the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Dispersal of bacteria and stimulation of permafrost decomposition by Collembola
Janine Mariën
Eveline J. Krab
Contrary to most soils, permafrost soils have the atypical feature of being almost entirely deprived of soil fauna. Abiotic constraints on the fate of permafrost carbon after thawing are increasingly understood, but biotic constraints remain scarcely investigated. Incubation studies, essential to estimate effects of permafrost thaw on carbon cycling, typically measure the consequences of permafrost thaw in isolation from the topsoil and thus do not account for the effects of altered biotic interactions because of e.g. colonization by soil fauna. Microarthropods facilitate the dispersal of microorganisms in soil, both on their cuticle (ectozoochory) and through their digestive tract (endozoochory), which may be particularly important in permafrost soils, considering that microbial community composition can strongly constrain permafrost biogeochemical processes.
Here we tested how a model species of microarthropod (the Collembola Folsomia candida) affected aerobic CO2 production of permafrost soil over a 25 d incubation. By using Collembola stock cultures grown on permafrost soil or on an arctic topsoil, we aimed to assess the potential for endo- and ectozoochory of soil bacteria, while cultures grown on gypsum and sprayed with soil suspensions would allow the observation of only ectozoochory.
The presence of Collembola introduced bacterial amplicon sequence variants (ASVs) absent in the no-Collembola control, regardless of their microbiome manipulation, when considering presence–absence metrics (unweighted UniFrac metrics), which resulted in increased species richness. However, these introduced ASVs did not induce changes in bacterial community composition as a whole (accounting for relative abundances, weighted UniFrac), which might only become detectable in the longer term.
CO2 production was increased by 25.85 % in the presence of Collembola, about half of which could be attributed to Collembola respiration based on respiration rates measured in the absence of soil. We argue that the rest of the CO2 being respired can be considered a priming effect of the presence of Collembola, i.e. a stimulation of permafrost CO2 production in the presence of active microarthropod decomposers. Overall, our findings underline the importance of biotic interactions in permafrost biogeochemical processes and the need to explore the additive or interactive effects of other soil food web groups of which permafrost soils are deprived.
- Article
(2400 KB) - Full-text XML
- BibTeX
- EndNote
Carbon fluxes from soils are largely governed by the rate of decomposition of organic matter. Soil fauna is a crucial component in organic matter decomposition (García-Palacios et al., 2013; Griffiths et al., 2021), both directly through mechanical degradation of litter and corpses into smaller pieces and indirectly through feeding habits controlling the abundances of other decomposer groups such as fungi, microbial eukaryotes or bacteria (Hanlon and Anderson, 1979; Kaneda and Kaneko, 2008; Frouz et al., 2020; Potapov et al., 2020). In arctic soils, the scarcity of macrofaunal decomposers (e.g. earthworms – Blume-Werry et al., 2020) coupled with the high abundance of microbivorous microarthropods such as Collembola (Potapov et al., 2022) results in a particularly strong impact of Collembola on decomposition through microbial population control (Koltz et al., 2018; Crowther et al., 2012; Seastedt and Crossley, 1984).
Contrary to other important groups of soil fauna, such as earthworms, millipedes (Golovatch and Kime, 2009; Berman et al., 2015) or woodlice (Sfenthourakis and Hornung, 2018), Collembola are ubiquitous in arctic soils, where they can reach high densities – up to 130 000 individuals per square metre in high-Arctic Greenland (Sørensen et al., 2006). However, Collembola are mostly abundant in the topsoil and to our best knowledge have never been observed in the perennially frozen subsoil, the permafrost. Permafrost soils are a prominent feature of Arctic landscapes, and the huge carbon stock they represent and positive feedback to climate change that their thawing will likely induce cause concern (Masson-Delmotte et al., 2021). The frozen conditions over long periods of time have eliminated most fauna and non-microbial life from these environments, and despite the possibility of resuscitating organisms such as plants (Yashina et al., 2012), nematodes (Shatilovich et al., 2018) or rotifers (Shmakova et al., 2021), newly thawed permafrost soils generally harbour an extremely simplified soil food web entirely deprived of metazoans. Despite this particularity, the absence of Collembola and the consequence of their possible introduction into newly thawed permafrost on its biogeochemical cycling have been mostly overlooked.
In contrast to Collembola, some microorganisms survive and/or thrive in permafrost conditions, and microbial adaptations to frozen conditions have been studied (Mackelprang et al., 2011, 2017; Hultman et al., 2015; Bottos et al., 2018). However, not all microbes survive these conditions, and the combination of environmental constraints exerted over long periods of time (Mackelprang et al., 2017) and strong dispersal limitations (Bottos et al., 2018) results in microbial communities that can be deprived of some functions (Knoblauch et al., 2018; Monteux et al., 2020; Barbato et al., 2022). The re-introduction of such functions can result in drastic changes in permafrost processes, and sizable impacts on greenhouse gas production have been observed in vitro for CH4 and CO2 (Knoblauch et al., 2018; Monteux et al., 2020) and confirmed in situ for N2O (Marushchak et al., 2021). Upon thawing, this re-introduction of missing functions or ecological rescue (Calderón et al., 2017) requires microorganisms to migrate into this newly available habitat, which could happen for instance laterally through airborne dispersal (Harding et al., 2011) – e.g. for permafrost exposed to the air in abrupt thaw processes (Inglese et al., 2017) – or vertically through percolation in the soil column when the active layer becomes deeper (Monteux et al., 2018; Johnston et al., 2019).
