the Creative Commons Attribution 3.0 License.
the Creative Commons Attribution 3.0 License.
Distribution and drivers of symbiotic and free-living diazotrophic cyanobacteria in the western tropical South Pacific
Andrea Caputo
Carlo Berg
Sophie Bonnet
Rachel A. Foster
The abundance and distribution of cyanobacterial diazotrophs were quantified in two regions (Melanesian archipelago, MA; and subtropical gyre, SG) of the western tropical South Pacific using nifH quantitative polymerase chain reaction (qPCR) assays. UCYN-A1 and A2 host populations were quantified using 18S rRNA qPCR assays including one newly developed assay. All phylotypes were detected in the upper photic zone (0–50 m), with higher abundances in the MA region. Trichodesmium and UCYN-B dominated and ranged from 2.18 × 102 to 9.41 × 106 and 1.10 × 102 to 2.78 × 106 nifH copies L−1, respectively. Het-1 (symbiont of Rhizosolenia diatoms) was the next most abundant (1.40 × 101–1.74 × 105 nifH copies L−1) and co-occurred with het-2 and het-3. UCYN-A1 and A2 were the least abundant diazotrophs and were below detection (bd) in 63 and 79, respectively, of 120 samples. In addition, in up to 39 % of samples in which UCYN-A1 and A2 were detected, their respective hosts were bd. Pairwise comparisons of the nifH abundances and various environmental parameters supported two groups: a deep-dwelling group (45 m) comprised of UCYN-A1 and A2 and a surface group (0–15 m) comprised of Trichodesmium, het-1 and het-2. Temperature and photosynthetically active radiation were positively correlated with the surface group, while UCYN-A1 and A2 were positively correlated with depth, salinity, and oxygen. Similarly, in a meta-analysis of 11 external datasets, all diazotrophs, except UCYN-A were correlated with temperature. Combined, our results indicate that conditions favoring the UCYN-A symbiosis differ from those of diatom diazotroph associations and free-living cyanobacterial diazotrophs.
- Article
(2892 KB) - Full-text XML
-
Supplement
(667 KB) - BibTeX
- EndNote
Biological di-nitrogen (N2) fixation is considered a major source of new nitrogen (N) to oceanic ecosystems (Karl et al., 1997). N2 fixation is an energetically expensive process, in which N2 gas is reduced to bioavailable ammonia (Howard and Rees, 1996), and is performed by a small but diverse group of bacteria and archaea. The nitrogenase enzyme, which is encoded by a suite of nif genes, mediates N2 fixation (Jacobson et al., 1989; Young, 2005). Nitrogenase has a high iron (Fe) requirement (Howard and Rees, 1996), and often N2 fixers, or diazotrophs, are Fe limited (Kustka et al., 2003; Raven, 1988). Nitrogenase is also sensitive to oxygen (O2), which has been shown to negatively influence N2 fixation efficiency (Meyerhof and Burk, 1928; Stewart, 1969). Thus, autotrophic diazotrophs (e.g., cyanobacteria) have evolved strategies, such as temporal and spatial separation of the fixation process, to protect their nitrogenase from O2 evolution during photosynthesis (Berman-Frank et al., 2001; Haselkorn, 1978; Mitsui et al., 1986). N2 fixation is widespread and occurs in marine, limnic, and terrestrial habitats. In marine ecosystems it mainly occurs in the photic zone, closest to the surface; however, more recently, evidence has shown activity in deeper depths below the photic zone, including oxygen minimum zones (Benavides et al., 2016; Bonnet et al., 2013; Fernandez et al., 2011; Halm et al., 2009; Löscher et al., 2015).
N2 fixation in the photic zone is often attributed to a diverse group of cyanobacteria. Traditionally, marine, photic-zone-dwelling diazotrophs are divided into two groups based on cell diameter, e.g., > 10 and < 10 µm size fractions. Diatom diazotroph associations (DDAs), symbioses between heterocystous cyanobacteria and a variety of diatom genera, and large filamentous non-heterocystous Trichodesmium spp. compose the larger size fraction (> 10 µm). Trichodesmium spp. occur as free filaments or often in two morphologies of colonies: tufts/rafts and puffs. There are three defined lineages of symbionts of DDAs based on their nifH phylogeny: het-1 and het-2 refer to the two Richelia intracellularis lineages that associate with diatom genera Rhizosolenia and Hemiaulus, respectively, while the third lineage, het-3, associates with the heterocystous Calothrix rhizosoleniae and Chaetoceros compressus diatoms (Foster et al., 2010; Foster and Zehr, 2006).
The unicellular diazotrophic cyanobacterial groups are divided into UCYN-A, UCYN-B, and UCYN-C groups and are representatives of the < 10 µm size fraction. The UCYN-A (Candidatus Atelocyanobacterium thalassa) group can be further delineated into six sublineages (Thompson et al., 2014; Turk-Kubo et al., 2017); two (UCYN-A1, UCYN-A2) are identified as symbiotic with small prymnesiophyte microalgae (reviewed by Farnelid et al., 2016; see references within). The UCYN-B group has its closest cultured relative as Crocosphaera watsonii and lives freely, colonially, and also in symbiosis with the diatom Climacodium frauenfeldianum (Bench et al., 2013; Carpenter and Janson, 2000; Webb et al., 2009; Zehr et al., 2001). Often overlooked is the observation that UCYN-B, when colonial or symbiotic, could also be associated with the > 10 µm size fraction. Less is known about the UCYN-C group, and given that its nifH nucleotide sequence is 90 % similar (Foster et al., 2007) to Cyanothece spp. ATCC51142, it is assumed to be analogous and thus co-occurring with the other < 10 µm size fraction. A diverse group of free-living heterotrophic bacteria (e.g., gamma proteobacteria) (Berthelot et al., 2015; Bombar et al., 2016; Halm et al., 2012; Langlois et al., 2005) and archaea (Zehr et al., 2005) are also within the < 10 µm size fraction.
The distribution and activity of diazotrophs in open-ocean ecosystems are governed by different ambient environmental factors, including macronutrient availability (Moutin et al., 2008; Sañudo-Wilhelmy et al., 2001) and temperature (Messer et al., 2016; Moisander et al., 2010). There are also simultaneous influences by several factors (i.e., co-limitation of nutrients; Mills et al., 2004). Moreover, most oceanic models of N2 fixation assume that all diazotrophs are equally controlled by the same environmental parameters (Deutsch et al., 2007; Hood et al., 2004; Landolfi et al., 2015), despite well-recognized differences in genetic repertoires for assimilating dissolved nutrient pools (e.g., dissolved organic phosphate; Dyhrman et al., 2006; Dyhrman and Ruttenberg, 2006), life histories (free, symbiotic, colonial), and cell sizes (micrometers to millimeters). A better understanding of these differences would benefit oceanic models of N2 fixation, our understanding of marine N2 budgets, and the impact of N2 fixation in oceanic surface waters and their communities.
The tropical South Pacific Ocean is considered one of the most oligotrophic regions in the world's oceans (Claustre and Maritorena, 2003), with a widespread N deficiency (Deutsch et al., 2007; Raimbault et al., 2008), and the central SP gyre has some of the lowest reported concentrations of dissolved Fe (Blain et al., 2008). One exception is the western tropical South Pacific (WTSP), which harbors many islands with Fe-rich sediments adding to an island mass effect (Shiozaki et al., 2014) and is influenced by multiple ocean currents, both surface and subsurface, that drive the distribution of dissolved nutrients, micronutrients, and the biota (Fitzsimmons et al., 2014; Gourdeau et al., 2008; Marchesiello and Estrade, 2010; Wells et al., 1999). The structure of these currents also promotes shearing instabilities and strong eddies (Qiu et al., 2009). Moreover, Van Den Broeck et al. (2004) suggested that the WTSP is phosphate limited, while Law et al. (2011) hypothesized that primary production and N2 fixation in the WTSP follow the seasonality of cyclones, which in their wake, enrich surface waters with phosphate and fuel primary and new production. An earlier investigation along a transect in the western equatorial Pacific estimated that 74 % of the total N2 fixation could be attributed to the < 10 µm size fraction, although abundances of unicellular cyanobacteria were moderate (17 cells mL−1) (Bonnet et al., 2009). However, diazotroph quantification is lacking further south in tropical waters, despite being recently recognized as a hot spot of N2 fixation, with average rates of ∼ 570 µmol N m−2 d−1 (Bonnet et al., this issue), i.e., in the upper range (100–1000 µmol N m−2 d−1) of rates gathered in the global N2 fixation MAREDAT database (Luo et al., 2012).
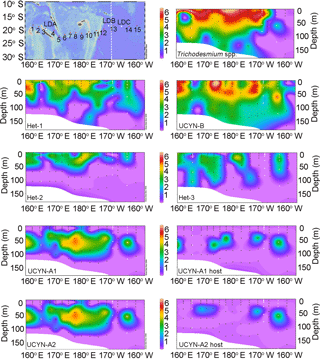
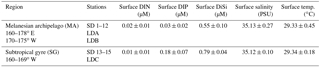
Figure 1Sampling locations and the horizontal and vertical distributions of diazotrophs and the UCYN-A1 and UCYN-A2 hosts in the study area. Sampling depths are indicated as black dots (white for LD stations) and the abundances are the log nifH gene copies L−1 for the diazotrophs and 18S rRNA gene copies L−1 for the UCYN-A host lineages. The boundary between the Melanesian archipelago and subtropical gyre in the transect map is marked with a dotted line.
The primary aim of this study was to quantify diazotroph abundance and distribution in the WTSP, with an emphasis on symbiotic N2-fixing populations, both by at-sea and lab-based quantitative approaches. Since earlier expeditions to the SW Pacific identified it as a potential hotspot for diazotrophy (Bonnet et al., 2015, 2016, 2017; Moisander et al., 2010), the at-sea quantitative polymerase chain reaction (qPCR) was performed in order to make informed decisions about the presence, absence, and relative abundance of diazotrophs so that stations could be selected for other assays and characterization of the hydrographic conditions (e.g., incubation experiments, sediment and drifter deployments; see Bonnet et al., 2018). For a more comprehensive investigation of the symbiotic diazotrophs, we developed a new primer and probe set for quantification of the UCYN-A1 host. We also identified key environmental parameters, both biotic and abiotic, which influenced the distribution of diazotrophs in the WTSP and tested the congruency of these parameters in an additional 11 publicly available datasets. We hypothesized that the distribution and the environmental drivers of the diazotrophic symbioses should differ due to the major differences in host taxonomy (e.g., diatom vs. prymnesiophyte), size (1–2 to hundreds of micrometers), and life history (free vs. symbiotic, chain forming). For comparison and for similarly divergent characteristics (symbiotic vs. free; colonial vs. single), several free-living (UCYN-B, Trichodesmium spp., and UCYN-C) cyanobacterial diazotrophs were also included.
2.1 Sampling
Sampling was conducted on a transect in the WTSP during austral summer (19 February–5 April 2015), onboard the R/V L'Atalante (Fig. 1a). The cruise transect and stations were selected according to the following: regions with low advection to better estimate biogeochemical budgets, contrasting diazotrophic community (e.g., Trichodesmium spp. dominated vs. UCYN dominated), and along a trophic and N2 fixation gradient from west to east (Moutin et al., 2017). The cruise is divided into two geographic regions: Melanesian archipelago (MA: SD 1–12, LD A and LD B stations, 160–178∘ E and 170–175∘ W) and the subtropical gyre (SG: SD 13–15 and LD C stations, 160–169∘ W). Long-duration (LD) stations of 8 days were chosen based on hydrographic conditions, satellite imagery, microscopic analyses of > 10 µm cyanobacterial diazotrophs, and the results of our at-sea qPCR analyses of four unicellular diazotrophic targets (UCYN-A1, UCYN-A2, UCYN-B, and UCYN-C) (see below).
2.1.1 Nucleic acids
Seawater (2.5 L) was collected from 18 stations (LD A–C; SD 1–15) into clean (10 % bleach rinsed) 2.75 L polycarbonate bottles from six to seven discrete depths based on surface incident light intensity (100, 75, 54, 36, 10, 1, and 0.1 %) once per station at both short-duration (SD) and LD stations using Niskin bottles (12 L) arranged on a conductivity temperature depth (CTD; Seabird 911) rosette.
After collection from the CTD rosette, 2.5 L seawater was immediately filtered onto a 0.2 µm pore size Supor filter (Pall Corporation, Pall Norden AB; Lund, Sweden) held within a 25 mm diameter Swinnex filter holder (Merck Millipore; Solna, Sweden) using a peristaltic pump (Cole-Parmer, Masterflex, Easy-load II; USA). In case the filters clogged with too much material, the remaining volume was measured and noted for later calculations. The filters were placed in pre-sterilized bead beater tubes (Biospec; Bartlesville, OK, USA) containing 30 µL of a 0.1 and 0.5 mm glass bead mixture, flash frozen in liquid nitrogen, and archived at −80 ∘C. Four additional DNA samples were collected from four discrete depths, (75, 50, 36, 10 % light), at 11 of the 18 stations, for the at-sea qPCR (see below) and filtered as described above. The at-sea samples were extracted and processed for qPCR immediately after collection (see below).
2.1.2 Cell abundances and microscopy observations
At the LD stations, 5 L of seawater was collected at the same depths in parallel with the nucleic acid samples from the CTD rosette. Two sets of samples, one set each day, were taken on days 1 and 3 at each LD station and immediately filtered onto a 47 mm diameter Poretics (Millipore) membrane filter with a pore size of 5 µm using a peristaltic pump.
