the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Effects of sea animal colonization on the coupling between dynamics and activity of soil ammonia-oxidizing bacteria and archaea in maritime Antarctica
Qing Wang
Yanling Zheng
Tao Bao
Lijun Hou
The colonization by a large number of sea animals, including penguins and seals, plays an important role in the nitrogen cycle of the tundra ecosystem in coastal Antarctica. However, little is known about the effects of sea animal colonization on ammonia-oxidizing archaea (AOA) and bacteria (AOB) communities involved in nitrogen transformations. In this study, we chose active seal colony tundra soils (SSs), penguin colony soils (PSs), adjacent penguin-lacking tundra soils (PLs), tundra marsh soils (MSs), and background tundra soils (BSs) to investigate the effects of sea animal colonization on the abundance, activity, and diversity of AOA and AOB in maritime Antarctica. Results indicated that AOB dominated over AOA in PS, SS, and PL, whereas AOB and AOA abundances were similar in MS and BS. Penguin or seal activities increased the abundance of soil AOB amoA genes but reduced the abundance of AOA amoA genes, leading to very large ratios (1.5×102 to 3.2×104) of AOB to AOA amoA copy numbers. Potential ammonia oxidation rates (PAORs) were significantly higher (P=0.02) in SS and PS than in PL, MS, and BS and were significantly positively correlated (P<0.001) with AOB amoA gene abundance. The predominance of AOB over AOA and their correlation with PAOR suggested that AOB play a more important role in the nitrification in animal colony soils. Sequence analysis for gene clones showed that AOA and AOB in tundra soils were from the Nitrososphaera and Nitrosospira lineages, respectively. Penguin or seal activities led to a predominance of AOA phylotypes related to Nitrososphaera cluster I and AOB phylotypes related to Nitrosospira clusters I and II but very low relative abundances in AOA phylotypes related to cluster II, and AOB phylotypes related to clusters III and IV. The differences in AOB and AOA community structures were closely related to soil biogeochemical processes under the disturbance of penguin or seal activities: soil C : N alteration and sufficient input of –N and phosphorus from animal excrements. The results significantly enhanced the understanding of ammonia-oxidizing microbial communities in the tundra environment of maritime Antarctica.
- Article
(4317 KB) - Full-text XML
-
Supplement
(1218 KB) - BibTeX
- EndNote
Nitrification, the oxidation of ammonia to nitrate through nitrite, plays a pivotal role in the global biogeochemical nitrogen cycle (Nunes-Alves, 2016). As the first and rate-limiting step of nitrification, ammonia oxidation (the aerobic oxidation of ammonia to nitrite) is performed by phylogenetically and physiologically distinct groups of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) (Belser and Schmidt, 1978; Könneke et al., 2005). AOA and AOB have been investigated using the amoA gene as a functional marker in a wide variety of environments, including soils (Di et al., 2009; Gubry-Rangin et al., 2017; Leininger et al., 2006; Ouyang et al., 2016; Shen et al., 2012), sediments (Li et al., 2015; Zheng et al., 2013), estuaries (Dang et al., 2008; Mosier et al., 2008; Santoro et al., 2011), the oxic and suboxic marine water column (Baker et al., 2012; Bouskill et al., 2012), plateau permafrost (Zhang et al., 2009; Zhao et al., 2017), and in subarctic and arctic soils (Alves et al., 2013; Daebeler et al., 2017). Results indicated that the relative abundance and functional importance of AOA vs. AOB vary greatly in natural ecosystems. Environmental drivers, including substrate concentration, oxygen availability, pH, and salinity, might be responsible for the different AOA and AOB abundances and distribution (Alves et al., 2013; Bouskill et al., 2012; Le Roux et al., 2008; Wang et al., 2015). The abundance, diversity, and activity of ammonia oxidizers have been explored in tundra soils of the Antarctic Peninsula (Jung et al., 2011; Yergeau et al., 2007) and the Antarctic Dry Valleys (Ayton et al., 2010; Magalhães et al., 2014; Richter et al., 2014) and in Antarctic coastal waters (Kalanetra et al., 2009; Tolar et al., 2016). However, there is still a large gap in our understanding of factors that control AOA vs. AOB prominence, and the relationships between nitrification rates and ammonia-oxidizer dynamics need to be explored in Antarctica.
In maritime Antarctica, a large number of sea animals, such as penguins or seals, settle on coastal ice-free tundra patches. Tundra vegetation including mosses, lichens, and algae, penguin colonies, and their interactions form a special ornithogenic tundra ecosystem (Tatur et al., 1997). The soil biogeochemistry of an ornithogenic tundra ecosystem has become a research hotspot under penguin-activity disturbance (Otero et al., 2018; Riddick et al., 2012; Simas et al., 2007; Zhu et al., 2013, 2014). Previous studies indicated that sea animals significantly affect the tundra N and P cycles (Lindeboom et al., 1984; Simas et al., 2007; Zhu et al., 2011), and the total N and P excreted by seabird breeders and chicks are 470 Gg N yr−1 and 79 Gg P yr−1 in Antarctica and the Southern Ocean, accounting for 80 % of the N and P from total global seabird excreta (Otero et al., 2018). Uric acid is the dominant N compound in penguin guano, and during its mineralization, different N forms, such as NH3, , and , can be produced via ammonification, nitrification, and deposition, following the changes in soil pH and the C : N ratio (Blackall et al., 2007; Otero et al., 2018; Riddick et al., 2012). The alteration of soil biogeochemistry under the sea-animal-activity disturbance might have an impact on the abundance and diversity of the AOA and AOB involved in the nitrogen cycle. Increased bacterial abundance, diversity, and activity have been detected in penguin or seal colony soils (Ma et al., 2013; Zhu et al., 2015). Penguin or seal colonies have been confirmed as strong sources for greenhouse gas N2O (Zhu et al., 2008, 2013), a by-product of microbial ammonia oxidation (Santoro et al., 2011). However, the effects of sea animal colonization on AOA and AOB community structures have not been thoroughly investigated in the maritime Antarctic tundra.
In the present study, we investigated the abundance, potential activity, and diversity of soil AOA and AOB in five tundra patches, including a penguin colony, a seal colony, the adjacent animal-lacking tundra, tundra marsh, and background tundra, where soil biogeochemical properties were subjected to the differentiating effects of sea animal activities. Our objectives were (a) to examine the abundance, diversity, and community structure of soil AOA and AOB using the amoA gene as a functional marker; (b) to investigate potential links between amoA gene abundance, AOA and AOB community structures, potential activity, and environmental variables; and (c) to assess the relative contribution of these two distinct ammonia-oxidizing groups to nitrification.
2.1 Study area
The study area is located on the Fildes Peninsula and Ardley Island in the southwest of King George Island (Fig. 1), having oceanic climate characteristics. The mean annual air temperature is about −2.5 ∘C, with a range of daily mean temperature from −26.6 to 11.7 ∘C, and mean annual precipitation is about 630 mm, mainly in the form of snow. The Fildes Peninsula (about 30 km2 area) is a host to important sea animal colonies. Based on annual statistical data, a total of over 10 700 sea animals colonize this peninsula in the austral summer. On the western coast there are established seal colonies including elephant seal (Mirounga leonine), Weddell seal (Leptonychotes weddellii), fur seal (Arctocephalus gazella), and leopard seal (Hydrurga leptonyx) (Sun et al., 2004). Ardley Island, with an area of 2.0 km in length and 1.5 km in width, is connected to the Fildes Peninsula via a sand dam. This island belongs to an important ecological reserve for penguin populations in western Antarctica. A great majority of breeding penguins, including Adélie penguins (Pygoscelis adeliae), gentoo penguins (Pygoscelis papua), and chinstrap penguins (Pygoscelis antarcticus), colonize the east of this island in the austral summer. Seal excrements or penguin droppings rich in nitrogen and phosphorus are transported into local tundra soils by ice and snow melting water during the breeding period (Sun et al., 2000, 2004). Mosses and lichens dominate local vegetation. However, the vegetation is almost absent in penguin or seal colonies because of over-manuring and animal trampling. A more detailed description of the study area can be found in Zhu et al. (2013).
