the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Dark septate endophytic fungi associated with pioneer grass inhabiting volcanic deposits and their functions in promoting plant growth
Han Sun
Tomoyasu Nishizawa
Hiroyuki Ohta
Kazuhiko Narisawa
Growth of the pioneer grass Miscanthus condensatus, one of the first vegetation types to be established on volcanic deposits, is promoted by root-associated fungi, particularly dark septate endophytes (DSEs). Fungal taxa within DSEs colonize the root of Miscanthus condensatus in oligotrophic Andosol, and their function in plant growth promotion remains largely unknown. We, therefore, comprehensively assessed the composition of the DSE community associated with Miscanthus condensatus root in volcanic ecosystems using the approaches of both metabarcoding (next-generation sequencing) and isolation (culturing). Also, the promotion effects of DSEs on plant growth (rice as a proxy) were evaluated by inoculation of core isolates to rice roots. Here, we found the following: (i) 70 % of culturable fungi that colonized Miscanthus condensatus phylogenetically belonged to DSEs; (ii) seven orders were identified by both sequencing and culturing methods; and (iii) inoculation of DSE isolates (Phialocephala fortinii, P. helvetica, and Phialocephala sp.) validated their effects on rice growth, particularly under an extremely low pH condition (compared to the control without inoculation, rice biomass was enhanced 7.6-fold after inoculation of P. fortinii). This study helps improve our understanding of the community of Miscanthus condensatus-associated DSE fungi and their functions in promoting plant growth.
- Article
(2050 KB) - Full-text XML
- BibTeX
- EndNote
Numerous studies have demonstrated that symbiotic fungi play a significant role in the establishment of pioneer vegetation in harsh environments or agricultural soils with extremely low pH. The association of these fungal micro-organisms that promote plant colonization is significant in extreme conditions, as these fungal symbionts help plant survival mainly by improving host nutrient uptake (Usuki and Narisawa, 2007; Yadav et al., 2009), defending against pathogens (Busby et al., 2016), promoting tolerance to abiotic stress (Rodriguez et al., 2008; Gill et al., 2016), and modifying trophic interactions (Clay, 1996; Omacini et al., 2001; Bultman et al., 2003).
One of the most common groups of monocotyledonous root endophytes is dark septate endophytes (DSEs), which usually colonize intracellularly and intercellularly in the tissues of more than 600 living herbaceous and woody plant species (Jumpponen and Trappe, 1998). DSEs, which are characterized by their morphology of melanized septate hyphae and structure like microsclerotia, also confer the ability to improve plant performance through enhanced nutrient uptake and have increased ability to withstand adverse environmental conditions (Khastini and Jannah, 2021). Increasing evidence shows that DSEs gradually become the most prevalent root colonizers under extreme environmental conditions of different ecosystems (Haruma et al., 2021; Yu et al., 2021). For example, Huusko et al. (2017) reported DSE-dominated colonization in Deschampsia flexuosa roots along a postglacial land uplift gradient. Gonzalez Mateu et al. (2020) reported that DSE-inoculated Phragmites australis had higher aboveground biomass under mesohaline conditions. DSEs, e.g., Phialocephala fortinii, promote host plant growth and adaptation to the hostile environment by (i) increasing resistance to heavy-metal contamination and heat and/or drought stress via producing melanized cell walls (Li et al., 2018; Haruma et al., 2021) and (ii) facilitating uptake of nutrients such as nitrogen and phosphorous (Jumpponen et al., 1998; Surono and Narisawa, 2017).
Wild plant species may live in symbiosis with mycoflora that may have been lost during breeding of the cultivars used in agriculture (Yuan et al., 2010), whilst some of symbiotic fungi that can assist plants in adapting to a given stress in a natural habitat might increase tolerance of crop species to that stress in an agricultural system. Thus, from an agricultural point of view, the plant symbiotic fungi could be seen as an extended source for crop adaptation and growth in agronomy. In attempts to domesticate “wild” symbiotic fungi (associated with genetically wild type plants), some of these DSE species in natural systems have been successfully transferred to agricultural species from their original host, providing benefits to the inoculated crops (Toju et al., 2018).
Rice (Oryza sativa) is the principal food grain crop (one of the four major food crops) for more than 3 billion people, and its consumption exceeds 100 kg per capita annually in many Asian countries (Yuan et al., 2010). During the last several decades, there have been major climatic events, including global warming, soil acidification, etc., that have influenced the agricultural productivity of rice around the world. Soil pH is a highly sensitive factor to determine plant survival, distribution, and interactions with micro-organisms, which are vital for the availability of essential nutrients and plant growth (Luo et al., 2013). About 13 % of the world's rice is produced in acid soil. Compared with other crops, rice has relatively strong Al toxic resistance (Famoso et al., 2010) and is also the most complex cereal crop with Al resistance genes (Ma et al., 2002). Nevertheless, as for other crops, heavy-metal toxicity in acid soil limits rice growth and nutrient uptake and subsequently reduces grain yield (Chen et al., 2020). The optimal pH range for rice growth is 5.0–8.5, which shows a likely reduction in yield in the soil with the extended pH range (Ma et al., 2002). To improve this soil acidity, liming is often used but is practically difficult and unsustainable.
Micro-organism inoculation is a sustainable approach to potentially promote plant resistance to acidic stress. For instance, plant-associated fungi, such as arbuscular mycorrhizal fungi (AM fungi), reportedly play a key role in the protection of plants in acidic soils (Toju et al., 2018). Yet, high concentrations of H+ and Al3+ can inhibit hyphal growth and spore germination in AM fungi, thereby decreasing the possibility of colonizing plant roots (Clark, 1997; Van Aarle et al., 2002; Postma et al., 2007). Comparably, DSEs show marked potential to help host plants resist acidity because of their higher H+ tolerance than other colonizing fungi (Postma et al., 2007). Still, there is a lack of reports of DSEs improving host crop (e.g., rice) growth under acidic conditions, especially in extremely acidic conditions (pH 3.0).