At the SD stations, the same collection was implemented; however a 25 mm diameter Poretics membrane filter was used. The high densities of cells on the latter made it impossible to properly enumerate the various cyanobacterial diazotrophs and as such these samples were used only for qualitative observations (see below). Immediately after filtration, samples were fixed in 1 % paraformaldehyde (v∕v) for 30 min prior to storing at −20 ∘C. For enumeration, the filter was mounted on a glass slide and examined under an Olympus BX60 microscope equipped with a filter for blue (460–490 nm) and green (545–580 nm) excitation wavelengths. Three areas (area = 0.94 mm2) per filter were counted separately and values were averaged. When abundances were low, the entire filter (area = 1734 mm2) was observed and cells enumerated. Due to weak fluorescence, only Trichodesmium colonies and free filaments could be accurately estimated with microscopy and in addition, the larger cell diameter Trichodesmium, hereafter referred to as Katagynemene pelagicum, was enumerated separately as these were often present albeit at lower cell densities. Other cyanobacterial diazotrophs, e.g., C. watsonii-like cells, C. rhizosoleniae (het-3), and R. intracellularis (het-1, het-2), were also present on the larger 47 mm diameter samples; however fluorescence was weak and therefore difficult to enumerate. Pico-eukaryote populations, identified as round 1–3 µm diameter cells, with red excitation under the blue filter set, were also observed. For the latter populations, qualitative observations of presence and some details on cell integrity (e.g., fluorescence, diatom frustule, free-living or symbiotic form) are included.
2.2 Nutrient analyses
Seawater for nutrient analyses was collected from each station using the CTD rosette at the same depths as those collected for the nucleic acids. Seawater for inorganic nutrient analysis was collected in 20 mL high-density polyethylene HCL-rinsed bottles and poisoned with HgCl2 to a final concentration of 20 µg L−1 and stored at 4 ∘C until analysis. The samples were frozen and returned to the laboratory for analysis within 4 months. Dissolved nitrate and nitrite (NO + NO, DIN), phosphate (PO, DIP), and silicate (Si (OH)4, DiSi) concentrations were determined with standard colorimetric techniques using a segmented flow analyzer according to Aminot and Kérouel (2007) on a SEAL Analytical AA3 HR system (Seal Analytical, Serlabo Technologies; Entraigues Sur La Sorgue, France). Quantification limits for nitrate, phosphate, and silicate were all 0.05 µmol L−1.
2.3 DNA extraction
The DNA from the 120 archived samples was extracted as described in Moisander et al. (2008), with a 30 s reduction in the agitation step in a Fast Prep cell disrupter (Thermo, model FP120; Qbiogene, Inc.; CEDEX, France) and an elution volume of 70 µL. The nucleic acid samples collected for the at-sea qPCR were extracted immediately after filtration using a modified version of the DNeasy plant kit (Qiagen) total DNA extraction protocol. The modifications were an initial 2 min agitation step using a bead beater (Biospec Mini-BeadBeater-16, model 607EUR; Biospec), 10 min proteinase K incubation, and final elution volume of 25 µL.
2.4 Oligonucleotide design
A new primer and probe set was designed to amplify the UCYN-A1 host and was based on a published 18S rRNA sequence (accession number JX291893) reported from the North Pacific gyre (station ALOHA) (Thompson et al., 2012). The design utilized the same 96 bp target region of the 18S rRNA used to amplify UCYN-A2 hosts described in Thompson et al. (2014) (Table S1 in the Supplement). The primers and probe for the UCYN-A1 host 18S rRNA gene assay are as follows: forward, 5′AGGTTTGCCGGTCTGCCGAT-3′; reverse, 5′GAGCGGGTGTCGGAGACGGAT-3′; probe, 5′-FAM-CTGGTAGAACTGTCCT-TAMRA-3′. The forward, reverse, and probe contain two to four, one, and five mismatches, respectively, to UCYN-A2 host sequences (accession number KF771248-KF771254) and the following closely related sequences (98–100 %): uncultured eukaryote clones (station ALOHA: EU50069; Cariaco Basin: GU824119) Chrysochromulina parkeae: AM490994; Braarudosphaera bigelowii TP056a: AB250784; B. bigelowii Furue-15: AB478413; B. bigelowii Funahama T3: AB478413; and B. bigelowii Yastushiro-1 AB478414. The UCYN-A1 host oligonucleotides specificity was tested de nova against the following closely related sequences derived from uncultured eukaryotic clonal sequences (accession numbers: EU500067-68, FJ537341, EU500138-39, EF695227, EU500141, EU499958, EF695229, EF695220). Only one mismatch was found in the forward probe for one sequence (EU500138) (Table S1). The nucleotide sequence identity for UCYN-A1 and UCYN-A2 host sequences is 97.95 %. Finally, a cross-reactivity test between the newly designed UCYN-A1 host oligonucleotides and a dilution series of the UCYN-A2 host template was run (see below).
2.5 Quantitative PCR
Abundances of nifH gene copies from selected diazotrophic groups (UCYN-A1, UCYN-A2, UCYN-B, UCYN-C, het-1, het-2, het-3, and Trichodesmium spp.) and the 18S rRNA of UCYN-A1 and A2 hosts were quantified using previously published oligonucleotides and TaqMan assays (Church et al., 2005; Foster et al., 2007; Moisander et al., 2010; Thompson et al., 2014) and the newly designed UCYN-A1 host oligonucleotides (Table S1). The qPCRs were conducted in a StepOnePlus system (Applied Biosystems, Life Technologies; Stockholm, Sweden) in fast (> 40 min) mode with the following parameters: 95 ∘C for 20 s, followed by 45 cycles of 95 ∘C for 1 s, and 60 ∘C for 20 s.
Cross-reactivity tests were run on two of the heterocystous symbiont (het-1 and het-2) oligonucleotides, the UCYN-A1 and UCYNA-2 oligonucleotides, and the newly designed UCYN-A1 host oligonucleotides and UCYN-A2 host primer and probe set. Cross-reactivity tests for het-3 were omitted as previously published tests (Foster et al., 2007) showed no cross-reaction with het-1 or het-2. The standard curve for a particular target was run in reactions with the other primers and probe sets. For example, the UCYN-A1 TaqMan host primers and probes were run in reactions with UCYN-A2 template DNA. The cross-reactivity for the het-1 and het-2 primer and probe sets has been previously reported (Foster et al., 2007), however only when the assay is run in standard mode. Standard mode runs the holding, denaturation, and annealing stages at the following longer intervals than in Fast mode: 11 min and 40 s, 14 s, and 40 s, respectively. Hence, we tested the cross-reactivity for the het primers and probes when run in fast mode, as the fast mode was used in our study. Similarly, the cross-reactivity between UCYN-A1 and UCYN-A2 was tested in fast mode at two annealing temperatures, 60 and 64 ∘C; 64 ∘C is the recommended annealing temperature for the UCYN-A2 assay (Thompson et al., 2014).
Reaction volume was 20 µL in all qPCRs and consisted of 10 µL of 2X TaqMan fast buffer (Applied Biosystems), 5.5 µL of nuclease free water (Sigma Aldrich Sweden AB; Stockholm Sweden), 1 µL each of the forward and reverse primers (10 µM), 0.5 µL of fluorogenic probe (10 µM), and 2 µL of DNA extract. For standard mode runs, the latter master mix was identical, with the exception of replacing the fast 2X buffer with the standard 2X buffer. For reactions quantifying Trichodesmium spp. nifH copies, SD 9 was excluded and 1 µL of DNA template was used for the remaining stations due to low template volume, and total reaction volume was adjusted by the addition of 1 µL of nuclease free water. Reactions were performed in duplicates for the at-sea qPCR and in triplicates for the archived samples and lab-based qPCR. For the at-sea qPCR, only four targets (UCYN-A1, UCYN-A2, UCYN-B, and UCYN-C) were quantified and only at the SD stations. No assays were processed at SD 5–6, 10–12, and 14 for the at-sea qPCR due to no potential LD station selection for these sites. Two microliters of nuclease free water was used as template in no-template controls; no nifH copies were detected in the no-template controls.
Gene copy abundance was calculated from the mean cycle threshold (Ct) value of the three replicates and the standard curve for the appropriate oligonucleotides in the lab-based qPCRs. For the at-sea qPCR, a mean Ct value of two replicates was used to maximize the number of samples run on one amplification plate (96 well). In samples in which one or two out of three replicates produced an amplification, signals were noted as detectable but not quantifiable (dnq) and no amplification was noted as below detection (bd).
2.6 Standard curves and PCR efficiency
Standard curves were plotted and analyzed in Excel for each target based on the qPCR Ct values from known dilutions of synthesized target gene fragments (gBlocks®; Integrated DNA Technologies; Leuven, Belgium) (359 bp nifH and 733 bp 18S rRNA for UCYN-A hosts). We made 10-fold dilutions starting with 108 to 101 gene copies L−1. The PCR efficiency, for identification of possible interfering contaminants in our samples, was determined as previously described (Short et al., 2004) for 12 samples run on the het-1, het-2, and het-3 primers and probe tests. The qPCR efficiency ranged from 90 to 99 % with an average of 94 % efficiency for the diazotroph targets het-1, het-2, and het-3.
2.7 Statistics and data analysis
Skewness and normal distribution tests by descriptive statistics were performed in IBM SPSS (version 23) on the following parameters recorded during sample collection in the WTSP from the CTD package: depth (m), oxygen (mL L−1), temperature (∘C), chlorophyll fluorescence (µg L−1), photosynthetically active radiation (PAR; µmol photons m−2 s−1), salinity (PSU), and gene copy abundances determined by qPCR. Since fewer samples and targets were processed in the at-sea qPCR, only the lab-based qPCR results from the archived samples was used for statistical analysis. Significant skew was noted when skewness, divided by its standard deviation, exceeded 1.95. All but three targets (het-1, UCYN-B, and Trichodesmium spp.) and three environmental parameters (temperature, salinity, and oxygen) were significantly skewed (not normally distributed) even after LOG10 transformation. Therefore, a nonparametric Spearman's rank correlation was conducted to test possible correlations between the targets and environmental parameters, where we assume that the het groups and UCYN-A clade are symbiotic, while UCYN-B is free living. The resulting correlation matrices were visualized in the form of a heat map of hierarchical clustering in R (version 3.2.2) using packages “hmisc” and “gplots”. Multivariate statistics by redundancy analysis (RDA) was conducted using the R package “vegan”. T tests, in IBM SPSS (version 23), were performed to characterize the MA and SG regions and used the mean concentrations of nutrients at each station as input. For meta-analysis on the external data from 11 publically available datasets, sampled in the Atlantic and Pacific oceans and the South China Sea, data were acquired from the PANGAEA database and previous publications (Benavides et al., 2016; Bombar et al., 2011; Church et al., 2005, 2008; Foster et al., 2007, 2009; Goebel et al., 2010; Kong et al., 2011; Langlois et al., 2008; Moisander et al., 2008, 2010). We included only datasets with a minimum of 10 data points on the previously mentioned diazotrophic targets. Note that in all datasets the two UCYN-A phylotypes (A1 and A2) were not distinguished, and het-3 was excluded since it was rarely quantified. The meta-analysis was conducted using the software OpenMEE (Wallace et al., 2017) (based on R package “metafor”), where correlation coefficients from Spearman's rank were z-transformed (Fisher's) and tested using weighted random effect models. Graphical visualization of the mean abundances of the most numerous diazotrophs across the cruise transect was also performed in IBM SPSS (version 23).
3.1 Hydrographic conditions
Near-surface (0–5 m) DIN concentrations were below the quantification limit (bq) in both the MA and SG regions, while the mean surface DIP and DiSi concentrations were bq or low across all stations in the MA (bq-0.08 µM and 0.54–0.56 µM, respectively) and significantly (p<0.001; t test) higher (0.18 ± 0.07 µM and 0.79 ± 0.04 µM, respectively) at the stations in the SG (Table 1). The upper 25–30 m of the euphotic zone had stable temperatures of 29–30 ∘C, and the thermocline was deeper in the SG compared to the MA. The depth of the deep chlorophyll maximum (DCM) was between 70 and 165 m throughout the cruise, except for LD B (DCM at 35 m), which was sampled during a degrading surface phytoplankton bloom. A 30-day composite of the surface chlorophyll a (Chl a) confirmed the decreasing level of surface fluorescence measured by the CTD package at LD B (data not shown).
3.2 Cross-reactivity tests
No amplification was detected for the newly designed UCYN-A1 host oligonucleotides run with the UCYN-A2 as template DNA and vice versa (Fig. S1a–b in the Supplement).
Running the het assay in fast mode showed a lower cross-reactivity between the het-1 assay and the het-2 template than vice versa (the het-2 assay and het-1 template) (Fig. S1e–f). In fact, no amplification was detected in the last two template additions and the Ct differences were > 9 when het-1 assay was run with het-2 templates. The UCYN-A2 assay detected the UCYN-A1 template in all but the last template addition and with Ct differences > 3 (1 order of magnitude) while there was a 18–20 difference in Ct value (less gene copies) when the UCYN-A1 assay was run in fast mode with UCYN-A2 templates at either annealing temperature (60 or 64 ∘C) and only the first three template additions (108–106 nifH copies µL−1) were detected (Fig. S1c–d).