Collembola are another possible vector for the dispersal of microorganisms into newly thawed permafrost (Buse et al., 2014). With the deepening of the active layer, Collembola migration into newly thawed permafrost is unlikely since they mostly reside in topsoil layers, but it will likely occur in soil-mixing events (Väisänen et al., 2020) such as thermokarst, active-layer detachment or thaw slump processes where newly thawed permafrost is exposed to surface conditions. Like all large organisms, Collembola host a variety of microorganisms, their microbiome (Agamennone et al., 2015; Leo et al., 2021). Because microarthropods, such as Collembola, can move across large distances compared to fungi or bacteria, the microbial species in collembolan microbiomes might be among the first to colonize and establish in newly thawed permafrost when Collembola access it. This could occur through a combination of two main processes: ectozoochory, where the microbiome of the cuticle disperses into the new habitat, and endozoochory, where microorganisms disperse after transiting through the gut of the animals. To our best knowledge, whether Collembola affect the biogeochemical functioning of newly thawed permafrost and whether and how they can serve as a vector for microbial colonization have not been explored yet.
We incubated permafrost from the Yedoma domain, which represents a large carbon stock in parts of Siberia and North America (Strauss et al., 2017) and has previously been shown to lack certain microbial functions (Monteux et al., 2020), in the presence or absence of a model species of microarthropod (Collembola Folsomia candida), and assessed CO2 production and bacterial community composition over a 25 d aerobic incubation. We used Folsomia candida Collembola from a stock culture, as well as Collembola subjected to manipulation of their cuticle microbiome or cuticle and gut microbiomes to test the following hypotheses:
-
Collembola presence in permafrost will alter bacterial community composition, through their grazing.
-
Collembola more closely exposed to topsoil bacteria will change permafrost bacterial community composition further than those exposed less intricately. In other words, we expect a gradual change between clean Collembola, Collembola with cuticle microbiome manipulated (ectozoochory), and Collembola with both gut and cuticle microbiomes manipulated (endozoochory).
-
Collembola presence in permafrost will increase CO2 production, through both Collembola respiration and a stimulation of microbial activity (“priming effect”).
-
The gradual introduction of distinct bacterial communities hypothesized above (2) will result in increased CO2 production, owing to the functional limitations of the Yedoma permafrost microbial community in terms of CO2 production.
2.1 Experimental design
2.1.1 Soils
The Yedoma sediment used to assess the impacts of Collembola on permafrost CO2 production and bacterial community composition originated from the Cold Regions Research and Engineering Laboratory (CRREL) permafrost tunnel (Fox, Alaska, USA). The sediment was sampled from the upper silt unit and is an Upper Pleistocene silty deposit, previously described in detail (Shur et al., 2004; Mackelprang et al., 2011, 2017; Monteux et al., 2020). This sediment was chosen due to its microbial communities being vulnerable to invasions and exhibiting functional limitations, allowing the discernment of impacts of introduced microorganisms on broad proxies such as CO2 production (Monteux et al., 2020). Approximately 35 g (fresh weight) of homogenized sediment was set in 200 mL glass jars, sealed with parafilm to allow for gas but not moisture or microorganism exchange, and pre-incubated at 10 ∘C for 11 d before inoculation.
To manipulate the Collembola microbiome and make it more similar to that found in natural settings, we collected a topsoil (0–15 cm depth) from a subarctic meadow (Kärkevagge, 30 km west of Abisko, northern Sweden; 68∘24′23.8′′ N, 18∘18′51.6′′ E) in September 2019 and kept it frozen until the cultivation of Collembola. We could not obtain topsoil from the same location as the Yedoma permafrost due to practical constraints during the Covid-19 pandemic.
2.1.2 Collembola
A strain of the Collembola species Folsomia candida was obtained from Vrije Universiteit Amsterdam and cultured 6 months prior to the onset of the experiment. Folsomia candida is a parthenogenetic ground-dwelling Collembola, which has been routinely used as a model organism in soil ecology. Stock cultures were maintained on a gypsum and coal medium and fed baker's yeast; traces of mould were removed; fresh yeast and water were added once to twice a week, and fresh stock cultures were started monthly.
2.1.3 Collembola inoculation treatments
Two months prior to the onset of the experiment, separate stock cultures were established on gypsum and coal medium supplemented with a 2–3 cm layer of topsoil to obtain Collembola whose skin and gut microbiome were both colonized with topsoil microorganisms (“topsoil stock culture”). In parallel, similar stock cultures using permafrost sediment were established as an additional control (“permafrost stock culture”). These stock cultures with soil or sediment were supplemented with yeast and water like the gypsum stock culture to maintain high adult population densities prior to the experiment. One day before inoculating the incubation jars, soil suspensions were made from the topsoil and permafrost sediment (5 and 10 g, respectively, in 100 mL ddH2O), shaken at 150 rpm for 1 h and filtered (Ahlstrom-Munksjö grade 006, 1.5 µm pore size) to manipulate the skin microbiome of Collembola in the ectozoochory treatments.