Re-vegetation in volcanic soil, characterized by a dominance of biological processes, is difficult due to (i) strong acidity of volcanic deposits, (ii) high concentration of toxic elements, and (iii) deficiencies in essential nutrients. Miscanthus sinensis, an unique pioneer grass plant during recovery after volcanic eruption, is the first to be established on volcanic deposits and is frequently found as primary vegetation in lahar deposited by volcanic eruptions (Watanabe et al., 2006; Hirata et al., 2007; An et al., 2008; Ezaki et al., 2008). This is because M. sinensis can tolerate a wide range of environmental stresses due to the trait of C4 photosynthesis, leading to high productivity and a low nutrient requirement (Stewart et al., 2009). Apart from Miscanthus traits that adapt to the volcanic soil, the root-associated fungal communities are widely reported to benefit the growth of host plants and promote their adaptation to stress, such as aridity (Wu and Xia, 2006), salinity (Porcel et al., 2012), and oligotrophic conditions (Jeewani et al., 2021). A better understanding of plant–microbe interactions, therefore, can help improve our understanding of vegetation recovery and plant growth promotion including in agricultural application scenarios. The isolation and culture of fungal species, therefore, are indispensable as they complement taxonomic databases and validate taxa revealed by sequencing. Bai et al. (2015) established Arabidopsis root-derived bacterial culture collections representing the majority of species that were reproducibly detectable by culture-independent community sequencing. Laval et al. (2021) investigated fungal and bacterial communities in soils receiving wheat and oilseed rape residues, and they confirmed the feasibility of combined culture–unculture approaches that revealed consistent community profiles. The role of keystone taxa revealed by the sequencing-data-based co-occurrence network can be further validated by culturing and subsequent inoculation. For example, isolation was used to test whether the interaction between micro-organisms predicted by metagenomic sequencing actually occurs (Laval et al., 2021). By isolation and inoculation, Zheng et al. (2021) identified the strong decomposition ability of keystone taxa such as the genera Chryseobacterium (bacteria) and Fusarium, Aspergillus, and Penicillium (fungi), which are consistent with the keystone taxa revealed by the co-occurrence network. Combining sequencing and culturing methods, therefore, is powerful for the identification of putative taxa (either individually or via the creation of synthetic communities). Yet studies on DSEs in volcanic ecosystems by culture–unculture approaches are lacking, and inoculation to validate the function in rice growth still awaits further investigation.
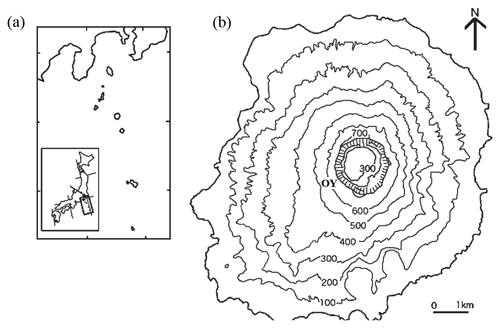
Figure 1(a) DEM map showing the location of Miyake-jima on the western rim of the Pacific Ocean. (b) DEM map showing study site OY near the summit crater in Miyake-jima.
For this purpose, both culture-dependent and culture-independent approaches were adopted to comprehensively reveal the fungal communities of Miscanthus-associated fungi, particularly DSEs, of volcanic ecosystems. Their functions in promoting plant growth (via isolation–inoculation) in different-pH soils were further evaluated. This study, therefore, aimed to (i) reveal the fungal taxa associated with the roots of M. condensatus during vegetation recovery by a combination of sequencing and culturing approaches and (ii) inoculate the major food crop, i.e., rice, with these indigenous isolates (overlapped with sequencing-revealed taxa) to evaluate their effects on rice growth, in particularly under low-pH conditions. We hypothesized that prevalent colonization by DSE fungi occurs in the pioneer grass M. condensatus inhabiting volcanic deposits near the crater of Miyake-jima, due to DSE traits of preferential colonization under oligotrophic and acidic conditions.
2.1 Study site description and root sampling
Miyake-jima (55.5 km2 in area; highest point, 775 m), a basaltic volcanic island (34∘05′ N, 139∘31′ E; Fig. 1), belongs to the Fuji volcanic southern zone in the east Japan volcanic belt. As a model active basaltic volcanic island with an eruption in 2000, it ejected large amounts of volcanic ash and gases such as sulfur dioxide and hydrogen sulfide (60 % of vegetation on the island was affected) (Yamanishi et al., 2003; Guo et al., 2014). As a result of SO2 gas exposure, volcanic ash deposits were acidified due to SO absorption. They were characterized by strong acidity, with high levels of exchangeable Ca2+ and Al3+ (Fujimura et al., 2016). Mount Ōyama is an active volcano, located in the center of the island. A large amount of volcanic SO2 gas (∼54 kt d−1) was ejected immediately from a newly created summit caldera after the latest eruption in 2000 (Fujimura et al., 2016). The SO2 gas exposure declined slowly after the eruption, and as a result of this exposure, the volcanic ash deposits were acidified due to SO absorption. They were characterized by strong acidity (pH (H2O) 3.1–4.0), with high levels of exchangeable Ca2+ (33.5–115 cmol kg−1) and Al3+ (0.8–10.2 cmol kg−1) (Fujimura et al., 2016). At 18 years after the eruption, the patchy vegetation of a pioneer grass, Miscanthus condensatus, was established at the OY site near the Miyake-jima summit crater (34∘04′ N, 139∘31′ E; 553 m a.s.l.; Fig. 1). The rhizosphere soils of Miscanthus condensatus were collected at the OY site in November 2017 and March and September 2018. From each period, three healthy specimens of M. condensatus were collected, kept in sterile plastic bags, and immediately stored on ice. Samples were divided into two portions: (1) the first kept at 4 ∘C and processed within 48 h of collection for isolation and (2) the second kept at −20 ∘C until DNA extraction and molecular analysis.
2.2 Root surface sterilization and culturable endophytic fungal isolation
In order to remove adhering soil and free-living microbes, root surface sterilization was performed by modifying the method of Sahu et al. (2022). Root samples were gently rinsed with tap water. Individual roots were severed aseptically in 1 cm long sections with a sterile scalpel and put into 50 mL conical centrifuge tubes. Then they were superficially sterilized with 0.005 % Tween 20 and rinsed with sterilized distilled water before the aseptic stepwise sterilization process was carried out. Root sections were treated with 70 % ethanol for 1 min; with a further step in the above process, 1 % sodium hypochlorite was added and sections were sterilized for 5 min. Finally, sections were rinsed with sterilized distilled water three times (Sahu et al., 2022).
After surface sterilization, the final wash was spread-plated onto cornmeal agar medium (cornmeal, Difco 25 g L−1, Bacto agar, Difco 15 g L−1) to confirm the disinfection and incubated for 2 weeks at 23 ∘C to check for the presence of a growth colony. Root sections were dried with sterile filter paper overnight and then placed onto cornmeal agar medium containing 0.1 mg kg−1 streptomycin and incubated at 23 ∘C for 2 weeks. When endophytic fungal growth was observed, the mycelia were immediately transferred to a new plate. An isolate was transferred only when the probability of a good pure culture was considered high. Thus, when the strains originated very close to each other and, in later stages, when they overgrew earlier strains, they were left untransferred and were not calculated in the total number of the isolates. After incubation, pure cultures were obtained by transferring single hyphae to cornmeal malt yeast agar medium (CMMY; malt extract 10 g L−1, yeast extract 2 g L−1, cornmeal 8.5 g L−1, Bacto agar 7.5 g L−1).