3.3 Comparison of at-sea and lab-based qPCR
In order to expedite the sample processing for the at-sea qPCR, a shortened and modified DNA extraction protocol was performed, four depths were sampled, and four targets were run (UCYN groups). In total, 44 samples can be compared with results from the parallel archived samples processed in the lab. Moreover, we considered the differences to be noteworthy only when there was at least 1 order of magnitude of difference in detection. A summary of the comparison, including the difference in nifH copy abundance is provided in Table S2. In addition, a graphical comparison shows a 1 : 1 relationship between the nifH gene copies detected in the at-sea and archived samples (Fig. S3).
In general, the at-sea and lab-based qPCR values were similar in quantifying the targets. Discrepancies were noted in 7, 8, and 11 samples, which had higher detection in the at-sea analyses for UCYN-A1, UCYN-A2, and UCYN-B, respectively. There were fewer instances (three, four, and five, respectively) of samples processed in the lab with the full extraction that had higher nifH gene abundances for UCYN-A1, UCYN-A2, and UCYN-B, respectively.
3.4 Horizontal and vertical distributions
Trichodesmium and UCYN-B were the most abundant diazotrophs and nifH gene abundances ranged from 104 to 106 nifH copies L−1 at multiple depths (four to six depths) in the upper water column (0–35 m) (Figs. 1–2; Table S3). Trichodesmium represented 80–99 % of total nifH genes detected at 9 out of 17 stations with highest detection in the MA and low to bd in the SG. Microscopy observations and abundances of Trichodesmium spp. confirmed a high abundance of free filaments of Trichodesmium and C. watsonii-like cells at LD B, while colonies were in general rarely observed (Table S5).
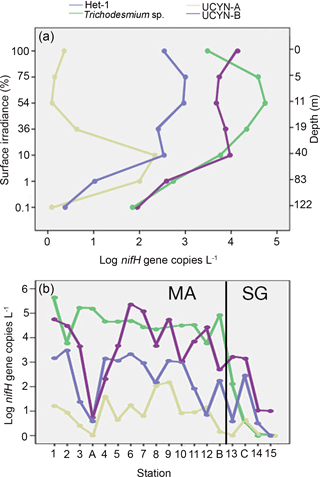
Figure 2(a–b) LOG10 transformed mean abundances for four of the diazotrophs across the transect: het-1 (blue), Trichodesmium (green), UCYN-A1 (yellow), and UCYN-B (purple). The mean nifH abundance values are shown as a function of (a) percent (%) of surface irradiance and corresponding depth (m) on the secondary y axis and (b) at each station. The corresponding depths for the percent of surface irradiance varied little; hence values were pooled and the mean is plotted. The solid black line in (b) designates the transition between the Melanesian archipelago (MA) and the subtropical gyre (SG).
At stations where Trichodesmium was not the most abundant diazotroph (e.g., SD 2, 6, 7, 14, 15, and LD C), UCYN-B had the highest depth-integrated nifH copy abundance. UCYN-B was also the most consistently detected diazotroph and was quantifiable from all stations sampled, accounting for 81–100 % of the total detected nifH gene copies in the SG. There was also a depth dependency for maximum nifH gene abundance such that the average depth maxima of Trichodesmium (1.88 × 106 nifH copies L−1) and UCYN-B (5.03 × 105 nifH copies L−1) at the stations in the MA were 10 and 25 m, respectively. In the SG, the average depth maximum for UCYN-B (1.50 × 105 nifH copies L−1) was the same (25 m), while the average depth of the Trichodesmium maximum (1.30 × 104 nifH copies L−1) deepened to 31 m.
Of the three heterocystous cyanobacterial symbiont lineages (het-1, het-2, and het-3), het-1 was the most dominant (60 % detection in total samples, 72 of 120 samples), and similar to Trichodesmium had higher detection in the stations of the MA region. For example, at stations SD 2, 4, and 9, het-1 represented 10–15 % of the total nifH genes quantified in the depth profiles, but in the total nifH genes quantified across the entire transect, het-1 only represented 1.5 %. Abundances for het-1 ranged between 103 and 105 nifH copies L−1 (15 of the 18 stations) at multiple depths (0–90 m) and the average depth maximum at MA stations (3.11 × 104 nifH copies L−1) was closer to the surface (15 m) compared to the SG stations (1.63 × 103 nifH copies L−1; 60 m) (Fig. 1; Table S3). Het-2 and het-3 co-occurred with het-1, however at lower abundances (102–104 nifH copies L−1) and often bd. For example, het-2 and het-3 were bd at all depths sampled at 1 and 3 stations, respectively, located in the SG. The average depth of maximum abundance (17 m) for het-2 (3.89 × 103 nifH copies L−1) was similar to het-1 (15 m), while het-3 (1.53 × 103 nifH copies L−1) was deeper at 33 m (considering only the MA stations). Microscopy observations confirmed the presence of R. intracellularis at five SD stations of the MA and LD B and absence at the SD stations and LD C of the SG. Noticeable was the co-occurrence of free filaments of R. intracellularis and degrading diatom cells (mainly belonging to the genus Rhizosolenia), especially at the SD 5, 6, and 7.
The unicellular symbiotic groups, UCYN-A1 and A2 (and their respective hosts), were the least detected targets. For example, UCYN-A1 was bd in 53 % (63 of 120 samples) and UCYN-A2 was bd in 66 % (79 of 120 samples) of samples. UCYN-A1 and A2 represented < 0.4 % of total nifH genes detected and UCYN-A symbionts were bd in the SG, except at LD C. When detected, average nifH gene abundance for UCYN-A1 and A2 was 8.60 × 104 and 4.60 × 104 nifH copies L−1, respectively, and usually accounted for < 1.0–1.5 % of the total nifH gene copies enumerated per station. One exception was at LD C in the SG, when UCYN-A1 and A2 accounted for 4 and 11 %, respectively, of the total nifH gene copies and were the second most abundant diazotroph (3.19 × 104 and 8.53 × 104 nifH copies L−1). The average depth of maximum nifH gene abundance for the UCYN-A1 (1.60 × 104 nifH copies L−1) and A2 (5.76 × 103 nifH copies L−1) symbionts was consistently recorded at deeper depths (55 and 58 m, respectively; 10 % light level).
The detection of the UCYN-A1 and A2 hosts mirrored the detection of their respective symbionts. However, in 22 and 15 samples, respectively, the UCYN-A1 and A2 symbionts were quantified while their hosts were bd. The UCYN-A hosts were never detected in samples in which their respective symbionts were bd or dnq. When both UCYN-A host and symbiont were present, the abundances of the host's 18S rRNA genes were always 1 order of magnitude less than their respective symbiont's nifH genes, with the exception of two samples for UCYN-A1 symbionts in which their respective host 18S rRNA gene abundances were half, or nearly equal in abundance. UCYN-C was the least abundant unicellular diazotroph and was only quantified in the at-sea qPCR for which detection was poor and limited to the MA region (3 of 11 stations: one to three of four depths sampled) and abundances never exceeded 102 nifH copies L−1 (Table S3).
3.5 Diazotroph and UCYN-A host covariation
Several significant correlations between the target diazotrophs and hosts were identified (Fig. 3; Table S4a). The nifH gene abundances of Trichodesmium and UCYN-B were significantly positively correlated with each other (N=108, p<0.01). In addition, UCYN-B nifH gene copy abundance was significantly positively correlated with that of both UCYN-A symbionts (A1 and A2; N=120, p<0.01) and UCYN-A2 host abundance (N=118, p<0.04). Abundances of UCYN-A1 and A2 nifH genes were significantly positively correlated with each other, and in addition, with their respective host 18S rRNA gene abundances (N=118–120, p<0.01). Lastly, the nifH gene abundances of het-1, het-2, and het-3 were significantly positively correlated with one another and with the nifH gene abundances of Trichodesmium and UCYN-B (N=108–120, p<0.01). The only correlations that were not significant were between the UCYN-A (including their hosts) and Trichodesmium and the het groups (with the exception of het-3, which correlated with the UCYN-A2 host (N=118, p<0.04)).
3.6 Influence of environmental conditions on diazotroph and UCYN-A host abundances in the WTSP
The nifH gene abundances of UCYN-A1 and A2 were significantly positively correlated with salinity (N=107, p<0.02 and N=107, p<0.03, respectively) and depth (N=118, p<0.01 and N=118, p<0.002, respectively) (Fig. 3; Table S4b). However, except for het-3 (regarding depth), all other diazotrophs were significantly negatively correlated with salinity (N=97–108, p<0.006) and depth (N=108–120, p<0.001). Moreover, Trichodesmium, UCYN-B, and the het group (except het-3) were significantly positively correlated with PAR (N=63–72, p<0.003) and temperature (N=97–108, p<0.001) while UCYN-A1 and A2 were significantly negatively correlated (N=72–108, p<0.02) with the latter parameters. All diazotrophic targets, except UCYN-A1, UCYN-A2, and het-3, were significantly negatively correlated with DIN concentration (N=90–94, p<0.02). All diazotrophs, except UCYN-A2, were significantly negatively correlated with DIP concentration (N=96–100, p<0.02), and all diazotrophs except UCYN-A1, A2, and het-3 were significantly negatively correlated with DiSi concentration (N=97–102, p<0.001). The gene copy abundances for the UCYN-A hosts, UCYN-A1 and UCYN-A2, and UCYN-B were significantly correlated with dissolved oxygen (UCYN-A and hosts, N=106–108, p<0.003; UCYN-B, N=108, p<0.03). In general, the correlations between nifH gene abundances and several hydrographic parameters divided the diazotrophs into two groups: the UCYN-A symbionts (and respective hosts) and all other target diazotrophs.
Figure 3Hierarchical clustering heat map of Spearman's rho results. The histogram shows negative (blue) and positive (green) values of correlation strength between parameters. Stars within cells mark significant correlations (p<0.05). In brackets are the two distinct groups in the WTSP.
Hierarchical clustering based on the Spearman's rank analyses resulted in two major groups: (1) a shallow group and (2) a deep group. The clustering in the euphotic zone was inferred from the negative and positive correlations for shallow and deep groups, respectively, with depth (Fig. 3). For example, Trichodesmium and the symbiotic het-1 and het-2 lineages characterize an upper water column (shallow) group with significant clustering and positive correlations with temperature (N=97–108, p<0.001) and PAR (N=63–72, p<0.003), while only UCYN-A1 and A2 symbionts and their respective hosts represent the deep group. UCYN-B was unique in an overlapping distribution and resulted in positive significant correlations with both the shallow and deep group euphotic zone diazotrophs (e.g., Trichodesmium, N=108, p<0.001; UCYN-A1, N=120, p<0.004). Diazotroph targets in the deep-dwelling group significantly clustered and correlated positively with oxygen, depth, salinity, and fluorescence (N=108–120, p<0.03, except for UCYN-A2 and fluorescence, N=108, p=0.053). Despite clustering with the shallow group, het-3 was less robust in a negative correlation with salinity (N=108, p=0.005).
The results from the Spearman's rank correlations were further confirmed and visualized in the RDA bi-plot (Fig. 4a), which explains parameter importance (Fig. 4b). Correlations with nutrients and PAR were omitted due to the limited number of data points. Fluorescence, depth, and salinity correlated positively with each other and negatively with temperature, while oxygen was not significantly correlated with any other environmental parameters. The response variables UCYN-A1 and A2 and their respective hosts clustered with the explanatory variables, fluorescence, salinity, and depth, with a dependency towards oxygen. Conversely, the shallow euphotic group (response variables Trichodesmium, het-1, and het-2) clustered closer to explanatory variable temperature. In addition, most of the observed variance is explained by the two axes RDA1 (72 %) and RDA2 (22 %), indicative of depth and temperature, respectively, as the most important environmental parameters for diazotroph abundance in our study. Together they form a depth–temperature gradient (RDA1) where Trichodesmium occupies the warmest and shallowest waters and UCYN-A1 and A2 occupy the coldest and deepest waters, among the investigated cyanobacterial diazotrophs.
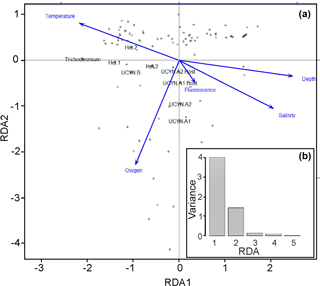
Figure 4(a–b) Multivariate RDA bi-plot (a) depicts the variance explained by the environmental parameters and qPCR data in several dimensions. (b) The majority of the variance (y axis: 0–4) in the dataset is explained by the RDA1 and RDA2 axes, meaning that most of the variance observed is explained by the environmental parameters. The arrows are the constrained explanatory vectors with the dots representing the superimposed unconstrained response variables. PAR and nutrients (DIP and DIN) were omitted due to limited data points.
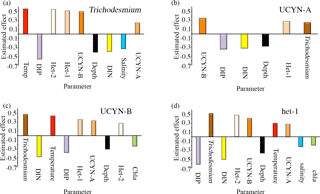
Figure 5(a–d) Meta-analysis bar graphs based on Table S6 for (a) Trichodesmium, (b) UCYN-B, (c) UCYN-A, and (d) het-1. Only the significant (p<0.05) parameters are included and arranged with the strongest effect to the left and weakest to the right (either positive or negative). Each parameter is color coded, and the cyanobacterial diazotrophs are shades of orange and brown. Red: temperature; blue: salinity; black: depth; green: chlorophyll a; yellow: DIN; and purple: DIP.