All incubation jars containing permafrost were randomly assigned a treatment on the day of inoculation from among the following, replicating each treatment across six jars (Fig. 1). The treatments consisted in adding to the jars with permafrost
-
no Collembola (hereafter, “no-Collembola control”),
-
Collembola grown on gypsum stock culture (“Collembola”),
-
Collembola grown on gypsum stock culture and sprayed with topsoil suspension (“ectozoochory”),
-
Collembola grown on gypsum stock culture and sprayed with permafrost suspension (“ectozoochory control”),
-
Collembola grown on topsoil stock culture (“endozoochory”),
-
Collembola grown on permafrost stock culture (“endozoochory control”).
To obtain a baseline allowing the quantification of the effect of Collembola on permafrost CO2 production and bacterial community composition, Yedoma permafrost was incubated in the absence of Collembola (no-Collembola control). For all other treatments, the permafrost was supplemented with adult Collembola isolated from the stock cultures using a handheld vacuum cleaner mounted with a 10 mL pipet tip. The Collembola were transferred into a black plastic tray, allowing them to be spread and the adults to be picked out, excluding juveniles as much as possible. From this tray, a similar number of Collembola (30–80 individuals, in the range of values used in the literature, e.g. by Kaneda and Kaneko, 2008) was sampled with the vacuum cleaner and then inoculated into each jar by pouring them into a plastic funnel, with the Collembola provenance or manipulation depending on the treatment. Prior to transferring them into incubation jars, Collembola used in the ectozoochory treatment and its control were sprayed with the corresponding soil suspensions. To limit cross-contamination, separate funnels, as well as separate 10 mL pipet tips on the vacuum cleaner, were used for the different treatments. A picture of the inside of each jar was taken to count the exact number of Collembola, then the jars were closed with rubber septa and flushed with moisturized CO2-free air and incubated. To estimate Collembola respiration per individual, Collembola were also incubated in six jars under identical conditions, except that the soil was replaced with a few drops of autoclaved ddH2O to prevent dehydration (hereafter, no-soil calibration).
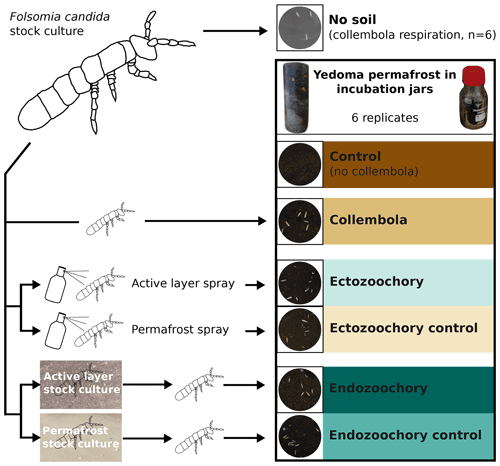
Figure 1Summary of the experimental design. Folsomia candida Collembola were grown in a stock culture on gypsum and fed with yeast and then subjected to different treatments before being inoculated into jars filled with Yedoma permafrost sediment (30–80 individuals per jar). Ectozoochory was assessed by spraying the Collembola with soil suspensions, while endozoochory was assessed by growing Collembola in stock cultures supplemented with soil. The jars were incubated in the dark for 25 d at 10 ∘C; CO2 production was measured throughout the incubation period, and bacterial community composition was determined at the end of the incubation.
2.1.4 Incubation
We dark-incubated all flasks for 25 d under aerobic conditions at 10 ∘C. This incubation temperature is similar to summer active-layer temperatures in permafrost-affected areas and within the thermal tolerance range of psychrophilic microorganisms (D'Amico et al., 2006). We used a short (25 d) incubation period to ensure a relatively stable Collembola population level, by limiting uncertain numbers of newly hatched Collembola individuals, since the eggs of Folsomia candida take 18–20 d to hatch at 16 ∘C (Marshall and Kevan, 1962) and presumably longer than 25 d at 10 ∘C.
2.2 Measurements
2.2.1 CO2 production
Headspace air was sampled with a syringe to measure CO2 concentrations (EGM-5 IRGA, PP Systems, Amesbury, Massachusetts, USA) at intervals ensuring CO2 concentrations remained below 20 000 ppm to prevent a toxic CO2 build-up (i.e. after 3, 7, 14 and 25 d). After each measurement, the jars were flushed with 0.45 µm filtered CO2-free air moisturized by bubbling it through two 5 L bottles of ddH2O, for 3 min at 1 to 2 L min−1, i.e. with at least 15 times the volume of the jar. CO2 concentrations were adjusted for changes in temperature and atmospheric pressure to calculate CO2 production rates (τ) as follows:
where (Δt)i is the time interval between measurement (subscript i) and previous flushing, Pi atmospheric pressure at measurement time, V the headspace volume, R the ideal gas constant, and Ti the temperature. To calculate cumulative CO2 production over the entire incubation, we summed up the quantity of CO2 present in the headspace at each sampling, within each jar.
