the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
The pH-based ecological coherence of active canonical methanotrophs in paddy soils
Jun Zhao
Yuanfeng Cai
Zhongjun Jia
Soil pH is considered one of the main determinants of the assembly of globally distributed microorganisms that catalyze the biogeochemical cycles of carbon (C) and nitrogen (N). However, direct evidence for niche specialization of microorganisms in association with soil pH is still lacking. Using methane-oxidizing bacteria (methanotrophs) as a model system of C cycling, we show that pH is potentially the key driving force selecting for canonical γ (type I) and α (type II) methanotrophs in rice paddy soils. DNA-based stable isotope probing (DNA-SIP) was combined with high-throughput sequencing to reveal the taxonomic identities of active methanotrophs in physiochemically contrasting soils from six different paddy fields across China. Following microcosm incubation amended with 13CH4, methane was primarily consumed by Methylocystis-affiliated type II methanotrophs in soils with a relatively low pH (5.44–6.10), whereas Methylobacter- or Methylosarcina-affiliated type I methanotrophs dominated methane consumption in soils with a high pH (7.02–8.02). Consumption of 13CH4 contributed 0.203 % to 1.25 % of soil organic C, but no significant difference was observed between high-pH and low-pH soils. The fertilization of ammonium nitrate resulted in no significant changes in the compositions of 13C-labeled methanotrophs in the soils, although significant inhibition of methane oxidation activity was consistently observed in low-pH soils. Mantel analysis further validated that soil pH, rather than other parameters tested, had significant correlation to the variation in active methanotrophic compositions across different rice paddy soils. These results suggest that soil pH might have played a pivotal role in mediating the niche differentiation of ecologically important aerobic methanotrophs in terrestrial ecosystems and imply the importance of such niche specialization in regulating methane emissions in paddy fields following increasingly intensified input of anthropogenic N fertilizers.
- Article
(1267 KB) - Full-text XML
-
Supplement
(884 KB) - BibTeX
- EndNote
Rice paddy fields are one of the major sources of the potent greenhouse gas methane, contributing approximately 10 %–25 % of global methane emissions (Kögel-Knabner et al., 2010). Constantly produced through methanogenesis from the anaerobic compartment of inundated paddy fields, methane can diffuse into the oxic–anoxic interface of the soil and reach high concentrations of above 5 mM, which is approximately equivalent to 50 000 ppmv (Eller and Frenzel, 2001; Nouchi et al., 1990, 1994). It is estimated that up to 80 % of methane gas is consumed by soil aerobic methanotrophs (i.e., methane-oxidizing bacteria, MOB) before release into the atmosphere (Conrad and Rothfuss, 1991; Frenzel et al., 1992). Therefore, methanotrophs in rice paddy fields are considered a crucial biological filter attenuating potential methane emissions, as well as an important contributor to the soil organic C, because a substantial amount of CH4 was used for MOB growth and biomass synthesis (Bridgham et al., 2013).
Accumulating evidence has indicated that the specialized MOB clades designated as upland soil clusters α and γ (USCα and USCγ, respectively) have high CH4 affinity and catalyze atmospheric methane oxidation in unsaturated soils (Holmes et al., 1999; Knief et al., 2003; Roslev and Iversen, 1999; Tveit et al., 2019), while the canonical α and γProteobacteria MOB (known as type II and type I methanotrophs) are considered to adapt better to high-concentration methane and thus regulate the methane oxidation in ecosystems with constant methane production, such as wetlands. Despite coexistence of type I and II methanotrophs in these ecosystems, their activity and contribution to methane oxidation has often been observed to vary largely depending on different environmental conditions and the predominant activity of either type I or type II methanotrophs (Chen et al., 2008a; Daebeler et al., 2014; He et al., 2012; Liebner and Wagner, 2007; Lin et al., 2004). This might be due to some major physiological differences that exist between these two groups. For instance, type I methanotrophic strain is more competitive under relatively lower methane and higher oxygen concentrations compared to type II methanotroph (Graham et al., 1993). Additionally, adaptation to slightly acidic pH values (growth optima 5.0–6.0) is characteristic for type IIb (Methylocella and Methylocapsa) and some Methylocystis type Ia MOB strains (Belova et al., 2013; Dedysh et al., 2000, 2007). These and other physiological traits of type I and II methanotrophs may be important in partitioning their specialized niches in different ecosystems.
Increasing lines of evidence from ecological studies have suggested that soil pH, among other environmental variables, might be one of the most important determinants in the emergence and maintenance of microbial communities across a wide variety of environments (Lauber et al., 2009; Tripathi et al., 2018). Recent studies have provided compelling evidence for niche specialization of biogeochemically important guilds associated with pH variation and consequent distinct patterns of soil resource utilization. For example, the biogeographical distribution of ammonia oxidizers is more strongly associated with soil pH than other parameters tested in soils (Aigle et al., 2019; Gubry-Rangin et al., 2011), as is that of denitrifiers (Liu et al., 2010). It is implied that type I and II methanotrophs might also be selectively favored under different pH conditions in natural wetland system, despite no systematic comparison has yet been made. Dominance of type II methanotrophs has been observed in many natural acidic peatlands (Chen et al., 2008a, b; Dedysh, 2009; Gupta et al., 2012; Kip et al., 2012), whereas in neutral–alkaline wetlands, type I methanotrophs appear to be more active (Gupta et al., 2012; Lin et al., 2004; Morris et al., 2002). The similar pattern, however, has not been obtained in anthropogenically flooded rice paddy fields, possibly due to lack of studies on methanotrophic activity in such soils with a wide range of pH. In fact, type I methanotrophs were considered to dominantly catalyze methane at high concentrations in the rice paddy soils (Ma et al., 2013; Qiu et al., 2008; Reim et al., 2012; Shrestha et al., 2008), while the activity of type II methanotrophs and their contribution to methane oxidation in rice soils remain unclear (Leng et al., 2015; Semrau et al., 2011). Since these findings all came from neutral–alkaline soils, it is necessary to investigate the active methanotrophs in more acidic soils, which likely have different community compositions (Shiau et al., 2018).
In this study, we selected six rice paddy soils with a pH gradient ranging from 5.45 to 8.02 collected from six geographically different rice fields in China and used DNA-SIP to identify active methanotrophs under unfertilized conditions and following a simulated fertilization. We predict distinct compositions of methanotrophic phylotypes of active type I and type II MOB in the soils that are associated with soil pH. Other environmental factors pertinent to methane oxidation were also tested to further elaborate the importance of soil pH in selection of active methanotrophs in the rice paddies.
2.1 Site description and soil sampling
The soils were collected from six different rice fields located in the main rice production areas across the south, east and center of China, including Yuxi (YX), Yingtan (YT), Taoyuan (TY), Ziyang (ZY), Changshu (CS) and Leizhou (LZ) (Fig. S1 in the Supplement). All sites have a subtropical monsoon climate and a history of rice cultivation for >50 years. The fields usually receive annual fertilization of 250 to 350 kg N ha−1. Soil sampling was performed at a 0–20 cm depth by mixing at least three randomly collected soil cores. The composite soil samples were air-dried as previously described (Mohanty et al., 2006; Murase and Frenzel, 2007; Noll et al., 2008) and passed through a sieve with a 2.0 mm pore size before the construction of microcosms.