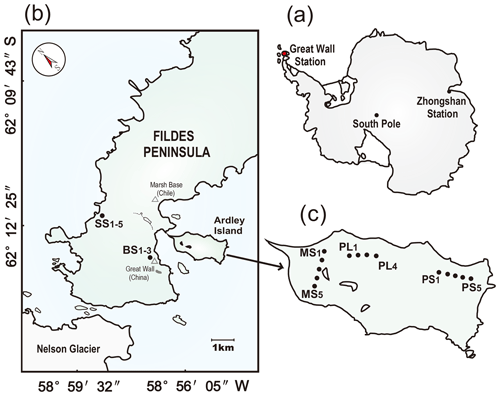
Figure 1Study area and soil sampling sites. Panel (a): the red dot indicates the location of the investigation area in maritime Antarctica. Panel (b): location of the sampling sites on the Fildes Peninsula. The sampling soils from tundra patches included the active seal colony tundra soils SS (SS1–5) in the western coast of the Fildes Peninsula and the background tundra soils on the upland areas (BS1–3). Panel (c): the location of the sampling sites on Ardley Island. The sampling soils from tundra patches included the western tundra marsh soils (MS1–5), the eastern active penguin colony tundra soils PS (PS1–5), and the adjacent penguin-lacking tundra soils PL (PL1–4). Note: the map was drawn using CorelDRAW X7 software (http://www.corel.com/cn/, last access: 20 September 2019).
2.2 Tundra soil collection
In the summer of 2014/2015, soil samples were collected from the following tundra patches, as illustrated in Fig. 1.
- i.
Penguin colony and penguin-lacking tundra sites: the tundra on Ardley Island was categorized into three areas from east to west according to the distance to the penguin nesting sites (i.e., the intensity of penguin activity) – the eastern active penguin colony with nesting sites, PS (i.e., high penguin-activity area), where penguins have the highest density and a high-frequency presence during the breeding period; the adjacent penguin-lacking tundra areas, PLs (i.e., low penguin-activity areas) in the middle of Ardley Island, where penguins occasionally wander and have a typically low density; and the western tundra marsh, MS, moderately far from penguin nesting sites (i.e., a slight penguin-activity area), where penguins rarely frequent the sites. In total, 14 soil samples were collected from Ardley Island to study the effects of penguin colonization on the abundance, activity, and community structures of soil AOA and AOB. Specifically, samples PS1–PS5 were collected sequentially from the center of the colony in the PS. Samples PL1–PL4 and MS1–MS5 were randomly collected in the PL and MS.
- ii.
The seal colony and its adjacent tundra sites, SSs: these sites are on the western coast of the Fildes Peninsula. According to the distance to seal wallows (i.e., the intensity of seal activity), samples SS1–SS5 were collected in sequence to investigate the effects of seal colonization. Site SS1 was closest to the seal colony (i.e., a high seal-activity site), whereas SS5 was the farthest from the seal colony (i.e., a low seal-activity site).
- iii.
Background tundra sites, BSs: three soil samples were collected from an upland tundra at about 40 m a.s.l. and with no sea animals around. The tundra surface is covered with mosses or lichens with a 10–15 cm organic clay layer (Zhu et al., 2013).
At each sampling site, soil was collected aseptically using a clean scoop from the top 5–10 cm at the four corners of a 1 m2 subarea, and combined into one sample. Appropriate precautions were taken to avoid cross-site or human-made contamination. Immediately after collection, each sample was divided into two portions: one was stored in sterile plastic containers at −80 ∘C for the analysis of the microbial community structures, and the other portion was stored at close to the in situ temperature to determine the geochemical characteristics and potential ammonia oxidation rates. All of the analyses were conducted within 1 month.
2.3 General analysis of soil characteristics
Soil pH was determined by mixing the soil and 1 M KCl solution (1:3 ratio). Soil moisture was measured by oven drying at 105 ∘C to a constant weight. Total carbon (TC), total nitrogen (TN), and total sulfur (TS) contents in the soils were determined through a CNS (carbon, nitrogen, sulfur) analyzer (vario MACRO, Elementar, Germany). The samples were digested in Teflon tubes using HNO3-HCl-HF-HClO4 digestion at 190 ∘C, and total phosphorus (TP) was determined using ICP-OES (inductively coupled plasma optical emission spectrometer; Perkin Elmer 2100DV, Waltham, MA, USA). The -N, -N, and -N concentrations were determined through a continuous-flow analyzer (Skalar, Netherlands) (Gao et al., 2018; Zhu et al., 2011).
2.4 Measurement of soil potential ammonia oxidation rate
The potential ammonia oxidation rate (PAOR) in tundra soil was determined using the chlorate inhibition method (Kurola et al., 2005; Xia, 2007). Sodium chlorate was used to inhibit from being oxidized into . Briefly, 5 g fresh tundra soil was incubated in 20 mL of 1 mM phosphate-buffered saline with 1 mM of (NH4)2SO4 and NaClO3 in the dark at 15 ∘C. After moderately shaking for 24 h, the 5 mL of 2 M KCl was used to extract the nitrite. The optical density for the supernatant after centrifugation was determined spectrophotometrically at 540 nm. The standard curve obtained from NaNO2 (0–2.5 µmol L−1) was used to calculate the PAOR in the tundra soils.
2.5 DNA extraction and gene amplification (polymerase chain reaction, PCR)
Genomic DNA was extracted from 0.25 g of homogenized tundra soils using a PowerSoil™ DNA Isolation Kit (Mo Bio, Carlsbad, CA, USA) as described in the manufacturer's protocol. The extracted DNA was eluted in 50 µL of elution buffer, quantified by a Nanodrop-2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), and stored at −20 ∘C. AOA amoA gene fragments (635 bp) were amplified using the primers Arch-amoAF (5′-STAATGGTCTGGCTTAGACG-3′) and Arch-amoAR (5′-GCGGCCATCCATCTGTATGT-3′) (Francis et al., 2005). The amoA gene fragment (491 bp) of β-proteobacterial AOB, which represents known AOB in soil, was amplified using the primer set composed of amoA-1F (5′-GGGGTTTCTACTGGTGGT-3′) and amoA-2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′) (Rotthauwe et al., 1997). All PCRs were performed using Taq PCR Master Mix (Sangon Biotech, Shanghai, China) in a total volume of 50 µL. PCRs were carried out with a thermal profile of 5 min at 95 ∘C; 35 cycles of 94 ∘C for 30 s, 56 ∘C for AOA or 55 ∘C for AOB for 45 s, 72 ∘C for 1 min; and a final 5 min extension cycle at 72 ∘C (Y. L. Zheng et al., 2014). Subsequently, the amplification products were visualized by electrophoresis on 1.0 % agarose gels.
2.6 Sequencing and phylogenetic analysis
The amplification products were sent to Sangon Company (Shanghai, China) for purification, cloning, and sequencing (Y. L. Zheng et al., 2014). The sequences were edited using DNAstar (DNASTAR, Madison, WI, USA) and then aligned by MUSCLE (Edgar, 2004) using the UPGMB (unweighted pair group method with arithmetic mean) clustering method with the ClustalX program. The sequences with 97 % identity were grouped into one OTU (operational taxonomic unit) using the mothur program (version 1.23.0; Schloss et al., 2009) by the furthest-neighbor approach (Y. L. Zheng et al., 2014). The closest reference sequences were identified at NCBI (http://www.ncbi.nlm.nih.gov/BLAST/, last access: 5 August 2018) using the BLASTn tool (Madden, 2002), and phylogenetic trees were constructed by the neighbor-joining method using the Molecular Evolutionary Genetics Analysis (MEGA) software (version 5.03, https://www.megasoftware.net/, last access: 5 August 2018). The sequences reported in this study have been deposited in GenBank under accession numbers MH318029 to MH318568 and MH301331 to MH302505.
2.7 Quantitative real-time PCR
The AOB and AOA amo A gene copy numbers for tundra soils were determined in triplicate using quantitative real-time PCR (qPCR) on an ABI 7500 Sequence Detection System (Applied Biosystems). The specific details were given by Y. L. Zheng et al. (2014). The strong linear inverse relationship confirmed the consistency of the qPCR assay between the threshold cycle and the log value of gene copy numbers (R2=0.997 for AOA; R2=0.999 for AOB). The amplification efficiencies for AOA and AOB were 99.8 % and 90.4 %, respectively. Melting curve analysis had only one observable peak at a melting temperature (Tm) (84.9 ∘C for AOA and 89.6 ∘C for AOB) (Fig. S1 in Supplement). Negative controls were subjected to exclude any possible carryover or contamination in all experiments.
2.8 Statistical analysis
The Shannon–Wiener index, Simpson index, and the richness estimator Chao 1 were calculated by the mothur program (version 1.23.0; Schloss et al., 2009). The coverage was the percentage of the number of observed OTUs divided by the Chao 1 (Table S1 in the Supplement). The Kruskal–Wallis test and Wilcoxon signed rank test were conducted for the comparison between amoA gene abundance and PAOR from five tundra patches using SPSS Statistics 17 (IBM Corp, Armonk, NY, USA). Correlations between ammonia-oxidizer gene abundance, PAOR and environmental variables were obtained by Spearman correlation analysis. The relationships between the ammonia-oxidizer community structure and environmental variables were explored using canonical correspondence analysis (CCA) in the software Canoco for windows (version 4.5; Microcomputer Power, Ithaca, NY, USA) because the maximum gradient length of both AOA and β-AOB was longer than four SD (AOA: 4.406; AOB: 18.326). All environmental parameter values were transformed into ln(x+1) before statistical analyses. The OTU richness (defined at 3 % distance) served as the species input, and several simulations of manual forward selection were performed with 499 Monte Carlo permutations to build the optimal models. The scaling in the final CCA biplots was focused on inter-sample relations.