2.3 Identification of fungal isolates and phylogenetic analysis
Genomic DNA from each fungal isolate was extracted from mycelium using PrepMan Ultra Sample Preparation Reagent protocol (Applied Biosystems, California, USA). The universal primer pairs of ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) hybridize at the end of 18S rDNA, and the primer ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) that hybridizes at the beginning of 28S rDNA was used to amplify fungal isolates (Mahmoud and Narisawa, 2013). PCR amplification was carried out in a 50 µL reaction mixture containing 1 µL of fungal genomic DNA, 2.5 µL of each primer, 5 µL of 10× Ex Taq buffer, 4 µL of dNTP, 0.25 µL of Ex Taq DNA polymerase, and 34.75 µL of sterilized Milli-Q water under thermal conditions of 4 min at 94 ∘C, 35 cycles of 94 ∘C for 35 s, 52 ∘C for 55 s, and 72 ∘C for 2 min, with a final extension of 72 ∘C for 10 min using a TaKaRa PCR Thermal Cycler Dice (TaKaRa Bio Inc., model TP600, Japan). The PCR products were purified and sequenced using an Applied Biosystems 3130xl DNA sequencer. All sequences obtained were compared with similar DNA sequences retrieved from the GenBank database using the NCBI BLASTN program. The nucleotide sequences from three dominant DSE isolates (NH1121, NH1221, and NH1331) were deposited in the NCBI GenBank database under accession numbers OR743765–OR743767.
2.4 Illumina MiSeq sequencing for culture-independent identification
Roots of samples which were collected in November were added to 10 mL aliquots of sterile distilled water and macerated with a pestle and mortar for DNA extraction with a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. DNA was purified using an UltraClean DNA Purification Kit (MO BIO, Carlsbad, California, USA). Then DNA was eluted in 50 µL of tris and EDTA buffer. A NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, Delaware, USA) was used to quantify the DNA concentration. Finally, DNA samples were stored at −80 ∘C before molecular analysis. The second nuclear ribosomal internal transcribed spacer (ITS2) region of the rRNA operon was targeted using the fungal-specific primer pairs ITS3 (5′-GCATCGATGAAGAACGCAGC-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Chen et al., 2021). PCR amplification was carried out in triplicate with 50 µL reactions containing 25 µL of Premix Taq (TaKaRa, Shiga, Japan), 23 µL of sterilized Milli-Q water, 0.5 µL of both forward and reverse primers (125 pmol), and 1 µL of template DNA. The PCR program had the following thermocycling conditions: 35 cycles of denaturation at 94 ∘C for 30 s, annealing at 54 ∘C for 30 s, 72 ∘C for 45 s, and a final extension of 72 ∘C for 10 min. PCR products were pooled, and their relative quantity was estimated by running 5 µL of amplicon DNA on 1.5 % agarose gel, and products were purified with a QIAquick PCR Purification Kit (Qiagen, Shenzhen, China). The purified mixture was diluted and denatured to obtain an 8 pmol amplicon library and mixed with an equal volume of 8 pmol PhiX (Illumina) following the manufacturer's recommendations in the Illumina MiSeq reagent kit preparation guide (Illumina, San Diego, California, USA). Finally, 600 µL of the amplicon mixtures was loaded with read 1, read 2, and the index sequencing primers. The paired-end sequencing (each 250 bp) was completed on a MiSeq platform (Illumina). The sequencing data were processed using the UPARSE pipeline (http://drive5.com/usearch/manual/uparse_pipeline.html, last access: 1 March 2019). The raw sequences were subjected to quality control. The singleton and chimeric sequences were removed after dereplication, and the remaining sequences were categorized into operational taxonomic units (OTUs) with 97 % similarity and then assigned taxonomy using the UNITE database (https://unite.ut.ee/, last access: 1 March 2019).
2.5 Inoculation
The experiment was conducted as a complete randomized factorial design with two factors. The first factor had four levels: non-inoculation control or inoculation with three dominant isolates (Phialocephala fortinii, P. helvetica, and Phialocephala sp.); and the second factor had three levels: pH 3, 4, and 5. Each treatment consisted of four replicates with two plants per pot, thus totaling 48 experimental pots in the study. Fungal inoculates were prepared by aseptically growing three dominant DSE isolates on Petri dishes with oatmeal agar medium (10 g L−1 oatmeal and 15 g L−1 Bacto agar enriched with nutrients: 1 g L−1 MgSO4.7H2O, 1.5 g L−1 KH2PO4, and 1 g L−1 NaNO3). Due to the host non-specific character of DSEs, rice was chosen as a host plant in this study mostly for its important role in consumed cereal in the world and because it is from the same family as Miscanthus. Rice seeds were surface-sterilized by immersion in 70 % ethanol for 2 min and a solution of 1 % sodium hypochlorite for 5 min with agitation. The sterilized seeds were gently rinsed several times with sterilized distilled water, then dried overnight, and plated onto 1 % water agar medium in Petri dishes for germination at 30 ∘C. Following pre-germination, 2 d old seedlings (two seedlings per plate) were transplanted as growing fungal colonies on the medium at pH 3, 4, or 5. For DSE inoculation, two 5 mm plugs excised from the edge of an actively growing colony on culture medium were inoculated at a 1 cm range close to the rice seedlings. Seedlings transplanted onto non-inoculated medium were used as controls. The whole set was placed into sterile plastic culture bottles and incubated for 3 weeks at room temperature with an 18 h : 6 h (L : D (light : dark))) regimen and intensity of 180 . Assessed plants were harvested and oven-dried at 40 ∘C for 72 h. The shoot and root dry weights of treated plants were measured and compared with the control.
2.6 DSE root colonization observations
Root colonization by DSE fungal isolates was observed to confirm whether the selected DSEs colonized the inner roots endophytically. Roots were harvested from plants after 3 weeks of cultivation. Root systems were washed thoroughly under running tap water to remove adhering agar, then rinsed with distilled water, and used for root staining. The root samples were cleared with 10 % () potassium hydroxide in a water bath at 80 ∘C for 20 min. Subsequently, roots were acidified with 1 % hydrochloric acid at room temperature for 5 min and then stained with 50 % acetic acid solution containing 0.005 % cotton blue at room temperature overnight. Root fragments were placed on a slide glass and covered with a cover glass. Fungal colonization was observed using a light microscope equipped with an Olympus DP25 digital camera.
2.7 Statistical analyses
All statistical analyses were performed in the R environment (version V4.1.2). Homoscedasticity was checked using Levene's test and normality using the Shapiro–Wilk test. The differences in mean dry biomass between the analyzed traits of the seedlings in different treatments in this study were calculated and analyzed statistically with two-way analysis of variance (ANOVA) and Tukey's honestly significant difference test at P values <0.05.
3.1 The core fungal taxa identified by both culture-dependent and culture-independent methods
This study compared the culture-dependent isolates with the fungal taxa revealed by culture-independent methods. Based on 97 % sequence similarity, all reads were clustered into 224 OTUs, and the valid sequences were classified into five phyla, including two major dominant phyla of Ascomycota (71.5 %) and Basidiomycota (17.1 %), followed by Mortierellomycota, Mucoromycota, and Calcarisporiellomycota, while the cultivable endophytic fungi were classified into two different phyla of Ascomycota (97.5 %) and Basidiomycota (2.50 %). Fifteen and four classes were detected by culture-independent and culture-dependent approaches, respectively. Specifically, classes Sordariomycetes and Leotiomycetes (both belonging to phylum Ascomycota) were the major classes in terms of the number of OTUs. These data were in agreement with a previous study showing that Leotiomycetes and Sordariomycetes were the major classes of endophytic fungi associated with plants (regardless of plant species, associated host tissue) in acidic, oligotrophic ecosystems and nutrient-limiting boreal and arctic areas (Arnold et al., 2007; Yuan et al., 2010; Ghimire et al., 2011; Luo et al., 2014; Knapp et al., 2019).
