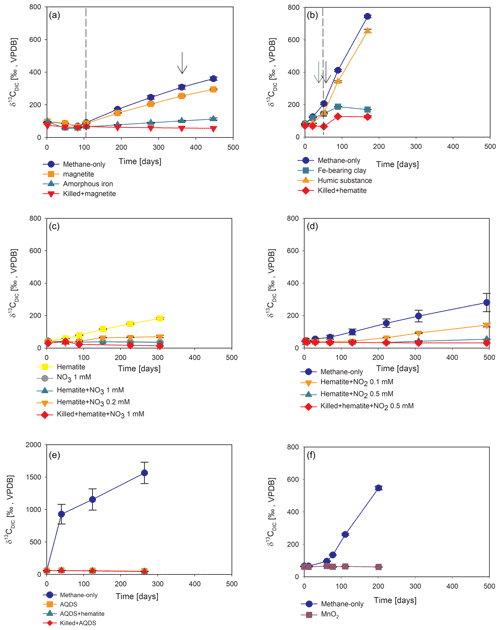
Figure 4Potential of different electron acceptors for AOM in Lake Kinneret in the two-stage long-term slurry experiments (at the second stage of the 1:3 ratio of sediment to porewater) with 13C-labeled methane and the following treatments: (a) with and without the addition of magnetite and amorphous iron (Fe(OH)3) – the dashed line represents the specific time of 13C-labeled methane addition, and the black arrow represents the addition of Na-molybdate as an inhibitor for sulfate reduction. (b) With clay and natural humic substance – the green arrow represents the time clay was added to the relevant bottles; the dashed line represents the time the headspace of each bottle was flushed again with N2; and the black arrow represents the second injection of 1 mL of 13C-labeled methane. (c) With the addition of hematite and two different concentrations of nitrate. (d) With the addition of hematite and two different concentrations of nitrite. (e) With the addition of AQDS. (f) With and without the addition of 13C-labeled methane to all the bottles (see Table 1 for specific experimental details). Error bars represent the average deviations of the data points from their means of duplicate/triplicate bottles.
3.1.1 Metals as electron acceptors
Iron and manganese oxides were added as potential electron acceptors to the second-stage long-term slurries. The addition of hematite to three different experiments increased the δ13CDIC values over time to 694 ‰, which is similar to the behavior of the methane-only controls and in a different pattern than the fresh experiments (Fig. 3). The average AOM rate in those two-stage treatments was 1.0 ± 0.3 nmol g−1 d−1 (Table 2). Magnetite amendments resulted in a minor increase in δ13CDIC values compared to the methane-only controls (200 ‰ and 265 ‰, respectively, Fig. 4a) with an AOM rate of 1.8 nmol g−1 d−1. Amorphous iron amendments resulted in only a 22 ‰ increase in δ13CDIC and a lower AOM rate (0.1 nmol g−1 d−1, Fig. 4a and Table 2). The addition of iron-bearing clay nontronite did not cause any increase in the δ13CDIC values (Fig. 4b), but the concentration of dissolved Fe(II) increased compared to the natural methane-only control (Fig. 5). Based on δ13CDIC estimates, no AOM was detected 200 d after the addition of MnO2, whereas the δ13CDIC values of the methane-only controls increased to over 500 ‰ (Fig. 4f).
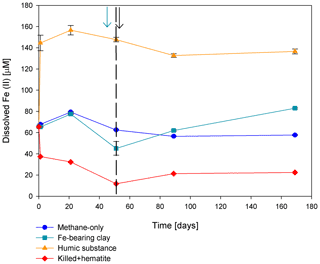
Figure 5Change in dissolved Fe(II) in the second stage of experiment no. 7 containing clay and natural humic acid. The green arrow represents the time at which clay was added to the specific bottles and those bottles were flushed with N2; the dashed line represents the time at which the rest of the bottles were flushed; and the black arrow represents the time at which 13C-labeled methane was added again. Error bars represent the average of the absolute deviations of the data points from their means.
3.1.2 Sulfate as an electron acceptor
The involvement of sulfate in the AOM in the incubations was tested, even in the absence of detectable sulfate in the methanogenic sediments. This is because sulfate could theoretically still be a short-lived intermediate for the AOM process in an active cryptic sulfur cycle (Holmkvist et al., 2011). It was quantified directly by adding Na-molybdate to the methane-only controls and the magnetite-amended treatments in the second-stage long-term incubations (Fig. 4a). The addition of Na-molybdate did not affect the increasing trend of δ13CDIC with time, and therefore, the AOM rates remained unchanged, which is similar to the observation in the fresh-batch incubations (Bar-Or et al., 2017).
3.1.3 Nitrate and nitrite as electron acceptors
Nitrate and nitrite involvement in the AOM was tested for the feasibility of an active cryptic nitrogen cycle, even in the absence of detectable amounts of nitrate and nitrite in the sediments (Nüsslein et al., 2001; Sivan et al., 2011). Nitrate was added at two different concentrations (0.2 and 1 mM, Fig. 4c) to the second-stage long-term slurries amended with hematite, as these concentrations were shown previously to promote AOM in other settings (Ettwig et al., 2010). The addition of hematite alone increased the δ13CDIC values by ∼ 200 ‰ during the 306 d of the experiment. The δ13CDIC in the bottles with the addition of 1 mM of nitrate, with and without hematite (Fig. 4c; the data points of the two treatments are on top of each other), decreased from 43 ‰ at the beginning of the experiment to 35 ‰ after 306 d. The δ13CDIC in the bottles with the addition of 0.2 mM of nitrate and hematite increased by 27 ‰ at the end of the experiment. Following the addition of 0.5 mM of nitrite, we observed no increase in δ13CDIC values during the first 222 d (Fig. 4d), after which they increased from 34 ‰ to 54 ‰ by the end of the experiment. The AOM rate of the high-nitrite-concentration treatment was 0.2 nmol g−1 d−1 (Table 2). Following the addition of 0.1 mM of nitrite, δ13CDIC increased only after 130 d to 158 ‰ on day 493. The AOM rate of the low-nitrite-concentration treatment was 0.5 nmol g−1 d−1. In the methane-only controls, the δ13CDIC value reached a maximum of 330 ‰.
3.1.4 Organic compounds as electron acceptors
Two of the second-stage long-term incubation experiments were amended with synthetic and natural organic electron acceptors to test the potential of organic electron acceptors. The addition of AQDS to slurries with and without hematite caused a decrease in δ13CDIC values over the entire duration of the experiment (Fig. 4e). Dissolved Fe(II) increased by 50 µM in these treatments, while in those without AQDS, it exhibited an increase of 20 µM (Fig. S3). We further tested the effect of naturally occurring humic substances by using those isolated from a different natural lake. The results show that the δ13CDIC values did not change at the beginning of the experiments (Fig. 4b), while a steep increase of ∼ 90 µM in their Fe(II) concentration was observed (Fig. 5). After 20 d, the δ13CDIC values of these slurries started to increase dramatically from 84 ‰ to 150 ‰ with an AOM rate of 1.2 nmol g−1 d−1 (Fig. 4b, Table 2). Dissolved Fe(II) concentrations mirrored the trend of δ13CDIC with a steep increase during the first 20 d followed by a decrease of 37 µM (Fig. 5).
3.1.5 Metabolic pathways
To elucidate which metabolic processes drive AOM, we analyzed δ13CDIC following the addition of inhibitors to the second-stage long-term slurries: (i) BES, a specific inhibitor for methanogenesis (Nollet et al., 1997), and (ii) acetylene, a non-specific inhibitor for methanogenesis and methanotrophy (Oremland and Capone, 1988). In both cases and similarly to the killed control, labeled 13C-DIC production was completely inhibited following the addition (Fig. 6). Though acetylene can also inhibit nitrogen cycling in some cases, it has been shown to result in the production of ethylene (Oremland and Capone, 1988). In our case, however, no ethylene was detected, supporting the conclusion that only the methanogenesis activity was inhibited.