2.2.2 Bacterial community
Microcentrifuge tubes (1.5 mL) were filled with soil and snap-frozen in dry ice to analyse microbial communities from the jars harvested at the end of the incubation. The frozen tubes were kept at −20 ∘C for up to 4 months before freeze-drying and then homogenized by bead beating (Precellys CK68 15 mL tubes, 2×30 s at 4500 rpm). DNA was extracted from 183 to 285 mg of homogenized freeze-dried soil using the DNeasy PowerSoil Pro Kit (Qiagen) according to the manufacturer's instructions, and DNA concentrations in the extracts were measured on a Qubit 1.0 fluorometer.
The V4–V5 region of the 16S ribosomal RNA gene was targeted in PCR amplification using primers 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 926R (5′-CCGYCAATTYMTTTRAGTTT-3′) with Illumina sequencing adapters, using 12.5 µL Phusion Taq Green PCR Master Mix (Thermo Scientific), 0.25 µM of each primer, 2 µL of DNA extract diluted to 5 ng µL−1 and nuclease-free water in 25 µL reaction volume. PCR conditions were as follows: initial denaturation (98 ∘C, 3 min), 25 cycles of denaturation (98 ∘C, 15 s), annealing (50 ∘C, 30 s) and elongation (72 ∘C, 40 s), and a final elongation (72 ∘C, 10 min), after which PCR products were checked by electrophoresis on 1 % agarose SB gel. A total of 20 µL of each PCR product was cleaned, and the products' DNA concentrations were normalized using a SequalPrep Normalization Plate Kit (Invitrogen), according to manufacturer's instructions. Three DNA extraction blanks and two PCR blanks were included as negative controls, as well as a mock community as the positive control (ZymoBIOMICS, diluted to 5 and 0.5 ng µL−1 in two replicates each).
A second PCR step was performed to add Nextera dual-indexing barcodes, using 30 µL reaction volume, 1 µM of each primer and 5 µL of cleaned PCR product. PCR conditions were as follows: initial denaturation (98 ∘C, 3 min), eight cycles of denaturation (98 ∘C, 30 s), annealing (55 ∘C, 30 s) and elongation (72 ∘C, 40 s), and a final elongation (72 ∘C, 10 min), after which PCR products were checked by electrophoresis on 1 % agarose SB gel. A total of 25 µL of each PCR product was cleaned, and the products' DNA concentrations were normalized using a SequalPrep Normalization Plate Kit (Invitrogen). Serial elution across columns was used to increase concentration of the pooled products, i.e. using only 8×20 µL elution buffer instead of 96×20 µL. The eluted DNA was pooled; its concentration was measured on a Qubit fluorometer, and the size distribution of the amplicons was measured by automated electrophoresis (Agilent 2100 Bioanalyzer). The library was then sent for sequencing on an Illumina MiSeq with V3 chemistry (2×300 bp, 15 % PhiX spike-in) at the SNP&SEQ Technology Platform in Uppsala. Demultiplexing was performed by the sequencing facility, and data were deposited at ENA with accession number PRJEB51992.
2.3 Data analysis
2.3.1 Bioinformatics
All bioinformatics and statistics were performed in R v4.1.3 (R Core Team, 2022), unless specified otherwise. The whole analysis pipeline is found at the Bolin Centre Code Repository (Monteux, 2022) and the processed data and figure-generating script at Zenodo (Monteux et al., 2022). In short, amplicon sequence variants (ASVs) were created with DADA2 1.18.0 (pseudo-pooling; Callahan et al., 2016) after removing primers and adapters with Cutadapt (v3.10; Martin, 2011). Taxonomy was assigned to ASVs with the RDP naïve Bayesian classifier (v1.8; Wang et al., 2007), and ASVs resolved to the genus rank were further assigned a species rank by the exact string-matching algorithm implemented in DADA2 (assignSpecies), using SILVA v138.1 reference data (Quast et al., 2013). Putative contaminant ASVs were manually selected from those identified in silico using the decontam algorithm (Davis et al., 2018) with combined prevalence- and frequency-based methods using the default threshold of 0.1 and separate prevalence- and frequency-based methods with a threshold of 0.05. Eight contaminant ASVs amounting to up to 0.024 % of the total reads were removed. Appropriateness of the bioinformatics analysis parameters was judged by visually assessing the composition of mock communities (microbial community DNA standard, ZymoBIOMICS) at the genus level, leading to ASVs amounting to up to fewer than 10 reads and/or present in fewer than 3 samples being removed from the dataset. Sequencing depth was not associated with experimental treatments (ANOVA 0.701, P= 0.627); therefore read numbers were converted to proportional abundances within samples to normalize sample sizes.
2.3.2 Diversity analyses
The effect of the different treatments on bacterial communities was visualized using principal coordinates analysis (PCoA) and tested with permutational multivariate analyses of variance (PERMANOVAs, adonis function in the vegan package; Oksanen et al., 2022) after verifying homoscedasticity (betadisper). Two distance matrixes were computed for that purpose, using weighted and non-weighted UniFrac distances (Lozupone et al., 2011) to distinguish between compositional effects accounting for bacterial relative abundances and for only presence–absence, respectively. Pairwise contrasts were subsequently computed using the wrapper provided in the pairwiseAdonis R package (Arbizu, 2022).