Table 1Soil properties, potential ammonia oxidation rates, and ammonia-oxidizer populations for the soil samples (n=22) that span a penguin colony, a seal colony, and their adjacent animal-lacking tundra across Ardley Island and the Fildes Peninsula in maritime Antarctica.
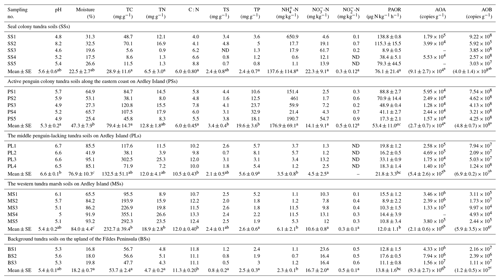
Note: ND indicated that the soil sample was not determined. The average value with different superscript letters (a, b or c) indicates a significant difference at P<0.05 between the different patches.
3.1 Soil chemistry and sea animal activities
Almost all the tundra soils were slightly acidic, and the mean pH ranged from 5.3 to 6.6 at each tundra patch (Table 1). In PS and SS, soil properties including TC, TN, TS, TP, -N, and -N levels showed high heterogeneity due to the deposition of penguin or seal excreta. In the seal colony tundra soils, the highest TC, TN, TP, TS, and -N levels occurred at the sites (SS1–2) close to the seal wallows. In the tundra soils on Ardley Island, the highest TP, TS, and -N levels occurred in the soils close to the eastern penguin nesting sites (PS1-5). PS and SS had generally lower C : N ratios than PL, MS, and BS. Soil mean TN, TS, and –N levels were higher in PS, SS, PL, and MS than in BS. Soil –N contents were 1–2 orders of magnitude higher in PS and SS than in PL, MS, and BS, with means of 176.9 and 137.6 mg -N kg−1, respectively. The highest -N contents occurred in SS. Phosphorus levels were significantly greater (p<0.05) in PS (10.6–32.9 mg g−1) than in other types of tundra soils (mean < 6.0 mg g−1). Overall, penguin or seal activities altered the local soil biogeochemical properties through the deposition of their excreta, leading to generally low C : N ratios in tundra soils.
3.2 Gene abundances under sea animal colonization
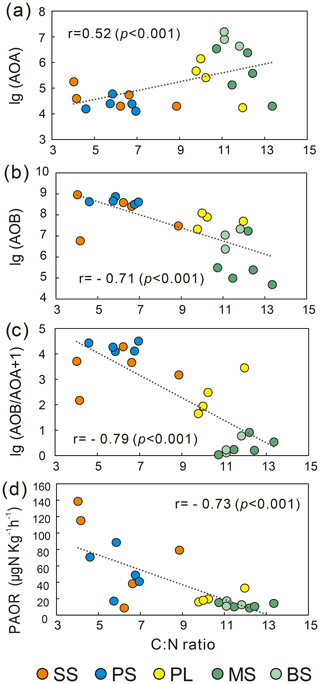
AOB amoA gene abundances were significantly higher (by approximately 2–4 orders of magnitude) than AOA amoA gene abundances (Wilcoxon test, n=22, P=0.002) in the penguin and seal colony and the adjacent tundra soils, PS, SS, and PL. However, amoA gene abundances were similar in the MS and BS soils (Fig. 2a). Overall, the abundances of AOB and AOA amoA genes were significantly negatively correlated (, P=0.002) across all the tundra patches (Fig. S2). The AOA amoA gene abundances showed a heterogeneous distribution in the abundances among the different tundra patches, and they were 2 orders of magnitude lower in PS and SS relative to those in BS and MS. Maximum AOA amoA gene abundance appeared in BS, followed by MS and PL, whereas the PS and SS soils had the lowest AOA amoA gene abundances. The log values of soil AOA amoA gene abundances showed a significant positive correlation (r=0.52, P<0.001) with C : N ratios (Fig. 3a), but their abundances showed a significant negative correlation with -N contents (, P=0.013) (Table 2).
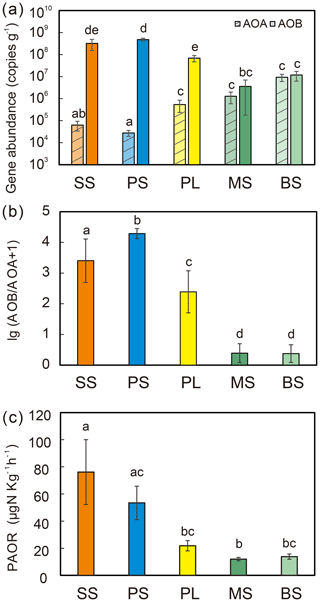
Figure 2Comparisons of soil AOA and AOB amoA gene copy numbers (a), log ratio of AOB : AOA abundances (b), and potential ammonia oxidation rates (PAORs) (c) between five tundra patches. The error bars indicate standard deviations of the means.
Table 2Spearman correlations (n=22) among ammonia-oxidizer populations, the ratios of AOA : AOB abundances, potential ammonia oxidation rates (PAOR), and environmental variables in the soils of maritime Antarctic tundra.
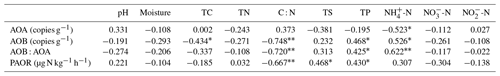
Note: significant correlations are indicated by * at the P=0.05 level and at the P=0.01 level.
Unlike AOA amoA gene abundances, AOB amoA genes showed the opposite distribution pattern. AOB amoA gene abundances were significantly higher (by approximately 2–3 orders of magnitude) in PS and SS compared with those in MS and BS (Fig. 2a). The log values of soil AOB amoA gene abundances showed a significant negative correlation with C : N ratios (, P<0.001) (Fig. 3b), but their abundances showed a significant positive correlation with -N (r=0.53, P<0.05) and TP (r=0.47, P<0.05) (Table 2). The ratios of AOB to AOA amoA copy numbers were strongly affected by animal activities and were much higher in PS and SS than in PL, MS, and BS (Fig. 2b; Kruskal–Wallis test, χ2=18.2, P=0.01). Their ratios showed a significant positive correlation with -N contents (r=0.62; P<0.01) and TP (r=0.43, P<0.05) (Table 2) but a significant negative correlation with the C : N ratios (; P<0.001) (Fig. 3c). Overall, penguin or seal activities, which were indicated by soil C : N ratios, significantly increased the abundance of soil AOB amoA genes but reduced the abundance of AOA amoA genes, leading to very large ratios (1.5×102 to 3.2×104) of AOB to AOA amoA copy numbers in PS and SS. However, the ratios varied only from 0.1 to 7.2 in BS and MS.
3.3 Potential ammonia oxidation rates under sea animal colonization
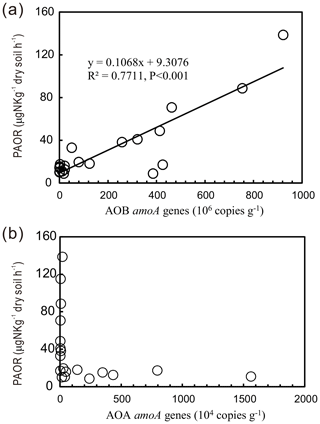
PAORs ranged from 8.9 to 138.8 µg N kg−1 h−1 in all the soil samples (Table 1). The PAOR was slightly higher in SS (mean 76.1 µg N kg−1 h−1) than in PS (mean 64.7 µg N kg−1 h−1) but significantly higher than in PL, MS, and BS (mean 12.0–21.8 µg N kg−1 h−1). Overall the PAOR was significantly higher in animal colony soils (mean 70.4 µg N kg−1 h−1 for SS and PS) than in non-animal colony soils (mean 15.7 µg N kg−1 h−1 for PL, MS, and BS; Kruskal–Wallis test, χ2=11.6, P=0.02) (Fig. 2c). The greatest PAOR occurred at the sites PS1 nearest the penguin nests (88.8±2.7 µg N kg−1 h−1) and SS1 close to seal wallows (138.8±0.8 µg N kg−1 h−1). The PAOR followed the distribution changes of AOB amoA gene abundances but showed the opposite trend to the AOA amoA gene abundances. A significant positive correlation (r2=0.77, P<0.001) was observed between the PAOR and the AOB amoA gene abundance when the data from all the tundra patches were combined, whereas no correlation occurred between PAOR and AOA amoA gene abundance (Fig. 4). The higher abundance of AOB compared to AOA in PS, SS, and PL and their correlation with the PAOR suggested that AOB populations might contribute more to the PAOR than the AOA populations in penguin or seal colonies. In addition, PAOR significantly negatively correlated with soil C : N ratios (, P<0.001) (Fig. 3d) but significantly positively correlated with TS contents (r=0.47, P<0.05) and TP contents (r=0.43, P<0.05) (Table 2).