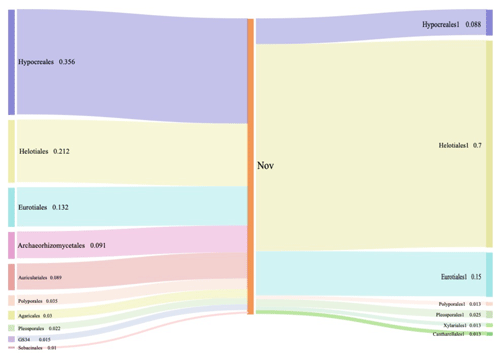
Figure 2Composition and relative abundance of endophytic fungi at the order level by culture-independent (left) and culture-dependent (right) methods.
While looking at the lower level, 27 orders were found by Illumina-based sequencing analysis, and 10 of them had an average abundance over 1 %. Among these orders detected by sequencing, seven orders were identified via culture-dependent methods as well (Fig. 2). Significantly higher proportions of Hypocreales (35.6 %), Helotiales (21.2 %), and Eurotiales (13.2 %) were observed by Illumina-based analysis (Fig. 2). Through culture-dependent methods, an abundance of Helotiales (70.0 %) occupied the whole community, followed by Eurotiales (15.0 %) and Hypocreales (8.75 %). In general, the abundant orders of fungal isolates also showed abundance in the OTU table generated by high-throughput sequencing. The overlapping of taxa (Hypocreales and Helotiales) identified by both approaches suggests their significance and dominance in Miscanthus condensatus-associated fungal communities. Similarly, the key fungal and bacterial communities in soils amended with wheat and oilseed residues were identified via culture and non-culture approaches (Laval et al., 2021). Several other studies have also confirmed the feasibility of revealing major microbial taxa and have shown the marked potential of adopting the combination of both culture and non-culture approaches to identify putative taxa (Laval et al., 2021; Bai et al., 2015; Zheng et al., 2021). Undoubtedly a combination of culture-dependent and culture-independent methods might provide a powerful strategy to identify and obtain novel endophytes.
Table 1Summary of the endophytic fungal isolates among 3 months of sampling in Miscanthus condensatus.
The overlapping order Helotiales identified by both culture-dependent methods was abundant in the Miscanthus condensatus-associated fungal community (Fig. 2). The isolates including P. fortinii, P. helvetica, and Phialocephala sp. belonged to Helotiales species, which are highly conserved and found to be co-occurring species in the root symbiont communities based on Sanger sequencing (Walker et al., 2011; Bruzone et al., 2015). This study also found these fungi, i.e., Phialocephala sp., P. helvetica, and P. fortinii, in all samples irrespective of the sampling period (Table 1). Previous studies isolated P. fortinii from the root of Pinus resinosa (Wang and Wilcox, 1985), Vaccinium vitis-idaea, Betula platyphylla var. japonica, Luetkea pectinata (Addy et al., 2000), Picea abies, Betula pendula (Menkis et al., 2004), Rhododendron sp. (Grünig et al., 2008), Chamaecyparis obtusa, and Rubus sp. (Surono and Narisawa, 2017). Yet, the phylogeny and ecological effects of P. fortinii on plant quality still remain largely unknown (Tedersoo et al., 2009). For example, P. fortinii itself is genotypically diverse and composed of at least 21 morphologically indistinguishable but genetically isolated cryptic species (CSP) (Grünig et al., 2008). Up to seven isolates belonging to P. fortinii have been formally described as CSP (Grünig et al., 2008). Phialocephala helvetica (sub-species of P. fortinii), associated with the root of Picea abies (Stroheker et al., 2021) and Pinus sylvestris (Landolt et al., 2020), is regarded as one of the most common CSP. Yet, its functions in promoting plant growth remain largely unknown.
3.2 Colonization of DSE fungal isolates in plant root
Isolating and characterizing micro-organisms could provide insights into their phylogenetic identification, physiological properties, and metabolic potential, which will help in understanding the formation, persistence, adaptation mechanisms, and ecological functions of microbial communities (Li et al., 2019). Therefore, these three most promising isolates of Phialocephala sp., P. fortinii, and P. helvetica, as typical DSEs, were further examined regarding their effects on growth-promoting activity for plants. Based on the inoculation test, all rice seedlings exhibited healthy growth throughout the experimental period by fungal isolate × agar pH interaction (Fig. 3).

Figure 3Growth and development of rice plants inoculated with DSE fungal isolates under different pH conditions.
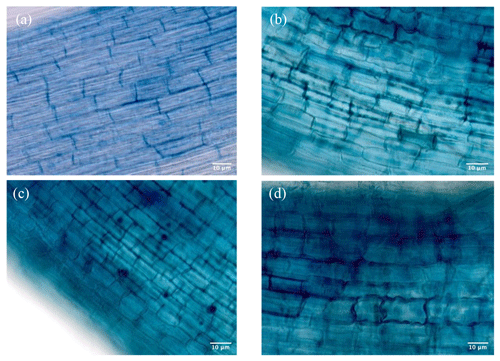
Figure 4(a) Non-treated DSEs as control roots. (b) Phialocephala sp. (NH1121)-treated roots. (c) Phialocephala helvetica (NH1221)-treated roots. (d) Phialocephala fortinii (NH1331)-treated roots.
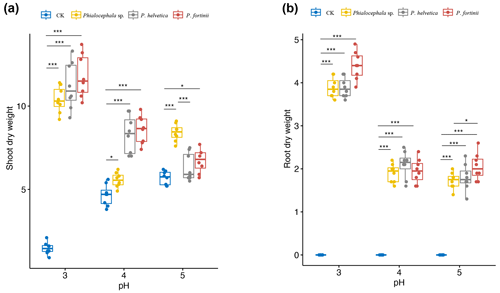
Figure 5Shoot and root dry weights of rice seedlings inoculated with NH1121 (Phialocephala sp.), NH1221 (Phialocephala helvetica), and NH1331 (Phialocephala fortinii) after 3 weeks of growth on oatmeal agar at pH 3, 4, or 5 (acidic conditions). There are biological replicates (n=8). Median values are lines across the box, with lower and upper boxes indicating the 25th and 75th percentiles, respectively. Whiskers represent the maximum and minimum values. Significance was determined by ANOVA. * P<0.05. ** P<0.01. *** P<0.001. “CK” means control.