2.2 Soil physiochemical properties
Soil pH was assessed by a Mettler Toledo 320-S pH meter (Mettler Toledo Instruments, China) with a water-to-soil ratio of 2.5. Soil inorganic N (ammonium and nitrate) was measured with a 2 M KCl solution and quantified using a Skalar San Plus segmented flow analyzer (Skalar, the Netherlands). The soil organic matter (SOM) content was determined using the dichromate oxidation method. Total organic carbon (TOC) and total nitrogen (TN) were determined by a vario Max CN Element Analyzer (Elementar, Germany). The available soil copper content was determined using an OPTIMA 8000 inductively coupled plasma optical emission spectroscope (ICP-OES) (PerkinElmer, USA) after extraction with buffered 5 mM Diethylenetriaminepentaacetic acid (DTPA) solution. The soil oxidation capacity (OXC) was determined as detailed previously (Zhang et al., 2009). The results of these properties were shown in Table 1.
Table 1The sampling locations and chemical characteristics of the rice paddy soils.
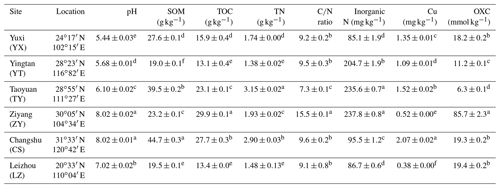
Abbreviations: SOM, soil organic matter; TOC, total organic carbon; OXC, soil oxidation capacity. Different letters (a–f) in each column of chemical properties indicate a significant difference between soils (P<0.05).
2.3 Stable isotope probing microcosms
Three treatments were established in triplicate, including “control” (under natural atmospheric conditions), “13CH4” (incubated with 5 % v∕v 13CH4 supplementation) and “13CH4+N” (incubated with 5 % v∕v 13CH4 supplementation plus N fertilization). Soil equivalent to 6.0 g dry weight was incubated at approximately 60 % maximum water-holding capacity and at 28 ∘C in the dark in a 120 mL serum bottle sealed with a butyl stopper. For the 13CH4 and 13CH4+N treatments, 6 mL of the headspace air in the bottles was replaced with the same volume of 13CH4 gas (>99 % 13C atom purity, Cambridge Isotope Laboratories, USA) to make an initial methane mixing ratio of approximately 5 % in the headspace. For the 13CH4+N treatment, ammonium nitrate (NH4NO3) solution instead of distilled water was added to the soil microcosm in a dropwise manner to produce a supplement of 200 µg inorganic N g−1 dry soil. A 2 d preincubation was performed before applying 13C-labeled methane and N fertilizer.
For the 13CH4 and 13CH4+N treatments, the headspace methane mixing ratios were measured every 2 d by an Agilent 7890A Gas Chromatograph (Agilent Technologies, USA) to assess the methane oxidation rate in SIP microcosms. Each microcosm incubation completed when approximately 90 % of the methane gas was consumed, i.e., the headspace methane concentration dropped to below 5000 ppmv, or after 6 weeks if the headspace methane concentration was still higher than 5000 ppmv. Soils were then collected and stored at −80 ∘C for further analyses.
2.4 Soil 13C atom abundance assay
The synthesis of biomass C derived from 13C-CH4 was assessed by determination of 13C atom abundance in soil organic matter. Approximately 1.5 g of each frozen soil sample was vacuum freeze-dried using an Alpha 1–2 LDplus freeze dryer (Christ, Germany). The relative 13C atom ratio was assessed by a FLASH 2000 elemental analyzer coupled to a Delta V Advantage isotope ratio mass spectrometer (Thermo Scientific, USA), and the TOC content was then measured by a vario Max CN Element Analyzer (Elementar) using the desiccated soil samples.
2.5 DNA extraction and SIP gradient fractionation
Total DNA was extracted using 0.5 g of each soil by a FastDNA spin kit for soil (MP Biomedicals, USA) according to the manufacturer's instructions. The quantity and quality of DNA extracts were assessed using a NanoDrop ND-1000 UV-visible light spectrophotometer (NanoDrop Technologies, USA).
The isopycnic density gradient centrifugation was employed to separate 13C-DNA from 12C-DNA in the total DNA extract. In brief, approximately 2.5 µg of the extracted DNA was mixed with a CsCl solution to achieve a final volume of 5.5 mL with a buoyant density of 1.725 g mL−1, followed by ultracentrifugation at 177 000 g and 20 ∘C for 44 h in a Vti65.2 vertical rotor (Beckman Coulter, USA). The DNA fractions for each sample were collected and CsCl density was measured as previously described (Wang et al., 2015; Zhao et al., 2015). The fractionated DNA was purified with 70 % ethanol after polyethylene glycol 6000 precipitation and dissolved in 30 µL of sterile water.
2.6 Real-time quantitative PCR of biomarker pmoA genes
To determine the changes in abundance of methanotrophic communities and to assess the 13CH4 labeling of methanotrophs, the copy number of pmoA genes in the total DNA extracts as well as in the DNA gradient fractions (fractions 3–13) were determined by real-time quantitative polymerase chain reaction (qPCR) using a CFX96 Optical Real-Time detection system (Bio-Rad Laboratories, USA). The PCR primers A189f and mb661r were used (Costello and Lidstrom, 1999; Holmes et al., 1995) following the conditions shown in Table S1. The standards were generated using plasmid DNA from one representative clone containing bacterial pmoA genes, and a dilution series of standard template from 102 to 108 per assay was used. In addition, the total DNA extracts were diluted in a series to assess possible PCR inhibition by soil humic substances, and DNA extracts were diluted 20-fold for subsequent analysis. The amplification efficiencies ranged from 92 % to 103 %, with R2 values of 0.994 to 0.999. Melting curve analysis and standard agarose gel electrophoresis were always performed at the end of a PCR run to verify the amplification specificity.
2.7 MiSeq sequencing of 16S rRNA and pmoA genes
Illumina MiSeq sequencing was employed to investigate the community compositions of methanotrophs in the soils. The total microbial communities were analyzed in all soil microcosms using universal primers for 16S rRNA genes to investigate the proportional changes in methanotrophs relative to the total microbial communities in soils. In addition, the 13C-DNA retrieved from “heavy” CsCl fractions (with a density of 1.738–1.740 g mL−1) in 13CH4-labeled microcosms was subjected to amplicon-based sequencing targeting both the 16S rRNA and pmoA genes. The “light” DNA fractions (with a density of 1.719–1.726 g mL−1) from the 12C control samples were also used for 16S rRNA gene sequencing to reveal the background microbial community compositions. The PCR primer pairs were 515F and 907R (Stubner, 2002) for 16S rRNA genes and A189f and mb661r for pmoA genes, with each forward primer fused with a unique bar code sequence. The PCR primers and conditions are detailed in Table S1. The resultant PCR products were gel purified and combined in equimolar ratios in a single tube. The sequencing samples were prepared using a TruSeq DNA kit (Illumina, USA), and the purified library was diluted, denatured, re-diluted and mixed with PhiX as described in the Illumina library preparation protocols. Paired-end sequencing (2×300 bp) was conducted using the MiSeq system (Illumina).