In addition, the same effect was tested with the more robust and sensitive manyglm approach (Wang et al., 2012), which avoids certain pitfalls of distance-based methods (Warton et al., 2012). The manyglm models were fitted with negative binomial distribution, after visually checking that assumptions were met, on the non-normalized ASV count data using the default PIT-trap resampling with 1999 bootstrap permutations and a likelihood-ratio testing method. An analysis of deviance was carried out (anova.manyglm), subsequently using the provided wrapper for pairwise comparisons with free step-down P-value adjustment.
2.3.3 CO2 production analysis
A repeated-measures ANOVA was carried out, using Greenhouse–Geisser ϵ correction to degrees of freedom to account for the violation of the sphericity assumption and thus to assess the interactive effect of the Collembola manipulation treatments over time. Since this interactive effect was not statistically significant (P= 0.56, Table A1), we removed the time dimension and used cumulative CO2 production at the end of the incubation period in further analyses.
To assess our hypothesis that Collembola presence would increase CO2 production we used two-sample t tests with unequal variances to compare the cumulative CO2 production at the end of the incubation between the no-Collembola control and all other jars.
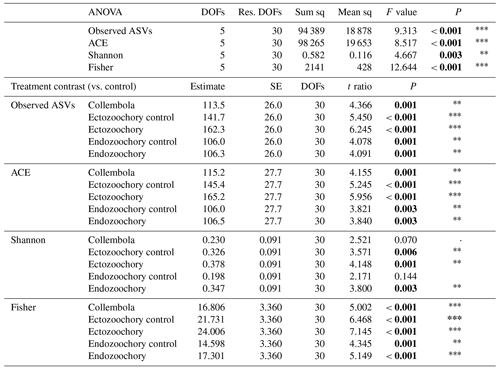
To explore differences between the Collembola microbiome manipulations, we used a one-way ANOVA followed by treatment contrasts to assess the difference from the control (i.e. all treatment and controls compared to the no-Collembola control) and selected orthogonal contrasts to assess the effects of each treatment (i.e. each treatment compared to its own control) using the “emmeans” package (Lenth, 2016).
2.3.4 Estimated Collembola respiration
Based on photographs taken upon inoculating the incubation jars with Collembola and 3 d after inoculating, we counted the exact number of live Collembola (i.e. ignoring Collembola which had apparently not moved between when the two pictures were taken). Only a few animals did not survive the transfer, and no dead animals were observed at the end of the incubation. The number of Collembola per jar varied across treatments ( 3.19, P= 0.030), although no pairs of treated jars significantly differed from each other (when including the no-soil calibration jars, 4.048, P= 0.006; Fig. B1); we therefore needed to account for differing Collembola numbers. We averaged the respiration per individual in the no-soil calibration set for each of the CO2 concentration measurement times and multiplied this amount by the number of Collembola present in each jar to estimate the amount of CO2 produced by Collembola basal respiration in treatment jars.
2.3.5 Response-ratio calculations
A response ratio of CO2 production in Collembola treatments was calculated by dividing cumulative CO2 production by the average of the no-Collembola control. This was performed both for the net CO2 production after subtracting the estimated Collembola basal respiration (RRsoil) and for the gross CO2 production (RRgross). This allows the partition of the difference in altered CO2 production between what was respired by the Collembola and a putative priming effect on soil CO2 production.
3.1 Bacterial communities
Bacterial community composition was overall largely unaffected by the presence of Collembola. Taking relative abundances into account, no effect of the different treatments was identified with PERMANOVA (P > 0.05, Fig. 2a), but a significant treatment effect was observed using the presence–absence distance metrics (PERMANOVA 2.91, P= 0.001, Fig. 2b). More specifically, treatments containing Collembola differed from the no-Collembola control (Fig. 2b) and to some extent from each other (Table C1). Using manyglm, a significant treatment effect was observed (manyglm analysis of deviance P < 0.001; Table 1). The ectozoochory treatment and its control (Collembola sprayed with topsoil or permafrost soil suspension, respectively) differed significantly from the no-Collembola control in the post hoc test (0.01 < P < 0.05), while the other treatments including Collembola tended to differ from the no-Collembola control (0.05 < P < 0.1, Table 1). Similarly to the presence–absence analysis, treatments that included Collembola did not significantly differ from each other in the manyglm analysis. Overall, soil with Collembola exhibited higher alpha-diversity metrics (richness estimators Chao1, abundance-based coverage estimator – ACE, number of observed ASVs, as well as Shannon and Fisher diversity indexes; Fig. 3, Table C2) than the no-Collembola control, but those did not differ among the Collembola treatments.