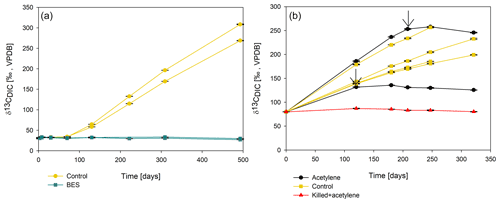
Figure 6Change in δ13CDIC values over time in the second-stage long-term sediment slurry incubations amended with hematite and 13C-labeled methane. (a) With/without BES and (b) with/without acetylene. Black arrows represent the time at which acetylene was injected into the experiment bottle. The error bars are smaller than the symbols.
3.2 Microbial dynamics
Analyses of taxonomy and coverage of metagenome-assembled genomes suggest that in the pre-incubated two-stage slurries, Bathyarchaeia are the dominant archaea, together with putative methanogens such as Methanofastidiales (Thermococci), Methanoregulaceae (Methanomicrobia) and Methanotrichales (Methanosarcina) (Supplement table). Bona fide ANMEs (ANME-1) were detected with substantial coverage of approximately 1 (the 27th most abundant from among the 195 MAGs detected) in all of the treatments. Among the bacteria, the sulfate reducers Desulfobacterota and Thermodesulfovibrionales (Nitrospirota) were prominent together with the GIF9 Dehalococcoidia lineage, which is known to metabolize chlorinated compounds in lake sediments (Biderre-Petit et al., 2016). Some Methylomirabilales (NC10) were found (average coverage of 0.32 ± 0.06), and no Methanoperedens were detected. Methylococcales methanotrophs were found in the natural sediments and the fresh-batch and bioreactor incubations (average of 0.34 ± 0.02), in contrast to their average coverage of 0.09 ± 0.04 in the long-term incubations. Methylococcales comprised the Methyloterricola, Methylomonas and Methylobacter genera (Supplement table). The methylotrophic partners of aerobic methanotrophs, Methylotenera, were found in fresh-batch and bioreactor incubations, where Methylomonas was found, findings that are in line with those of previous studies that showed their association (Beck et al., 2013). Principal component analysis shows the grouping of long-term pre-incubated slurries; semi-aerobic bioreactor incubations; and fresh-batch experiments (Fig. 7), emphasizing the microbial dynamics over time.
3.3 Lipid analysis
The δ13C values of the archaeol-derived isoprenoid phytane were between −5 ‰ and −17 ‰ in the long-term pre-incubated samples and thus showed 13C enrichment of 15 ‰–27 ‰ relative to the original sediment. This is indicative of methane-derived carbon assimilation by archaea (Table 3). Acyclic biphytane, derived mainly from caldarchaeol, exhibited a less pronounced 13C enrichment of 5 ‰–10 ‰. For bacterial-derived fatty acids, δ13C values similarly shifted by up to 10 ‰ relative to the original sediment. Nonetheless, one would have expected much higher values if aerobic methanotrophs were active, as was previously indicated by strong 13C enrichments of up to 1650 ‰ in C16:1ω5c observed in freshly incubated batch samples (Bar-Or et al., 2017).
Table 3The δ13C values (in ‰) of fatty acids and isoprenoid hydrocarbons from different experiments compared to values obtained from the original sediment in the methanogenic zone.
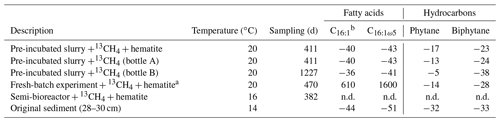
a Bar-Or et al. (2017). n.d.: not detected. b A mixture of three fatty acid double isomers at positions 9, 8 and 7.
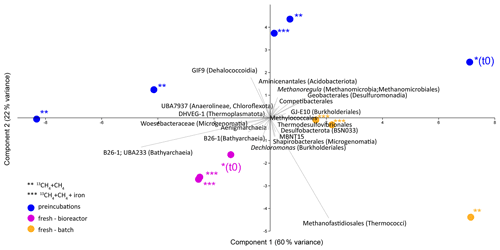
Figure 7Principal component analysis comparison of three types of samples: long-term pre-incubated slurries (blue – experiment A), semi-continuous bioreactor (pink – experiment B) and fresh-batch experiments (orange – experiment C). One asterisk represents t0; two asterisks denote methane-only treatments; three asterisks represent hematite treatment.
4.1 Anaerobic oxidation of methane in the methanogenic sediment incubation experiments
The in situ geochemical and microbial diversity profiles (Bar-Or et al., 2015) and the geochemical (Sivan et al., 2011; Bar-Or et al., 2017; Fig. 3) and metagenomic (Elul et al., 2021) analyses of batch incubations with fresh sediments provided strong support for the occurrence of Fe-AOM in sediments of the methanogenic zone below 20 cm. Such profiles and alongside incubations showed an unexpected presence of aerobic bacterial methanotrophs together with anaerobic microorganisms, such as methanogens and iron reducers (Adler et al., 2011; Sivan et al., 2011; Bar-Or et al., 2015, 2017; Elul et al., 2021). These findings suggested that both mcr-gene-bearing archaea and aerobic bacterial methanotrophs mediate methane oxidation. In the current study, we have supportive evidence of considerable AOM in the long-term incubations, even after the two treatment stages and considering the low abundance of the microbial populations.
The data from the two-stage incubations show a similar increasing trend in the δ13CDIC values of both the natural (methane-only) and the hematite-amended treatments (Fig. 3). This deviates from our observations during experiments B and C with fresh sediment, wherein higher δ13CDIC values were obtained after the addition of hematite than in the methane-only treatment (Fig. 3 and Bar-Or et al., 2017). This was particularly dramatic in the batch slurries (experiment C), but it was also observed in the semi-continuous bioreactor (experiment B). We assume that the observed difference in the bioreactors would have been more pronounced if methane concentrations had been higher, but it is still a relevant finding. We also note that the difference between the bioreactor results may also be due to the fact that each bioreactor community developed separately. The results of the type-A experiments (compared to those of types B and C) suggest that either hematite lacks the potential to stimulate the AOM activity during the two-stage experiments or there is enough natural Fe(III) in the sediments to sustain the maximum potential of Fe-AOM. Below we characterize the AOM process in the long-term two-stage incubation experiments.
4.2 Potential electron acceptors for AOM in the long-term two-stage incubation experiments
4.2.1 Metal oxides as electron acceptors
Measurements of δ13CDIC show that the additions of magnetite, amorphous iron, clays and manganese oxide in the second-stage incubations resulted in a less pronounced increase in the δ13CDIC values compared to those of the methane-only controls (Fig. 4). A possible explanation for the latter may be that these metal oxides inhibit AOM, either directly or via a preference for organoclastic iron reduction over Fe-AOM, which adds a natural, more negative carbon isotope signal from the organic materials rather than the heavy carbon from the 13C-labeled methane. Using mass balance estimations in the methane-only and in the amorphous iron treatments and considering the DIC concentrations and δ13CDIC values of the methane-only treatments at the beginning of the experiment (6 mM and 60 ‰, respectively) and the values at the end (6.5 mM and 360 ‰, respectively), about 0.5 mM of the DIC was added by the AOM of methane with δ13C of ∼ 4000 ‰. The DIC and δ13CDIC values of the amorphous iron treatment at the beginning of the experiment were 5.4 mM and 60 ‰, respectively, and by the end were 6.1 mM and 120 ‰, respectively. Assuming the same δ13C of the added methane of 4000 ‰ and a δ13CTOC of −30 ‰ (Sivan et al., 2011), 0.1 mM of the DIC should derive from AOM and 0.6 mM from organoclastic metabolism. This means that adding amorphous iron to the system encouraged iron reduction that was coupled to the oxidation of organic compounds other than methane. Intrinsic microbes, particularly the commonly detected ex-deltaproteobacterial lineages such as Geobacterales, may catalyze Fe(III) metal reduction, regardless of AOM (Xu et al., 2021). Manganese oxides are found in very low abundance in Lake Kinneret sediments (0.1 %; Table S1 and Sivan et al., 2011). Thus, their role in metal AOM is likely minimal.