After harvesting, the roots were stained with 0.05 % cotton blue to determine the endophytism of DSE isolates. Microscopic observation revealed that all DSE isolates successfully colonized hair roots of rice seedlings. The hair roots were coated with loose wefts of fungal hyphae. This feature was identical to that previously described for typical DSEs; i.e., they are characterized by microsclerotia and thick and darkly pigmented septate hyphae in the hair roots. Non-inoculated plants as a control showed no DSE colonization. The root colonization pattern was similar in P. fortinii and P. helvetica, but the degree of fungal colonization of Phialocephala sp. was the lowest compared with those two isolates. The images show the dense networks of hyphae of DSE inter- and intracellularly colonizing rice roots (Fig. 4). Very few studies, however, have investigated the role and ecological significance of isolated DSEs underlying plant growth.
3.3 The role of isolated DSEs in rice growth promotion
As rice is the most important of the four major food crops for most Asian people, to domesticate these isolated wild DSEs can benefit agricultural production. Thus we transferred these DSE isolates from their original hosts of Miscanthus condensatus to agricultural species (rice). DSEs are widely reported to be characterized by non-host-specific traits, but different hosts (cross-family) may have different responses (in terms of morphology) to inoculated isolates. For example, P. fortinii is frequently reported in roots and forms typical ectomycorrhizae with members of the Pinaceae plants (Jumpponen et al., 1998). In contrast, for other family plants, P. fortinii is often found to be an endophytic fungi. In addition, Cladophialophora chaetospira was reported to be able to develop and form spiral structures resembling ericoid mycorrhizae within the roots of ericaceous plants (Usuki and Narisawa, 2005), whilst C. chaetospira colonizing in other host families is characterized by the formation of microsleclerotia aggregations of irregularly lobed hyphae and dark septate hyphae growing inter- and intracellularly. Considering that both plants used in this study as a host (e.g., Miscanthus and rice) belong to the same family of grass with similar host responses to DSEs (showing non-host-specific traits), we aimed to test the effects of these isolated DSEs in crop (rice as a proxy) growth promotion. Here, we found that the shoot biomass of rice inoculated with DSE isolates increased up to 7.6-fold compared with non-inoculated controls (Fig. 5). The greatest shoot dry weight was recorded in plants treated with P. fortinii, followed by P. helvetica and Phialocephala sp. Similarly, P. fortinii isolates were used to inoculate asparagus plants and promote plant growth; e.g., shoot biomass increased by up to 53.5 % (Surono and Narisawa, 2017). The beneficial effects of P. fortinii on enhancing plant yield have been reported (Jumpponen et al., 1998; Jumpponen and Trappe, 1998).
This improvement in plant growth may be related to the ability of these isolates to use organic nitrogen sources under nitrogen-deficient conditions. Low nitrogen uptake by plants is associated with soil acidity. The presence of P. fortinii associated with plant tissues demonstrated its ability to produce a variety of extracellular enzymes that break down complex forms of organic matter containing nitrogen and phosphorus (Jumpponen et al., 1998). For example, Cladophialophora chaetospira activates soil nitrogen and promotes aboveground transfer in Chinese cabbage (Usuki and Narisawa, 2007). Therefore, the most abundant DSEs identified by both culture and non-culture approaches, acting as an important mycorrhizal symbiont via melanized septate hyphae formation that removes resource limitation, might promote plant growth. A labeled nitrogen study is required to validate this mechanism.
Rice growth was markedly different depending on the combination of DSE isolates and pH. Differences in the dry weight of DSE-inoculated rice compared with non-inoculated rice grown at pH 3.0 (as high as 7.6-fold) were significantly greater than for that DSE-inoculated rice grown at pH 4.0 and 5.0 (as high as 1.6-fold and 1.2-fold, respectively). In particular, the root dry weight of P. fortinii-treated seedlings was the highest at pH 3.0 with respect to that of the control. Also, we observed that inoculated species of Phialocephala effectively promoted plant growth, particularly under acidic conditions. The enhanced shoot biomass via DSE isolate inoculation was most marked in acidic environments, e.g., with 7.6, 1.6, and 1.2 times greater shoot biomass at pH 3, 4, and 5, respectively. Less promotion of plant growth by inoculation with Phialocephala at pH 5 compared with pH 4 and 3 agar indicated that these DSE isolates likely promote plant tolerance to soil acidity.
Many researchers have reported relatively narrow ranges of pH for the presence or activity of mycorrhizal fungi in soils (Clark, 1997; Postma et al., 2007). This is consistent with the observation that most colonized isolates associated with plants were found in acidic agar. Similarly, the colonization of investigated plants with DSEs significantly decreased with increasing soil pH (Postma et al., 2007). The mechanisms underlying the promotion of plant growth by DSE fungal have been addressed. DSE fungal might help adaptability of crop to acid stress, i.e., low soil pH, and subsequent support of plant growth. The relatively high abundance of DSEs supports host survival in stress habitats mainly via high chitin contents and forming melanized septate hyphae and microsclerotia in plant roots (Likar and Regvar, 2013). Also, it might increase the concentration of Mg, known to ameliorate Al toxicity, in the roots of M. sinensis to decrease Al activity (Haruma et al., 2021).
Here, we validated the effects of these DSE isolates on rice growth, particularly under an extremely low pH condition; e.g., compared to control without inoculation, rice biomass enhanced 7.6-fold after inoculation of P. fortinii. DSEs show great potential to help host crop resist acidity and thus enable crop cultivation, especially in acidic soil (Postma et al., 2007). Acidic soils occupy up to 50 % of the arable area worldwide, and around 13 % of paddy is acid soil. While soil acidification can be a problem for crop yield, these DSE isolates might be used as a management strategy to reduce acidic harm to crops. This, however, awaits field investigation.
Taken together, our findings help improve understanding of the community of Miscanthus condensatus-associated DSE fungi and their functions. Our results suggest that DSEs have the ability to support rice growth under extremely acidic conditions, and the formation of melanized septate hyphae and microsclerotia-associated rice tissues might promote increases in rice growth and root biomass via removing stress and resource limitations, and thus they show marked potential in not only re-vegetation of pioneer plants in post-volcanic ecosystems but also promotion of rice growth.
The present study provided detailed insights into the diversity and function of the endophytic fungal community in Miscanthus condensates, using both culture-dependent and culture-independent approaches. Here, we showed that the fungal community was dominated by isolates of Phialocephala, which were abundant and widely distributed in the volcanic deposits. Additionally, we validated the functions of these DSEs in rice growth, particularly under acidic conditions, by adopting the approach of isolation–inoculation. Considering that these fungal isolates promote plant adaptation to acidic soil, the identified DSEs, e.g., Phialocephala fortinii, P. helvetica, and Phialocephala sp., might be potential candidates as plant-growth-promoting fungi for either restoring vegetation or promoting rice growth under extreme conditions.
Sequences of the fungal communities presented in this work were deposited at the NCBI Sequence Read Archive (SRA) with the project number BioProject ID PRJNA1046139 and BioSample submission ID SUB13935530. Fungal ITS sequences have the accession numbers OR743765–OR743767 and are available from GenBank. Raw data and code can be obtained by contacting the author.