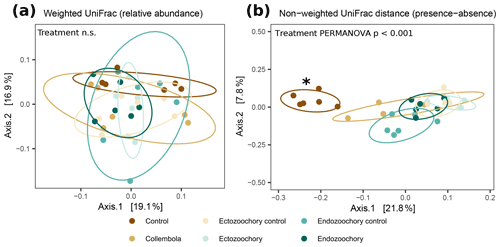
Figure 2Response of permafrost bacterial communities to the addition of Collembola undergoing different microbiome manipulation treatments. (a) Principal coordinates analysis (PCoA) of normalized abundance of bacterial ASVs (amplicon sequence variants) based on weighted UniFrac phylogenetic distance (i.e. accounting for relative abundances); (b) PCoA of normalized abundance of bacterial ASVs based on non-weighted UniFrac phylogenetic distance (i.e. presence–absence only). Ellipses indicate the 80 % confidence interval around treatment centroids, and axes are scaled to the percentage of explained variance. The asterisk indicates that the no-Collembola control (dark brown) in (b) significantly differs from all other treatments (pairwise PERMANOVA P < 0.05) and endo- and ectozoochory treatments did not differ from the Collembola treatment. Pairwise comparisons are summarized in Table C1.
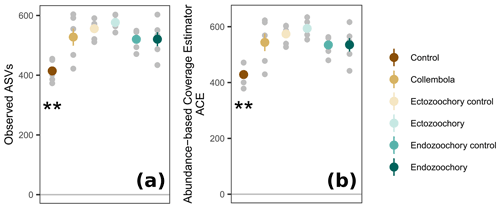
Figure 3Response of permafrost bacterial alpha diversity to the addition of Collembola undergoing different microbiome manipulation treatments. (a) Number of observed ASVs (amplicon sequence variants) per sample; (b) abundance-based coverage estimator (ACE). Coloured symbols and error bars are means and standard errors within a treatment (n=6); small grey symbols are individual samples. Error bars are only shown when exceeding the symbols' size. Asterisks denote that the no-Collembola control differs from all other treatments (estimated marginal means pairwise comparison, 0.001 < Holm-adjusted P < 0.01).
3.2 CO2 production
Overall, the Collembola addition resulted in higher CO2 production than the no-Collembola control, but the ectozoochory and endozoochory treatments did not result in higher CO2 production than their respective controls (using gross CO2 production, as in RRgross; Table A2, Fig. A1). When accounting for Collembola basal respiration (as in RRsoil), only the ectozoochory treatment (P= 0.005) and to a lesser extent its control (P= 0.069) differed from the no-Collembola control (Table A2, Fig. A1).
We hypothesized increased CO2 production in the zoochory treatments through an effect on bacterial communities; however that effect was absent. We therefore tested for the overall effect of Collembola presence across all treatments compared to the no-Collembola control (Fig. 4). Using a t test with unequal variances, we observed a 25.85 % increase in CO2 production in the presence of Collembola (RRgross; Fig. 4a). When subtracting estimated Collembola basal respiration, this increase was on average 13.22 % (RRsoil; Fig. 4b); thus roughly half of the observed increase in CO2 production could be attributed to Collembola basal respiration.
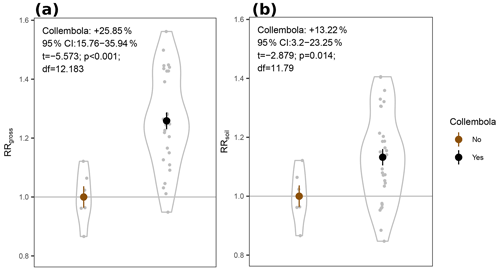
Figure 4Response ratios of permafrost cumulative CO2 production rates to addition of Collembola. (a) RRgross: measured CO2 production rates. (b) RRsoil: measured CO2 production rates with estimated Collembola basal respiration subtracted. All values are divided by the mean of the no-Collembola control; small grey circles are individual values; large symbols are means; error bars are standard errors of the mean (n= 6 and 30 for brown and black symbols, respectively); violin plots denote the shape of the distributions with the maximum width proportional to the number of observations. Statistics are derived from two-sample t tests with unequal variances.
We showed that Collembola stimulated CO2 production from permafrost sediment through “priming” while subtly altering the presence of certain bacterial ASVs, irrespectively of the microbiome manipulation imposed on the Collembola. Colonization of permafrost by Collembola therefore has the potential to result in bacterial community changes and to increase C turnover from tundra soils.
4.1 Introduction of bacteria by Collembola
Collembola introduced new bacteria into thawed permafrost soil, resulting in an increase in alpha diversity and significant differences in bacterial community composition, on a presence–absence basis. This pattern was consistent in all treatments where Collembola were added but did not depend on the microbiome manipulation treatment assigned to the Collembola (Figs. 2b, 3). Despite their effects on the presence of certain bacteria, Collembola did not impose broader community changes as bacterial communities were not affected when considering relative abundances as we had hypothesized (Fig. 2a, Table 1). However, given the subtle effect of Collembola on bacteria during this short incubation, this suggests that on longer timescales Collembola may modify bacterial communities more dramatically.