4.2.2 Sulfate as an electron acceptor
Sulfate concentrations in the methanogenic Lake Kinneret sediments have been below the detection limit in years past, similarly to their representation in the natural sediments we used for the incubations (<5 µM; Bar-Or et al., 2015; Elul et al., 2021). Sulfide concentrations have also been reported to be minor (<0.3 µM; Sivan et al., 2011). However, sulfate could theoretically still be a short-lived intermediate for the AOM process, as pyrite and FeS precipitate in the top sediments and cryptic cycling via pyrite or FeS may replenish the sulfate, thus rendering it available for AOM (Bottrell et al., 2000). The addition of Na-molybdate to the second-stage slurries, including those amended with and without magnetite, did not change the δ13CDIC dynamics, which remained similar to those from before the addition of the inhibitor (Fig. 4a). This finding is in line with that in fresh-batch sediment slurries (Bar-Or et al., 2017) and suggests that sulfate is not a potent electron acceptor for AOM in this environment. Furthermore, although sulfate-reducing bacteria were abundant, none of the reducers belonged to the known clades of ANME-2d partners, which were connected previously to the Fe–S–CH4-coupled AOM (Su et al., 2020; Mostovaya et al., 2021).
4.2.3 Nitrogen species as electron acceptors
Nitrate and nitrite concentrations are also undetectable in the porewater of Lake Kinneret sediments (Nüsslein et al., 2001; Sivan et al., 2011) but again may appear as short-lived intermediate products of ammonium oxidation that is coupled to iron reduction (Tan et al., 2021; Ding et al., 2014; Shrestha et al., 2009; Clement et al., 2005). We thus assessed the roles of nitrate and nitrite as electron acceptors in the two-stage slurries. Our results indicate that the addition of nitrate did not promote AOM, likely due to the absence of ANME-2d, which is known to use nitrate (Arshad et al., 2015; Haroon et al., 2013). In the case of nitrite, even low concentrations appeared to delay the increase in δ13CDIC values, suggesting that organoclastic denitrification outcompetes AOM, and despite the occurrence of Methylomirabilia, the role of nitrite-AOM is not prominent in the two-stage incubations (Fig. 4c, d).
4.2.4 Humic substances as electron acceptors
Humic substances may promote AOM by continuously shuttling electrons to metal oxides (Valenzuela et al., 2019). Though humic substances were not measured directly in Lake Kinneret sediments, the DOC concentrations in the methanogenic depth porewater were previously found to be high (∼ 1.5 mM; Adler et al., 2011), suggesting that they may play a role in AOM. Compared to the methane-only treatments, the treatment with the synthetic humic analog AQDS caused an increase in dissolved Fe(II) concentrations, but it did not cause 13C-DIC enrichment. This may be explained by the behavior of AQDS as a strong electron shuttle in organoclastic iron reduction (Lovley et al., 1996), which produces isotopically more negative carbon that masks the AOM signal (Figs. 4e, S3). Yet, as was done by Valenzuela et al. (2017), the addition of natural humic substances did promote AOM, compared to the rest of the electron acceptors tested, and may thus support AOM (Fig. 4b). In our incubations, the natural humic substances promoted first the oxidation of organic matter by iron reduction, probably by shuttling electrons from the broad spectrum of organic compounds to natural iron oxides (Figs. 4b and 5). When the availability of the iron oxides or the organic matter decreased, humic substances likely took over to facilitate the AOM (Fig. 4b).
Overall, the results of our long-term two-stage experiments indicate that sulfate, nitrate, nitrite and manganese oxides do not support AOM in the methanogenic sediments of Lake Kinneret. The candidate electron acceptors for AOM in the long-term experiments are natural humic substances and/or naturally abundant iron minerals. Future experiments can simulate iron limitation and the involvement of iron oxides in the AOM by removing natural iron oxides from the sediments.
4.3 Main microbial players in the long-term two-stage slurries
Methane oxidation in the pre-incubated Lake Kinneret sediments is likely mediated by either ANMEs or methanogens, as the addition of BES and acetylene immediately stopped the AOM (Fig. 6), similarly to the results of the killed bottles and the BES treatment in the fresh-batch experiment (Bar-Or et al., 2017). Apart from being methane-metabolizing, acetylene can inhibit nitrogen cycling, which results in ethylene production (Oremland and Capone, 1988). This was not the case in our incubations as no ethylene was produced. The increase in δ13C values in phytane and biphytane (Table 3) also indicates the presence of active archaeal methanogens or ANMEs (Wegener et al., 2008; Kellermann et al., 2012; Kurth et al., 2019).
Using the isotopic compositions of specific lipids and metagenomics, we identified a considerable abundance of aerobic methanotrophs and methylotrophs in the fresh sediments but not in the long-term slurries (Table 3, Fig. 7). In the natural sediments, micro-levels (nanomolar) of oxygen could be trapped in clays and slowly released to the porewater (Wang et al., 2018). However, if such micro-levels of oxygen still existed during the time of the pre-incubation, they were probably already exhausted. Indeed, the results of our specific lipids and metagenomics analyses suggest that the aerobic methanotrophs lineages play only a minor role in the long-term slurries, probably due to complete depletion of the oxygen. The metagenomic data (Fig. 7, Supplement table) also indicate that Bathyarchaeia, which may be involved in methane metabolism (Evens et al., 2015), were enriched in the bioreactor incubations, yet their role in Lake Kinneret AOM remains to be evaluated. We also observed changes in the abundance of bacterial degraders of organic matter and necromass: for example, GIF9 Dehalococcoidia, which can metabolize complex organic materials under methanogenic conditions (Cheng et al., 2019; Hug et al., 2013), were most abundant in the long-term incubations (Fig. 7, Supplement table). Though ANME-1 archaea are likely mediators of AOM in these sediments, methane oxidation via reverse methanogenesis is feasible for some methanogens in Lake Kinneret sediments (Elul et al., 2021).