2.8 Bioinformatic analyses
All raw sequence files were processed using the Quantitative Insights Into Microbial Ecology (QIIME) pipeline (Caporaso et al., 2010). Paired-end reads were first assembled using Fast Length Adjustment of Short Reads (FLASH) with a minimum overlap parameter value of 10 bp (Magoč and Salzberg, 2011). The quality control procedure removed reads with average quality score <20 containing mismatched primers and ambiguous bases. Chimeras were eliminated using USEARCH program. For pmoA genes, putative frame-shifting reads were removed using the FrameBot program (Wang et al., 2013). Subsequently, a total of 2 871 893 high-quality 16S rRNA and 421 696 pmoA sequences were retained for further analyses. The high-quality sequences were then clustered into operational taxonomic units (OTUs) at 97 % (16S rRNA gene) or 93 % (pmoA gene) sequence similarities by the UPARSE algorithm (Edgar, 2013). To cluster pmoA genes at 93 % similarity, the “otu_radius_pct” option (default of 3) was changed to 7 when performing the “cluster_otus” command, and the id option (default of 0.97) was modified to 0.93 for the “usearch_global” command. Clustering pmoA genes at 93 % similarity rather than the commonly used 86 %–87 % similarity (Degelmann et al., 2010; Wen et al., 2016) is useful to detect spatial patterning in microbial assemblage, as suggested previously (van de Kamp et al., 2019). The representative sequences of all 16S rRNA OTUs were taxonomically classified using the Ribosomal Database Project (RDP) classifier (Wang et al., 2007). For pmoA reads, representative OTU sequences were classified using a naïve classifier implemented with the mothur “classify.seq” command, as described previously (Dumont et al., 2014). For the major 13C-labeled pmoA OTUs retrieved from heavy CsCl fractions (containing ≥2 % of pmoA gene sequences in at least one of the samples), a representative sequence was selected for phylogenetic analysis by comparison with known sequences from GenBank. A heatmap was generated from the relative abundances of the major 13C-labeled pmoA OTUs across different microcosms by HemI version 1.0 (Deng et al., 2014), and hierarchical clustering of samples was performed with the calculated Pearson distance.
2.9 Statistical analysis
Mantel tests were used to test for significant correlations between methanotrophic community distance and different soil physiochemical properties, namely, pH, TOC, TN, C : N ratio, SOM, inorganic N, soil copper content and OXC. The tests were performed in the R environment with the vegan package.
One-way analysis of variance with Tukey's post hoc test was used for comparisons among different treatments for each soil. Student's t test was conducted to assess the possibility of significant differences between two groups. Analyses were conducted using the SPSS version 13.0 package (SPSS, Inc.). P<0.05 was regarded as statistically significant.
3.1 Conversion of CH4 to soil organic matter
During SIP microcosm's incubation, a fraction of 13C-CH4 was converted to SOM by MOB through cell biomass synthesis, and it was assessed as the changes in the 13C atom percent of the soil TOC. The average background 13C-atom abundance in soils under natural atmospheric conditions was 1.08±0.01 %, and all soils showed statistically significant increases in the 13C-atom abundance of TOC up to 1.74±0.41 % after consumption of 13CH4 (Fig. 1a). There was no significant difference in conversion ratio of 13CH4 into soil organic matters between low-pH and high-pH soils, and fertilizing soils with inorganic N did not result in higher 13C incorporation into organic matter than that in unfertilized soils (Table S2). Based on the changes in methane concentrations, soil organic C contents and 13C-atom percent during incubation of SIP microcosms, it was theoretically estimated that 10.4 %–38.1 % of 13C-CH4 was converted to soil organic C during the microcosm incubations (Table S2), and 13CH4-derived C contributed 0.203 %–1.25 % of total soil organic C after incubation (Table S2).
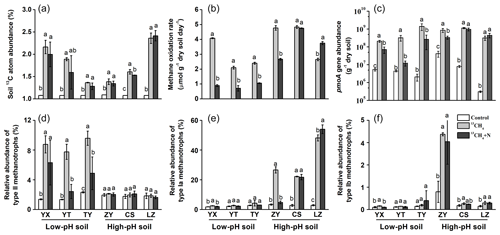
Figure 1Changes in soil 13C-atom abundances, methane oxidation rates, abundances and compositions of methanotrophs following soil microcosm incubation. (a) Soil 13C-atom abundance was measured to assess methane assimilation in soil microcosms amended with 5 % 13CH4. (b) The soil methane oxidation rate was compared between soil microcosms incubated with or without NH4NO3 fertilization. (c) The pmoA gene copy numbers of methanotrophs were estimated using real-time qPCR. Illumina sequencing targeting 16S rRNA genes was performed at the whole microbial community level, and the relative abundance of type II (d), type Ia (e) and type Ib (f) methanotrophs was expressed as the percentage of affiliated gene reads relative to the total 16S rRNA gene reads in each microcosm. “Control” indicates soil under natural atmospheric conditions; “13CH4” and “13CH4+N” refer to soil microcosms incubated with 5 % v∕v 13CH4 without and with extra NH4NO3 fertilization, respectively. The designations below the x axis represent the soil sampling sites of rice paddy fields. All treatments were conducted in triplicate. The error bars represent the standard errors of the means. Different letters above the columns indicate a significant difference between different treatments in a given soil (P<0.05).
3.2 Methane oxidation rates
Assuming linear kinetics, the methane oxidation rates were 0.71–4.08 µmol CH4 g−1 dry weight soil (d.w.s.) d−1 in soils of YX, YT and TY (with a low pH) and 2.65–4.83 µmol CH4 g−1 d.w.s. d−1 in soils of ZY, CS and LZ (with a high pH) (Fig. 1b). N fertilization led to significantly lower methane oxidation rates in low-pH soils, while varying effects were observed in high-pH soils (Fig. 1b). The lowered methane oxidation rate following fertilization might also suffer from decreased oxygen concentration at the later stage of the microcosm incubation, especially for the low-pH soil incubations that lasted more than 30 d. However, the temporal changes in the concentrations of headspace methane showed that inorganic N fertilization inhibited microbial methane oxidation in low-pH soils, leading to a prolonged period for consumption of the same amount of methane, particularly for YT soil (Fig. S2).
3.3 Change in abundance of methane-oxidizing bacteria
The absolute abundance of soil methanotrophs was estimated by qPCR using the biomarker pmoA gene at the microcosm incubation endpoints. The consumption of methane at high concentrations stimulated the growth of methanotrophs, represented by 20.4- to 1027-fold increases in the pmoA gene abundance following microcosm incubation of all six soils (Fig. 1c). Consistent with the changes in methane oxidation rates, N fertilization had a similar impact on the abundance of methanotrophic communities. Specifically, the pmoA gene abundances after N fertilization were significantly lower than those in the unfertilized low-pH soils (YX, YT and TY) (Fig. 1c).
Based on high-throughput sequencing of 16S rRNA genes, the percentages of type I and type II MOB in the total microbial communities were calculated to track the changes of methanotrophic communities during SIP incubations. The relative abundances of type II methanotrophs increased significantly in methane-amended microcosms with low-pH soils (YX, YT and TY) but did not change in the high-pH soils of ZY, CS and LZ (Fig. 1d). For type I methanotrophs, the reverse trend was observed. Specifically, type Ia methanotrophs showed an 8.0–16.9-fold increase in high-pH soils of ZY, CS and LZ, while no change or only a minor increase was observed in the low-pH soils (YX, YT and TY) following methane amendment (Fig. 1e). High enrichment of type Ib methanotrophs was also observed in the ZY soil (Fig. 1f).
3.4 Stable isotope probing of active methane-oxidizing bacteria
Following the isopycnic centrifugation of the total DNA extracted from 13CH4-SIP microcosms, real-time qPCR analysis of pmoA genes demonstrated active cell propagation and 13C assimilation in all six soils fueled by methane oxidation. A peak shift of relative abundances of pmoA genes towards heavy fractions was clearly observed in all soil microcosms amended with 13CH4 compared to the control treatments (Fig. 2). The pmoA genes in the 13CH4-amended microcosms accumulated in the heavy DNA fractions with a CsCl buoyant density of approximately 1.735–1.745 g mL−1, which was within the same range as in previous studies (Cai et al., 2016; Shiau et al., 2018), while the pmoA gene abundance in the control treatments peaked only in the light DNA fractions with a buoyant density of 1.717–1.726 g mL−1. Similar results were also obtained from SIP microcosms amended with inorganic N (Fig. 2). Notably, following N fertilization, the highest peak in the YT soil occurred in the light fraction, although the apparent labeling of pmoA gene-carrying methanotrophs was evidenced by increased abundances in the heavy fraction compared to control.