KN supervised the research and secured the funding. KN, HO, TN and HS planned and designed the research, analyzed and interpreted the data. HS performed the experiments, and wrote the manuscript. All authors critically reviewed and edited the manuscript.
The contact author has declared that none of the authors has any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
This research was supported by a Grant-in-Aid for Scientific Research (B) (grant no. 21H02191) from the Japan Society for the Promotion of Science (JSPS) and a Grant-in-Aid for Challenging Exploratory Research (grant no. 22K19164) from JSPS.
This research has been supported by the Japan Society for the Promotion of Science (Grant-in-Aid for Scientific Research (B) (grant no. 21H02191) and Grant-in-Aid for Challenging Exploratory Research (grant no. 22K19164)).
This paper was edited by Luo Yu and reviewed by two anonymous referees.
Addy, H. D., Hambleton, S., and Currah, R. S.: Distribution and molecular characterization of the root endophyte Phialocephala fortinii along an environmental gradient in the boreal forest of Alberta, Mycol. Res., 104, 1213–1221, https://doi.org/10.1017/S0953756200002896, 2000.
An, G.-H., Miyakawa, S., Kawahara, A., Osaki, M., and Ezawa, T.: Community structure of arbuscular mycorrhizal fungi associated with pioneer grass species Miscanthus sinensis in acid sulfate soils: Habitat segregation along pH gradients, Soil Sci. Plant Nutr., 54, 517–528, https://doi.org/10.1111/j.1747-0765.2008.00267.x, 2008.
Arnold, A. E., Henk, D. A., Eells, R. L., Lutzoni, F., and Vilgalys, R.: Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR, Mycologia, 99, 185–206, https://doi.org/10.1080/15572536.2007.11832578, 2007.
Bai, Y., Müller, D. B., Srinivas, G., Garrido-Oter, R., Potthoff, E., Rott, M., Dombrowski, N., Münch, P. C., Spaepen, S., Remus-Emsermann, M., Hüttel, B., McHardy, A. C., Vorholt, J. A., and Schulze-Lefert, P.: Functional overlap of the Arabidopsis leaf and root microbiota, Nature, 528, 364–369, https://doi.org/10.1038/nature16192, 2015.
Bruzone, M. C., Fontenla, S. B., and Vohník, M.: Is the prominent ericoid mycorrhizal fungus Rhizoscyphus ericae absent in the Southern Hemisphere's Ericaceae? A case study on the diversity of root mycobionts in Gaultheria spp. from northwest Patagonia, Argentina, Mycorrhiza, 25, 25–40, https://doi.org/10.1007/s00572-014-0586-3, 2015.
Bultman, T. L., McNeill, M. R., and Goldson, S. L.: Isolate-dependent impacts of fungal endophytes in a multitrophic interaction, Oikos, 102, 491–496, https://doi.org/10.1034/j.1600-0706.2003.11477.x, 2003.
Busby, P. E., Ridout, M., and Newcombe, G.: Fungal endophytes: modifiers of plant disease, Plant Mol. Biol., 90, 645–655, https://doi.org/10.1007/s11103-015-0412-0, 2016.
Chen, J., Akutse, K. S., Saqib, H. S. A., Wu, X., Yang, F., Xia, X., Wang, L., Goettel, M. S., You, M., and Gurr, G. M.: Fungal Endophyte Communities of Crucifer Crops Are Seasonally Dynamic and Structured by Plant Identity, Plant Tissue and Environmental Factors, Front. Microbiol., 11, 1519, https://doi.org/10.3389/fmicb.2020.01519, 2020.
Chen, Y., Liu, F., Kang, L., Zhang, D., Kou, D., Mao, C., Qin, S., Zhang, Q., and Yang, Y.: Large-scale evidence for microbial response and associated carbon release after permafrost thaw, Glob Change Biol., 27, 3218–3229, https://doi.org/10.1111/gcb.15487, 2021.
Clark, R. B.: Arbuscular mycorrhizal adaptation, spore germination, root colonization, and host plant growth and mineral acquisition at low pH, Plant Soil., 192, 15–22, https://doi.org/10.1023/A:1004218915413, 1997.
Clay, K.: Interactions among fungal endophytes, grasses and herbivores, Res. Popul. Ecol., 38, 191–201, https://doi.org/10.1007/BF02515727, 1996.
Ezaki, B., Nagao, E., Yamamoto, Y., Nakashima, S., and Enomoto, T.: Wild plants, Andropogon virginicus L. and Miscanthus sinensis Anders, are tolerant to multiple stresses including aluminum, heavy metals and oxidative stresses, Plant Cell Rep., 27, 951–961, https://doi.org/10.1007/s00299-007-0503-8, 2008.
Famoso, A. N., Clark, R. T., Shaff, J. E., Craft, E., McCouch, S. R., and Kochian, L. V.: Development of a Novel Aluminum Tolerance Phenotyping Platform Used for Comparisons of Cereal Aluminum Tolerance and Investigations into Rice Aluminum Tolerance Mechanisms, Plant Physiol., 153, 1678–1691, https://doi.org/10.1104/pp.110.156794, 2010.
Fujimura, R., Kim, S.-W., Sato, Y., Oshima, K., Hattori, M., Kamijo, T., and Ohta, H.: Unique pioneer microbial communities exposed to volcanic sulfur dioxide, Sci. Rep., 6, 19687, https://doi.org/10.1038/srep19687, 2016.
Ghimire, S. R., Charlton, N. D., Bell, J. D., Krishnamurthy, Y. L., and Craven, K. D.: Biodiversity of fungal endophyte communities inhabiting switchgrass (Panicum virgatum L.) growing in the native tallgrass prairie of northern Oklahoma, Fungal Divers., 47, 19–27, https://doi.org/10.1007/s13225-010-0085-6, 2011.
Gill, S. S., Gill, R., Trivedi, D. K., Anjum, N. A., Sharma, K. K., Ansari, M. W., Ansari, A. A., Johri, A. K., Prasad, R., Pereira, E., Varma, A., and Tuteja, N.: Piriformospora indica: Potential and Significance in Plant Stress Tolerance, Front. Microbiol., 7, 332, https://doi.org/10.3389/fmicb.2016.00332, 2016.
Gonzalez Mateu, M., Baldwin, A. H., Maul, J. E., and Yarwood, S. A.: Dark septate endophyte improves salt tolerance of native and invasive lineages of Phragmites australis, ISME J., 14, 1943–1954, https://doi.org/10.1038/s41396-020-0654-y, 2020.
Grünig, C. R., Queloz, V., Sieber, T. N., and Holdenrieder, O.: Dark septate endophytes (DSE) of the Phialocephala fortinii s.l. – Acephala applanata species complex in tree roots: classification, population biology, and ecology, Botany, 86, 1355–1369, https://doi.org/10.1139/B08-108, 2008.