4.4 Mechanism of methane oxidation in the long-term two-stage incubations
Our results indicate net methanogenesis in the two-stage incubation experiments with an average rate of 25 nmol g−1 d−1 (Fig. 2 and Table S2), which is similar to those from fresh incubation experiments (Bar-Or et al., 2017). This is despite the overall trend of increasing δ13CDIC values, a result representing potential methane turnover (Figs. 3 and 4). A likely explanation for the presence of both signals is an interplay between methane production and oxidation, which is possibly triggered by reversal of the methanogenesis pathway in bona fide ANMEs or certain methanogens (Hallam et al., 2004; Timmers et al., 2017). Due to the overall production of methane and the lack of intense stimulation of AOM by any electron acceptor added, the increase in δ13CDIC values could theoretically result from the occurrence of carbon back flux during methanogenesis, which is feasible in environments that are close to thermodynamic equilibrium (Gropp et al., 2021). To test this, we used DIC mass balance calculations to determine the strength of back flux in our incubations. Based on Eqs. (1) and (2), the observed level of 13C enrichment indicates that 3 %–8 % of the 13C methane should be converted into DIC. These estimates are orders of magnitude higher than the previously reported values of 0.001 %–0.3 % for methanogenesis back flux in cultures (Zehnder and Brock, 1979; Moran et al., 2005), but they are in the same range as the back flux of 3.2 % to 5.5 % observed in ANME enrichment cultures (Holler et al., 2011). For the latter, however, modeling approaches from AOM-dominated marine sediment samples and associated ANME enrichment cultures indicated the absence of net methanogenesis (Yoshinaga et al., 2014; Chuang et al., 2019; Meister et al., 2019; Wegener et al., 2021). Thus, it seems unlikely that back flux alone can account for the methane-to-DIC conversion in Lake Kinneret sediments. Moreover, the occurrence of back flux alone in marine methanogenic sediments with similar net methanogenesis rates and abundant methane-metabolizing archaea did not yield considerable 13C enrichment in the DIC pool following sediment incubations (Sela-Adler et al., 2015; Vigderovich et al., 2019; Yorshansky, 2019) (Table S3). It is, therefore, less likely that the observed DIC values in our study were sustained by methanogenesis back flux alone (without an external electron acceptor) than by active AOM, which, in this case, is probably performed by ANME-1 archaea or by methanogens, with the latter performing reverse methanogenesis to some extent.
Previous results of geochemical and microbial profiles as well as incubations with fresh sediments from Lake Kinneret constitute evidence of the occurrence of Fe-AOM in the methanogenic zone. The process is performed by anaerobic archaeal methanogens and bacterial methanotrophs, which remove about 10 %–15 % of the methane produced in the lake's sediment. In the current study, we found that after two incubation stages and intensive purging for a prolonged duration, AOM was still significant, consuming 3 %–8 % of the methane produced. However, the abundance of aerobic methanotrophs decreased and anaerobic archaea (ANME-1 or specific methanogens) appeared to be solely responsible for methane turnover. AOM could be a result of carbon back flux as the methanogenic/AOM pathway is reversible; however, the high δ13CDIC signal points to a metabolic reaction. Terminal electron acceptors or electron shuttles stimulating Fe-AOM are hematite and/or humic substances. The role of the aerobic methanotrophs of the order Methylococcales, which were found in the freshly collected sediment experiments, remains to be examined.
The markdown file is available at https://doi.org/10.6084/m9.figshare.19585933.v1 (Rubin-Blum, 2022).
Metagenomic reads were submitted to NCBI Sequence Read Archive, BioProject PRJNA826318, and the geochemical data sets can be accessed at https://doi.org/10.5281/zenodo.6489360 (Vigderovich, 2022)
The supplement related to this article is available online at: https://doi.org/10.5194/bg-19-2313-2022-supplement.
HV, WE and OS designed the research. HV, ME, MRB and ME analyzed the samples and the data. WE and OS supervised HV and provided resources and funding. HV and OS synthesized the data and wrote the original draft. HV wrote the paper with contributions from all the co-authors.
The contact author has declared that neither they nor their co-authors have any competing interests.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
We would like to thank Benni Sulimani and Oz Tzabari from the Yigal Allon Kinneret Limnological Laboratory for their onboard technical assistance. We thank all of O. Sivan's lab members for their help during sampling and express especially heartfelt thanks to Noam Lotem for the invaluable assistance with the mass balance calculations and the fruitful discussions and to Efrat Eliani-Russak for her technical assistance. Many thanks to Kai Hachmann from Marcus Elvert's lab for his help during lipid analysis and to Jonathan Gropp for insightful discussions about the back flux. This work was supported by ERC Consolidator (818450) and Israel Science Foundation (857-2016) grants awarded to Orit Sivan. Funding for Marcus Elvert was provided by the Deutsche Forschungsgemeinschaft (DFG) under Germany's Excellence Initiative/Excellence Strategy through the clusters of excellence EXC 309 “The Ocean in the Earth System” (project no. 49926684) and EXC 2077 “The Ocean Floor – Earth's Uncharted Interface” (project no. 390741601). Funding for Maxim Rubin-Blum was provided by the Israel Science Foundation (913/19), the U.S.-Israel Binational Science Foundation (2019055), and the Israel Ministry of Science and Technology (1126). Hanni Vigderovich was supported by a student fellowship from the Israel Water Authority.
This research has been supported by the H2020 European Research Council (MERIR (grant no. 818450)), the Israel Science Foundation (grant nos. 857-2016 and 913/19), the Deutsche Forschungsgemeinschaft (project no. 49926684), the U.S.-Israel Binational Science Foundation (grant no. 2019055), and the Israel Ministry of Science and Technology (grant no. 1126).
This paper was edited by Tina Treude and reviewed by four anonymous referees.
Adler, M., Eckert, W., and Sivan, O.: Quantifying rates of methanogenesis and methanotrophy in Lake Kinneret sediments (Israel) using porewater profiles, Limnol. Oceanogr., 56, 1525–1535, https://doi.org/10.4319/lo.2011.56.4.1525, 2011.
Aepfler, R. F., Bühring, S. I., and Elvert, M.: Substrate characteristic bacterial fatty acid production based on amino acid assimilation and transformation in marine sediments, FEMS Microbiol. Ecol., 95, 1–15, https://doi.org/10.1093/femsec/fiz131, 2019.
Arshad, A., Speth, D. R., De Graaf, R. M., Op den Camp, H. J. M., Jetten, M. S. M., and Welte, C. U. A: Metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like archaea, Front. Microbiol., 6, 1–14, https://doi.org/10.3389/fmicb.2015.01423, 2015.
Bai, Y. N., Wang, X. N., Wu, J., Lu, Y. Z., Fu, L., Zhang, F., Lau, T. C., and Zeng, R. J.: Humic substances as electron acceptors for anaerobic oxidation of methane driven by ANME-2d, Water Res., 164, 114935, https://doi.org/10.1016/j.watres.2019.114935, 2019.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., Lesin, V. M., Nicolenko, S. I., Pham, S., Prjibelski, A. D., Sirotkin, A. V., Vyahhi, N., Tesler, G., Aleksyev, A. M., and Pevzner, P. A.: SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing, J. Comput. Biol., 19, 455–477, https://doi.org/10.1089/cmb.2012.0021, 2012.
Bar-Or, I., Ben-Dov, E., Kushmaro, A., Eckert, W., and Sivan, O.: Methane-related changes in prokaryotes along geochemical profiles in sediments of Lake Kinneret (Israel), Biogeosciences, 12, 2847–2860, https://doi.org/10.5194/bg-12-2847-2015, 2015.
Bar-Or, I., Elvert, M., Eckert, W., Kushmaro, A., Vigderovich, H., Zhu, Q., Ben-Dov, E., and Sivan, O.: Iron-Coupled Anaerobic Oxidation of Methane Performed by a Mixed Bacterial-Archaeal Community Based on Poorly Reactive Minerals, Environ. Sci. Technol., 51, 12293–12301, https://doi.org/10.1021/acs.est.7b03126, 2017.
Bastviken D.: Methane, in: Encyclopedia of Inland waters, Oxford, Elsevier, 783–805, https://doi.org/10.1016/B978-012370626-3.00117-4, 2009.
Beck, D. A. C., Kalyuzhnaya, M. G., Malfatti, S., Tringe, S. G., del Rio, T. G., Ivanova, N., Lidstorm, M. E., and Chistoserdova, L.: A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between the Methylococcaceae and the Methylophilaceae, PeerJ, 1, 1–23, https://doi.org/10.7717/peerj.23, 2013.