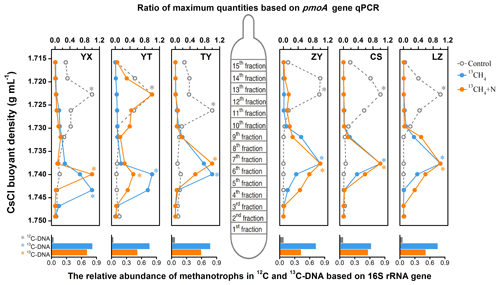
Figure 2The distribution of 13C-labeled methanotrophs based on qPCR of pmoA and sequencing of 16S rRNA genes. The 13C-labeled methanotrophs were determined by quantitative distribution of pmoA genes across the entire buoyant density gradient of the DNA fractions from soil microcosms incubated with 13CH4 compared to the controls. The normalized data are the ratios of the gene abundance in each DNA gradient to the maximum number for each treatment. The columns beneath display the relative abundance of methanotroph-affiliated reads in all 16S rRNA genes in the 12C-DNA from the control and 13C-DNA from 13CH4-amended soil microcosms, respectively. * The DNA fractions selected for Illumina sequencing.
High-throughput sequencing of the 16S rRNA genes in the heavy DNA fractions at the whole-community level further showed significant increase in relative abundance of methanotrophs in 13C-labeled microcosms. The methanotroph-affiliated 16S rRNA genes accounted for 61.9 % to 81.2 % of the total microbial communities in the 13C-DNA fractions, while in the control treatment, the methanotrophs constituted only 3.1 %–7.2 % of the total microbial communities (bottom columns of Fig. 2).
3.5 Linking soil physiochemical properties with active methanotrophs
Phylogenetic analysis of 13C-labeled pmoA genes from heavy DNA fractions demonstrated that Methylocystis related type II organisms dominated the 13C-labeled methanotrophs in the YX, YT and TY soils with low pH values (Fig. 3), which was confirmed by taxonomic classification of 13C-labeled 16S rRNA gene classification (Figs. S3 and S4). In the high-pH soils (ZY, CS and LZ), 13C-labeled methanotrophs were predominated by type Ia organisms. The 13C-pmoA genes were related to the type Ia methanotroph Methylobacter sp. (Fig. 3), but the 13C-labeled 16S rRNA genes suggested the methanotrophs might be closer to Methylosarcina sp. (Fig. S4). Notably, in the ZY soil, 17 %–30 % of the 13C-labeled sequences were phylogenetically related to type Ib methanotroph Methylocaldum sp. (Fig. 3). The community compositions of the 13C-labeled methanotrophs were deeply branched between high- and low-pH soils (Fig. 3).
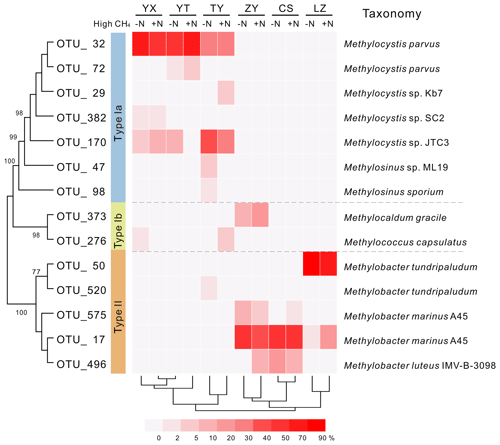
Figure 3Heat map of relative abundances of major 13C-labeled methanotrophic OTUs based on pmoA gene sequencing. “−N” and “+N” indicate 13CH4-amended microcosms without and with N fertilization, respectively. Hierarchical clustering of samples was performed, and phylogenetic relations between different OTUs are shown by the topology, with bootstrap values >60 % indicated at branch nodes. Each OTU representative sequence is taxonomically annotated to a defined strain from GenBank with the closest phylogenetic relation.
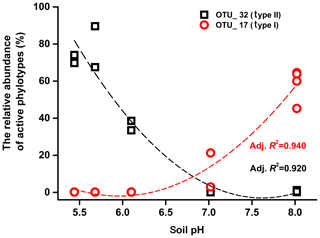
Mantel tests showed that only the pH, out of all the eight soil characteristics tested, was significantly correlated with variation in the active methanotrophic communities (according to 13C-labeled pmoA gene sequencing) between different soils (P<0.05) (Table S3). Regression analysis further revealed a significantly positive relationship between soil pH and the relative abundances of the primary 13C-labeled type I methanotrophic cluster (OTU_17), while pH was negatively related to the relative abundances of the dominant type II cluster (OTU_32) under high methane concentrations (Fig. 4).
Our results provide strong evidence for the important roles of pH-based selection of type I and type II methanotrophs in methane oxidation and assimilation in paddy soils. In soils amended with 13CH4 gas, the incorporation of 13C into genomic DNA occurred in methanotrophic communities that directly utilized methane-derived C for growth. Therefore, the relative gene abundances of type I versus type II methanotrophs in 13C-labeled DNA fractions can reflect their relative contributions to actual methane uptake and oxidation. In this study, the ratios of 13C-labeled type II to type I methanotrophs in ZY, CS and LZ soils with high pH values were very low, ranging from 0.002 to 0.014 (Table S4), suggesting that the type I methanotrophs in these three soils were more active than their type II counterparts. However, the ratios of 13C-labeled type II to type I methanotrophs were 20.0 to 101 in the low-pH soils of YX, YT and TY (Table S4). Assuming that one cell contained two copies of pmoA genes (Kolb et al., 2003), type II methanotrophs could reach a cell-specific methane oxidation rate of 0.2–9.6 fmol CH4 h−1 cell−1 in these three soils (calculated in Table S4), being consistent with previous report of 0.2–15 fmol CH4 h−1 cell−1 obtained from both pure cultures and complex soils (Hanson and Hanson, 1996). These results thus suggest that aerobic methanotrophy was mostly sustained by the growth of type II methanotrophs in these low-pH paddy soils.
Although we cannot rule out the possibility that there were some unlabeled pmoA genes “drifting” to the heavy fractions during the process of DNA fractionation, since the absence of 12CH4 amended microcosms for background calibration prevents precise calculation of relative abundance of 13C-labeled microbes, we can confidently conclude that the dominant phylotypes revealed in the heavy fractions in this study represented the properly labeled and most active methanotrophs. Dominant type II methanotrophs identified in low-pH soils were affiliated to Methylocystis based on 13C-labeled pmoA genes (Fig. 3), which was also congruent with phylogenetic analysis of 16S rRNA genes (Fig. S4). The analysis of pmoA genes indicated that 13C-methanotrophs in high-pH soils could be most closely related to Methylobacter of type Ia methanotrophs. However, phylogenetic analysis of the 13C-labeled 16S rRNA genes suggested that in high-pH soils the dominant methanotrophs could cluster closely with Methylosarcina species (Fig. S4). We assume this discrepancy might be attributed to the lack of whole genome information in the present study, and the presence of phylogenetic incongruence between pmoA and 16S rRNA genes, which could be better resolved with the increasing genome availability of methanotrophs in the future. Nevertheless, our results (both from pmoA and 16S rRNA genes) indicated that type Ia methanotrophs dominated methane oxidation activities in the high-pH paddy soils.