3.7 Global drivers of diazotrophic abundance
We found consistency between our results in the WTSP and the correlations identified in the 11 external datasets using the nonparametric correlation analyses and meta-analyses (Fig. 5a–d; Table S6). For example, in three of the external datasets, nifH gene abundances of Trichodesmium spp., UCYN-B, and het-1 were significantly positively correlated with temperature and negatively correlated with the same three parameters as in our study in the WTSP: salinity, DIP, and DIN. The latter correlations were identified in two regions of the WTSP (tropical and subtropical) and in the northern South China Sea (NSCS). In contrast to a significant positive correlation between UCYN-A nifH gene abundance and depth reported here in the WTSP, UCYN-A nifH gene abundance was negatively correlated with depth in 4 of the 11 external datasets (two regions of the WTSP, tropical Atlantic (TA), and NSCS). Moreover, and consistent with several of the other diazotrophs (Trichodesmium, UCYN-B, het-1), UCYN-A nifH gene abundance was negatively correlated with DIP and DIN concentrations (five and three additional external datasets, respectively) (Table S6).
Meta-analysis revealed similar groupings (e.g., shallow and deep) as observed in the WTSP; however, the significance was less robust (Fig. 5a–d; Table S6). For example nifH gene abundances of Trichodesmium and het-1 and het-2 were significantly positively correlated with temperature and negatively correlated with salinity (p<0.05). No significance was found for UCYN-A nifH gene abundance for the latter parameters, and UCYN-B nifH gene abundance was not significantly correlated with salinity, but was significantly positively correlated with temperature (p<0.05). In addition, UCYN-A was the only diazotroph that was not significantly correlated with het-2, while all other diazotrophs had a significant positive correlation with het-2 (p<0.05). Similar to our findings reported for the WTSP, all diazotrophs, except UCYN-A, correlated significantly negatively with depth and DIP and DIN concentrations (p<0.05) (except het-2 with DIP, which was not significant). Finally, UCYN-B and het-1 nifH gene abundances were significantly negatively correlated with Chl a (p<0.05), while Trichodesmium, UCYN-A, and het-2 were not.
4.1 Environmental conditions in the WTSP
The tropical South Pacific Ocean is one of the most oligotrophic regions of the world's oceans with chronically low dissolved nutrient concentrations, especially DIN, and is thus considered an area primed for N2 fixation. Likewise, we encountered surface hydrographic conditions in the WTSP that were consistently low in dissolved nutrient concentrations and similar to earlier reports for the equatorial Pacific (Bonnet et al., 2009; Dufour et al., 1999; Moutin et al., 2008; Van Den Broeck et al., 2004). The conservative tracers of temperature and salinity remained constant in the surface between the MA and SG regions; hence the elevated nutrient concentrations in the SG are likely not related to an eddy intrusion. The deviation away from a 16 : 1 relationship (Redfield ratio) (data not shown) in the upper 125 m in both regions (MA and SG) was indicative of DIN limitation. The low DIP concentrations in MA waters suggest utilization of DIP by diazotrophs in the absence of DIN; however, other sources of nitrogen were likely available to the phytoplankton community, e.g., dissolved organic nitrogen and N2 fixation (Karl et al., 2001).
4.2 Detection of diazotrophs and application of at-sea qPCR
Trichodesmium, UCYN-B, and the het group are easily identifiable with standard epi-fluorescence microscopy, and so these populations can readily be observed at sea. However, UCYN-A1 and UCYN-A2, and their respective hosts, require a lengthy fluorescent in situ hybridization (FISH) protocol that is difficult to implement in the field. Conversely, modern oceanographers have a suite of other molecular genetic tools, some of which are also seagoing and autonomous (e.g., Robidart et al., 2014; Ottesen et al., 2013; Preston et al., 2011), thereby making quantification of microscopically unidentified microorganisms tangible by quantifying their genes, simultaneous with collection of hydrographic data. Here, we describe a rather effective, steadfast (within 3 h of sample collection) seagoing nucleic acid extraction and qPCR technique to quantify diazotrophs by their nifH gene, which was used in real time during the OUTPACE cruise to help locate the LD stations for the purpose of the project (see Moutin et al., 2017). The comparisons of the at-sea assays to the lab-based full extraction protocol and qPCR on archived samples indicated that the assays were consistent (Fig. S3a–c), and surprisingly the shortened DNA extraction performed at sea had higher abundances for all three targets (UCYN-A1, UCYN-A2, and UCYN-B) in 16–25 % of the samples processed, depending on the target diazotroph.
The disparity between the enumerations made at sea and in the lab could result from technical and/or sampling differences. For example, we do not know the DNA extraction efficiency of either method used (at-sea vs. lab-based) or if extraction efficiency varies for a particular target, e.g., UCYN-A or UCYN-B. Variations between samples or replicates could be attested to different DNA extractions varying in their efficiencies based on target organism (Boström et al., 2004). Some strains of UCYN-B are known to secrete thick extracellular matrix (Sohm et al., 2011), while others form dense colonies, and some are symbiotic with diatoms (Carpenter and Janson, 2000). Moreover, the UCYN-A symbioses are considered fragile and easily dismantled (Thompson et al., 2012). The latter observations could potentially result in differences in extraction efficiency, which is supported by significant disparities in all samples for which the UCYN-A hosts were enumerated. However we find no clear pattern in higher detection for one method (at-sea vs. lab-based assays) or one particular target. Despite taking the samples from the same Niskin bottle for the archived and at-sea assays, we cannot discount the natural heterogeneity of plankton and differences in settling and that perhaps some samples were more enriched than others. Seldom are replicates processed in qPCR studies as those presented here, and so the disparity reported here highlights the need to address variation derived from sampling and/or extraction efficiencies. Nonetheless, at-sea (and lab-based) qPCRs could also be appended with a multiplexing approach to both increase and broaden the number of metabolic pathways (e.g., narB, rbcL, nirS) and/or phylotypes quantified simultaneously.
4.3 Abundance and vertical distribution of diazotrophs in the WTSP
Earlier work based on N isotope ratios (delta 15N) of suspended particulate matter and dissolved organic N in the WTSP suggested that new production is likely fueled by N2 fixation in this region (Hansell and Feely, 2000; Yoshikawa et al., 2005). The SP is also an area in which high nifH gene abundances of the unicellular diazotrophs, in particular UCYN-A and UCYN-B, have been previously reported (Biegala and Raimbault, 2008; Bonnet et al., 2009, 2015; Moisander et al., 2010) and account for a significant (74 %) portion of the areal N2 fixation (Bonnet et al., 2009). Hence, it was likely to encounter the presence of diazotrophic populations.
Recently UCYN-A and its various lineages have been highlighted as some of the most widespread and abundant diazotrophs (Farnelid et al., 2016, and references therein), which has led to the dramatic shift in the canonical paradigm of Trichodesmium as the only significant diazotroph. Surprisingly, here we report nifH gene abundances of the UCYN-A1 and UCYN-A2 lineages that are comparatively lower than earlier reports. In fact, UCYN-A1 and A2 were the least detected diazotrophs of our targets. Consistent with higher UCYN-A1 and A2 biomass (3.2 × 104 and 8.5 × 104 nifH copies L−1, respectively) at depth (60 m) at LD C were microscopy observations of high abundances of pico-eukaryotes similar in size and shape previously reported for the UCYN-A hosts (Krupke et al., 2013). The vertical distribution of UCYN-A1 (and A2) was similar to that reported by Moisander et al. (2010) and others, including earlier studies in the North Pacific Ocean (NP) and North Atlantic (NA), where maximum abundances of UCYN-A are common to deeper depths in the euphotic zone (below 45 m) (e.g., Bonnet et al., 2015; Foster et al., 2007; Goebel et al., 2010; Needoba et al., 2007). Likewise, we also observed, like others (Cabello et al., 2016), that the UCYN-A1 and A2 symbioses co-occur and typically have decreased abundance towards the DCM and maximum abundances slightly above the nitracline.
Unlike UCYN-A phylotypes, high nifH gene abundances of Trichodesmium were common in the upper 10 m of the MA region and included both surface slicks and free filaments, which was consistent with earlier observations of high surface densities further north in the SP (Moisander et al., 2010; Shiozaki et al., 2014), as well as regional satellite observations of blooms (Dupouy et al., 2011). Surface slicks have also been reported elsewhere, e.g., the NA (Goebel et al., 2010; Langlois et al., 2005). The depth of maximum nifH gene abundance for Trichodesmium deepened from the MA (10 m) region to the open gyre (SG, 31 m), which was similar to earlier reports in the equatorial Pacific (Bonnet et al., 2009). A niche partitioning has been suggested for Trichodesmium and unicellular diazotrophs in the SP (Bonnet et al., 2015; Moisander et al., 2010) and elsewhere (Goebel et al., 2010; Langlois et al., 2005; Messer et al., 2015). However, here in the WTSP, Trichodesmium abundance was positively correlated with UCYN-B, which instead is consistent with previous studies in other ocean basins, e.g., the Atlantic Ocean (Foster et al., 2007, 2009; Langlois et al., 2008), and the South China Sea (Moisander et al., 2008). UCYN-B co-occurred with Trichodesmium in the surface samples, although at lower nifH gene abundances, and more often UCYN-B had subsurface maxima (35–70 m) in both regions (MA and SG) of the transect. The latter is also consistent with Moisander et al. (2010), who observed maximum nifH gene abundances of UCYN-B north of the Fijian islands at 37 m.
All three heterocystous symbiont phylotypes co-occurred and were widespread in the MA, with het-1 as the most abundant and most highly detected het group (70 % detection or 84 of 120 samples). The early work of Moisander et al. (2010) detected het-1 at all but one of 26 stations sampled (56 % detected, or 56 of 100 samples), and highest nifH gene densities were reported northeast of our cruise transect. Moreover, Bonnet et al. (2015) detected het-1 and het-2 at the surface of one out of 10 stations west (approximately 10∘ W) of our cruise transect. Het-2 and het-3 were not quantified by Moisander et al. (2010) and het-3 was not quantified by Bonnet et al. (2015). Therefore, our study is among the first to report on the abundances and distributions for all three heterocystous diazotrophs in a large expanse of the SP. The three het phylotypes were however recently reported from a mesocosm (enclosed design) experiment in the Noumea lagoon, a low-nutrient low-chlorophyll region located along the New Caledonian coast (Turk-Kubo et al., 2015). In fact, het-1 and het-2 were among the most abundant diazotrophs in the first half of the experiment (Turk-Kubo et al., 2015). Two additional earlier studies have also reported microscopic observations of free-living Richelia in the same lagoon (Biegala and Raimbault, 2008; Garcia et al., 2007).
The highest densities (104–106 nifH copies L−1) of the Richelia phylotypes (het-1 and het-2) were restricted to the western region of the MA, and in the upper 12 m, which is shallower than the subsurface maximum (e.g., ≥ 25 m) previously reported for het-1 (and het-2) in the western tropical North Atlantic (WTNA) and NP (Church et al., 2005; Foster et al., 2007; Goebel et al., 2010). Our microscopy observations from SD 5 to 7 and LD A indicated that near-surface Rhizosolenia populations were in a moribund state since frustules were broken and free filaments of Richelia were observed. Our observations also coincide with a region of high backscattering measurements in the upper water column (5–30 m) (Dupouy et al., 2018). Het-1 nifH genes were 4 orders of magnitude higher in abundance in the moored sediment traps of LD A (325 m: 2.0 × 107 nifH copies L−1) and LD B (325 and 500 m: 5.8 × 106 and 1.10 × 107 nifH copies L−1, respectively) (Caffin et al., 2018) than the nifH genes detected in the overlying waters (3.11 × 103 nifH copies L−1 and 4.1 × 102 nifH copies L−1, respectively). Combined, the latter observations suggest that a higher density of the het-1 population was likely present prior to our sampling and perhaps derived from a “seed” population originating in the coastal regions of New Caledonia and that they play an important role for export production in this region, as has previously been shown in the NP (Karl et al., 2012), for example.
The UCYN-C phylotype was poorly detected in the at-sea assays (61 % samples were bd and maximum abundance was 5.0 × 102 nifH copies L−1), and as such was not enumerated in the archived samples. The low detection of UCYN-C is consistent with Taniuchi et al. (2012), who estimated that UCYN-C only represented a small portion of diazotrophs detected in the western Pacific (Kuroshio Current). However, a recent study reported relatively high UCYN-C abundances in the open waters of the Solomon Sea (north of the MA) (Berthelot et al., 2017). UCYN-C has also been observed in the New Caledonian lagoon (Turk-Kubo et al., 2015), where it was the most dominant diazotroph in the first part of the aforementioned mesocosm experiment (Turk-Kubo et al., 2015). Moreover, Turk-Kubo et al. (2015) reviewed the specificity of the UCYN-C assay used in our study (Foster et al., 2007) and concluded that it does quantify the majority of UCYN-C phylotypes. Hence, like most plankton, abundances can be patchy, as was observed with UCYN-C in our study.