Biderre-Petit, C., Dugat-Bony, E., Mege, M., Parisot, N., Adrian, L., Moné, A., Denonfoux, J., Peyretaillade, E., Debroas. D., Boucher, D., and Peyret, P.: Distribution of Dehalococcoidia in the anaerobic deep water of a remote meromictic crater lake and detection of Dehalococcoidia-derived reductive dehalogenase homologous genes, Plos One, 11, 1–19, https://doi.org/10.1371/journal.pone.0145558, 2016.
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gieseke, A., Amann, R., Jørgensen, B. B., Witte, U., and Pfannkuche, O.: A marine microbial consortium apparently mediating AOM, Nature, 407, 623–626, https://doi.org/10.1038/35036572, 2000.
Bottrell, S. H., Parkes, R. J., Cragg, B. A., and Raiswell, R.: Isotopic evidence for anoxic pyrite oxidation and stimulation of bacterial sulphate reduction in marine sediments, J. Geol. Soc., 157, 711–714, https://doi.org/10.1144/jgs.157.4.711, 2000.
Cabrol, L., Thalasso, F., Gandois, L., Sepulveda-Jauregui, A., Martinez-Cruz, K., Teisserenc, R., Tananaev, N., Tveit, A., Svenning, M. M., and Barret, M.: Anaerobic oxidation of methane and associated microbiome in anoxic water of Northwestern Siberian lakes, Sci. Total Environ., 736, 139588, https://doi.org/10.1016/j.scitotenv.2020.139588, 2020.
Cai, C., Leu, A. O., Xie, G.-J., Guo, J., Feng, Y., Zhao, J.-X., Tyson, G. W., Yuan, Z., and Hu, S.: A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(II) reduction, ISME J., 12, 1929–1939, https://doi.org/10.1038/s41396-018-0109-x, 2018.
Cheng, L., Shi, S. bao, Yang, L., Zhang, Y., Dolfing, J., Sun, Y., Liu, L., Li, Q., Tu, B., Dai, L., Shi, Q., and Zhang, H.: Preferential degradation of long-chain alkyl substituted hydrocarbons in heavy oil under methanogenic conditions, Org. Geochem., 138, 103927, https://doi.org/10.1016/j.orggeochem.2019.103927, 2019.
Chuang, P. C., Yang, T. F., Wallmann, K., Matsumoto, R., Hu, C. Y., Chen, H. W., Lin, S., Sun, C.-H., Li, H.-C., Wang, Y., and Dale, A. W.: Carbon isotope exchange during anaerobic oxidation of methane (AOM) in sediments of the northeastern South China Sea, Geochim. Cosmochim. Ac., 246, 138–155, https://doi.org/10.1016/j.gca.2018.11.003, 2019.
Clement, J.-C., Shrestha, J., Ehrenfeld, J. G., and Jaffe, P. R.: Ammonium oxidation coupled to dissimilatory iron reduction under anaerobic conditions in wetland soils, Soil biology and biochemistry, 37, 2323–2328, https://doi.org/10.1016/j.soilbio.2005.03.027, 2005.
Conrad, R.: The global methane cycle: Recent advances in understanding the microbial processes involved, Environ. Microbiol. Rep., 1, 285–292, https://doi.org/10.1111/j.1758-2229.2009.00038.x, 2009.
Crowe, S. A., Katsev, S., Leslie, K., Sturm, A., Magen, C., Nomosatryo, S., Pack, M. A., Kessler, J. D., Reeburgh, W. S., Roberts, J. A., González, L., Douglas Haffner, G., Mucci, A., Sundby, B., and Fowle, D. A.: The methane cycle in ferruginous Lake Matano, Geobiology, 9, 61–78, https://doi.org/10.1111/j.1472-4669.2010.00257.x, 2011.
Damgaard, L. R., Revsbech, N. P., and Reichardt, W.: Use of an oxygen-insensitive microscale biosensor for methane to measure methane concentration profiles in a rice paddy, Appl. Environ. Microbiol., 64, 864–870, https://doi.org/10.1128/aem.64.3.864-870.1998, 1998.
Dershwitz, P., Bandow, N. L., Yang, J., Semrau, J. D., McEllistrem, M. T., Heinze, R. A., Fonseca, M., Ledesma, J. C., Jennett, J. R., DiSpirito, A. M., Athwal, N. S., Hargrove, M. S., Bobik, T. A., Zischka, H., and DiSpirito, A. A.: Oxygen Generation via Water Splitting by a Novel Biogenic Metal Ion-Binding Compound, Appl. Environ. Microbiol., 87, 1–14, https://doi.org/10.1128/aem.00286-21, 2021.
Ding, L. J., An, X. L., Li, S., Zhang, G. L., and Zhu, Y. G.: Nitrogen loss through anaerobic ammonium oxidation coupled to iron reduction from paddy soils in a chronosequence, Environ. Sci. Technol., 48, 10641–10647, https://doi.org/10.1021/es503113s, 2014.
Elul, M., Rubin-Blum, M., Ronen, Z., Bar-Or, I., Eckert, W., and Sivan, O.: Metagenomic insights into the metabolism of microbial communities that mediate iron and methane cycling in Lake Kinneret iron-rich methanic sediments, Biogeosciences, 18, 2091–2106, https://doi.org/10.5194/bg-18-2091-2021, 2021.
Elvert, M., Boetius, A., Knittel, K., and Jørgensen, B. B.: Characterization of specific membrane fatty acids as chemotaxonomic markers for sulfate-reducing bacteria involved in anaerobic oxidation of methane, Geomicrobiol. J., 20, 403–419, https://doi.org/10.1080/01490450303894, 2003.
Ettwig, Katharina F, Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M. M., Schreiber, F., Dutilh, B. E., Zedelius, J., de Beer, D. Gloerich, J., Wessels, H. J. C. T., van Alen, T., Luesken, F., Wu, M. L., van de Pas-Schoonen K. T., Op den Camp, H. J. M., Jansen-Megens, E. M., Francojs, K. J., Stunnenberg, H., Weissenbach, J., Jetten, M. S. M., and Strous, M.: Nitrite-driven anaerobic methane oxidation by oxygenic bacteria, Nature, 464, 543–548, https://doi.org/10.1038/nature08883, 2010.
Ettwig, K. F., Zhu, B., Speth, D., Keltjens, J. T., Jetten, M. S. M., and Kartal, B.: Archaea catalyze iron- dependent anaerobic oxidation of methane, P. Natl. Acad. Sci. USA, 113, 12792–12796, https://doi.org/10.1073/pnas.1609534113, 2016.
Evans, P. N., Parks, D. H., Chadwick, G. L., Robbins, S. J., Orphan, V. J., Golding, S. D., and Tyson, G. W.: Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics, Science, 350, 434–438, https://doi.org/10.1126/science.aac7745, 2015.
Fan, L., Dippold, M. A., Ge, T., Wu, J., Thiel, V., Kuzyakov, Y., and Dorodnikov, M.: Anaerobic oxidation of methane in paddy soil: Role of electron acceptors and fertilization in mitigating CH4 fluxes, Soil Biol. Biochem., 141, 107685, https://doi.org/10.1016/j.soilbio.2019.107685, 2020.
Froelich, P. N., Klinkhammer, G. P., Lender, M. L., Luedtke, N. A., Heath, G. R., Cullen, D., Dauphin, P., Hammond, D., Hartman, B., and Maynard, V.: Geochem. Cosmochim. Ac., 43, 1075–1090, https://doi.org/10.1016/0016-7037(79)90095-4, 1979.
Gropp, J., Iron, M. A., and Halevy, I.: Theoretical estimates of equilibrium carbon and hydrogen isotope effects in microbial methane production and anaerobic oxidation of methane, Geochem. Cosmochim. Ac., 295, 237–264, https://doi.org/10.1016/j.gca.2020.10.018, 2021.