It is technically challenging to track the in situ activities of microbes, especially in ecosystems that are exposed to constantly fluctuating environments such as rice paddy fields. Agricultural management might influence the population dynamics of methanotrophs through irrigation, fertilization and plantation. In view of this variability, results based on microcosms cannot entirely represent the in situ conditions. However, the incubation of SIP microcosms was conducted under the same conditions, but the labeling of distinct methanotrophs indeed occurred, which might be the result of long-term ecological and evolutionary adaption of methane oxidizers in paddy soils with contrasting physiochemical variables. Therefore, our results might reflect what is largely occurring under in situ inundated conditions, in which high methane emissions occur, particularly in regard to the relative activities of type I versus type II methanotrophs that were likely controlled by intrinsic biotic and abiotic factors in the paddy soils.
The active methanotrophic compositions in the six paddy soils were strongly associated with only soil pH, based on tests of potential correlations with eight key soil physiochemical properties. Specifically, rice paddy soils with higher type II methanotrophic activities were all acido-neutral (YX, YT and TY, with a pH of 5.44–6.10), while the more alkaline soils (ZY, CS and LZ, with a pH of 7.02–8.02) displayed stronger type I methanotrophic activity. Previous culture-dependent and ecophysiological studies, where type II methanotrophs were described as stress tolerators (Ho et al., 2013), provided strong support for low pH as the potential driving force for selection of type II over type I methanotrophs in paddy soils. Cultivated acidophilic or acid-tolerant methanotrophs are by far mostly type II strains (Belova et al., 2013; Dedysh et al., 2000, 2007), which are phylogenetically close to the dominant 13C-methanotrophs in our low-pH soils (Fig. 3). Our results are similar to some observations from natural wetland systems, in which the activity of type II methanotrophs appeared to be more prevalent under low-pH than high-pH conditions (Chen et al., 2008a, b; Dedysh, 2009; Gupta et al., 2012; Kip et al., 2012). It thus seems plausible that oxidation of methane at high concentrations by type II methanotrophs more frequently occurs in acidic than alkaline rice paddy soils. However, the consistency of our conclusion of the pH-based ecological coherence of methanotrophs in other ecosystems should be further investigated, since previous studies also revealed codominance of type I and II methanotrophs in low-pH natural peatlands (Deng et al., 2016; Esson et al., 2016). In addition, the possibility cannot be ruled out that other untested abiotic or biotic variables might have stronger forces in shaping the community structure of methanotrophs in rice paddy soils.
Our results demonstrated that chemical N fertilization did not alter the dominant community compositions of active methanotrophs in paddy soils (Fig. 3), implying that the growth of methanotrophs in these soils was not N-limited, or that N availability was not the key factor for selection of distinct methanotrophs in rice paddy soils. In this context, we speculate that N input could have stimulated plant growth and increased the levels of exudates and that litter decomposition served as precursors for methanogenesis to enhance methane production. Despite no consistent pattern being observed with respect to the effects of N fertilization on methane oxidation rates in this study, it appears that N fertilization had an inhibitory effect on the methane oxidation rates in the low-pH soils, which were dominated by type II methanotrophic activity (Figs. 1b and S2). Consistently, the methanotrophic abundances following N fertilization were significantly lower than those in unfertilized low-pH soils (Fig. 1c). These results agreed well with previous findings that high N input suppressed the activity and growth rates of type II methanotrophs from pure cultures (Graham et al., 1993) and in complex soil samples (Bodelier et al., 2000; Mohanty et al., 2006; Noll et al., 2008). Meanwhile it is noteworthy that contradictory results were obtained in high-pH paddy soils. N fertilization stimulated the methane oxidation rate in only one of the high-pH soils (LZ), which had a unique dominant type Ia cluster (OTU_50, as shown in Fig. 3) when compared to the other two high-pH soils (dominated by OTU_17). Our study therefore implied that the contradicting effects of N fertilization on methane oxidation (inhibition or stimulation) frequently reported in different soils (Alam and Jia, 2012; Bodelier et al., 2000; Cai and Yan, 1999; Zheng et al., 2014) might be determined by the dominant methanotrophic phylotypes, but a larger-scale sampling with activity-based molecular analysis (e.g., RNA- or SIP-based tools) is required to test this hypothesis in the future.
Despite different compositions of active methanotrophs, there was no fundamental difference of methane oxidation rate between low-pH and high-pH paddy soils, indicating similar methane oxidation rate of type II and I methanotrophs in rice paddy fields. The culture-dependent studies have showed similar kinetic traits between these two groups of methanotrophs, including substrate affinity and specific cell activity (Bedard and Knowles, 1989; Calhoun and King, 1998; Hanson and Hanson, 1996), which might explain the similar methane oxidation rates in different soils in the present study. The significant 13C enrichment of soil organic C indicated that 13CH4-derived microbial biomass contributed significantly to the turnover of soil C. Meta-analysis indicated microbial biomass represents 0.6 %–1.1 % of total soil organic C (Fierer et al., 2009), but the contribution of methane-driven microbial food web to soil fertility and quality remained largely unknown (Murase and Frenzel, 2007). Our results showed no statistically significant difference in net soil 13C input between low-pH and high-pH soils during methane oxidation, although type I and type II methanotrophs employed different strategies for C metabolisms (Trotsenko and Murrell, 2008). For instance, 5 %–15 % of cell biomass C in type I methanotrophs could be derived from CO2 (Trotsenko and Murrell, 2008), and a recent study suggests the proportion up to be 60 % in type II methanotrophs (Yang et al., 2013). It is noteworthy that the fresh input of 13CH4-derived biomass accounted for up to 1.25 % of total organic C in LZ soil (Table S2), implying that the amount of 13CH4-C incorporated into trophic networks comprised a substantial fraction of soil microbial biomass. Quantitative assessment of soil microbial biomass pools and the relative contribution of phylogenetically distinct methanotrophs to active C pool in soil would be essential for deciphering the underlying metabolism of methane oxidizers and their ecological and agricultural importance in paddy fields.
This study provides evidence for niche differentiation of type I and II methanotrophs strongly associated with pH variation in rice paddy soils. Low pH could have likely selected for type II methanotrophs in paddy soils, while type I was favored in high-pH soils. The incorporation of CH4-derived C into biomass contributed up to 1.25 % of total organic C in paddy soil. The fresh input of new C from aerobic methanotrophy could have played a vital role in the turnover of soil microbial biomass and subsequent cycling of soil nutrients in support of agricultural sustainability. N fertilization changed methane oxidation rates in five of the soils tested, but the composition of active methanotrophs was not significantly affected. These results provide a mechanistic basis for better understandings of community assembly mechanisms of ecologically important microbial guilds and their possible roles in agricultural sustainability.
The raw Illumina sequencing data have been deposited in the European Nucleotide Archive (ENA) under accession nos. PRJEB37235 (https://www.ebi.ac.uk/ena/data/view/PRJEB37235, Zhao et al., 2017a) for 16S rRNA genes and PRJEB40045 (https://www.ebi.ac.uk/ena/data/view/PRJEB40045, Zhao et al., 2017b) for pmoA gene sequences. The sequences of dominant 13C-labeled pmoA OTUs were deposited to GenBank with accession numbers MK613983-MK613993 (https://www.ncbi.nlm.nih.gov/genbank/, Zhao, 2019a) and MK621911-MK621913 (https://www.ncbi.nlm.nih.gov/genbank/, Zhao, 2019b).
The supplement related to this article is available online at: https://doi.org/10.5194/bg-17-1451-2020-supplement.
JZ and ZJ designed the experiments and JZ carried them out. JZ analyzed all the data, and YC assisted in the bioinformatic analyses. JZ and ZJ wrote the manuscript.