4.4 UCYN-A and host (co-)occurrence
Earlier and recent work has suggested a high host dependency (e.g., smaller and streamlined genomes) and selectivity in the UCYN-A based symbioses (Cabello et al., 2016; Cornejo-Castillo et al., 2016; Farnelid et al., 2016; Krupke et al., 2013, 2014; Thompson et al., 2012; Tripp et al., 2010). Moreover, the UCYN-A partnerships are also considered mutualistic, where the host and symbiont both benefit by exchange of metabolites (e.g., reduced C and N, respectively) (Krupke et al., 2014; Thompson et al., 2012), and based on streamlined genome content of UCYN-A, it has a photo-fermentative metabolism dependent on other organisms (Tripp et al., 2010; Zehr et al., 2008). Hence one would expect parallel distributions for both partners. Some have argued that the partnership is also obligatory since few observations of free-living hosts have been reported and abundances of free symbionts assumed to be derived from disruption during sample preparation are always correlated with their hosts (Cabello et al., 2016; Krupke et al., 2014; Thompson et al., 2012). Thus, by use of our newly designed oligonucleotides for the UCYN-A1 host and previously designed oligonucleotides for the UCYN-A2 host (Thompson et al., 2014), we unexpectedly found that both UCYN-A1 and A2 were often (89 and 59 %, respectively; not considering dnq) detected in the absence (or bd) of their respective hosts, while the hosts, when detected, always coincided with increased UCYN-A abundance. Our observations could result if the UCYN-A lineages live in either a loose association, or perhaps with a wider diversity of hosts than detected by the UCYN-A host assays.
The number of cells per partner lineage is considered specific as well, such that 1–2 UCYN-A1 cell(s) is associated with a prymnesiophyte partner (UCYN-A1 host) and the larger B. bigelowii (UCYN-A2 host) host associates with multiple and variable numbers of UCYN-A2 cells to compensate for its higher N requirement (Cornejo-Castillo et al., 2016). We consistently observed higher abundances of the nifH genes for the UCYN-A1 and A2 symbionts compared to their respective hosts' 18S rRNA genes (2–10 and 6–34 times more abundant, for A1 and A2, respectively). Similar results were found by Thompson et al. (2014), who observed symbiont ∕ host ratios of 0.2–11. However, a higher detection of the UCYN-A 1 (and A2) in absence of the host detection would result if there is a broader diversity of hosts associated with the UCYN-A symbionts. Ultimately a CARD-FISH (CAtalyzed Reporter Deposition – Fluorescence In Situ Hybridization) approach using the same 18S rRNA oligonucleotide as in the qPCR would help distinguish the discrepancy between the partner gene copy abundances.
A symbiosome-like compartment has also been described attached to the UCYN-A2 host or residing freely (Cornejo-Castillo et al., 2016). Thus, another plausible explanation for the higher abundances of the UCYN-A2, in particular, in the absence of their respective host, could result if our assays quantified UCYN-A2 residing in a dislodged free-floating symbiosome, or an overestimate of the UCYN-A2 due to cross-reactivity with UCYN-A3 lineage as expected by in silico tests (Farnelid et al., 2016). It is less likely that the UCYN-A2 was overestimated due to cross-reaction with UCYN-A1 templates since our cross-reactivity tests showed a weak cross-reaction (see below).
4.5 Environmental influence on diazotroph abundances and distributions
The annual N inputs through biological N2 fixation in the oceans is considered high, ranging from 100 to 200 Tg N (Eugster and Gruber, 2012; Luo et al., 2012), yet large uncertainties remain in what factor(s) influence the abundance, distribution, and activity of marine diazotrophs. Initially, we hypothesized that the conditions favoring a particular cyanobacterial diazotroph would be unique given the contrasting life histories of different diazotrophic groups (free-living, colonial, and symbiotic). Moreover, we also suspected that the conditions promoting DDAs would differ from those influencing the UCYN-A-based symbioses given the vast differences in the symbionts and hosts (e.g., genome content of symbiont, cell size of symbiont and hosts in the two systems; expected number of symbionts per host; host phylogeny: diatom vs. prymnesiophyte). Thus, determining the condition or sets of conditions that drive cyanobacterial diazotroph distribution, abundance, and activity is of great interest.
Hydrographic conditions and dissolved nutrient concentrations measured at the time of sampling were used to correlate diazotrophic abundances with various environmental parameters. Consistently, in two independent statistical tests, two groups emerged in the WTSP: (1) UCYN-A1 and A2 and their hosts (2) het-1, het-2, het-3, UCYN-B and Trichodesmium. Thus, conditions favoring the UCYN-A-based symbioses differ from the conditions for DDAs and free-living cyanobacterial diazotrophs.
Temperature is often cited as the most important driver of diazotroph abundance and distribution (Messer et al., 2016; Moisander et al., 2010), and recently marine nitrogen fixation was shown to be controlled by surface irradiance and dissolved oxygen concentration (Luo et al., 2014). As shown earlier in the WTSP, both Trichodesmium spp. and UCYN-B were most abundant in warmer surface waters (> 27 ∘C) in the north, while UCYN-A dominated in the cooler (24–26 ∘C) southern waters of WTSP (Bonnet et al., 2015; Moisander et al., 2010). Likewise, we found similar abundances and temperature optima for the latter three diazotrophs and significant correlations between the various diazotrophs and temperature. In fact, all diazotrophs, except the UCYN-A lineages, were significantly positively correlated with temperature in the WTSP. In addition to temperature, the environmental parameters of PAR, salinity, and depth also significantly influenced nifH gene abundance and distribution. Moreover, the latter two variables separated the abundances of the UCYN-A symbioses (A1 and A2) from the rest of the diazotrophs in the WTSP, including both free-living phylotypes and the symbiotic heterocystous lineages.
The maximum abundances at depth for UCYN-A1 and A2 were slightly above or at the nitracline and coincided with higher measures of fluorescence from the CTD. The latter is consistent with observations of high UCYN-A abundances in coastal habitats (Bombar et al., 2014), estuaries (Messer et al., 2015), or in waters that have been recently entrained with new nutrients (Moisander et al., 2010). Increased nifH copies and/or nifH gene expression for UCYN-A have also been reported from bioassay experiments amended with nutrients, including DIN, phosphate, and iron (Krupke et al., 2015; Langlois et al., 2012; Moisander et al., 2012). The latter is in contrast with the data reported here in the WTSP (including the meta-analysis) and several of the external datasets (e.g., WTSP, TA, NA, NSCS), which find a negative correlation between DIN and DIP concentrations and nifH gene abundance of most of the diazotrophs, including UCYN-A. Across the cruise transect, conditions instead favored diatoms in symbiosis with diazotrophs since surface DIP and DiSi concentrations were considered not limiting (Thierry Moutin, personal communication, 2017), while DIN was below detection, and as reported here, resulted in higher abundances of het-1 nifH gene copies and observations of Rhizosolenia hosts in the MA. In the WTNA, waters with high DiSi concentration and low N : P ratios, driven by a disproportionate utilization of N relative to P, results in consistent and widespread blooms of the Hemiaulus–Richelia symbioses (het-2) (Foster et al., 2007; Subramaniam et al., 2008).
All the diazotrophs described here are either photoautotrophic or associated with photoautotrophic partners (UCYN-A, het-group). Therefore, light irradiance (e.g., PAR) and availability will impact the abundance and distribution of the diazotrophic populations. Results from CARD-FISH observations of the UYCN-A1 and A2 symbioses have reported a strong dependency on light intensity, which results in higher abundances nearer to the surface (Cabello et al., 2016). Presence in shallower waters is also thought to be strategic for avoiding competition (Cabello et al., 2016). However, in the WTSP, in 11 of the 14 stations at which UCYN-A1 and A2 were detected at subsurface depth maxima, the same lineages (and corresponding hosts) were undetected at the surface and displayed a negative correlation with PAR. Microscopy observations also confirmed higher numbers of pico-eukaryotes at depth. Hence, it would appear that low light correlates with high abundances of UCYN-A, while the other free-living diazotrophs and symbiotic het-1 and het-2 were positively correlated with PAR and had maxima closer to the surface with higher PAR. Interestingly, we found no significant correlation between PAR and the UCYN-A host lineages, which is unexpected since the host partners require light for photosynthesis.
In an attempt to identify the consistency in the correlation patterns identified in the WTSP with other regions of the world's ocean, the same statistical analyses were performed on 11 publically available datasets and subsequently run through a meta-analysis. Our statistical analyses provided coefficients and p values for easy evaluation and comparisons between data sets for the influence of environmental parameter(s) and diazotroph abundance. The analyses confirmed that UCYN-A indeed stands out from the other diazotrophs in terms of environmental parameter influence, mainly by not being significantly correlated with temperature, which for all other diazotrophs was a significant positive correlation. In terms of temperature, UCYN-A has the lowest temperature optimum among the target cyanobacterial diazotrophs, which will influence their distributions and subsequent correlation with temperature both regionally and vertically. The studies in the meta-analysis include a wide geographical range and, independent of ocean basin, the majority of the datasets found no significant correlation between UCYN-A and temperature. For most other environmental parameters the pattern for UCYN-A in the WTSP does not hold true in the meta-analysis. For example a significant negative correlation with depth was found for UCYN-A and perhaps related to high surface temperatures, given that Moisander et al. (2010) observed UCYN-A closer to the surface in the southern SP where surface temperatures were typically cooler. However, for the other target diazotrophs, depth and salinity follow the same pattern as observed in the WTSP (except for UCYN-B, which was not significantly correlated with salinity). Interestingly, UCYN-B seems to be mostly detected in the Pacific, where it was also found to be significantly negatively correlated with salinity, while in the Atlantic, perhaps due to lower detections or riverine impact, it was not significantly correlated, resulting in no significant correlation in the meta-analysis overall. Furthermore, what did unify all diazotrophs in the meta-analysis were their consistent significant positive correlations with each other and significant negative correlations between abundance and concentrations of DIP and DIN, which was also observed in the WTSP, and again UCYN-A was the exception.
In summary, the correlations observed in the WTSP were not always consistent with the meta-analysis of the external datasets. We attribute the inconsistencies in part to seasonal differences in sample collections and the impact of an individual environmental parameter or sets of parameters on a local and regional scale, particularly for coastal studies that make it difficult to unambiguously explain the abundance and distribution patterns. Determining the condition or sets of conditions favoring one diazotroph or life history strategy (free-living vs. symbiotic) is complex and likely not all diazotrophs are influenced by the same condition in time and space.
4.6 Estimation of diazotrophs by nifH qPCR
When interpreting abundance estimates by qPCR there are a few assumptions to keep in mind. A limitation of qPCR assays is that gene copies do not equal cell abundances. Recent evidence in filamentous and heterocystous cyanobacteria reports evidence of polyploidy dependent on cell cycle (Griese et al., 2011; Sargent et al., 2016; Sukenik et al., 2012). Moreover, Trichodesmium may contain up to 100 genome copies per cell (Sargent et al., 2016), thus a potential for overestimation. Conversely, underestimation of gene copies by qPCR is also plausible if one considers that DNA extraction efficiency is not 100 % and can vary among species and DNA extraction kits (Mumy and Findlay, 2004), and if high probe specificity favors exclusion of closely related phylotypes for a particular target or lineage.
A final consideration with qPCR, as shown here, is the degree of cross-reactivity in assays targeting closely related lineages (e.g., UCYN-A and het). Oligonucleotide specificity as a source of underestimation of the UCYN-A lineages was recently reviewed by a de nova analysis (Farnelid et al., 2016) showing the potential to underestimate UCYN-A sublineages since the widely used oligonucleotides for UCYN-A1 contain several mismatches to the other UCYN-A sublineages. The latter becomes important when the sublineages co-occur. Here, however, we highlight the potential to overestimate. For example, UCYN-A2 oligonucleotides amplified the UCYN-A1 templates, indicating a tendency to overquantify UCYN-A2 in the presence of A1. Moreover, when the annealing temperature was set to 64 ∘C, to distinguish between UCYN-A1 and A2 as recommended by Thompson et al. (2014), the assay still failed to separate the two sub-lineages when run in fast mode. Thus, the fast mode feature has a shortcoming that could influence a wider range of targets than the ones presented here. We observed the same cross-reactivity reported earlier (Foster et al., 2007) for het-1 and het-2 when run in fast mode and highlight the potential to overestimate het-2 if het-1 co-occurs at densities of approximately 106 nifH copies L−1. The latter observation has never been reported.
Consistent with earlier observations in the WTSP, we found diazotrophic cyanobacteria to be abundant, with the most abundant cyanobacterial diazotrophs being UCYN-B, Trichodesmium, and the symbiotic Richelia lineage het-1. Although the cell integrity and detection of het-1 in water column samples and those from depth (e.g., sediment traps) indicated that the populations were in a senescent state, our work represents one of the first documentations of the three DDA populations in a wide expanse of the WTSP. In contrast to earlier work in the SP and other recent reports from global ocean surveys (Farnelid et al., 2016; Martínez-Pérez et al., 2016), we observed low abundances and poor detection of both UCYN-A (A1 and A2) lineages. According to our qPCR results, UCYN-A was also enumerated when its respective hosts were below detection, which contrasts to the assumed high fidelity and dependency in the partnerships; however, we cannot discount that the disparity in host–symbiont detection was not a result of qPCR oligonucleotide assay bias and/or overestimations indicated by our cross-reactivity tests.