3.4 Community structure of AOA and AOB under sea animal colonization
The PCR products were insufficient to construct the clone libraries for the AOA amoA gene from SS and PS because of the low AOA abundance in the soils, as was the case with the AOB amoA gene from MS and BS. Overall, 10 AOA and 14 AOB amoA gene clone libraries were successfully constructed. The 543 AOA sequences and 1175 AOB quality sequences were generated from the respective sites. Within each individual site, 1–6 AOA OTUs and 6–15 AOB OTUs were identified, as defined by < 3 % divergence in nucleotides. The AOA and AOB OTU numbers for each library are presented in Table S1. These numbers might be higher if more clones were sequenced, based on the rarefaction curves (Figs. S3 and S4). AOB amoA gene diversity was generally higher compared to AOA, based on the indices of Shannon–Wiener and Simpson. Specifically, AOA amoA gene diversity was higher in PL and MS than in BS, whereas AOB amoA gene diversity was higher in SS and PS compared with that in adjacent animal-lacking tundra soils (Table S1).
The 543 AOA amoA gene sequences had 76 %–100 % sequence similarity to each other and 95 %–100 % identity with the corresponding top hit amoA sequences deposited in GenBank. Phylogenetic analysis showed that the AOA amoA sequences were grouped into 16 unique OTUs, representing 100 % of all the AOA amoA OTUs identified, and these sequences were affiliated with two Nitrososphaera clusters (Fig. 5a): cluster I contained 11 OTUs and 264 clones, and 57.9 % of AOA amoA sequences were from PL, 41.3 % from SS and only 0.8 % from MS. In cluster II, there are five unique OTUs and 279 clones, and 58.8 % of them were from BS, 38.3 % from MS, and only 2.9 % from PL. Almost all the AOA phylotypes retrieved from PL and SS were related to Nitrososphaera cluster I, whereas the AOA phylotypes retrieved from MS and BS were distributed in cluster II (Fig. S5a). Seal or penguin activities led to the predominant existence of AOA phylotypes related to cluster I but very low relative abundances in AOA phylotypes related to cluster II, which were almost completely excluded in SS and PL. Almost all AOA phylotypes in BS and MS were related to Nitrososphaera cluster II, whereas the relative abundances of AOA phylotypes related to cluster I were very low or undetectable.
The 1175 AOB amoA gene sequences shared 87 %–100 % sequence identity to each other and 93 %–100 % identity with the closest matched GenBank sequences. Phylogenetic analysis showed that the AOB amoA sequences could be grouped into 38 unique OTUs, representing 58.5 % of all the AOB amoA OTUs identified, and they were grouped into four Nitrosospira clusters according to the evolutionary distance of the phylogenetic tree (Fig. 5b): cluster I contained 11 OTUs and 226 clones, and 67.7 % of AOB amoA sequences were from PS, 23.5 % from SS, 8.4 % from PL, and only 0.4 % from MS. Clusters II and III contained 17 unique OTUs and 521 clones. The sources of the OTUs in cluster II were similar to those of cluster I, with 69.8 % from PS, 29.9 % from SS, and 0.3 % from PL. For cluster III, 79.2 % of the sequences were from PL, 19.8 % from SS, and 1.0 % from MS. Cluster IV contained nine unique OTUs and 370 clones from PL (50.0 %), SS (36.8 %), and MS (13.2 %). All the AOB phylotypes retrieved from PS were related to dominant Nitrosospira clusters I and II, whereas AOB phylotypes related to clusters III and IV were completely excluded because of penguin colonization (Fig. S5b). The AOB phylotypes retrieved from SS were distributed in clusters I, II, III, and IV (16 %–38 % for each cluster). Almost all the AOB phylotypes retrieved from PL and MS were related to Nitrosospira clusters III and IV.
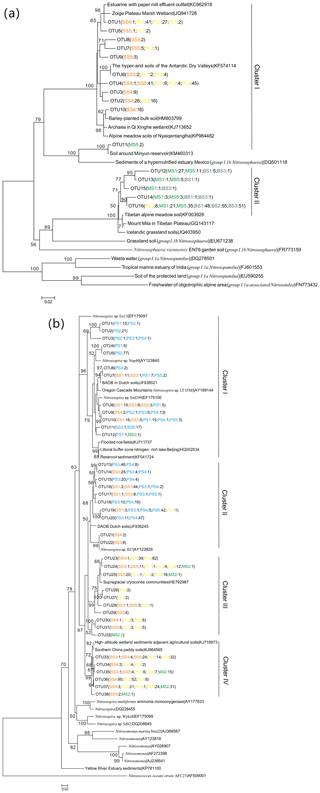
Figure 5Neighbor-joining phylogenetic tree of AOA amoA (a) and AOB amoA (b). The phylogeny is based on nucleotide sequences. Bootstrap values ≥50 % (of 1000 iterations) are shown near the nodes. GenBank accession numbers are shown for sequences from other studies. OTUs were defined at 97 % similarity. Numbers in parentheses following each OTU indicate the number of sequences recovered from each sampling site.
3.5 Relationships of the ammonia-oxidizer community structure with environmental variables
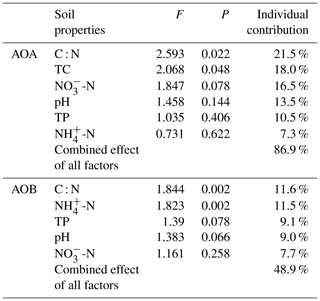
The relationships of the AOA and AOB communities with environmental variables were analyzed using CCA. The environmental variables explained 62.1 % of the total variance in the AOA amoA genotype compositions and 71.5 % of the cumulative variance of the genotype-environment relationships in the first two CCA dimensions (Fig. 6a). Overall, the AOA community structures significantly correlated with C : N (F=2.59, P=0.022) and TC (F=2.07, P=0.048) in tundra soils (Table 3), and the combination of the two factors explained 39.6 % of the variation. High soil C : N and TC concentrations increased the AOA richness in MS and BS. Although other environmental parameters, including TP, pH, -N, and -N were not statistically significant (P>0.05), these variables additionally explained 47.3 % of the variation. As illustrated in Fig. 6b, the first two dimensions explained 26.6 % of the total variance in the AOB compositions and 54.3 % of the cumulative variance of the AOB genotype–environment relationships. The composition and distribution of AOB communities correlated significantly with C : N ratios (F=1.844, P=0.002) and -N (F=1.823, P=0.002), and the two factors combined yielded 21.9 % of total CCA explanatory power. The others including TP, -N, and pH accounted for 27.1 % of the variance. Penguin or seal activities significantly increased the AOB richness in SS and PS through higher -N and P input from sea animal excrement, whereas AOB richness was closely related to the soil C : N in PL and MS.
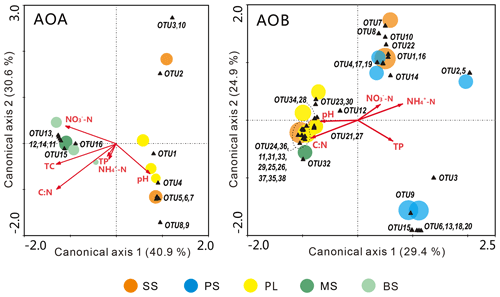
Figure 6Canonical correspondence analysis (CCA) ordination plots for the relationship between the AOA and AOB community structures with environmental variables. The circles with different colors represent the various sampling sites. The size of the circles corresponds to the OTU richness in individual samples. The black triangles represent amoA phylotypes. Environmental variables are represented by red arrows. The percentage of species–environment relation variance explained by the two principal canonical axes is represented close to the axes.