4.2 Differences between ecto- and endozoochory treatments
Although Collembola introduced new bacteria into permafrost soil, our results did not support that they did so differently through ecto- and/or endozoochory. This suggests that Collembola presence mostly introduced bacteria that were part of the core Collembola microbiome or that the way in which their microbiome changed was not well represented by our treatments. For example, spraying may not have altered the cuticle microbiome as Collembola cuticle is particularly hydrophobic (Hensel et al., 2013a, b; Nickerl et al., 2013). This should however not have been an issue for the endozoochory treatment, in which Collembola were in contact with the topsoil for a prolonged (> 1 month) period of time. Despite this prolonged exposure, no additional effect of the endozoochory treatment on bacterial community composition was observed, compared to ectozoochory or Collembola-only treatments. Collembola typically harbour a gut microbiome reflecting their diet and direct environment (Xiang et al., 2019; Leo et al., 2021), and observing soil-coloured faeces in the topsoil and permafrost stock cultures suggests that soil bacteria were present in the gut of Collembola used in the endozoochory treatments. In contrast with the permafrost stock cultures, where the yeast was necessary as an additional food source to maintain stable populations, Collembola populations remained stable in topsoil stock cultures similar to those used in the endozoochory treatment even in the absence of yeast, further indicating their consumption of soil particles and associated microorganisms. Therefore, the absence of the effect of the ectozoochory treatment on permafrost bacterial communities may indicate that Collembola cuticle does not serve as a vector for bacterial dispersal, possibly due to its omniphobic structure. Further, the lack of difference between the effects of Collembola and endozoochory treatments suggests that few to none of the bacteria present in this topsoil are able to establish in – or survive transit through – collembolan guts. This suggests against generalist “hitch-hiking” bacteria using microfauna guts as a means of dispersal, emphasizing instead that such hitch-hiking bacteria are similar regardless of the feeding context of the Collembola.
It is not unlikely that the observed limited response of the bacterial community to zoochory treatments was transient and that Collembola presence would have eventually affected community composition also in terms of relative abundances. A longer incubation period may have resulted in stronger effects on bacterial community composition, as observed by Coulibaly et al. (2019), but this would have been at the expense of controlling the number of Collembola in the jars and thus of being able to account for their basal respiration. Using an RNA-based approach to target the “active” bacterial community may be another way to detect such transient effects. It should also be noted that our semi-quantitative approach to bacterial community composition did not allow us to assess putative changes in absolute abundances, although such changes are not necessarily observed in similar studies (Kaneda and Kaneko, 2008).
Collembola effects on fungal communities may have indirectly affected bacterial communities and thus represent a potential additional mechanism by which Collembola have affected the presence of certain bacteria. It is not unlikely that fungi would have responded more strongly to the presence of mostly fungivorous Folsomia candida. Using fungi as the focus organism in this study, however, may have had its difficulties. For example, the use of Candida albicans as a food source for the Collembola stock cultures may have strongly distorted fungal community composition, thus requiring questionable bioinformatic workarounds. Further, assessing the effects on fungal communities may not allow for distinguishing between the consequences of Collembola as a vector for microbial migration and as a grazing consumer. Although assessing responses of the whole soil food web may have given better insights into potential mechanisms by which Collembola affect thawed permafrost soil, we here chose to focus on their effects on bacteria given their preponderant role on realizing soil functions in this system (Monteux et al., 2020).
4.3 Collembola effects on CO2 production
Our hypothesis stating that soil CO2 production would be increased in the presence of Collembola was supported by our results. However, since no differences in bacterial community composition were detected between the different Collembola treatments, the rationale for our follow-up hypothesis (attributing an increased influence on CO2 production to the treatments exhibiting stronger changes in bacterial community) was not met. We therefore did not deem it relevant to formally evaluate this hypothesis. Our results suggest that it would be rejected (Fig. A1), but we could not say whether that is because the rationale is wrong or because the manipulation of bacterial communities did not yield the expected effect.
Overall, Collembola presence increased respiration by 25 %, half of which was attributed to respiration of Collembola themselves. We interpret the remaining half (13.22 %) as a priming effect, i.e. a stimulation of soil-organic-matter-derived CO2 production induced by the presence of Collembola. The mechanisms behind this priming effect remain unclear and could for instance relate to stimulation of microbial activity by the input of more labile substrates by the Collembola (digestion by-products). Overall, our results add to a list of contrasting findings on the direction and magnitude of priming of soil-organic-matter decomposition by Collembola presence in soils in general. Earlier studies that compared CO2 production in the presence or absence of Collembola or other faunal groups have mostly focused on litter (e.g. Hanlon and Anderson, 1979). The scarce studies on soil CO2 production including Collembola reported contrasting findings, with some studies observing increases in soil CO2 emissions in the presence of Collembola by up to +400 % (Addison and Parkinson, 1978; Bakonyi, 1989; Kaneda and Kaneko, 2008; Wang et al., 2017), while others found no significant changes (Theenhaus et al., 1999; Lubbers et al., 2020; Lucas et al., 2020). The Collembola effect on CO2 production in litter can be species-specific (Hanlon and Anderson, 1979), and our results could support this as we found a similar increase to those in three studies where Folsomia candida was used (Bakonyi, 1989; Kaneda and Kaneko, 2008; Wang et al., 2017). Kaneda and Kaneko (2008) reported a density-dependent increase in CO2 production with the addition of F. candida, which showed no effect at the density used in our study but an increase with 5–10 times higher density (i.e. 400 individuals in 30 g of soil compared to our 30–80). Accordingly, we did not find any trend supporting a density-dependent response in our data. Nevertheless, we suggest that permafrost soils colonized by Collembola in particular, as well as by soil fauna in general, will likely see an increase in their rates of decomposition, in CO2 production in particular, due to both priming and the basal respiration of the newly established food web.