Our initial hypothesis was that the condition or sets of conditions, which promote the distribution of one diazotroph, would differ. Moreover, the parameters for symbiotic diazotrophs should also differ from those of free-living phylotypes, and given the vast difference in hosts (diatoms and prymnesiophyte) and genome content for the het and UCYN-A symbionts, we further hypothesized divergent conditions favoring one symbiosis over another. In the WTSP, the same conditions favored abundances of both the free-living phylotypes and the diatom (het groups) symbioses. However, the same conditions impacted the abundance of UCYN-A-based symbiosis negatively, hence somewhat supporting our initial hypothesis that conditions for one symbiosis type would differ. In the external datasets, however, we observed differences in environmental conditions favoring abundances of the investigated diazotrophs compared to the WTSP, which underscores that diazotrophs are not similarly influenced by the same condition in time and space.
Multivariate approaches on numerous parameters and with high spatial resolution are required to understand the complex and often indirect effects that govern species distribution. Finally, this study highlights the application and considerations of at-sea qPCR for steadfast quantification of microbial populations. Moreover, a meta-analysis was used to determine the influence of environmental parameters on the distribution and abundance of diazotrophs in the WTSP region and in other ocean basins. However, it is of great interest to know if the same parameters influence gene expressions (e.g., nifH), and ultimately N2 fixation rates, in the same manner, thus understanding the weight of environmental parameters influencing diazotrophic abundance and distribution. Given the global significance of N2 fixation as a major new source of N to the oceans, the meta-analysis presented here could be directly applicable to improving parameter constraints on model-based approaches for predicting areas prone to diazotrophy.
Underlying data are reported in the Supplement, and all cruise and meta-data can also be accessed at http://www.obs-vlfr.fr/proof/php/outpace/outpace.php. The qPCR data will also be made available at https://pangaea.de.
The supplement related to this article is available online at: https://doi.org/10.5194/bg-15-1559-2018-supplement.
The authors declare that they have no conflict of interest.
This article is part of the special issue “Interactions between planktonic organisms and biogeochemical cycles across trophic and N2 fixation gradients in the western tropical South Pacific Ocean: a multidisciplinary approach (OUTPACE experiment)”. It is not associated with a conference.
The participation (Marcus Stenegren and Andrea Caputo), sample processing,
and work presented here is supported by the Knut and Alice Wallenberg
foundation (to Rachel A. Foster). This is a contribution to the OUTPACE
(Oligotrophy from Ultra-oligoTrophy PACific Experiment) project funded by the
French research national agency (ANR-14-CE01-0007-01), the LEFE-CYBER program
(CNRS-INSU), the GOPS program (IRD), and the CNES (BC T23, ZBC 4500048836).
The OUTPACE cruise (https://doi.org/10.17600/15000900) was managed by Thierry Moutin
and Sophie Bonnet from the MIO (Mediterranean Institute of Oceanography).
Thierry Moutin is specifically thanked for invaluable input and feedback on
this paper. The authors thank the crew of the R/V L'Atalante for
outstanding shipboard operation. Gilles Rougier and Marc Picheral are thanked
for their efficient help in CTD rosette management and data processing, as is
Catherine Schmechtig for the LEFE-CYBER database management. We especially
acknowledge Andreas Krupke (formerly of WHOI), Kyle Frischkorn (WHOI),
Mar Benavides (MIO/IRD), Hugo Berthelot (MIO), and Mathieu Caffin (MIO).
Olivier Grosso (MIO) and Sandra Hélias (MIO) are acknowledged for
nutrient analyses. Additionally we thank Lotta Berntzon for assisting in
sample processing and Konrad Karlsson for assistance with the multivariate
statistics. Lastly we thank Bethany Jenkins, Anders Bignert and two anonymous
referees for invaluable feedback and suggestions on our paper.
Edited by: Gerhard Herndl
Reviewed by: two anonymous referees
Aminot, A. and Kérouel, R.: Dosage automatique des nutriments dans les eaux marines: méthodes en flux continu, in: Méthodes d'analyse en milieu marin, Ifremer, p. 188, available at: http://cat.inist.fr/?aModele=afficheN&cpsidt=18706171 (last access: 1 December 2016), 2007.
Benavides, M., Bonnet, S., Hernández, N., Martínez-Pérez, A. M., Nieto-Cid, M., Álvarez-Salgado, X. A., Baños, I., Montero, M. F., Mazuecos, I. P., Gasol, J. M., Osterholz, H., Dittmar, T., Berman-Frank, I., and Arístegui, J.: Basin-wide N2 fixation in the deep waters of the Mediterranean Sea, Global Biogeochem. Cy., 30, 952–961, https://doi.org/10.1002/2015GB005326, 2016.
Bench, S. R., Heller, P., Frank, I., Arciniega, M., Shilova, I. N., and Zehr, J. P.: Whole genome comparison of six Crocosphaera watsonii strains with differing phenotypes, J. Phycol., 49, 786–801, https://doi.org/10.1111/jpy.12090, 2013.
Berman-Frank, I., Lundgren, P., Chen, Y.-B., Küpper, H., Kolber, Z., Bergman, B., and Falkowski, P.: Segregation of Nitrogen Fixation and Oxygenic Photosynthesis in the Marine Cyanobacterium Trichodesmium, Science, 294, 1534–1537, https://doi.org/10.1126/science.1064082, 2001.
Berthelot, H., Moutin, T., L'Helguen, S., Leblanc, K., Hélias, S., Grosso, O., Leblond, N., Charrière, B., and Bonnet, S.: Dinitrogen fixation and dissolved organic nitrogen fueled primary production and particulate export during the VAHINE mesocosm experiment (New Caledonia lagoon), Biogeosciences, 12, 4099–4112, https://doi.org/10.5194/bg-12-4099-2015, 2015.
Berthelot, H., Benavides, M., Moisander, P. H., Grosso, O., and Bonnet, S.: High-nitrogen fixation rates in the particulate and dissolved pools in the Western Tropical Pacific (Solomon and Bismarck Seas), Geophys. Res. Lett., 44, 8414–8423, https://doi.org/10.1002/2017GL073856, 2017.
Biegala, I. C. and Raimbault, P.: High abundance of diazotrophic picocyanobacteria (< 3 µm) in a Southwest Pacific coral lagoon, Aquat. Microb. Ecol., 51, 45–53, https://doi.org/10.3354/ame01185, 2008.
Blain, S., Sarthou, G., and Laan, P.: Distribution of dissolved iron during the natural iron-fertilization experiment KEOPS (Kerguelen Plateau, Southern Ocean), Deep-Sea Res. Pt. II, 55, 594–605, https://doi.org/10.1016/j.dsr2.2007.12.028, 2008.
Bombar, D., Moisander, P. H., Dippner, J. W., Foster, R. A., Voss, M., Karfeld, B., and Zehr, J. P.: Distribution of diazotrophic microorganisms and nifH gene expression in the Mekong River plume during intermonsoon, Mar. Ecol.-Prog. Ser., 424, 39–52, https://doi.org/10.3354/meps08976, 2011.
Bombar, D., Heller, P., Sanchez-Baracaldo, P., Carter, B. J., and Zehr, J. P.: Comparative genomics reveals surprising divergence of two closely related strains of uncultivated UCYN-A cyanobacteria, ISME J., 8, 2530–2542, https://doi.org/10.1038/ismej.2014.167, 2014.
Bombar, D., Paerl, R. W., and Riemann, L.: Marine Non-Cyanobacterial Diazotrophs: Moving beyond Molecular Detection, Trends Microbiol., 24, 916–927, https://doi.org/10.1016/j.tim.2016.07.002, 2016.
Bonnet, S., Biegala, I. C., Dutrieux, P., Slemons, L. O., and Capone, D. G.: Nitrogen fixation in the western equatorial Pacific: Rates, diazotrophic cyanobacterial size class distribution, and biogeochemical significance, Global Biogeochem. Cy., 23, GB3012, https://doi.org/10.1029/2008GB003439, 2009.
Bonnet, S., Dekaezemacker, J., Turk-Kubo, K. A., Moutin, T., Hamersley, R. M., Grosso, O., Zehr, J. P., and Capone, D. G.: Aphotic N2 Fixation in the Eastern Tropical South Pacific Ocean, PLOS ONE, 8, e81265, https://doi.org/10.1371/journal.pone.0081265, 2013.
Bonnet, S., Rodier, M., Turk-Kubo, K. A., Germineaud, C., Menkes, C., Ganachaud, A., Cravatte, S., Raimbault, P., Campbell, E., Quéroué, F., Sarthou, G., Desnues, A., Maes, C., and Eldin, G.: Contrasted geographical distribution of N2 fixation rates and nifH phylotypes in the Coral and Solomon Seas (southwestern Pacific) during austral winter conditions, Global Biogeochem. Cy., 29, 1874–1892, https://doi.org/10.1002/2015GB005117, 2015.
Bonnet, S., Berthelot, H., Turk-Kubo, K., Cornet-Barthaux, V., Fawcett, S., Berman-Frank, I., Barani, A., Grégori, G., Dekaezemacker, J., Benavides, M., and Capone, D. G.: Diazotroph derived nitrogen supports diatom growth in the South West Pacific: A quantitative study using nanoSIMS, Limnol. Oceanogr., 61, 1549–1562, https://doi.org/10.1002/lno.10300, 2016.
Bonnet, S., Caffin, M., Berthelot, H., and Moutin, T.: Hot spot of N2 fixation in the western tropical South Pacific pleads for a spatial decoupling between N2 fixation and denitrification, P. Natl. Acad. Sci. USA, 114, E2800–E2801, https://doi.org/10.1073/pnas.1619514114, 2017.
Bonnet, S., Caffin, M., Berthelot, H., Grosso, O., Benavides, M., Helias-Nunige, S., Guieu, C., Stenegren, M., and Foster, R. A.: In depth characterization of diazotroph activity across the Western Tropical South Pacific hot spot of N2 fixation, Biogeosciences Discuss., https://doi.org/10.5194/bg-2017-567, in review, 2018.
Boström, K. H., Simu, K., Hagström, Å., and Riemann, L.: Optimization of DNA extraction for quantitative marine bacterioplankton community analysis, Limnol. Oceanogr.-Meth., 2, 365–373, 2004.
Cabello, A. M., Cornejo-Castillo, F. M., Raho, N., Blasco, D., Vidal, M., Audic, S., de Vargas, C., Latasa, M., Acinas, S. G., and Massana, R.: Global distribution and vertical patterns of a prymnesiophyte–cyanobacteria obligate symbiosis, ISME J., 10, 693–706, https://doi.org/10.1038/ismej.2015.147, 2016.
Caffin, M., Berthelot, H., Cornet-Barthaux, V., and Bonnet, S.: Transfer of diazotroph-derived nitrogen to the planktonic food web across gradients of N2 fixation activity and diversity in the Western Tropical South Pacific, Biogeosciences Discuss., https://doi.org/10.5194/bg-2017-572, in review, 2018.
Carpenter, E. J. and Janson, S.: Intracellular Cyanobacterial Symbionts in the Marine Diatom Climacodium Frauenfeldianum (bacillariophyceae), J. Phycol., 36, 540–544, https://doi.org/10.1046/j.1529-8817.2000.99163.x, 2000.
Church, M. J., Jenkins, B. D., Karl, D. M., and Zehr, J. P.: Vertical distributions of nitrogen-fixing phylotypes at Stn Aloha in the oligotrophic North Pacific Ocean, Aquat. Microb. Ecol., 38, 3–14, https://doi.org/10.3354/ame038003, 2005.
Church, M. J., Björkman, K. M., Karl, D. M., Saito, M. A., and Zehr, J. P.: Regional distributions of nitrogen-fixing bacteria in the Pacific Ocean, Limnol. Oceanogr., 53, 63–77, https://doi.org/10.4319/lo.2008.53.1.0063, 2008.
Claustre, H. and Maritorena, S.: The Many Shades of Ocean Blue, Science, 302, 1514–1515, https://doi.org/10.1126/science.1092704, 2003.
Cornejo-Castillo, F. M., Cabello, A. M., Salazar, G., Sánchez-Baracaldo, P., Lima-Mendez, G., Hingamp, P., Alberti, A., Sunagawa, S., Bork, P., Vargas, C. de, Raes, J., Bowler, C., Wincker, P., Zehr, J. P., Gasol, J. M., Massana, R., and Acinas, S. G.: Cyanobacterial symbionts diverged in the late Cretaceous towards lineage-specific nitrogen fixation factories in single-celled phytoplankton, Nat. Commun., 7, 11071, https://doi.org/10.1038/ncomms11071, 2016.
Deutsch, C., Sarmiento, J. L., Sigman, D. M., Gruber, N., and Dunne, J. P.: Spatial coupling of nitrogen inputs and losses in the ocean, Nature, 445, 163–167, https://doi.org/10.1038/nature05392, 2007.
Dufour, P., Charpy, L., Bonnet, S., and Garcia, N.: Phytoplankton nutrient control in the oligotrophic South Pacific subtropical gyre (Tuamotu Archipelago), Mar. Ecol.-Prog. Ser., 179, 285–290, https://doi.org/10.3354/meps179285, 1999.
Dupouy, C., Benielli-Gary, D., Neveux, J., Dandonneau, Y., and Westberry, T. K.: An algorithm for detecting Trichodesmium surface blooms in the South Western Tropical Pacific, Biogeosciences, 8, 3631–3647, https://doi.org/10.5194/bg-8-3631-2011, 2011.