4.1 Effects of sea animal colonization on AOA and AOB abundances
In this study, soil AOA amoA gene abundances were 2 orders of magnitude lower in PS and SS relative to BS and MS; however, AOB amoA gene abundances were approximately 2–3 orders of magnitude higher in PS and SS than in MS and BS, indicating that sea animal activities increased the AOB population size but decreased AOA abundances in tundra soils (Figs. 2 and 3). Overall, the AOA amoA gene abundances obtained here were similar to the abundance range reported in the soils of the Antarctic Dry Valleys and arctic tundra soils; however, the AOB amoA gene abundances were 2–3 orders of magnitude higher in PS and SS than in Antarctic Dry Valleys (Alves et al., 2013; Magalhães et al., 2014). In contrast to previous studies indicating that AOA were more abundant than AOB in some terrestrial or marine ecosystems (Beman et al., 2008; Lam et al., 2007; Wuchter et al., 2006; Yao et al., 2011) and in soils from the Antarctic Peninsula (Jung et al., 2011), our qPCR estimates showed that the AOB amoA copy numbers were much greater than those of AOA amoA in PS, SS, and PL because of sea animal activities. However, their abundances were very similar to each other in BS and MS. The ratios of AOB to AOA abundance were strongly affected by sea animal activities, which were indicated by soil C : N ratios (Fig. 2c). A shift in the relative abundance of AOA and AOB was recorded previously for the Antarctic Dry Valleys, with a greater abundance of AOB compared with that of AOA for Battleship Promontory and Miers Valley and the reverse for upper Wright Valley and Beacon Valley (Magalhães et al., 2014). The results for PS, SS, and PL are also in agreement with those detected in subglacial soils (Boyd et al., 2011).
The ratios of AOB to AOA showed significant correlations with C : N, -N, and TP when all the data were combined in the five tundra patches (Table 2). This suggested that C : N, -N, and TP are key factors in determining a predominance of AOB over AOA. In Antarctica, the productivity of terrestrial ecosystems is strongly limited because of the extremely low nitrogen levels (Park et al., 2007). However, the physiochemical properties for tundra soils were strongly influenced by the deposition of penguin or seal excreta under the effects of local microbes (Tatur et al., 1997). Sea animals provide considerable external N inputs for their colony soils and adjacent tundra soils through direct input of their excreta and atmospheric deposition via ammonia volatilization (Lindeboom, 1984; Sun et al., 2002; Blackall et al., 2007; Zhu et al., 2011; Riddick et al., 2012). In addition to ammonium, phosphorus can typically be found in penguin guano (Sun et al., 2000). Generally low C : N ratios and significantly elevated –N and TP concentrations occurred in PS and PL due to penguin or seal activities (Table 1). These conditions allow high abundance of AOB amoA genes, which explains the strong correlations between AOB abundances and C : N, -N, and TP in the sea animal colony soils (Table 2). This agreed with the high bacterial abundance previously documented in penguin or seal colony soils and ornithogenic sediments (Ma et al., 2013; Zhu et al., 2015).
The AOA abundance showed a significant negative correlation with -N levels in tundra patches (Table 2), indicating that AOA might better adapt to low and oligotrophic environments (Martens-Habbena et al., 2009; Stieglmeier et al., 2014). High -N concentrations might partially inhibit AOA populations (Hatzenpichler et al., 2008). This result is similar to that reported for some agricultural soils with increased fertilization and grassland soils with increased grazing (Fan et al., 2011; Prosser and Nicol, 2012; Pan et al., 2018), supporting the conclusion that AOA and AOB generally inhabit different niches in soil, distinguished by the concentration and availability (Verhamme et al., 2011; Wessén et al., 2011).
4.2 Effects of sea animal colonization on soil potential ammonia oxidation rates
The PAOR ranged from 9 to 139 µg N kg−1 h−1, lower than nitrification rates measured in most agricultural soils (83–1875 µg N kg−1 h−1) (Fan et al., 2011; Ouyang et al., 2016; Daebeler et al., 2017). One reason might be the selection of a 15 ∘C incubation temperature, which was lower than the incubation temperatures used in other studies. Generally, the gross nitrification rate and amoA abundance increased significantly when the incubation temperature was higher than 15 ∘C (Daebeler et al., 2017; Zhao et al., 2014). Our measurements indicated that there were significant differences (P=0.02) in the PAOR across different tundra patches, and the PAORs in SS and PS were about 10 times higher than those in BS and MS. A significant correlation was obtained between the PAOR and C : N, TP, and TS (Table 2). Overall, ammonia oxidation activity was modulated by soil biogeochemical processes under the disturbance of penguin or seal activities: generally low C : N ratios and sufficient input of the nutrients TP, TS, and –N from sea animal excrements.
The higher AOB abundances (Fig. 2b) and significant negative correlation of AOA abundance with –N levels (Table 2) indicated that AOB might play a more important role in nitrification in tundra soils. In agreement with these results, AOB dominated nitrification in the areas where it was easy to achieve nitrogen input, whereas the relative contribution of AOA to nitrification was higher in the areas where the ammonium concentration remained low (Fan et al., 2011; Sterngren et al., 2015). Moreover, the cell-specific activity for AOB was 10 times higher than that for AOA due to the bigger cell size of AOB (Hatzenpichler, 2012; Prosser and Nicol, 2012). Therefore, AOB might play a more important role in nitrification in SS, PS, and PL with the input of –N from penguin or seal excrements.
In addition, AOA might play a role that cannot be ignored in MS and BS, just like the prevalence of AOA among ammonia oxidizers in arctic soils (Alves et al., 2013; Daebeler et al., 2017). AOB groups were mostly undetectable in the analysis of MS and BS. Although unknown γ-AOB groups might not have been detected, the primer set used here covers the β-AOB groups typically found in soils (Alves et al., 2013). The BS and MS were moderately far away from penguin or seal colonies without the input of the nutrients from sea animal excrements, and their substrates can be provided only through the mineralization of organic matter from local tundra plants. The simple organic substrates and barren soil environment might favor AOA (Stopnišek et al., 2010; Habteselassie et al., 2013). Therefore, AOA showed relatively high abundance in MS and BS compared with PS and SS.
4.3 Effects of sea animal colonization on genotypic diversity of soil AOA and AOB
In this study, distinct AOA communities appear to inhabit different types of tundra patches, depending on sea animal activities (Fig. 5a). It was difficult to amplify the AOA amoA gene from SS and PS, whereas a high diversity of AOA amoA genes was observed in PL, MS, and BS. Phylogenetic analysis indicated that the AOA amoA sequences in cluster I were from PL and tundra soils close to seal wallows, while the sequences in cluster II were from BS and MS (Fig. S5). AOA in most extreme environments have lower levels of microbial diversity than in benign ecosystems because of the requirement for specific physiological adaptations which allow organisms to exploit the combination of physical and biochemical stressors (Cowan et al., 2015). Detected OTUs in cluster I had their closest matches mainly in the hyper-arid soils of Antarctic Dry Valleys (Magalhães et al., 2014), wetland soils (Y. K. Zheng et al., 2014), alpine meadow soils (Zhao et al., 2017), and some agricultural soils (Glaser et al., 2010). Cluster II was more prevalent in BS and MS, probably because of their stronger adaptation to barren soil environments. In cluster II, the sequences were affiliated with sequences recovered from cold environments, including the soils of the Tibetan Plateau (Xie et al., 2014) and Icelandic grassland soils (Daebeler et al., 2012). The compositions of soil AOA populations are likely not to be explained by individual physicochemical properties, and their community structures significantly correlated with tundra soil C : N, and TC, which was consistent with previous studies (Glaser et al., 2010; Wessén et al., 2011).
AOB amoA gene diversity was higher than that of AOA, similar to results in the Antarctic Dry Valley soils (Magalhães et al., 2014). A high diversity of AOB amoA genes occurred in SS, PS, and PL compared to BS, indicating that penguin or seal activities had important effects on AOB genotypic diversity. Phylogenetic analysis indicated that the sequences in clusters I and II were mainly from PS and SS (Fig. 5b), and the detected OTUs in cluster I had their closest matches in mixed community culture systems, a meadow-to-forest transect in Oregon Cascade Mountains (Mintie et al., 2003), and Dutch agricultural soils (M. C. Silva et al., 2012a) and reservoir sediments (A. F. Silva et al., 2012b). For clusters III and IV, the sequences were predominantly from PL and SS, and they were affiliated with sequences recovered from high-altitude wetland (Yang et al., 2014). Previous studies have shown that multiple environmental factors affected the AOB communities (Dang et al., 2008; Mosier and Francis, 2008). In this study, the C : N ratios and -N concentrations seemed to be the most important factors influencing the AOB community structure, which was in accordance with the results from different environments (Bouskill et al., 2012; Jung et al., 2011; Li et al., 2015). Moreover, the TP also affected the AOB amoA community compositions (Zheng et al., 2013). Therefore, the AOB community compositions were impacted by the biogeochemical factors related to sea animal activities, such as low C : N ratios and a sufficient supply of the nutrients –N and TP from sea animal excreta.