Hallam, S. J., Putnam, N., Preston, C. M., Detter, J. C., Rokhsar, D., Richardson, P. H., and DeLong, E. F.: Reverse methanogenesis: Testing the hypothesis with environmental genomics, Science, 305, 1457–1462, https://doi.org/10.1126/science.1100025, 2004.
Hammer, Ø., Harper, D. A. T., and Ryan, P. D.: Past: paleontological statistics software package for education and data analysis, Palaeontol. Electron., 4, 9 pp., 2001.
Haroon, M. F., Hu, S., Shi, Y., Imelfort, M., Keller, J., Hugenholtz, P., Yuan, Z., and Tyson, G. W.: Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage, Nature, 500, 567–570, https://doi.org/10.1038/nature12375, 2013.
Hoehler, T. M., Alperin, M. J., Albert, D. B., and Martens, C. S.: Field and laboratory, evidence for a methane-sulfate reducer consortium, Global Biogeochem. Cy., 8, 451–463, https://doi.org/10.1029/94GB01800, 1994.
Holler, T., Wegener, G., Niemann, H., Deusner, C., Ferdelman, T. G., Boetius, A., Brunner, B., and Widdel, F.: Carbon and sulfur back flux during anaerobic microbial oxidation of methane and coupled sulfate reduction, P. Natl. A. Sci. USA., 108, E1484–E1490, https://doi.org/10.1073/pnas.1106032108, 2011.
Holmkvist, L., Ferdelman, T. G., and Jørgensen, B. B.: A cryptic sulfur cycle driven by iron in the methane zone of marine sediment (Aarhus Bay, Denmark), Geochim. Cosmochim. Ac., 75, 3581–3599, https://doi.org/10.1016/j.gca.2011.03.033, 2011.
Hug, L. A., Castelle, C. J., Wrighton, K. C., Thomas, B. C., Sharon, I., Frischkorn, K. R., Williams, K. H., Tringe, S. G., and Banfield, J. F.: Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling, Microbiome, 1, 1–17, https://doi.org/10.1186/2049-2618-1-22, 2013.
Kang, D. D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., and Wang, Z.: MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies, PeerJ, 2019, 1–13, https://doi.org/10.7717/peerj.7359, 2019.
Kellermann, M. Y., Wegener, G., Elvert, M., Yoshinaga, M. Y., Lin, Y. S., Holler, T., Mollar, P. X., Knittel K., and Hinrichs, K. U.: Autotrophy as a predominant mode of carbon fixation in anaerobic methane-oxidizing microbial communities, P. Natl. A. Sci. USA, 109, 19321–19326, https://doi.org/10.1073/pnas.1208795109, 2012.
Kits, K. D., Klotz, M. G., and Stein, L. Y.: Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1, Environ. Microbiol., 17, 3219–3232, https://doi.org/10.1111/1462-2920.12772, 2015.
Knittel, K. and Boetius, A.: Anaerobic oxidation of methane: Progress with an unknown process, Annu. Rev. Microbiol., 63, 311–334, https://doi.org/10.1146/annurev.micro.61.080706.093130, 2009.
Kurth, J. M., Smit, N. T., Berger, S., Schouten, S., Jetten, M. S. M., and Welte C. U.: Anaerobic methanotrophic archaea of the ANME-2d clade feature lipid composition that differs from other ANME archaea, FEMS Microbiol. Ecol., 95, fiz082, https://doi.org/10.1093/femsec/fiz082, 2019.
Lin, Y. S., Lipp, J. S., Yoshinaga, M. Y., Lin, S. H., Elvert, M., and Hinrichs, K. U.: Intramolecular stable carbon isotopic analysis of archaeal glycosyl tetraether lipids, Rapid Commun. Mass Sp., 24, 2817–2826, https://doi.org/10.1002/rcm.4707, 2010.
Lovley, D. R. and Klug, M. J.: Sulfate reducers can outcompete methanogens at freshwater sulfate concentrations, Appl. Environ. Microbiol., 45, 187–192, https://doi.org/10.1128/aem.45.1.187-192.1983, 1983.
Lovley, D. R., Coates, J. D., Blunt-Harris, E. L., Phillips, E. J. P., and Woodward, J. C.: Humic substances as electron acceptors for microbial respiration, Nature, 382, 445–448, https://doi.org/10.1038/382445a0, 1996.
Lu, Y. Z., Fu, L., Ding, J., Ding, Z. W., Li, N., and Zeng, R. J.: Cr(VI) reduction coupled with anaerobic oxidation of methane in a laboratory reactor, Water Res., 102, 445–452, https://doi.org/10.1016/j.watres.2016.06.065, 2016.
Martinez-Cruz, K., Leewis, M., Charold, I., Sepulveda-Jauregui, A., Walter, K., Thalasso, F., and Beth, M.: Science of the Total Environment Anaerobic oxidation of methane by aerobic methanotrophs in sub-Arctic lake sediments, Sci. Total Environ., 607/608, 23–31, https://doi.org/10.1016/j.scitotenv.2017.06.187, 2017.
Meador, T. B., Gagen, E. J., Loscar, M. E., Goldhammer, T., Yoshinaga, M. Y., Wendt, J., Thomm, M., and Hinrichs, K. U.: Thermococcus kodakarensis modulates its polar membrane lipids and elemental composition according to growth stage and phosphate availability, Front. Microbiol., 5, 1–13, https://doi.org/10.3389/fmicb.2014.00010, 2014.
Meister, P., Liu, B., Khalili, A., Böttcher, M. E., and Jørgensen, B. B.: Factors controlling the carbon isotope composition of dissolved inorganic carbon and methane in marine porewater: An evaluation by reaction-transport modelling, J. Mar. Syst., 200, 103227, https://doi.org/10.1016/j.jmarsys.2019.103227, 2019.
Moran, J. J., House, C. H., Freeman, K. H., and Ferry, J. G.: Trace methane oxidation studied in several Euryarchaeota under diverse conditions, Archaea, 1(5), 303–309, https://doi.org/10.1155/2005/650670, 2005.
Mostovaya, A., Wind-Hansen, M., Rousteau, P., Bristow, L. A., and Thamdrup, B.: Sulfate- and iron- dependent anaerobic methane oxidation occuring side-by-side in freshwater lake sediments, Limnol. Oceanogr., 67, 231–246, https://doi.org/10.1002/lno.11988, 2021.
Nollet, L., Demeyer, D., and Verstraete, W.: Effect of 2-bromoethanesulfonic acid and Peptostreptococcus productus ATCC 35244 addition on stimulation of reductive acetogenesis in the ruminal ecosystem by selective inhibition of methanogenesis, Appl. Environ. Microbiol., 63, 194–200, https://doi.org/10.1128/aem.63.1.194-200.1997, 1997.
Norði, K. Á. and Thamdrup, B.: Nitrate-dependent anaerobic methane oxidation in freshwater sediment, Geochim. Cosmochim. Ac., 132, 141–150, https://doi.org/10.1016/j.gca.2014.01.032, 2014.
Norði, K Á., Thamdrup, B., and Schubert, C. J.: Anaerobic oxidation of methane in an iron-rich Danish freshwater lake sediment, Limnol. Oceanogr., 58, 546–554, https://doi.org/10.4319/lo.2013.58.2.0546, 2013.
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A., Korobeynikov, A., Lapidus, A., Prjibelsky, A., Pyshkin, A., Sirotkin, A., Sirotkin, Y, Stepanauskas, R., McLean, J., Lasken, R., Clingenpeel, S. R., Woyke, T., Tesler, G., Alekseyev, M. A., and Pevzner, P. A.: Assembling genomes and mini-metagenomes from highly chimeric reads, in: Research in Computational Molecular Biology, RECOMB 2013, Lecture Notes in Computer Science, Vol. 7821, edited by: Deng, M., Jiang, R., Sun, F., and Zhang, X., Springer, Berlin, Heidelberg, 158–170, https://doi.org/10.1007/978-3-642-37195-0, 2013.