Guo, Y., Fujimura, R., Sato, Y., Suda, W., Kim, S., Oshima, K., Hattori, M., Kamijo, T., Narisawa, K., and Ohta, H.: Characterization of Early Microbial Communities on Volcanic Deposits along a Vegetation Gradient on the Island of Miyake, Japan, Microb. Environ., 29, 38–49, https://doi.org/10.1264/jsme2.ME13142, 2014.
Haruma, T., Yamaji, K., and Masuya, H.: Phialocephala fortinii increases aluminum tolerance in Miscanthus sinensis growing in acidic mine soil, Lett. Appl. Microbiol., 73, 300–307, https://doi.org/10.1111/lam.13514, 2021.
Hirata, M., Hasegawa, N., Nogami, K., and Sonoda, T.: Evaluation of forest grazing as a management practice to utilize and control Miscanthus sinensis in a young tree plantation in southern Kyushu, Japan, Grassl. Sci., 53, 181–191, https://doi.org/10.1111/j.1744-697X.2007.00091.x, 2007.
Huusko, K., Ruotsalainen, A. L., and Markkola, A. M.: A shift from arbuscular mycorrhizal to dark septate endophytic colonization in Deschampsia flexuosa roots occurs along primary successional gradient, Mycorrhiza, 27, 129–138, https://doi.org/10.1007/s00572-016-0736-x, 2017.
Jeewani, P. H., Luo, Y., Yu, G., Fu, Y., He, X., Van Zwieten, L., Liang, C., Kumar, A., He, Y., Kuzyakov, Y., Qin, H., Guggenberger, G., and Xu, J.: Arbuscular mycorrhizal fungi and goethite promote carbon sequestration via hyphal-aggregate mineral interactions, Soil Biol. Biochem., 162, 108417, https://doi.org/10.1016/j.soilbio.2021.108417, 2021.
Jumpponen, A. and Trappe, J. M.: Dark septate endophytes: a review of facultative biotrophic root-colonizing fungi, New Phytol., 140, 295–310, https://doi.org/10.1046/j.1469-8137.1998.00265.x, 1998.
Jumpponen, A., Mattson, K. G., and Trappe, J. M.: Mycorrhizal functioning of Phialocephala fortinii with Pinus contorta on glacier forefront soil: interactions with soil nitrogen and organic matter, Mycorrhiza, 7, 261–265, https://doi.org/10.1007/s005720050190, 1998.
Khastini, R. O. and Jannah, R.: Potential Contribution of Dark-Septate Endophytic Fungus Isolated From Pulau Dua Nature Reserve, Banten on Growth Promotion of Chinese Cabbage, in: 2nd and 3rd International Conference on Food Security Innovation (ICFSI 2018–2019), Banten, Indonesia, https://doi.org/10.2991/absr.k.210304.015, 2021.
Knapp, D. G., Imrefi, I., Boldpurev, E., Csíkos, S., Akhmetova, G., Berek-Nagy, P. J., Otgonsuren, B., and Kovács, G. M.: Root-Colonizing Endophytic Fungi of the Dominant Grass Stipa krylovii From a Mongolian Steppe Grassland, Front. Microbiol., 10, 2565, https://doi.org/10.3389/fmicb.2019.02565, 2019.
Landolt, M., Stroheker, S., Queloz, V., Gall, A., and Sieber, T. N.: Does water availability influence the abundance of species of the Phialocephala fortinii s.l. – Acephala applanata complex (PAC) in roots of pubescent oak (Quercus pubescens) and Scots pine (Pinus sylvestris)?, Fungal Ecol., 44, 100904, https://doi.org/10.1016/j.funeco.2019.100904, 2020.
Laval, V., Kerdraon, L., Barret, M., Liabot, A.-L., Marais, C., Boudier, B., Balesdent, M.-H., Fischer-Le Saux, M., and Suffert, F.: Assessing the Cultivability of Bacteria and Fungi from Arable Crop Residues Using Metabarcoding Data as a Reference, Diversity, 13, 404, https://doi.org/10.3390/d13090404, 2021.
Li, A.-Z., Han, X.-B., Zhang, M.-X., Zhou, Y., Chen, M., Yao, Q., and Zhu, H.-H.: Culture-Dependent and -Independent Analyses Reveal the Diversity, Structure, and Assembly Mechanism of Benthic Bacterial Community in the Ross Sea, Antarctica, Front. Microbiol., 10, 2523, https://doi.org/10.3389/fmicb.2019.02523, 2019.
Li, X., He, X., Hou, L., Ren, Y., Wang, S., and Su, F.: Dark septate endophytes isolated from a xerophyte plant promote the growth of Ammopiptanthus mongolicus under drought condition, Sci. Rep., 8, 7896, https://doi.org/10.1038/s41598-018-26183-0, 2018.
Likar, M. and Regvar, M.: Isolates of dark septate endophytes reduce metal uptake and improve physiology of Salix caprea L., Plant Soil., 370, 593–604, https://doi.org/10.1007/s11104-013-1656-6, 2013.
Luo, J., Walsh, E., Naik, A., Zhuang, W., Zhang, K., Cai, L., and Zhang, N.: Temperate Pine Barrens and Tropical Rain Forests Are Both Rich in Undescribed Fungi, PLoS One., 9, e103753, https://doi.org/10.1371/journal.pone.0103753, 2014.
Luo, Y., Durenkamp, M., Nobili, M. D., Lin, Q., Devonshire, B. J., and Brookes, P. C.: Microbial biomass growth, following incorporation of biochars produced at 350 ∘C or 700 ∘C, in a silty-clay loam soil of high and low pH, Soil Biol. Biochem., 11, 513–523, https://doi.org/10.1016/j.soilbio.2012.10.033, 2013.
Ma, J. F., Shen, R., Zhao, Z., Wissuwa, M., Takeuchi, Y., Ebitani, T., and Yano, M.: Response of Rice to Al Stress and Identification of Quantitative Trait Loci for Al Tolerance, Plant Cell Physiol., 43, 652–659, https://doi.org/10.1093/pcp/pcf081, 2002.
Mahmoud, R. S. and Narisawa, K.: A New Fungal Endophyte, Scolecobasidium humicola, Promotes Tomato Growth under Organic Nitrogen Conditions, PLoS One, 8, e78746, https://doi.org/10.1371/journal.pone.0078746, 2013.
Menkis, A., Allmer, J., Vasiliauskas, R., Lygis, V., Stenlid, J., and Finlay, R.: Ecology and molecular characterization of dark septate fungi from roots, living stems, coarse and fine woody debris, Mycol. Res., 108, 965–973, https://doi.org/10.1017/S0953756204000668, 2004.
Omacini, M., Chaneton, E. J., Ghersa, C. M., and Müller, C. B.: Symbiotic fungal endophytes control insect host–parasite interaction webs, Nature, 409, 78–81, https://doi.org/10.1038/35051070, 2001.
Porcel, R., Aroca, R., and Ruiz-Lozano, J. M.: Salinity stress alleviation using arbuscular mycorrhizal fungi. A review, Agron. Sustain. Dev., 32, 181–200, https://doi.org/10.1007/s13593-011-0029-x, 2012.