The findings of this study concerning the abundance, potential activity, and diversity of tundra soil AOA and AOB provide insights into microbial mechanisms driving nitrification in maritime Antarctica. We confirmed the presence of AOA and AOB amoA genes in five different tundra patches and demonstrated that the spatial distribution heterogeneities of the tundra soil AOA and AOB communities were driven by penguin or seal activities. The soil AOB amoA copy numbers were generally higher than the AOA amoA copy numbers, following the higher PAOR in penguin or seal colonies and their adjacent tundra, compared with that in the background tundra and marsh tundra. Penguin or seal activities resulted in a significant shift in soil AOA and AOB community compositions. AOB amoA gene diversity was higher in SS and PS than in PL and MS, and the majority of the AOB sequences was closely related to Nitrosospira-like sequences. The AOA amoA gene had higher diversity in PL and MS than in BS, and it was associated with Nitrososphaera sequences recovered from barren soils. Soil AOB and AOA abundances, and their community compositions, were related to soil biogeochemical processes under the sea-animal-activity disturbance, such as soil C : N alteration and a sufficient supply of the nutrients –N, N and P from animal excreta. This study significantly enhances the understanding of ammonia-oxidizing microbial communities in the tundra environment of maritime Antarctica.
The final derived data presented in this study are available at https://doi.org/10.5281/zenodo.1260292 (Wang, 2018).
The supplement related to this article is available online at: https://doi.org/10.5194/bg-16-4113-2019-supplement.
RBZ, LJH, and QW conceived of the study together. QW conducted the laboratory analyses and statistical analyses with support from LJH and YLZ. TB retrieved samples during field work. The paper was written by QW with support by RBZ.
The authors declare that they have no conflict of interest.
This work was supported by the NSFC (grant nos. 41576181, 41776190). We are particularly grateful to the members of the Chinese Antarctic Research Expedition and the Polar Office of the National Ocean Bureau of China for their support and timely help.
This research has been supported by the National Natural Science Foundation of China (grant nos. 41576181, 41776190).
This paper was edited by Denise Akob and reviewed by Weidong Kong and Barbara Bayer.
Alves, R. J. E., Wanek, W., Zappe, A., Richter, A., Svenning, M. M., Schleper, C., and Urich, T.: Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidizing archaea, ISME J., 7, 1620–1631, https://doi.org/10.1038/ ismej.2013.35, 2013.
Ayton, J., Aislabie, J., Barker, G. M., Saul, D., and Turner, S.: Crenarchaeota affiliated with group 1.1 b are prevalent in coastal mineral soils of the Ross Sea region of Antarctica, Environ. Microbiol., 12, 689–703, https://doi.org/10.1111/j.14622920.2009.02111.x, 2010.
Baker, B. J., Lesniewski, R. A., and Dick, G. J.: Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume, ISME J., 6, 2269–2279, https://doi.org/10.1038/ismej.2012.64, 2012.
Belser, L. W. and Schmidt, E. L.: Diversity in the ammonia-oxidizing nitrifier population of a soil, Appl. Environ. Microbiol., 36, 584–588, 1978.
Beman, J. M., Popp, B. N., and Francis, C. A.: Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California,ISME J., 2, 429–441, 2008.
Blackall, T. D., Wilson, L. J., Theobald, M. R., Milford, C., Nemitz, E., Bull, J., and Sutton, M. A.: Ammonia emissions from seabird colonies, Geophys. Res. Lett., 34, L10801, https://doi.org/10.1029/2006GL028928, 2007.
Bouskill, N. J., Eveillard, D., Chien, D., Jayakumar, A., and Ward, B. B.: Environmental factors determining ammoniaoxidizing organism distribution and diversity in marine environments, Environ. Microbiol., 14, 714–729, https://doi.org/10.1111/j.1462-2920.2011.02623.x, 2012.
Boyd, E. S., Lange, R. K., Mitchell, A. C., Havig, J. R., Hamilton, T. L., Lafrenière, M. J., Shock, E. L., Peters, J. W., and Skidmore, M.: Diversity, abundance, and potential activity of nitrifying and nitrate-reducing microbial assemblages in a subglacial ecosystem, Appl. Environ. Microbiol., 77, 4778–4787, https://doi.org/10.1128/AEM.00376-11, 2011.
Cowan, D. A., Ramond, J. B., Makhalanyane, T. P., and De Maayer, P.: Metagenomics of extreme environments, Curr. Opin. Microbiol., 25, 97–102, https://doi.org/10.1016/j.mib.2015.05.005, 2015.
Daebeler, A., Abell, G. C., Bodelier, P. L., Bodrossy, L., Frampton, D. M., Hefting, M. M., and Laanbroek, H. J.: Archaeal dominated ammonia-oxidizing communities in Icelandic grassland soils are moderately affected by long-term N fertilization and geothermal heating, Front. Microbiol., 3, 352, https://doi.org/10.3389/fmicb.2012.00352, 2012.
Daebeler, A., Bodelier, P. L., Hefting, M. M., Rütting, T., Jia, Z., and Laanbroek, H. J.: Soil warming and fertilization altered rates of nitrogen transformation processes and selected for adapted ammonia-oxidizing archaea in sub-arctic grassland soil, Soil Biol. Biochem., 107, 114–124, https://doi.org/10.1016/j.soilbio.2016.12.013, 2017.
Dang, H., Zhang, X., Sun, J., Li, T., Zhang, Z., and Yang, G.: Diversity and spatial distribution of sediment ammonia-oxidizing crenarchaeota in response to estuarine and environmental gradients in the Changjiang Estuary and East China Sea, Microbiology, 154, 2084–2095, https://doi.org/10.1099/mic.0.2007/013581-0, 2008.
Di, H. J., Cameron, K. C., Shen, J. P., Winefield, C. S., O'callaghan, M., Bowatte, S., and He, J. Z.: Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils, Nat. Geosci., 2, 621–624, https://doi.org/10.1038/ngeo613, 2009.
Edgar, R. C.: MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucl. Acid. Res., 32, 1792–1797, 2004.
Fan, F., Yang, Q., Li, Z., Wei, D., Cui, X. A., and Liang, Y.: Impacts of organic and inorganic fertilizers on nitrification in a cold climate soil are linked to the bacterial ammonia oxidizer community, Microb. Ecol., 62, 982–990, https://doi.org/10.1007/s00248-011-9897-5, 2011.
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E., and Oakley, B. B.: Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean, P. Natl. Acad. Sci. USA, 102, 14683–14688, https://doi.org/10.1073/pnas.0506625102, 2005.
Gao, Y. S., Yang, L. J., Wang, J. J., Xie, Z. Q., Wang, Y. H., and Sun, L. G.: Penguin colonization following the last glacial-interglacial transition in the Vestfold Hills, East Antarctica, Palaeogeogr. Palaeocl., 490, 629–639, https://doi.org/10.1016/j.palaeo.2017.11.053, 2018.
Glaser, K., Hackl, E., Inselsbacher, E., Strauss, J., Wanek, W., Zechmeister-Boltenstern, S., and Sessitsch, A.: Dynamics of ammonia-oxidizing communities in barley-planted bulk soil and rhizosphere following nitrate and ammonium fertilizer amendment, FEMS Microbiol. Ecol., 74, 575–591, https://doi.org/10.1111/j.1574-6941.2010.00970.x, 2010.
Gubry-Rangin, C., Novotnik, B., Mandič-Mulec, I., Nicol, G. W., and Prosser, J. I.: Temperature responses of soil ammonia-oxidising archaea depend on pH, Soil Biol. Biochem., 106, 61–68, https://doi.org/10.1016/j.soilbio.2016.12.007, 2017.
Habteselassie, M. Y., Xu, L., and Norton, J. M.: Ammonia-oxidizer communities in an agricultural soil treated with contrasting nitrogen sources, Front. Microbiol., 4, 326, https://doi.org/10.3389/fmicb.2013.00326, 2013.
Hatzenpichler, R.: Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea, Appl. Environ. Microbiol., 78, 7501–7510, https://doi.org/10.1128/AEM.01960-12, 2012.
Hatzenpichler, R., Lebedeva, E. V., Spieck, E., Stoecker, K., Richter, A., Daims, H., and Wagner, M.: A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring, P. Natl. Acad. Sci. USA, 105, 2134–2139, https://doi.org/10.1073/pnas.0708857105, 2008.
Jung, J., Yeom, J., Kim, J., Han, J., Lim, H. S., Park, H., Hyun, S., and Park, W.: Change in gene abundance in the nitrogen biogeochemical cycle with temperature and nitrogen addition in Antarctic soils, Res. Microbiol., 162, 1018–1026, https://doi.org/10.1016/j.resmic.2011.07.007, 2011.
Kalanetra, K. M., Bano, N., and Hollibaugh, J. T.: Ammonia-oxidizing Archaea in the Arctic Ocean and Antarctic coastal waters, Environ. Microbiol., 11, 2434–2445, https://doi.org/10.1111/j.1462-2920.2009.01974.x, 2009.
Könneke, M., Bernhard, A. E., José, R., Walker, C. B., Waterbury, J. B., and Stahl, D. A.: Isolation of an autotrophic ammonia-oxidizing marine archaeon, Nature, 437, 543–546, https://doi.org/10.1038/nature03911, 2005.