Dupouy, C., Frouin, R., Tedetti, M., Maillard, M., Rodier, M., Lombard, F., Guidi, L., Picheral, M., Duhamel, S., Charrière, B., and Sempéré, R.: Diazotrophic Trichodesmium influence on ocean color and pigment composition in the South West tropical Pacific, Biogeosciences Discuss., https://doi.org/10.5194/bg-2017-570, in review, 2018.
Dyhrman, S. T. and Ruttenberg, K. C.: Presence and regulation of alkaline phosphatase activity in eukaryotic phytoplankton from the coastal ocean: Implications for dissolved organic phosphorus remineralization, Limnol. Oceanogr., 51, 1381–1390, https://doi.org/10.4319/lo.2006.51.3.1381, 2006.
Dyhrman, S. T., Chappell, P. D., Haley, S. T., Moffett, J. W., Orchard, E. D., Waterbury, J. B., and Webb, E. A.: Phosphonate utilization by the globally important marine diazotroph Trichodesmium, Nature, 439, 68–71, https://doi.org/10.1038/nature04203, 2006.
Eugster, O. and Gruber, N.: A probabilistic estimate of global marine N-fixation and denitrification, Global Biogeochem. Cy., 26, GB4013, https://doi.org/10.1029/2012GB004300, 2012.
Farnelid, H., Turk-Kubo, K., Muñoz-Marín, M. del C., and Zehr, J. P.: New insights into the ecology of the globally significant uncultured nitrogen-fixing symbiont UCYN-A, Aquat. Microb. Ecol., 77, 125–138, https://doi.org/10.3354/ame01794, 2016.
Fernandez, C., Farías, L., and Ulloa, O.: Nitrogen Fixation in Denitrified Marine Waters, PLOS ONE, 6, e20539, https://doi.org/10.1371/journal.pone.0020539, 2011.
Fitzsimmons, J. N., Boyle, E. A., and Jenkins, W. J.: Distal transport of dissolved hydrothermal iron in the deep South Pacific Ocean, P Natl Acad Sci USA, 111(47), 16654–16661, https://doi.org/10.1073/pnas.1418778111, 2014.
Foster, R. A. and Zehr, J. P.: Characterization of diatom–cyanobacteria symbioses on the basis of nifH, hetR and 16S rRNA sequences, Environ. Microbiol., 8, 1913–1925, https://doi.org/10.1111/j.1462-2920.2006.01068.x, 2006.
Foster, R. A., Subramaniam, A., Mahaffey, C., Carpenter, E. J., Capone, D. G., and Zehr, J. P.: Influence of the Amazon River plume on distributions of free-living and symbiotic cyanobacteria in the western tropical north Atlantic Ocean, Limnol. Oceanogr., 52, 517–532, https://doi.org/10.4319/lo.2007.52.2.0517, 2007.
Foster, R. A., Subramaniam, A., and Zehr, J. P.: Distribution and activity of diazotrophs in the Eastern Equatorial Atlantic, Environ. Microbiol., 11, 741–750, https://doi.org/10.1111/j.1462-2920.2008.01796.x, 2009.
Foster, R. A., Goebel, N. L., and Zehr, J. P.: Isolation of Calothrix Rhizosoleniae (cyanobacteria) Strain Sc01 from Chaetoceros (bacillariophyta) Spp. Diatoms of the Subtropical North Pacific Ocean1, J. Phycol., 46, 1028–1037, https://doi.org/10.1111/j.1529-8817.2010.00885.x, 2010.
Garcia, N., Raimbault, P., and Sandroni, V.: Seasonal nitrogen fixation and primary production in the Southwest Pacific: nanoplankton diazotrophy and transfer of nitrogen to picoplankton organisms, Mar. Ecol.-Prog. Ser., 343, 25–33, https://doi.org/10.3354/meps06882, 2007.
Goebel, N. L., Turk, K. A., Achilles, K. M., Paerl, R., Hewson, I., Morrison, A. E., Montoya, J. P., Edwards, C. A., and Zehr, J. P.: Abundance and distribution of major groups of diazotrophic cyanobacteria and their potential contribution to N2 fixation in the tropical Atlantic Ocean, Environ. Microbiol., 12, 3272–3289, https://doi.org/10.1111/j.1462-2920.2010.02303.x, 2010.
Gourdeau, L., Kessler, W. S., Davis, R. E., Sherman, J., Maes, C., and Kestenare, E.: Zonal Jets Entering the Coral Sea, J. Phys. Oceanogr., 38, 715–725, https://doi.org/10.1175/2007JPO3780.1, 2008.
Griese, M., Lange, C., and Soppa, J.: Ploidy in cyanobacteria, FEMS Microbiol. Lett., 323, 124–131, https://doi.org/10.1111/j.1574-6968.2011.02368.x, 2011.
Halm, H., Musat, N., Lam, P., Langlois, R., Musat, F., Peduzzi, S., Lavik, G., Schubert, C. J., Singha, B., LaRoche, J., and Kuypers, M. M. M.: Co-occurrence of denitrification and nitrogen fixation in a meromictic lake, Lake Cadagno (Switzerland), Environ. Microbiol., 11, 1945–1958, https://doi.org/10.1111/j.1462-2920.2009.01917.x, 2009.
Halm, H., Lam, P., Ferdelman, T. G., Lavik, G., Dittmar, T., LaRoche, J., D'Hondt, S., and Kuypers, M. M.: Heterotrophic organisms dominate nitrogen fixation in the South Pacific Gyre, ISME J., 6, 1238–1249, https://doi.org/10.1038/ismej.2011.182, 2012.
Hansell, D. A. and Feely, R. A.: Atmospheric Intertropical Convergence impacts surface ocean carbon and nitrogen biogeochemistry in the western tropical Pacific, Geophys. Res. Lett., 27, 1013–1016, https://doi.org/10.1029/1999GL002376, 2000.
Haselkorn, R.: Heterocysts, Ann. Rev. Plant Physio., 29, 319–344, https://doi.org/10.1146/annurev.pp.29.060178.001535, 1978.
Hood, R. R., Coles, V. J., and Capone, D. G.: Modeling the distribution of Trichodesmium and nitrogen fixation in the Atlantic Ocean, J. Geophys. Res., 109, C06006, https://doi.org/10.1029/2002JC001753, 2004.
Howard, J. B. and Rees, D. C.: Structural Basis of Biological Nitrogen Fixation, Chem. Rev., 96, 2965–2982, https://doi.org/10.1021/cr9500545, 1996.
Jacobson, M. R., Brigle, K. E., Bennett, L. T., Setterquist, R. A., Wilson, M. S., Cash, V. L., Beynon, J., Newton, W. E., and Dean, D. R.: Physical and genetic map of the major nif gene cluster from Azotobacter vinelandii, J. Bacteriol., 171, 1017–1027, 1989.
Karl, D., Letelier, R., Tupas, L., Dore, J., Christian, J., and Hebel, D.: The role of nitrogen fixation in biogeochemical cycling in the subtropical North Pacific Ocean, Nature, 388, 533–538, https://doi.org/10.1038/41474, 1997.
Karl, D. M., Björkman, K. M., Dore, J. E., Fujieki, L., Hebel, D. V., Houlihan, T., Letelier, R. M., and Tupas, L. M.: Ecological nitrogen-to-phosphorus stoichiometry at station ALOHA, Deep-Sea Res. Pt. II, 48, 1529–1566, https://doi.org/10.1016/S0967-0645(00)00152-1, 2001.
Karl, D. M., Church, M. J., Dore, J. E., Letelier, R. M., and Mahaffey, C.: Predictable and efficient carbon sequestration in the North Pacific Ocean supported by symbiotic nitrogen fixation, P. Natl. Acad. Sci. USA, 109, 1842–1849, https://doi.org/10.1073/pnas.1120312109, 2012.
Kong, L., Jing, H., Kataoka, T., Sun, J., and Liu, H.: Phylogenetic diversity and spatio-temporal distribution of nitrogenase genes (nifH) in the northern South China Sea, Aquat. Microb. Ecol., 65, 15–27, https://doi.org/10.3354/ame01531, 2011.
Krupke, A., Musat, N., LaRoche, J., Mohr, W., Fuchs, B. M., Amann, R. I., Kuypers, M. M. M., and Foster, R. A.: In situ identification and N2 and C fixation rates of uncultivated cyanobacteria populations, Syst. Appl. Microbiol., 36, 259–271, https://doi.org/10.1016/j.syapm.2013.02.002, 2013.
Krupke, A., Lavik, G., Halm, H., Fuchs, B. M., Amann, R. I., and Kuypers, M. M. M.: Distribution of a consortium between unicellular algae and the N2 fixing cyanobacterium UCYN-A in the North Atlantic Ocean, Environ. Microbiol., 16, 3153–3167, https://doi.org/10.1111/1462-2920.12431, 2014.
Krupke, A., Mohr, W., LaRoche, J., Fuchs, B. M., Amann, R. I., and Kuypers, M. M.: The effect of nutrients on carbon and nitrogen fixation by the UCYN-A–haptophyte symbiosis, ISME J., 9, 1635–1647, https://doi.org/10.1038/ismej.2014.253, 2015.
Kustka, A. B., Sañudo-Wilhelmy, S. A., Carpenter, E. J., Capone, D., Burns, J., and Sunda, W. G.: Iron requirements for dinitrogen- and ammonium-supported growth in cultures of Trichodesmium (IMS 101): Comparison with nitrogen fixation rates and iron:carbon ratios of field populations, Limnol. Oceanogr., 48, 1869–1884, https://doi.org/10.4319/lo.2003.48.5.1869, 2003.
Landolfi, A., Koeve, W., Dietze, H., Kähler, P., and Oschlies, A.: A new perspective on environmental controls of marine nitrogen fixation, Geophys. Res. Lett., 42, 4482–4489, https://doi.org/10.1002/2015GL063756, 2015.
Langlois, R., Mills, M. M., Ridame, C., Croot, P., and LaRoche, J.: Diazotrophic bacteria respond to Saharan dust additions, Mar. Ecol.-Prog. Ser., 470, 1–14, https://doi.org/10.3354/meps10109, 2012.
Langlois, R. J., LaRoche, J., and Raab, P. A.: Diazotrophic Diversity and Distribution in the Tropical and Subtropical Atlantic Ocean, Appl. Environ. Microbiol., 71, 7910–7919, https://doi.org/10.1128/AEM.71.12.7910-7919.2005, 2005.
Langlois, R. J., Hümmer, D., and LaRoche, J.: Abundances and Distributions of the Dominant nifH Phylotypes in the Northern Atlantic Ocean, Appl. Environ. Microb., 74, 1922–1931, https://doi.org/10.1128/AEM.01720-07, 2008.
Law, C. S., Woodward, E. M. S., Ellwood, M. J., Marriner, A., Bury, S. J., and Safi, K. A.: Response of surface nutrient inventories and nitrogen fixation to a tropical cyclone in the southwest Pacific, Limnol. Oceanogr., 56, 1372–1385, https://doi.org/10.4319/lo.2011.56.4.1372, 2011.
Löscher, C. R., LaRoche, J., and Schmitz, R. A.: Underestimation of Marine Dinitrogen Fixation: A Novel Method and Novel Diazotrophic Habitats, in Biological Nitrogen Fixation, edited by: de Bruijn, F. J., 891–896, John Wiley & Sons, Inc., available at: http://onlinelibrary.wiley.com/doi/10.1002/9781119053095.ch87/summary (last access: 29 November 2016), 2015.
Luo, Y.-W., Doney, S. C., Anderson, L. A., Benavides, M., Berman-Frank, I., Bode, A., Bonnet, S., Boström, K. H., Böttjer, D., Capone, D. G., Carpenter, E. J., Chen, Y. L., Church, M. J., Dore, J. E., Falcón, L. I., Fernández, A., Foster, R. A., Furuya, K., Gómez, F., Gundersen, K., Hynes, A. M., Karl, D. M., Kitajima, S., Langlois, R. J., LaRoche, J., Letelier, R. M., Marañón, E., McGillicuddy Jr., D. J., Moisander, P. H., Moore, C. M., Mouriño-Carballido, B., Mulholland, M. R., Needoba, J. A., Orcutt, K. M., Poulton, A. J., Rahav, E., Raimbault, P., Rees, A. P., Riemann, L., Shiozaki, T., Subramaniam, A., Tyrrell, T., Turk-Kubo, K. A., Varela, M., Villareal, T. A., Webb, E. A., White, A. E., Wu, J., and Zehr, J. P.: Database of diazotrophs in global ocean: abundance, biomass and nitrogen fixation rates, Earth Syst. Sci. Data, 4, 47–73, https://doi.org/10.5194/essd-4-47-2012, 2012.
Luo, Y.-W., Lima, I. D., Karl, D. M., Deutsch, C. A., and Doney, S. C.: Data-based assessment of environmental controls on global marine nitrogen fixation, Biogeosciences, 11, 691–708, https://doi.org/10.5194/bg-11-691-2014, 2014.
Marchesiello, P. and Estrade, P.: Upwelling limitation by onshore geostrophic flow, J. Mar. Res., 68, 37–62, https://doi.org/10.1357/002224010793079004, 2010.
Martínez-Pérez, C., Mohr, W., Löscher, C. R., Dekaezemacker, J., Littmann, S., Yilmaz, P., Lehnen, N., Fuchs, B. M., Lavik, G., Schmitz, R. A., LaRoche, J., and Kuypers, M. M. M.: The small unicellular diazotrophic symbiont, UCYN-A, is a key player in the marine nitrogen cycle, Nat. Microbiol., 1, 16163, https://doi.org/10.1038/nmicrobiol.2016.163, 2016.