Nüsslein, B., Chin, K. J., Eckert, W., and Conrad, R.: Evidence for anaerobic syntrophic acetate oxidation during methane production in the profundal sediment of subtropical Lake Kinneret (Israel), Environ. Microbiol., 3, 460–470, https://doi.org/10.1046/j.1462-2920.2001.00215.x, 2001.
Oremland, R. S. and Capone, D. G.: Use of “Specific” Inhibitors in Biogeochemistry and Microbial Ecology, in: Advances in microbial ecology, Vol. 10, edited by: Marshall, K. C., Plenum Press, New York and London, 285–362, https://doi.org/10.2307/4514, 1988.
Orphan, V. J., House, C. H., and Hinrichs, K.: Methane-Consuming Archaea Revealed by Directly Coupled Isotopic and Phylogenetic Analysis, Science, 293, 484–488, https://doi.org/10.1126/science.1061338, 2001.
Oswald, K., Milucka, J., Brand, A., Hach, P., Littmann, S., Wehrli, B., Albersten, M., Daims, H., Wagner, M., Kuypers, M. M. M., Schubert, C. J., and Milucka, J.: Aerobic gammaproteobacterial methanotrophs mitigate methane emissions from oxic and anoxic lake waters, Limnol. Oceanogr., 61, 101–118, https://doi.org/10.1002/lno.10312, 2016.
Parks, D. H., Chuvochina, M., Rinke, C., Mussig, A. J., Chaumeil, P.-A., and Hugenholtz, P.: GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy, Nucl. Acids Res., 202, 1–10, https://doi.org/10.1093/nar/gkab776, 2021.
Raghoebarsing, A. A., Pol, A., Van De Pas-Schoonen, K. T., Smolders, A. J. P., Ettwig, K. F., Rijpstra, W. I. C., Schouten, S., Sinninghe Damsté, J. S., Op den Camp, H. J. M., Jetten, M. S. M., and Strous, M.: A microbial consortium couples anaerobic methane oxidation to denitrification, Nature, 440, 918–921, https://doi.org/10.1038/nature04617, 2006.
Reeburgh, W. S.: Oceanic Methane Biogeochemistry, Chem. Rev., 38, 486–513, https://doi.org/10.1002/chin.200720267, 2007.
Rosentreter, J. A., Borges, A. V., Deemer, B. R., Holgerson, M. A., Liu, S., Song, C., Melack, J., Raymond, P. A., Duarte, C. M., Allen, G. H., Olefeldt, D., Poulter, B., Battin, T. I., and Eyre, B. D.: Half of global methane emissions come from highly variable aquatic ecosystem source, Nat. geosci., 14, 225-230, https://doi.org/10.1038/s41561-021-00715-2, 2021.
Rubin-Blum, M.: Vigderovich et al., 2022 markdown file, figshare [code], https://doi.org/10.6084/m9.figshare.19585933.v1, 2022.
Saunois, M., Stavert, A. R., Poulter, B., Bousquet, P., Canadell, J. G., Jackson, R. B., Raymond, P. A., Dlugokencky, E. J., Houweling, S., Patra, P. K., Ciais, P., Arora, V. K., Bastviken, D., Bergamaschi, P., Blake, D. R., Brailsford, G., Bruhwiler, L., Carlson, K. M., Carrol, M., Castaldi, S., Chandra, N., Crevoisier, C., Crill, P. M., Covey, K., Curry, C. L., Etiope, G., Frankenberg, C., Gedney, N., Hegglin, M. I., Höglund-Isaksson, L., Hugelius, G., Ishizawa, M., Ito, A., Janssens-Maenhout, G., Jensen, K. M., Joos, F., Kleinen, T., Krummel, P. B., Langenfelds, R. L., Laruelle, G. G., Liu, L., Machida, T., Maksyutov, S., McDonald, K. C., McNorton, J., Miller, P. A., Melton, J. R., Morino, I., Müller, J., Murguia-Flores, F., Naik, V., Niwa, Y., Noce, S., O'Doherty, S., Parker, R. J., Peng, C., Peng, S., Peters, G. P., Prigent, C., Prinn, R., Ramonet, M., Regnier, P., Riley, W. J., Rosentreter, J. A., Segers, A., Simpson, I. J., Shi, H., Smith, S. J., Steele, L. P., Thornton, B. F., Tian, H., Tohjima, Y., Tubiello, F. N., Tsuruta, A., Viovy, N., Voulgarakis, A., Weber, T. S., van Weele, M., van der Werf, G. R., Weiss, R. F., Worthy, D., Wunch, D., Yin, Y., Yoshida, Y., Zhang, W., Zhang, Z., Zhao, Y., Zheng, B., Zhu, Q., Zhu, Q., and Zhuang, Q.: The Global Methane Budget 2000–2017, Earth Syst. Sci. Data, 12, 1561–1623, https://doi.org/10.5194/essd-12-1561-2020, 2020.
Schubert, C. J., Vazquez, F., Lösekann-Behrens, T., Knittel, K., Tonolla, M., and Boetius, A.: Evidence for anaerobic oxidation of methane in sediments of a freshwater system (Lago di Cadagno), FEMS Microbiol. Ecol., 76, 26–38, https://doi.org/10.1111/j.1574-6941.2010.01036.x, 2011.
Segarra, K. E. A., Schubotz, F., Samarkin, V., Yoshinaga, M. Y., Hinrichs, K.-U., and Joye, S. B.: High rates of anaerobic methane oxidation in freshwater wetlands reduce potential atmospheric methane emissions, Nat. Commun., 6, 1–8, https://doi.org/10.1038/ncomms8477, 2015.
Sela-Adler, M., Herut, B., Bar-Or, I., Antler, G., Eliani-Russak, E., Levy, E., Makovsky, Y., and Sivan, O.: Geochemical evidence for biogenic methane production and consumption in the shallow sediments of the SE Mediterranean shelf (Israel), Cont. Shelf Res., 101, 117–124, https://doi.org/10.1016/j.csr.2015.04.001, 2015.
Shrestha, J., Rich, J. J., Ehrenfeld, J. G., and Jaffe, P. R.: Oxidation of ammonium to nitrite under iron-reducing conditions in wetland soils: Laboratory, field demonstrations, and push-pull rate determination, Soil Sci., 174, 156–164, https://doi.org/10.1097/SS.0b013e3181988fbf, 2009.
Sieber, C. M. K., Probst, A. J., Sharrar, A., Thomas, B. C., Hess, M., Tringe, S. G., and Banfield, J. F.: Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy, Nat. Microbiol., 3, 836–843, https://doi.org/10.1038/s41564-018-0171-1, 2018.
Sinke, A. J.C., Cornelese, A. A., Cappenberg, T. E., and Zehnder, A. J. B.: Seasonal variation in sulfate reduction and methanogenesis in peaty sediments of eutrophic Lake Loosdrecht, The Netherlands, Biogeochemistry, 16, 43–61, https://doi.org/10.1007/BF02402262, 1992.
Sivan, O, Adler, M., Pearson, A., Gelman, F., Bar-Or, I., John, S. G., and Eckert, W.: Geochemical evidence for iron-mediated anaerobic oxidation of methane, Limnol. Oceanogr., 56, 1536–1544, https://doi.org/10.4319/lo.2011.56.4.1536, 2011.
Stookey, L. L.: Ferrozine-a new spectrophotometric reagent for iron, Anal. Chem., 42, 779–781, https://doi.org/10.1021/ac60289a016, 1970.