Kurola, J., Salkinoja-Salonen, M., Aarnio, T., Hultman, J., and Romantschuk, M.: Activity, diversity and population size of ammoniaoxidising bacteria in oilcontaminated landfarming soil, FEMS Microbiol. Lett., 250, 33–38, https://doi.org/10.1016/j.femsle.2005.06.057, 2005.
Lam, P., Jensen, M. M., Lavik, G., McGinnis, D. F., Müller, B., Schubert, C. J., Amann, R., Thamdrup, B., and Kuypers, M. M.: Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea, P. Natl. Acad. Sci. USA, 104, 7104–7109, https://doi.org/10.1073/pnas.0611081104, 2007.
Le Roux, X., Poly, F., Currey, P., Commeaux, C., Hai, B., Nicol, G. W., Prosser, J. I., Schloter, M., Attard, E., and Klumpp, K.: Effects of aboveground grazing on coupling among nitrifier activity, abundance and community structure, ISME J., 2, 221–232, https://doi.org/10.1038/ismej.2007.109, 2008
Leininger, S., Urich, T., Schloter, M., Schwark, L., Qi, J., Nicol, G. W., Prosser, J. I., Schuster, S. C., and Schleper, C.: Archaea predominate among ammonia-oxidizing prokaryotes in soils, Nature, 442, 806–809, https://doi.org/10.1038/nature04983, 2006.
Li, J., Nedwell, D. B., Beddow, J., Dumbrell, A. J., McKew, B. A., Thorpe, E. L., and Whitby, C.: amoA gene abundances and nitrification potential rates suggest that benthic ammonia-oxidizing bacteria and not archaea dominate N cycling in the Colne Estuary, United Kingdom, Appl. Environ. Microbiol., 81, 159–165, https://doi.org/10.1128/AEM.02654-14, 2015.
Lindeboom, H. J.: The nitrogen pathway in a penguin rookery, Ecology, 65, 269–277, https://doi.org/10.2307/1939479, 1984.
Ma, D. W., Zhu, R. B., Ding, W., Shen, C. C., Chu, H. Y., and Lin, X. G.: Ex-situ enzyme activity and bacterial community diversity through soil depth profiles in penguin and seal colonies on Vestfold Hills, East Antarctica, Pol. Biol., 36, 1347–1361, https://doi.org/10.1007/s00300-013-1355-z, 2013.
Madden, T.: The BLAST Sequence Analysis Tool, in: The NCBI Handbook, edited by: McEntyre, J. and Ostell, J., Bethesda (MD), National Center for Biotechnology Information (US), Chap. 16, Bookshelf ID: NBK21097, 2002.
Magalhães, C. M., Machado, A., Frank-Fahle, B., Lee, C. K., and Cary, S. C.: The ecological dichotomy of ammonia-oxidizing archaea and bacteria in the hyper-arid soils of the Antarctic Dry Valleys, Front. Microbiol., 5, 515, https://doi.org/10.3389/fmicb.2014.00515, 2014.
Martens-Habbena, W., Berube, P. M., Urakawa, H., José, R., and Stahl, D. A.: Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria, Nature, 461, 976–979, https://doi.org/10.1038/nature08465, 2009.
Mintie, A. T., Heichen, R. S., Cromack Jr, K., Myrold, D. D., and Bottomley, P. J.: Ammonia-oxidizing bacteria along meadow-to-forest transects in the Oregon Cascade Mountains, Appl. Environ. Microbiol., 69, 3129–3136, https://doi.org/10.1128/AEM.69.6.3129-3136.2003, 2003.
Mosier, A. C. and Francis, C. A.: Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary, Environ. Microbiol., 10, 3002–3016, https://doi.org/10.1111/j.1462-2920.2008.01764.x, 2008.
Nunes-Alves, C.: Do it yourself nitrification, Nat. Rev. Microbiol., 14, 61, https://doi.org/10.1038/nrmicro.2015.20, 2016.
Otero, X. L., Peña-Lastra, S., Pérez-Alberti, A., Ferreira, T. O., and Huerta-Diaz, M. A.: Seabird colonies as important global drivers in the nitrogen and phosphorus cycles, Nat. Commun., 9, 246, https://doi.org/0.1038/s41467-017-02446-8, 2018.
Ouyang, Y., Norton, J. M., Stark, J. M., Reeve, J. R., and Habteselassie, M. Y.: Ammonia-oxidizing bacteria are more responsive than archaea to nitrogen source in an agricultural soil, Soil Biol. Biochem., 96, 4–15, https://doi.org/10.1016/j.soilbio.2016.01.012, 2016.
Pan, H., Xie, K. X., Zhang, Q. C., Jia, Z. J., Xu, J. M., Di, H. J., and Li, Y.: Archaea and bacteria respectively dominate nitrification in lightly and heavily grazed soil in a grassland system, Biol. Fert. Soils, 54, 41–54, https://doi.org/10.1007/s00374-017-1236-7, 2018.
Park, J. H., Day, T. A., Strauss, S., and Ruhland, C. T.: Biogeochemical pools and fluxes of carbon and nitrogen in a maritime tundra near penguin colonies along the Antarctic Peninsula, Pol. Biol., 30, 199–207, https://doi.org/10.1007/s00300-006-0173-y, 2007.
Prosser, J. I. and Nicol, G. W.: Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation, Trends Microbiol., 20, 523–531, https://doi.org/10.1016/j.tim.2012.08.001, 2012.
Richter, I., Herbold, C. W., Lee, C. K., McDonald, I. R., Barrett, J. E., and Cary, S. C.: Influence of soil properties on archaeal diversity and distribution in the McMurdo Dry Valleys, Antarctica, FEMS Microbiol. Ecol., 89, 347–359, https://doi.org/10.1111/1574-6941.12322, 2014.
Riddick, S. N., Dragosits, U., Blackall, T. D., Daunt, F., Wanless, S., and Sutton, M. A.: The global distribution of ammonia emissions from seabird colonies, Atmos. Environ., 55, 319–327, https://doi.org/10.1016/j.atmosenv.2012.02.052, 2012.
Rotthauwe, J. H., Witzel, K. P., and Liesack, W.: The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations, Appl. Environ. Microbiol., 63, 4704–4712, 1997.
Santoro, A. E., Buchwald, C., McIlvin, M. R., and Casciotti, K. L.: Isotopic signature of N2O produced by marine ammonia-oxidizing archaea, Science, 333, 1282–1285, https://doi.org/10.1126/science.1208239, 2011.
Schloss, P. D., Westcott, S., Ryabin, T., Hall, J., Hartmann, M., Hollister, E., Lesniewski, R. A., Oakley, B. B., Parks, D. H., Robinson, C. J., Sahl, J. W., Stres, B., Thallinger, G. G., Van Horn, D. J., and Weber, C. F.: Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities, Appl. Environ. Microbiol., 75, 7537–7541, https://doi.org/10.1128/AEM.01541-09, 2009.
Shen, J. P., Zhang, L. M., Di, H. J., and He, J. Z.: A review of ammonia-oxidizing bacteria and archaea in Chinese soils, Front. Microbiol., 3, 296, https://doi.org/10.3389/fmicb.2012.00296, 2012.
Simas, F. N. B., Schaefer, C. E. G. R. , Melo, V. F., Albuquerque-Filho, M. R. A., Michel, R. F. M., Pereira, V. V., Gomes, M. R. M., and da Costa, L. M.: Ornithogenic cryosols from Maritime Antarctica: Phosphatization as a soil forming process, Geoderma, 138, 191–203, https://doi.org/10.1016/j.geoderma.2006.11.011, 2007.
Silva, A. F., Carvalho, G., Oehmen, A., Lousada-Ferreira, M., van Nieuwenhuijzen, A., Reis, M. A., and Crespo, M. T. B.: Microbial population analysis of nutrient removal-related organisms in membrane bioreactors. Appl. Microbiol. Biotechnol., 93, 2171–2180, https://doi.org/10.1007/s00253-011-3499-5, 2012.
Silva, M. C., Poly, F., Guillaumaud, N., van Elsas, J. D., and Falcão Salles, J.: Fluctuations in ammonia oxidizing communities across agricultural soils are driven by soil structure and pH, Front. Microbiol., 3, 77, https://doi.org/10.3389/fmicb.2012.00077, 2012.
Sterngren, A. E., Hallin, S., and Bengtson, P.: Archaeal ammonia oxidizers dominate in numbers, but bacteria drive gross nitrification in N-amended grassland soil, Front. Microbiol., 6, 1350, https://doi.org/10.3389/fmicb.2015.01350, 2015.
Stieglmeier, M., Mooshammer, M., Kitzler, B., Wanek, W., Zechmeister-Boltenstern, S., Richter, A., and Schleper, C.: Aerobic nitrous oxide production through N-nitrosating hybrid formation in ammonia-oxidizing archaea, ISME J., 8, 1135–1146, 2014.