The authors declare that they have no conflict of interest.
We are grateful to Jing Ma, Yiming Wang and Lijun Bao at the Institute of Soil Science, CAS, for soil collection. We thank Baozhan Wang at the Institute of Soil Science for advice on the soil chemical characteristics and Zhiying Guo for technical support in the statistical analysis. We also thank the staff of the Analysis Center at the Institute of Soil Science for technical support, including Rong Huang and Zuohao Ma for Illumina MiSeq sequencing, Deling Sun for the 13C-atom abundance assay, Yufang Sun for the soil C and N content assay, Ruhai Wang for the ammonia and nitrate content assays, Guoxing Lu for the SOM assay, Hua Gong for the soil metal element measurements, and Li Gao for the content assay.
This research has been supported by the National Science Foundation of China (grant nos. 91751204, 41701302 and 41530857).
This paper was edited by Luo Yu and reviewed by Yongcui Deng and one anonymous referee.
Aigle, A., Prosser, J. I., and Gubry-Rangin, C.: The application of high-throughput sequencing technology to analysis of amoA phylogeny and environmental niche specialisation of terrestrial bacterial ammonia-oxidisers, Environ. Microbiomes, 14, 3, https://doi.org/10.1186/s40793-019-0342-6, 2019.
Alam, M. S. and Jia, Z.: Inhibition of methane oxidation by nitrogenous fertilizers in a paddy soil, Front. Microbiol., 3, 246, https://doi.org/10.3389/fmicb.2012.00246, 2012.
Bedard, C. and Knowles, R.: Physiology, biochemistry, and specific inhibitors of CH4, , and CO oxidation by methanotrophs and nitrifiers, Microbiol. Rev., 53, 68–84, 1989.
Belova, S. E., Kulichevskaya, I. S., Bodelier, P. L. E., and Dedysh, S. N.: Methylocystis bryophila sp. nov., a facultatively methanotrophic bacterium from acidic Sphagnum peat, and emended description of the genus Methylocystis (ex Whittenbury et al. 1970) Bowman et al. 1993, Int. J. Syst. Evol. Micr., 63, 1096–1104, https://doi.org/10.1099/ijs.0.043505-0, 2013.
Bodelier, P. L. E., Roslev, P., Henckel, T., and Frenzel, P.: Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots, Nature, 403, 421–424, https://doi.org/10.1038/35000193, 2000.
Bridgham, S. D., Cadillo-Quiroz, H., Keller, J. K., and Zhuang, Q.: Methane emissions from wetlands: Biogeochemical, microbial, and modeling perspectives from local to global scales, Glob. Change Biol., 19, 1325–1346, https://doi.org/10.1111/gcb.12131, 2013.
Cai, Y., Zheng, Y., Bodelier, P. L. E., Conrad, R., and Jia, Z.: Conventional methanotrophs are responsible for atmospheric methane oxidation in paddy soils, Nat. Commun., 7, 11728, https://doi.org/10.1038/ncomms11728, 2016.
Cai, Z. and Yan, X.: Kinetic model for methane oxidation by paddy soil as affected by temperature, moisture and N addition, Soil Biol. Biochem., 31, 715–725, https://doi.org/10.1016/S0038-0717(98)00170-9, 1999.
Calhoun, A. and King, G. M.: Characterization of root-associated methanotrophs from three freshwater macrophytes: Pontederia cordata, Sparganium eurycarpum, and Sagittaria latifolia, Appl. Environ. Microb., 64, 1099–1105, 1998.
Caporaso, J., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F., Costello, E., Fierer, N., Pena, A., Goodrich, J., Gordon, J., Huttley, G., Kelley, S., Knights, D., Koenig, J., Ley, R., Lozupone, C., McDonald, D., Muegge, B., Pirrung, M., Reeder, J., Sevinsky, J., Tumbaugh, P., Walters, W., Widmann, J., Yatsunenko, T., Zaneveld, J., and Knight, R.: QIIME allows analysis of high-throughput community sequencing data, Nat. Methods, 7, 335–336, https://doi.org/10.1038/nmeth.f.303, 2010.
Chen, Y., Dumont, M. G., McNamara, N. P., Chamberlain, P. M., Bodrossy, L., Stralis-Pavese, N., and Murrell, J. C.: Diversity of the active methanotrophic community in acidic peatlands as assessed by mRNA and SIP-PLFA analyses, Environ. Microbiol., 10, 446–459, https://doi.org/10.1111/j.1462-2920.2007.01466.x, 2008a.
Chen, Y., Dumont, M. G., Neufeld, J. D., Bodrossy, L., Stralis-Pavese, N., McNamara, N. P., Ostle, N., Briones, M. J. I., and Murrell, J. C.: Revealing the uncultivated majority: Combining DNA stable-isotope probing, multiple displacement amplification and metagenomic analyses of uncultivated Methylocystis in acidic peatlands, Environ. Microbiol., 10, 2609–2622, https://doi.org/10.1111/j.1462-2920.2008.01683.x, 2008b.
Conrad, R. and Rothfuss, F.: Methane oxidation in the soil surface layer of a flooded rice field and the effect of ammonium, Biol. Fert. Soils, 12, 28–32, https://doi.org/10.1007/BF00369384, 1991.
Costello, A. M. and Lidstrom, M. E.: Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments, Appl. Environ. Microb., 65, 5066–5074, 1999.
Daebeler, A., Bodelier, P. L. E., Yan, Z., Hefting, M. M., Jia, Z., and Laanbroek, H. J.: Interactions between Thaumarchaea, Nitrospira and methanotrophs modulate autotrophic nitrification in volcanic grassland soil, ISME J., 8, 2397–2410, https://doi.org/10.1038/ismej.2014.81, 2014.
Dedysh, S. N.: Exploring methanotroph diversity in acidic northern wetlands: Molecular and cultivation-based studies, Microbiology, 78, 655–669, https://doi.org/10.1134/S0026261709060010, 2009.
Dedysh, S. N., Liesack, W., Khmelenina, V. N., Suzina, N. E., Trotsenko, Y. A., Semrau, J. D., Bares, A. M., Panikov, N. S., and Tiedje, J. M.: Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine- pathway methanotrophs, Int. J. Syst. Evol. Micr., 50, 955–969, https://doi.org/10.1099/00207713-50-3-955, 2000.
Dedysh, S. N., Belova, S. E., Bodelier, P. L. E., Smirnova, K. V., Khmelenina, V. N., Chidthaisong, A., Trotsenko, Y. A., Liesack, W., and Dunfiel, P. F.: Methylocystis heyeri sp. nov., a novel type II methanotrophic bacterium possessing “signature” fatty acids of type I methanotrophs, Int. J. Syst. Evol. Micr., 57, 472–479, https://doi.org/10.1099/ijs.0.64623-0, 2007.
Degelmann, D. M., Borken, W., Drake, H. L., and Kolb, S.: Different atmospheric methane-oxidizing communities in european beech and norway spruce soils, Appl. Environ. Microb., 76, 3228–3235, https://doi.org/10.1128/aem.02730-09, 2010.
Deng, W., Wang, Y., Liu, Z., Cheng, H., and Xue, Y.: HemI: A toolkit for illustrating heatmaps, PLoS One, 9, e111988, https://doi.org/10.1371/journal.pone.0111988, 2014.
Deng, Y., Cui, X., and Dumont, M. G.: Identification of active aerobic methanotrophs in plateau wetlands using DNA stable isotope probing, FEMS Microbiol. Lett., 363, 1–8, https://doi.org/10.1093/femsle/fnw168, 2016.