Sturt, H. F., Summons, R. E., Smith, K., Elvert, M., and Hinrichs, K.-U.: Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry – New biomarkers for biogeochemistry and microbial ecology, Rapid Commun. Mass Sp., 18, 617–628, https://doi.org/10.1002/rcm.1378, 2004.
Su, G., Zopfi, J., Yao, H., Steinle, L., Niemann, H., and Lehmann, M. F.: Manganese/iron-supported sulfate-dependent anaerobic oxidation of methane by archaea in lake sediments, Limnol. Oceanogr., 65, 863–875, https://doi.org/10.1002/lno.11354, 2020.
Tamames, J. and Puente-Sánchez, F.: SqueezeMeta, A Highly Portable, Fully Automatic Metagenomic Analysis Pipeline, Front. Microbiol., 9, 3349, https://doi.org/10.3389/fmicb.2018.03349, 2019.
Tan, X., Xie, G. J., Nie, W. B., Xing, D-F., Liu, B. F., Ding, J., and Ren, N. Q.: Fe(III)-mediated anaerobic ammonium oxidation: A novel microbial nitrogen cycle pathway and potential applications, Crit. Rev. Environ. Sci. Technol., 0, 1–33, https://doi.org/10.1080/10643389.2021.1903788, 2021.
Timmers, P. H. A., Welte, C. U., Koehorst, J. J., Plugge, C. M., Jetten, M. S. M., and Stams, A. J. M.: Reverse Methanogenesis and Respiration in Methanotrophic Archaea, Archaea, 2017, 1654237, https://doi.org/10.1155/2017/1654237, 2017.
Treude, T., Niggemann, J., Kallmeyer, J., Wintersteller, P., Schubert, C. J., Boetius, A., and Jørgensen, B. B.: Anaerobic oxidation of methane and sulfate reduction along the Chilean continental margin, Geochim. Cosmochim. Ac., 69, 2767–2779, https://doi.org/10.1016/j.gca.2005.01.002, 2005.
Treude, T., Krause, S., Maltby, J., Dale, A. W., Coffin, R., and Hamdan, L. J.: Sulfate reduction and methane oxidation activity below the sulfate-methane transition zone in Alaskan Beaufort Sea continental margin sediments: Implications for deep sulfur cycling, Geochim. Cosmochim. Ac., 144, 217–237, https://doi.org/10.1016/j.gca.2014.08.018, 2014.
Valentine D. L.: Biogeochemistry and microbial ecology of methane oxidation in anoxic environments: A review, A. van Leeuw. J. Microb., 81, 271–282, https://doi.org/10.1023/A:1020587206351, 2002.
Valenzuela, E. I., Prieto-Davó, A., López-Lozano, N. E., Hernández-Eligio, A., Vega-Alvarado, L., Juárez, K., García-González, A. S., López, M. G., and Cervantes, F. J.: Anaerobic methane oxidation driven by microbial reduction of natural organic matter in a tropical wetland, Appl. Environ. Microbiol., 83, 1–15, https://doi.org/10.1128/AEM.00645-17, 2017.
Valenzuela, E. I., Avendaño, K. A., Balagurusamy, N., Arriaga, S., Nieto-Delgado, C., Thalasso, F., and Cervantes, F. J.: Electron shuttling mediated by humic substances fuels anaerobic methane oxidation and carbon burial in wetland sediments, Sci. Total Environ., 650, 2674–2684, https://doi.org/10.1016/j.scitotenv.2018.09.388, 2019.
Vigderovich, H.: Data sets for Vigderovich et al., 2022, Zenodo [data set], https://doi.org/10.5281/zenodo.6489360, 2022.
Vigderovich, H., Liang, L., Herut, B., Wang, F., Wurgaft, E., Rubin-Blum, M., and Sivan, O.: Evidence for microbial iron reduction in the methanic sediments of the oligotrophic southeastern Mediterranean continental shelf, Biogeosciences, 16, 3165–3181, https://doi.org/10.5194/bg-16-3165-2019, 2019.
Wang, L., Miao, X., Ali, J., Lyu, T., and Pan, G.: Quantification of Oxygen Nanobubbles in Particulate Matters and Potential Applications in Remediation of Anaerobic Environment, ACS Omega, 3, 10624–10630, https://doi.org/10.1021/acsomega.8b00784, 2018.
Wegener, G., Niemann, H., Elvert, M., Hinrichs, K.-U., and Boetius, A.: Assimilation of methane and inorganic carbon by microbial communities mediating the anaerobic oxidation of methane, Environ. Microbiol. 10, 2287–2298, https://doi.org/10.1111/j.1462-2920.2008.01653.x, 2008.
Wegener, G., Gropp, J., Taubner, H., Halevy, I., and Elvert, M.: Sulfate-dependent reversibility of intracellular reactions explains the opposing isotope effects in the anaerobic oxidation of methane, Sci. Adv., 7, 1–14, https://doi.org/10.1126/sciadv.abe4939, 2021.
Whiticar, M. J., Faber, E., and Schoell, M.: Biogenic methane formation in marine and freshwater environments: CO2 reduction vs. acetate fermentation-Isotope evidence, Geochim. Cosmochim. Ac., 50, 693–709, https://doi.org/10.1016/0016-7037(86)90346-7, 1986.
Wu, Y. W., Tang, Y.-H., Tringe, S. G., Simmons, B. A., and Singer, S. W.: MaxBin: an automated binning method to recover individual genomes from metagenomes using, Microbiome, 2, 4904–4909, 2014.
Wuebbles, D. J. and Hayhoe, K.: Atmospheric methane and global change, Earth-Sci. Rev., 57, 177–210, https://doi.org/10.1016/S0012-8252(01)00062-9, 2002.
Xu, Z., Masuda, Y., Wang, X., Ushijima, N., Shiratori, Y., Senoo, K., and Itoh, H.: Genome-Based Taxonomic Rearrangement of the Order Geobacterales Including the Description of Geomonas azotofigens sp. nov. and Geomonas diazotrophica sp. nov, Front. Microbiol., 12, 737531, https://doi.org/10.3389/fmicb.2021.737531, 2021.
Yorshansky, O.: Iron Reduction in Deep Marine Sediments of the Eastern Mediterranean Continental Shelf and the Yarqon Estuary, M.S. thesis, Ben Gurion University of the Negev, https://doi.org/10.5281/zenodo.6457287, 2019.
Yoshinaga, M. Y., Holler, T., Goldhammer, T., Wegener, G., Pohlman, J. W., Brunner, B., Kuypers, M. M. M., Hinrichs, K.-U., and Elvert, M.: Carbon isotope equilibration during sulphate-limited anaerobic oxidation of methane, Nat. Geosci., 7, 190–194, https://doi.org/10.1038/ngeo2069, 2014.
Zehnder, A. J. and Brock, T. D.: Methane formation and methane oxidation by methanogenic bacteria, J. Bacteriol., 137, 420–432, https://doi.org/10.1128/jb.137.1.420-432.1979 1979.
Zhang, X., Xia, J., Pu, J., Cai, C., Tyson, G. W., Yuan, Z., and Hu, S.: Biochar-Mediated Anaerobic Oxidation of Methane, Environ. Sci. Technol., 53, 6660–6668, https://doi.org/10.1021/acs.est.9b01345, 2019.
Zheng, Y., Wang, H., Liu, Y., Zhu, B., Li, J., Yang, Y., Qin, W., Chen, L., Wu, X., Chistoserdova, L., and Zhao, F.: Methane-Dependent Mineral Reduction by Aerobic Methanotrophs under Hypoxia, Environ. Sci. Technol. Lett., 7, 606–612, https://doi.org/10.1021/acs.estlett.0c00436, 2020.