Messer, L. F., Doubell, M., Jeffries, T. C., Brown, M. V., and Seymour, J. R.: Prokaryotic and diazotrophic population dynamics within a large oligotrophic inverse estuary, Aquat. Microb. Ecol., 74, 1–15, https://doi.org/10.3354/ame01726, 2015.
Messer, L. F., Mahaffey, C., Robinson, C. M., Jeffries, T. C., Baker, K. G., Bibiloni Isaksson, J., Ostrowski, M., Doblin, M. A., Brown, M. V., and Seymour, J. R.: High levels of heterogeneity in diazotroph diversity and activity within a putative hotspot for marine nitrogen fixation, ISME J., 10, 1499–1513, https://doi.org/10.1038/ismej.2015.205, 2016.
Meyerhof, O. and Burk, D.: Über die Fixation des Luftstickstoffs durch Azotobacter, Z. Phys. Chem. A-Chem. T., 139, 117–142, 1928.
Mills, M. M., Ridame, C., Davey, M., La Roche, J., and Geider, R. J.: Iron and phosphorus co-limit nitrogen fixation in the eastern tropical North Atlantic, Nature, 429, 292–294, https://doi.org/10.1038/nature02550, 2004.
Mitsui, A., Kumazawa, S., Takahashi, A., Ikemoto, H., Cao, S., and Arai, T.: Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically, Nature, 323, 720–722, https://doi.org/10.1038/323720a0, 1986.
Moisander, P. H., Beinart, R. A., Voss, M., and Zehr, J. P.: Diversity and abundance of diazotrophic microorganisms in the South China Sea during intermonsoon, ISME J., 2, 954–967, https://doi.org/10.1038/ismej.2008.51, 2008.
Moisander, P. H., Beinart, R. A., Hewson, I., White, A. E., Johnson, K. S., Carlson, C. A., Montoya, J. P., and Zehr, J. P.: Unicellular Cyanobacterial Distributions Broaden the Oceanic N2 Fixation Domain, Science, 327, 1512–1514, https://doi.org/10.1126/science.1185468, 2010.
Moisander, P. H., Zhang, R., Boyle, E. A., Hewson, I., Montoya, J. P., and Zehr, J. P.: Analogous nutrient limitations in unicellular diazotrophs and Prochlorococcus in the South Pacific Ocean, ISME J., 6, 733–744, https://doi.org/10.1038/ismej.2011.152, 2012.
Moutin, T., Karl, D. M., Duhamel, S., Rimmelin, P., Raimbault, P., Van Mooy, B. A. S., and Claustre, H.: Phosphate availability and the ultimate control of new nitrogen input by nitrogen fixation in the tropical Pacific Ocean, Biogeosciences, 5, 95–109, https://doi.org/10.5194/bg-5-95-2008, 2008.
Moutin, T., Doglioli, A. M., de Verneil, A., and Bonnet, S.: Preface: The Oligotrophy to the UlTra-oligotrophy PACific Experiment (OUTPACE cruise, 18 February to 3 April 2015), Biogeosciences, 14, 3207–3220, https://doi.org/10.5194/bg-14-3207-2017, 2017.
Mumy, K. L. and Findlay, R. H.: Convenient determination of DNA extraction efficiency using an external DNA recovery standard and quantitative-competitive PCR, J. Microbiol. Meth., 57, 259–268, https://doi.org/10.1016/j.mimet.2004.01.013, 2004.
Needoba, J. A., Foster, R. A., Sakamoto, C., Zehr, J. P., and Johnson, K. S.: Nitrogen fixation by unicellular diazotrophic cyanobacteria in the temperate oligotrophic North Pacific Ocean, Limnol. Oceanogr., 52, 1317–1327, https://doi.org/10.4319/lo.2007.52.4.1317, 2007.
Ottesen, E. A., Young, C. R., Eppley, J. M., Ryan, J. P., Chavez, F. P., Scholin, C. A., and DeLong, E. F.: Pattern and synchrony of gene expression among sympatric marine microbial populations, P. Natl. Acad. Sci. USA, 110, E488–E497, https://doi.org/10.1073/pnas.1222099110, 2013.
Preston, C. M., Harris, A., Ryan, J. P., Roman, B., Iii, R. M., Jensen, S., Everlove, C., Birch, J., Dzenitis, J. M., Pargett, D., Adachi, M., Turk, K., Zehr, J. P., and Scholin, C. A.: Underwater Application of Quantitative PCR on an Ocean Mooring, PLOS ONE, 6, e22522, https://doi.org/10.1371/journal.pone.0022522, 2011.
Qiu, B., Chen, S., and Kessler, W. S.: Source of the 70-Day Mesoscale Eddy Variability in the Coral Sea and the North Fiji Basin, J. Phys. Oceanogr., 39, 404–420, https://doi.org/10.1175/2008JPO3988.1, 2009.
Raimbault, P., Garcia, N., and Cerutti, F.: Distribution of inorganic and organic nutrients in the South Pacific Ocean – evidence for long-term accumulation of organic matter in nitrogen-depleted waters, Biogeosciences, 5, 281–298, https://doi.org/10.5194/bg-5-281-2008, 2008.
Raven, J. A.: The iron and molybdenum use efficiencies of plant growth with different energy, carbon and nitrogen sources, New Phytol., 109, 279–287, https://doi.org/10.1111/j.1469-8137.1988.tb04196.x, 1988.
Robidart, J. C., Church, M. J., Ryan, J. P., Ascani, F., Wilson, S. T., Bombar, D., Marin, R., Richards, K. J., Karl, D. M., Scholin, C. A., and Zehr, J. P.: Ecogenomic sensor reveals controls on N2-fixing microorganisms in the North Pacific Ocean, ISME J., 8, 1175–1185, https://doi.org/10.1038/ismej.2013.244, 2014.
Sañudo-Wilhelmy, S. A., Kustka, A. B., Gobler, C. J., Hutchins, D. A., Yang, M., Lwiza, K., Burns, J., Capone, D. G., Raven, J. A., and Carpenter, E. J.: Phosphorus limitation of nitrogen fixation by Trichodesmium in the central Atlantic Ocean, Nature, 411, 66–69, https://doi.org/10.1038/35075041, 2001.
Sargent, E. C., Hitchcock, A., Johansson, S. A., Langlois, R., Moore, C. M., LaRoche, J., Poulton, A. J., and Bibby, T. S.: Evidence for polyploidy in the globally important diazotroph Trichodesmium, FEMS Microbiol. Lett., 363, fnw244, https://doi.org/10.1093/femsle/fnw244, 2016.
Shiozaki, T., Kodama, T., and Furuya, K.: Large-scale impact of the island mass effect through nitrogen fixation in the western South Pacific Ocean, Geophys. Res. Lett., 41, 2907–2913, https://doi.org/10.1002/2014GL059835, 2014.
Short, S. M., Jenkins, B. D., and Zehr, J. P.: Spatial and Temporal Distribution of Two Diazotrophic Bacteria in the Chesapeake Bay, Appl. Environ. Microb., 70, 2186–2192, https://doi.org/10.1128/AEM.70.4.2186-2192.2004, 2004.
Sohm, J. A., Webb, E. A., and Capone, D. G.: Emerging patterns of marine nitrogen fixation, Nat. Rev. Micro., 9, 499–508, https://doi.org/10.1038/nrmicro2594, 2011.
Stewart, W. D. P.: Biological and Ecological Aspects of Nitrogen Fixation by Free-Living Micro-Organisms, P. Roy. Soc. Lond. B Bio., 172, 367–388, 1969.
Subramaniam, A., Yager, P. L., Carpenter, E. J., Mahaffey, C., Björkman, K., Cooley, S., Kustka, A. B., Montoya, J. P., Sañudo-Wilhelmy, S. A., Shipe, R., and Capone, D. G.: Amazon River enhances diazotrophy and carbon sequestration in the tropical North Atlantic Ocean, P. Natl. Acad. Sci. USA, 105, 10460–10465, https://doi.org/10.1073/pnas.0710279105, 2008.
Sukenik, A., Kaplan-Levy, R. N., Welch, J. M., and Post, A. F.: Massive multiplication of genome and ribosomes in dormant cells (akinetes) of Aphanizomenon ovalisporum (Cyanobacteria), ISME J., 6, 670–679, https://doi.org/10.1038/ismej.2011.128, 2012.
Taniuchi, Y., Chen, Y. L., Chen, H.-Y., Tsai, M.-L., and Ohki, K.: Isolation and characterization of the unicellular diazotrophic cyanobacterium Group C TW3 from the tropical western Pacific Ocean, Environ. Microbiol., 14, 641–654, https://doi.org/10.1111/j.1462-2920.2011.02606.x, 2012.
Thompson, A., Carter, B. J., Turk-Kubo, K., Malfatti, F., Azam, F., and Zehr, J. P.: Genetic diversity of the unicellular nitrogen-fixing cyanobacteria UCYN-A and its prymnesiophyte host, Environ. Microbiol., 16, 3238–3249, https://doi.org/10.1111/1462-2920.12490, 2014.
Thompson, A. W., Foster, R. A., Krupke, A., Carter, B. J., Musat, N., Vaulot, D., Kuypers, M. M. M., and Zehr, J. P.: Unicellular Cyanobacterium Symbiotic with a Single-Celled Eukaryotic Alga, Science, 337, 1546–1550, https://doi.org/10.1126/science.1222700, 2012.
Tripp, H. J., Bench, S. R., Turk, K. A., Foster, R. A., Desany, B. A., Niazi, F., Affourtit, J. P., and Zehr, J. P.: Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium, Nature, 464, 90–94, https://doi.org/10.1038/nature08786, 2010.
Turk-Kubo, K. A., Frank, I. E., Hogan, M. E., Desnues, A., Bonnet, S., and Zehr, J. P.: Diazotroph community succession during the VAHINE mesocosm experiment (New Caledonia lagoon), Biogeosciences, 12, 7435–7452, https://doi.org/10.5194/bg-12-7435-2015, 2015.
Turk-Kubo, K. A., Farnelid, H. M., Shilova, I. N., Henke, B., and Zehr, J. P.: Distinct ecological niches of marine symbiotic N2-fixing cyanobacterium Candidatus Atelocyanobacterium thalassa sublineages, J. Phycol., 53, 451–461, https://doi.org/10.1111/jpy.12505, 2017.
Van Den Broeck, N., Moutin, T., Rodier, M., and Bouteiller, A. L.: Seasonal variations of phosphate availability in the SW Pacific Ocean near New Caledonia, Mar. Ecol.-Prog. Ser., 268, 1–12, https://doi.org/10.3354/meps268001, 2004.
Wallace, B. C., Lajeunesse, M. J., Dietz, G., Dahabreh, I. J., Trikalinos, T. A., Schmid, C. H., and Gurevitch, J.: OpenMEE: Intuitive, open-source software for meta-analysis in ecology and evolutionary biology, Methods Ecol. Evol., 8, 941–947, https://doi.org/10.1111/2041-210X.12708, 2017.
Webb, E. A., Ehrenreich, I. M., Brown, S. L., Valois, F. W., and Waterbury, J. B.: Phenotypic and genotypic characterization of multiple strains of the diazotrophic cyanobacterium, Crocosphaera watsonii, isolated from the open ocean, Environ. Microbiol., 11, 338–348, https://doi.org/10.1111/j.1462-2920.2008.01771.x, 2009.
Wells, M. L., Vallis, G. K., and Silver, E. A.: Tectonic processes in Papua New Guinea and past productivity in the eastern equatorial Pacific Ocean, Nature, 398, 601–604, https://doi.org/10.1038/19281, 1999.
Yoshikawa, C., Nakatsuka, T., and Kawahata, H.: Transition of low-salinity water in the Western Pacific Warm Pool recorded in the nitrogen isotopic ratios of settling particles, Geophys. Res. Lett., 32, L14615, https://doi.org/10.1029/2005GL023103, 2005.
Young, J. P. W.: The Phylogeny and Evolution of Nitrogenases, in: Genomes and Genomics of Nitrogen-fixing Organisms, edited by: Palacios, R. and Newton, W. E., 221–241, Springer Netherlands, available at: http://link.springer.com/chapter/10.1007/1-4020-3054-1_14 (last access: 29 November 2016), 2005.
Zehr, J. P., Waterbury, J. B., Turner, P. J., Montoya, J. P., Omoregie, E., Steward, G. F., Hansen, A., and Karl, D. M.: Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean, Nature, 412, 635–638, https://doi.org/10.1038/35088063, 2001.
Zehr, J. P., Methe, B., and Foster, R.: New Nitrogen-Fixing Microorganisms from the Oceans: Biological Aspects and Global Implications, in: Biological Nitrogen Fixation, Sustainable Agriculture and the Environment, 361–365, Springer Netherlands, available at: http://link.springer.com/chapter/10.1007/1-4020-3570-5_87 (last access: 29 November 2016), 2005.
Zehr, J. P., Bench, S. R., Carter, B. J., Hewson, I., Niazi, F., Shi, T., Tripp, H. J., and Affourtit, J. P.: Globally Distributed Uncultivated Oceanic N2-Fixing Cyanobacteria Lack Oxygenic Photosystem II, Science, 322, 1110–1112, https://doi.org/10.1126/science.1165340, 2008.