Dumont, M. G., Lüke, C., Deng, Y., and Frenzel, P.: Classification of pmoA amplicon pyrosequences using BLAST and the lowest common ancestor method in MEGAN, Front. Microbiol., 5, 34, https://doi.org/10.3389/fmicb.2014.00034, 2014.
Edgar, R.: UPARSE: highly accurate OTU sequences from microbial amplicon reads, Nat. Methods, 10, 996–998, https://doi.org/10.1038/nmeth.2604, 2013.
Eller, G. and Frenzel, P.: Changes in activity and community structure of methane-oxidizing bacteria over the growth period of rice, Appl. Environ. Microb., 67, 2395–2403, https://doi.org/10.1128/aem.67.6.2395-2403.2001, 2001.
Esson, K. C., Lin, X., Kumaresan, D., Chanton, J. P., Murrell, J. C., and Kostka, E.: Alpha- and Gammaproteobacterial methanotrophs codominate the active methane-oxidizing communities in an acidic boreal peat bog, Appl. Environ. Microb., 82, 2363–2371, https://doi.org/10.1128/aem.03640-15, 2016.
Fierer, N., Strickland, M. S., Liptzin, D., Bradford, M. A., and Cleveland, C. C.: Global patterns in belowground communities, Ecol. Lett., 12, 1238–1249, https://doi.org/10.1111/j.1461-0248.2009.01360.x, 2009.
Frenzel, P., Rothfuss, F., and Conrad, R.: Oxygen profiles and methane turnover in a flooded rice microcosm, Biol. Fert. Soils, 14, 84–89, 1992.
Graham, D. W., Chaudhary, J. A., Hanson, R. S., and Arnold, R. G.: Factors affecting competition between type I and type II methanotrophs in two-organism, continuous-flow reactors, Microb. Ecol., 25, 1–17, https://doi.org/10.1007/BF00182126, 1993.
Gubry-Rangin, C., Hai, B., Quince, C., Engel, M., Thomson, B. C., James, P., Schloter, M., Griffiths, R. I., Prosser, J. I., and Nicol, G. W.: Niche specialization of terrestrial archaeal ammonia oxidizers, P. Natl. Acad. Sci. USA, 108, 21206–21211, https://doi.org/10.1073/pnas.1109000108, 2011.
Gupta, V., Smemo, K. A., Yavitt, J. B., and Basiliko, N.: Active methanotrophs in two contrasting North American peatland ecosystems revealed using DNA-SIP, Microb. Ecol., 63, 438–445, https://doi.org/10.1007/s00248-011-9902-z, 2012.
Hanson, R. S. and Hanson, T. E.: Methanotrophic bacteria, Microbiol. Rev., 60, 439–471, https://doi.org/10.1002/0471263397.env316, 1996.
He, R., Wooller, M. J., Pohlman, J. W., Quensen, J., Tiedje, J. M., and Leigh, M. B.: Diversity of active aerobic methanotrophs along depth profiles of arctic and subarctic lake water column and sediments, ISME J., 6, 1937–1948, https://doi.org/10.1038/ismej.2012.34, 2012.
Ho, A., Kerckhof, F. M., Luke, C., Reim, A., Krause, S., Boon, N., and Bodelier, P. L. E.: Conceptualizing functional traits and ecological characteristics of methane-oxidizing bacteria as life strategies, Env. Microbiol. Rep., 5, 335–345, https://doi.org/10.1111/j.1758-2229.2012.00370.x, 2013.
Holmes, A. J., Costello, A., Lidstrom, M. E., and Murrell, J. C.: Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related, FEMS Microbiol. Lett., 132, 203–208, https://doi.org/10.1016/0378-1097(95)00311-R, 1995.
Holmes, A. J., Roslev, P., McDonald, I. R., Iversen, N., Henriksen, K., and Murrell, J. C.: Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake, Appl. Environ. Microb., 65, 3312–3318, 1999.
Kip, N., Fritz, C., Langelaan, E. S., Pan, Y., Bodrossy, L., Pancotto, V., Jetten, M. S. M., Smolders, A. J. P., and Op den Camp, H. J. M.: Methanotrophic activity and diversity in different Sphagnum magellanicum dominated habitats in the southernmost peat bogs of Patagonia, Biogeosciences, 9, 47–55, https://doi.org/10.5194/bg-9-47-2012, 2012.
Knief, C., Lipski, A., and Dunfield, P. F.: Diversity and activity of methanotrophic bacteria in different upland soils, Appl. Environ. Microb., 69, 6703–6714, https://doi.org/10.1128/aem.69.11.6703-6714.2003, 2003.
Kögel-Knabner, I., Amelung, W., Cao, Z., Fiedler, S., Frenzel, P., Jahn, R., Kalbitz, K., Kölbl, A., and Schloter, M.: Biogeochemistry of paddy soils, Geoderma, 157, 1–14, https://doi.org/10.1016/j.geoderma.2010.03.009, 2010.
Kolb, S., Knief, C., Stubner, S., and Conrad, R.: Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays, Appl. Environ. Microb., 69, 2423–2429, https://doi.org/10.1128/aem.69.5.2423-2429.2003, 2003.
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N.: Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale, Appl. Environ. Microb., 75, 5111–5120, https://doi.org/10.1128/aem.00335-09, 2009.
Leng, L., Chang, J., Geng, K., Lu, Y., and Ma, K.: Uncultivated Methylocystis species in paddy soil include facultative methanotrophs that utilize acetate, Microbiol. Ecol., 70, 88–96, https://doi.org/10.1007/s00248-014-0540-0, 2015.
Liebner, S. and Wagner, D.: Abundance, distribution and potential activity of methane oxidizing bacteria in permafrost soils from the Lena Delta, Siberia, Environ. Microbiol., 9, 107–117, https://doi.org/10.1111/j.1462-2920.2006.01120.x, 2007.
Lin, J. L., Radajewski, S., Eshinimaev, B. T., Trotsenko, Y. A., McDonald, I. R., and Murrell, J. C.: Molecular diversity of methanotrophs in Transbaikal soda lake sediments and identification of potentially active populations by stable isotope probing, Environ. Microbiol., 6, 1049–1060, https://doi.org/10.1111/j.1462-2920.2004.00635.x, 2004.
Liu, B., Mørkved, P. T., Frostegård, Å., and Bakken, L. R.: Denitrification gene pools, transcription and kinetics of NO, N2O and N2 production as affected by soil pH, FEMS Microbiol. Ecol., 72, 407–417, https://doi.org/10.1111/j.1574-6941.2010.00856.x, 2010.
Ma, K., Conrad, R., and Lu, Y.: Dry/wet cycles change the activity and population dynamics of methanotrophs in rice field soil, Appl. Environ. Microb., 79, 4932–4939, https://doi.org/10.1128/aem.00850-13, 2013.
Magoč, T. and Salzberg, S. L.: FLASH: Fast length adjustment of short reads to improve genome assemblies, Bioinformatics, 27, 2957–2963, https://doi.org/10.1093/bioinformatics/btr507, 2011.
Mohanty, S. R., Bodelier, P. L., Floris, V., and Conrad, R.: Differential effects of nitrogenous fertilizers on methane-consuming microbes in rice field and forest soils, Appl. Environ. Microb., 72, 1346–1354, https://doi.org/10.1128/aem.72.2.1346-1354.2006, 2006.
Morris, S. A., Radajewski, S., Willison, T. W., and Murrell, J. C.: Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing, Appl. Environ. Microb., 68, 1446–1453, https://doi.org/10.1128/aem.68.3.1446-1453.2002, 2002.