Postma, J. W. M., Olsson, P. A., and Falkengren-Grerup, U.: Root colonisation by arbuscular mycorrhizal, fine endophytic and dark septate fungi across a pH gradient in acid beech forests, Soil Biol. Biochem., 39, 400–408, https://doi.org/10.1016/j.soilbio.2006.08.007, 2007.
Rodriguez, R. J., Henson, J., Van Volkenburgh, E., Hoy, M., Wright, L., Beckwith, F., Kim, Y.-O., and Redman, R. S.: Stress tolerance in plants via habitat-adapted symbiosis, ISME J., 2, 404–416, https://doi.org/10.1038/ismej.2007.106, 2008.
Sahu, P. K., Tilgam, J., Mishra, S., Hamid, S., Gupta, A., Verma, S. K., and Kharwar, R. N.: Surface sterilization for isolation of endophytes: Ensuring what (not) to grow, J. Basic Microbiol., 62, 647–668, https://doi.org/10.1002/jobm.202100462, 2022.
Stewart, J. R., Toma, Y., Fernández, F. G., Nishiwaki, A., Yamada, T., and Bollero, G.: The ecology and agronomy of Miscanthus sinensis , a species important to bioenergy crop development, in its native range in Japan: a review, GCB Bioenergy, 1, 126–153, https://doi.org/10.1111/j.1757-1707.2009.01010.x, 2009.
Stroheker, S., Dubach, V., Vögtli, I., and Sieber, T. N.: Investigating Host Preference of Root Endophytes of Three European Tree Species, with a Focus on Members of the Phialocephala fortinii–Acephala applanata Species Complex (PAC), JoF, 7, 317, https://doi.org/10.3390/jof7040317, 2021.
Surono and Narisawa, K.: The dark septate endophytic fungus Phialocephala fortinii is a potential decomposer of soil organic compounds and a promoter of Asparagus officinalis growth, Fungal Ecol., 28, 1–10, https://doi.org/10.1016/j.funeco.2017.04.001, 2017.
Tedersoo, L., Pärtel, K., Jairus, T., Gates, G., Põldmaa, K., and Tamm, H.: Ascomycetes associated with ectomycorrhizas: molecular diversity and ecology with particular reference to the Helotiales, Environ Microbiol., 11, 3166–3178, https://doi.org/10.1111/j.1462-2920.2009.02020.x, 2009.
Toju, H., Peay, K. G., Yamamichi, M., Narisawa, K., Hiruma, K., Naito, K., Fukuda, S., Ushio, M., Nakaoka, S., Onoda, Y., Yoshida, K., Schlaeppi, K., Bai, Y., Sugiura, R., Ichihashi, Y., Minamisawa, K., and Kiers, E. T.: Core microbiomes for sustainable agroecosystems, Nat Plants., 4, 247–257, https://doi.org/10.1038/s41477-018-0139-4, 2018.
Usuki, F. and Narisawa, K.: Formation of structures resembling ericoid mycorrhizas by the root endophytic fungus Heteroconium chaetospira within roots of Rhododendron obtusum var. kaempferi, Mycorrhiza, 15, 61–64, https://doi.org/10.1007/s00572-004-0333-2, 2005.
Usuki, F. and Narisawa, K.: A mutualistic symbiosis between a dark septate endophytic fungus, Heteroconium chaetospira, and a nonmycorrhizal plant, Chinese cabbage, Mycologia, 99, 175–184, https://doi.org/10.1080/15572536.2007.11832577, 2007.
Van Aarle, I. M., Olsson, P. A., and Söderström, B.: Arbuscular mycorrhizal fungi respond to the substrate pH of their extraradical mycelium by altered growth and root colonization, New Phytol., 155, 173–182, https://doi.org/10.1046/j.1469-8137.2002.00439.x, 2002.
Walker, J. F., Aldrich-Wolfe, L., Riffel, A., Barbare, H., Simpson, N. B., Trowbridge, J., and Jumpponen, A.: Diverse Helotiales associated with the roots of three species of Arctic Ericaceae provide no evidence for host specificity, New Phytol., 191, 515–527, https://doi.org/10.1111/j.1469-8137.2011.03703.x, 2011.
Wang, C. J. K. and Wilcox, H. E.: New Species of Ectendomycorrhizal and Pseudomycorrhizal Fungi: Phialophora finlandia, Chloridium paucisporum, and Phialocephala fortinii, Mycologia, 77, 951, https://doi.org/10.2307/3793308, 1985.
Watanabe, T., Jansen, S., and Osaki, M.: Al-Fe interactions and growth enhancement in Melastoma malabathricum and Miscanthus sinensis dominating acid sulphate soils, Plant Cell Environ., 29, 2124–2132, https://doi.org/10.1111/j.1365-3040.2006.001586.x, 2006.
Wu, Q.-S. and Xia, R.-X.: Arbuscular mycorrhizal fungi influence growth, osmotic adjustment and photosynthesis of citrus under well-watered and water stress conditions, J. Plant Physiol., 163, 417–425, https://doi.org/10.1016/j.jplph.2005.04.024, 2006.
Yadav, R. L., Shukla, S. K., Suman, A., and Singh, P. N.: Trichoderma inoculation and trash management effects on soil microbial biomass, soil respiration, nutrient uptake and yield of ratoon sugarcane under subtropical conditions, Biol. Fertil. Soils, 45, 461–468, https://doi.org/10.1007/s00374-009-0352-4, 2009.
Yamanishi, A., Kamijo, T., Tsunekawa, A., and Higuchi, H.: Monitoring vegetation damage caused by the year 2000 Miyake Island's volcanic eruption using satellite data, Journal of the Japanese Institute of Landscape Architecture, 66, 473–476, https://doi.org/10.5632/jila.66.473, 2003.
Yu, Y., Teng, Z., Mou, Z., Lv, Y., Li, T., Chen, S., Zhao, D., and Zhao, Z.: Melatonin confers heavy metal-induced tolerance by alleviating oxidative stress and reducing the heavy metal accumulation in Exophiala pisciphila, a dark septate endophyte (DSE), BMC Microbiol., 21, 40, https://doi.org/10.1186/s12866-021-02098-1, 2021.
Yuan, Z., Zhang, C., Lin, F., and Kubicek, C. P.: Identity, Diversity, and Molecular Phylogeny of the Endophytic Mycobiota in the Roots of Rare Wild Rice (Oryza granulate) from a Nature Reserve in Yunnan, China, Appl. Environ. Microb., 76, 1642–1652, https://doi.org/10.1128/AEM.01911-09, 2010.
Zheng, H., Yang, T., Bao, Y., He, P., Yang, K., Mei, X., Wei, Z., Xu, Y., Shen, Q., and Banerjee, S.: Network analysis and subsequent culturing reveal keystone taxa involved in microbial litter decomposition dynamics, Soil Biol. Biochem., 157, 108230, https://doi.org/10.1016/j.soilbio.2021.108230, 2021.