Stopnišek, N., Gubry-Rangin, C., Höfferle, Š., Nicol, G. W., Mandič-Mulec, I., and Prosser, J. I. : Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment, Appl. Environ. Microbiol., 76, 7626–7634, https://doi.org/10.1128/AEM.00595-10, 2010.
Sun, L. G., Liu, X. D., Yin, X. B., Zhu, R. B., Xie, Z. Q., and Wang, Y. H.: A 1,500-year record of Antarctic seal populations in response to climate change, Pol. Biol., 27, 495–501, https://doi.org/10.1007/s00300-004-0608-2, 2004.
Sun, L. G., Xie, Z. Q., and Zhao, J. L.: Palaeoecology: A 3,000-year record of penguin populations, Nature, 407, 858, https://doi.org/10.1038/35038163, 2000.
Sun, L., Zhu, R., Xie, Z., and Xing, G.: Emissions of nitrous oxide and methane from Antarctic tundra: role of penguin dropping deposition, Atmos. Environ., 36, 4977–4982, https://doi.org/10.1016/S1352-2310(02)00340-0, 2002.
Tatur, A., Myrcha, A., and Niegodzisz, J.: Formation of abandoned penguin rookery ecosystems in the maritime Antarctic, Pol. Biol., 17, 405–417, https://doi.org/10.1007/s003000050135, 1997.
Tolar, B. B., Ross, M. J., Wallsgrove, N. J., Liu, Q., Aluwihare, L. I., Popp, B. N., and Hollibaugh, J. T.: Contribution of ammonia oxidation to chemoautotrophy in Antarctic coastal waters, ISME J., 10, 2605, https://doi.org/10.1038/ismej.2016.61, 2016.
Verhamme, D. T., Prosser, J. I., and Nicol, G. W.: Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms, ISME J., 5, 1067–1071, https://doi.org/10.1038/ismej.2010.191, 2011.
Wang, B. Z., Zhao, J., Guo, Z. Y., Ma, J., Xu, H., and Jia, Z. J.: Differential contributions of ammonia oxidizers and nitrite oxidizers to nitrification in four paddy soils, ISME J., 9, 1062–1075, https://doi.org/10.1038/ismej.2014.194, 2015.
Wang, Q.: Table. Effects of sea animal colonization on the coupling between dynamics and activity of soil ammonia-oxidizing bacteria and archaea in maritime Antarctica, https://doi.org/10.5281/zenodo.1260292, 2018.
Wessén, E., Söderström, M., Stenberg, M., Bru, D., Hellman, M., Welsh, A., Thomsen, F., Klemedtson, L., Philippot, L., and Hallim, S.: Spatial distribution of ammonia-oxidizing bacteria and archaea across a 44-hectare farm related to ecosystem functioning, ISME J., 5, 1213–1225, https://doi.org/10.1038/ismej.2010.206, 2011.
Wuchter, C., Abbas, B., Coolen, M. J. L., Herfort, L., van Bleijswijk, J., Timmers, P., Strous, M., Teira, E., Herndl, G. J., Middelburg, J. J., Schouten, S., and Sinninghe Damsté, J. S.: Archaeal nitrification in the ocean, P. Natl. Acad. Sci. USA, 103, 12317–12322, https://doi.org/10.1073/pnas.0600756103, 2006.
Xia, Y., Zhu, Y. G., Gu, Q., and He, J. Z.: Does long-term fertilization treatment affect the response of soil ammonia-oxidizing bacterial communities to Zn contamination?, Plant Soil, 301, 245–254, https://doi.org/10.1007/s11104-007-9441-z, 2007.
Xie, Z., Le Roux, X., Wang, C., Gu, Z., An, M., Nan, H., Chen, B. Z., Li, F., Liu, Y. J., Du, G. Z., Feng, H. Y., and Ma, X. J.: Identifying response groups of soil nitrifiers and denitrifiers to grazing and associated soil environmental drivers in Tibetan alpine meadows, Soil Biol. Biochem., 77, 89–99, https://doi.org/10.1016/j.soilbio.2014.06.024, 2014.
Yang, Y., Shan, J., Zhang, J., Zhang, X., Xie, S., and Liu, Y.: Ammonia-and methane-oxidizing microorganisms in high-altitude wetland sediments and adjacent agricultural soils, Appl. Microbiol. Biotechnol., 98, 10197–10209, https://doi.org/10.1007/s00253-014-5942-x, 2014.
Yao, H. Y., Gao, Y. M., Nicol, G. W., Campbell, C. D., Prosser, J. I., Zhang, L. M., Han, W. Y., and Singh, B. K.: Links between ammonia oxidizer community structure, abundance, and nitrification potential in acidic soils, Appl. Environ. Microbiol., 77, 4618–4625, https://doi.org/10.1128/AEM.00136-11, 2011.
Yergeau, E., Kang, S., He, Z., Zhou, J., and Kowalchuk, G. A.: Functional microarray analysis of nitrogen and carbon cycling genes across an Antarctic latitudinal transect, ISME J., 1, 163–179, https://doi.org/10.1038/ismej.2007.24, 2007.
Zhang, L. M., Wang, M., Prosser, J. I., Zheng, Y. M., and He, J. Z.: Altitude ammonia-oxidizing bacteria and archaea in soils of Mount Everest, FEMS Microbiol. Ecol., 70, 208–217, https://doi.org/10.1111/j.1574-6941.2009.00775.x, 2009.
Zhao, K., Kong, W. D., Khan, A., Liu, J. B., Guo, G. X., Muhanmmad, S., Zhao, X. Z., and Dong, X. B.: Elevational diversity and distribution of ammonia-oxidizing archaea community in meadow soils on the Tibetan Plateau, Appl. Microbiol. Biotechnol., 101, 7065–7074, https://doi.org/10.1007/s00253-017-8435-x, 2017.
Zhao, X., Wang, S., and Xing, G.: Nitrification, acidification, and nitrogen leaching from subtropical cropland soils as affected by rice straw-based biochar: laboratory incubation and column leaching studies, J. Soil. Sediment., 14, 471–482, 2014.
Zheng, Y. K., Wang, X. B., Gu, Y. F., and Zhang, X. P.: Diversity of ammonia-oxidizing archaea in Tibetan Zoige plateau wetland, Acta Microbiol. Sin., 54, 1090–1096, 2014.
Zheng, Y. L., Hou, L. J., Liu, M., Lu, M., Zhao, H., Yin, G. Y., and Zhou, J. L.: Diversity, abundance, and activity of ammonia-oxidizing bacteria and archaea in Chongming eastern intertidal sediments, Appl. Microbiol. Biotechnol., 97, 8351–8363, https://doi.org/10.1007/s00253-012-4512-3, 2013.
Zheng, Y. L., Hou, L. J., Newell, S., Liu, M., Zhou, J. L., Zhao, H., You, L. L., and Cheng, X. L.:. Community dynamics and activity of ammonia-oxidizing prokaryotes in intertidal sediments of the Yangtze estuary, Appl. Environ. Microbiol., 80, 408–149, https://doi.org/10.1128/AEM.03035-13, 2014.
Zhu, R. B., Liu, Y. S., Xu, H., Ma, J., Zhao, S. P., and Sun, L. G.: Nitrous oxide emissions from sea animal colonies in the maritime Antarctic, Geophys. Res. Lett., 35, L09807, https://doi.org/10.1029/2007GL032541, 2008.
Zhu, R. B., Sun, J. J., Liu, Y. S., Gong, Z. J., and Sun, L. G.: Potential ammonia emissions from penguin guano, ornithogenic soils and seal colony soils in coastal Antarctica: effects of freezing-thawing cycles and selected environmental variables, Antarct. Sci., 23, 78–92, https://doi.org/10.1017/S0954102010000623, 2011.
Zhu, R. B., Liu, Y. S., Xu, H., Ma, D. W., and Jiang, S.: Marine animals significantly increase tundra N2O and CH4 emissions in maritime Antarctica, J. Geophys. Res.-Biogeo., 118, 1773–1792, https://doi.org/10.1002/2013JG002398, 2013.
Zhu, R. B., Wang, Q., Ding, W., Wang, C., Hou, L. J., and Ma, D. W.: Penguins significantly increased phosphine formation and phosphorus contribution in maritime Antarctic soils, Sci. Rep., 4, 7055, https://doi.org/10.1038/srep07055, 2014.
Zhu, R. B., Shi, Y., Ma, D. W., Wang, C., Xu, H., and Chu, H. Y.: Bacterial diversity is strongly associated with historical penguin activity in an Antarctic lake sediment profile, Sci. Rep., 5, 17231, https://doi.org/10.1038/srep17231, 2015.