Murase, J. and Frenzel, P.: A methane-driven microbial food web in a wetland rice soil, Environ. Microbiol., 9, 3025–3034, https://doi.org/10.1111/j.1462-2920.2007.01414.x, 2007.
Noll, M., Frenzel, P., and Conrad, R.: Selective stimulation of type I methanotrophs in a rice paddy soil by urea fertilization revealed by RNA-based stable isotope probing, FEMS Microbiol. Ecol., 65, 125–132, https://doi.org/10.1111/j.1574-6941.2008.00497.x, 2008.
Nouchi, I., Mariko, S., and Aoki, K.: Mechanism of methane transport from the rhizosphere to the atmosphere through rice plants, Plant Physiol., 94, 59–66, https://doi.org/10.1104/pp.94.1.59, 1990.
Nouchi, I., Hosono, T., Aoki, K., and Minami, K.: Seasonal variation in methane flux from rice paddies associated with methane concentration in soil water, rice biomass and temperature, and its modelling, Plant Soil, 161, 195–208, https://doi.org/10.1007/BF00046390, 1994.
Qiu, Q., Noll, M., Abraham, W., Lu, Y., and Conrad, R.: Applying stable isotope probing of phospholipid fatty acids and rRNA in a Chinese rice field to study activity and composition of the methanotrophic bacterial communities in situ, ISME J., 2, 602–614, https://doi.org/10.1038/ismej.2008.34, 2008.
Reim, A., Lu, C., Krause, S., Pratscher, J., and Frenzel, P.: One millimetre makes the difference: high-resolution analysis of methane-oxidizing bacteria and their specific activity at the oxic-anoxic interface in a flooded paddy soil, ISME J., 6, 2128–2139, https://doi.org/10.1038/ismej.2012.57, 2012.
Roslev, P. and Iversen, N.: Radioactive fingerprinting of microorganisms that oxidize atmospheric methane in different soils, Appl. Environ. Microb., 65, 4064–4070, 1999.
Semrau, J. D., DiSpirito, A. A., and Vuilleumier, S.: Facultative methanotrophy: false leads, true results, and suggestions for future research, FEMS Microbiol. Lett., 323, 1–12, https://doi.org/10.1111/j.1574-6968.2011.02315.x, 2011.
Shiau, Y. J., Cai, Y., Jia, Z., Chen, C. L., and Chiu, C. Y.: Phylogenetically distinct methanotrophs modulate methane oxidation in rice paddies across Taiwan, Soil Biol. Biochem., 124, 59–69, https://doi.org/10.1016/j.soilbio.2018.05.025, 2018.
Shrestha, M., Abraham, W. R., Shrestha, P. M., Noll, M., and Conrad, R.: Activity and composition of methanotrophic bacterial communities in planted rice soil studied by flux measurements, analyses of pmoA gene and stable isotope probing of phospholipid fatty acids, Environ. Microbiol., 10, 400–412, https://doi.org/10.1111/j.1462-2920.2007.01462.x, 2008.
Stubner, S.: Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen™ detection, J. Microbiol. Meth., 50, 155–164, https://doi.org/10.1016/S0167-7012(02)00024-6, 2002.
Tripathi, B. M., Stegen, J. C., Kim, M., Dong, K., Adams, J. M., and Lee, Y. K.: Soil pH mediates the balance between stochastic and deterministic assembly of bacteria, ISME J., 12, 1072–1083, https://doi.org/10.1038/s41396-018-0082-4, 2018.
Trotsenko, Y. A. and Murrell, J. C.: Metabolic aspects of aerobic obligate methanotrophy, Adv. App. Microbiol., 63, 183–229, https://doi.org/10.1016/S0065-2164(07)00005-6, 2008.
Tveit, A. T., Hestnes, A. G., Robinson, S. L., Schintlmeister, A., Dedysh, S. N., Jehmlich, N., von Bergen, M., Herbold, C., Wagner, M., Richter, A., and Svenning, M. M.: Widespread soil bacterium that oxidizes atmospheric methane, P. Natl. Acad. Sci. USA, 116, 8515–8524, https://doi.org/10.1073/pnas.1817812116, 2019.
van de Kamp, J., Hook, S. E., Williams, A., Tanner, J. E., and Bodrossy, L.: Baseline characterization of aerobic hydrocarbon degrading microbial communities in deep-sea sediments of the Great Australian Bight, Australia, Environ. Microbiol., 21, 1782–1797, https://doi.org/10.1111/1462-2920.14559, 2019.
Wang, B., Zhao, J., Guo, Z., Ma, J., Xu, H., and Jia, Z.: Differential contributions of ammonia oxidizers and nitrite oxidizers to nitrification in four paddy soils, ISME J., 9, 1062–1075, https://doi.org/10.1038/ismej.2014.194, 2015.
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R.: Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy, Appl. Environ. Microbiol., 73, 5261–5267, https://doi.org/10.1128/AEM.00062-07, 2007.
Wang, Q., Quensen, J. F. I., Fish, J. A., Lee, T. K., Sun, Y., Tiedje, J. M., and Cole, J. R.: Ecological patterns of nifH genes in four terrestrial climatic zones, MBio, 4, e00592-13, https://doi.org/10.1128/mBio.00592-13, 2013.
Wen, X., Yang, S., and Liebner, S.: Evaluation and update of cutoff values for methanotrophic pmoA gene sequences, Arch. Microbiol., 198, 629–636, https://doi.org/10.1007/s00203-016-1222-8, 2016.
Yang, S., Matsen, J. B., Konopka, M., Green-Saxena, A., Clubb, J., Sadilek, M., Orphan, V. J., Beck, D., and Kalyuzhnaya, M. G.: Global molecular analyses of methane metabolism in methanotrophic Alphaproteobacterium, Methylosinus trichosporium OB3b. Part II. metabolomics and 13C-labeling study, Front. Microbiol., 4, 70, https://doi.org/10.3389/fmicb.2013.00070, 2013.
Zhang, J., Cai, Z., Cheng, Y., and Zhu, T.: Denitrification and total nitrogen gas production from forest soils of Eastern China, Soil Biol. Biochem., 41, 2551–2557, https://doi.org/10.1016/j.soilbio.2009.09.016, 2009.
Zhao, J.: Methanotrophic bacterium clone, available at: https://www.ncbi.nlm.nih.gov/nuccore/MK613983-MK613993, last access: 9 March 2019a.
Zhao, J.: Uncultured methanotrophic bacterium clone, available at: https://www.ncbi.nlm.nih.gov/nuccore/MK621911-MK621913, last access: 12 March 2019b.
Zhao, J., Wang, B., and Jia, Z.: Phylogenetically distinct phylotypes modulate nitrification in a paddy soil, Appl. Environ. Microb., 81, 3218–3227, https://doi.org/10.1128/aem.00426-15, 2015.
Zhao, J., Cai, Y. F., and Jia, Z. J.: Type II and I methanotrophs in paddy soils, ENA-LAST-UPDATE, 2017-08-23, ENA, available at: https://www.ebi.ac.uk/ena/data/view/PRJEB37235, last access: 23 August 2017a.
Zhao, J., Cai, Y. F., and Jia, Z. J.: Methanotrophs in wetland paddies, ENA-LAST-UPDATE, 2017-08-23, ENA, available at: https://www.ebi.ac.uk/ena/data/view/PRJEB40045, last access: 23 August 2017b.
Zheng, Y., Huang, R., Wang, B. Z., Bodelier, P. L. E., and Jia, Z. J.: Competitive interactions between methane- and ammonia-oxidizing bacteria modulate carbon and nitrogen cycling in paddy soil, Biogeosciences, 11, 3353–3368, https://doi.org/10.5194/bg-11-3353-2014, 2014.