Our experimental setup did not allow us to distinguish further between soil and Collembola respiration. We estimated basal respiration, though this may not perfectly reflect reality, considering that Collembola in jars without soil may not have behaved similarly to those in jars with soil. Faced with a scarcity of food sources, they may have slowed their metabolism to endure that stress, which could result in decreased basal respiration and thus lead us to overestimate the magnitude of priming. We could not find reference values in the literature for Folsomia candida respiration rates, but Addison and Parkinson (1978) estimated the CO2 production of two high-Arctic collembolan species to 182.6 and 250.6 µL CO2 per gram live weight per hour at 10 ∘C, for Hypogastrura tullbergi and Folsomia regularis, respectively. Assuming a CO2 density of 1.977 kg m−3 and 170 µg per adult individual of Folsomia candida, our values range between 47 and 220 µL CO2 per gram live weight per hour. Addison and Parkinson (1978) refer to Mitchell (1973) for a description of the method used for estimating respiration rates, where the measurements appear to be carried out over a period of 20 h. When considering only the first measurement date in our data (3 d after inoculation), the rates are closer to the values reported by Addison and Parkinson (1978) with 157.5±18.5 µL CO2 per gram live weight per hour (mean ± SE, n= 6). In future studies of this mechanism, it would be interesting to use stable isotope methods to partition the Collembola-derived vs. soil-organic-matter-derived CO2 production more accurately.
Beyond the CO2 production increase due to priming by soil fauna, our Collembola addition treatments may have induced “microbial community priming”, in the form of the release of microbial functional limitations. The Yedoma sediment used in this study indeed lacks certain microbial taxa and functions, and the onset of an exotic microbial community can result in large increases in CO2 production (Monteux et al., 2020). Considering the limited to non-significant (depending on the distance metrics) effect of Collembola addition on bacterial communities, we do not think this mechanism explains our observed results, although we do not rule out this explanation.
Collembola presence modified bacterial communities in newly thawed permafrost and primed its CO2 production, highlighting the importance of non-microbial decomposers for the fate of soil organic matter in thawing permafrost. An emerging theme in permafrost research is the missing functions in permafrost microbial communities due to a lack of certain microbial groups, hampering the production of CO2, CH4 and N2O (Knoblauch et al., 2018; Monteux et al., 2020; Marushchak et al., 2021). It is also becoming clearer that the functionality of permafrost microbes can vary across space (Barbato et al., 2022); therefore the modalities of microbial dispersal into newly thawed permafrost likely affect the fate of the permafrost's organic matter and the rate of release of greenhouse gases. Our findings suggest that Collembola may accelerate the release of greenhouse gases, possibly in part through the introduction of microorganisms, although those did not seem to depend strongly on the preceding Collembola environment. In nature, thawing permafrost mostly occurs at the bottom of the seasonally thawing active layer, often below the water table and at depths far from the topsoil where most Collembola reside. Different soil fauna, such as earthworms, may access such newly thawed permafrost, but their current geographical range does not overlap much with the permafrost region. However, rapid thaw events such as hillslope thermokarst, active-layer detachments or retrogressive thaw slumps expose newly thawed permafrost to surface conditions, thus making it susceptible to colonization by Collembola. Fauna-induced dispersal of microorganisms is most likely to take place in these environments and should be further investigated in realistic field conditions.
It remains unclear whether the increase in CO2 production we attribute to priming stems from increased functionality of the microbial community due to bridging of functional limitations or from other mechanisms. Mechanical breakdown of organic material and digestion processes might provide microorganisms with more easily available substrates, while disruption of the soil pore structure may also result in increased microbial decomposition activity. Several studies indicate increases in CO2 production with the presence of soil fauna; however to our best knowledge no studies have specifically partitioned to which extent such increases were due to faunal respiration or to priming effects. We argue that elucidating this question would be an important next step towards opening the “black box” that soil systems still often represent, thus helping to mechanistically address the effects of global changes.
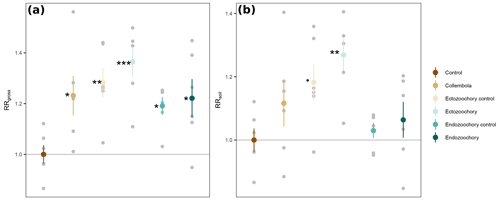
Figure A1Response ratios of permafrost cumulative CO2 production rates to addition of Collembola. (a) RRgross: measured CO2 production rates. (b) RRsoil: measured CO2 production rates with estimated Collembola basal respiration subtracted. All values are divided by the mean of the no-Collembola control; small grey circles are individual jar values; large symbols are means; error bars are standard errors of the mean (n= 6). Black symbols denote significant difference from the control; different symbols denote different statistical significance (estimated marginal means treatment contrasts with Holm adjustment for multiple comparisons, Appendix Table A2; dot (⋅) denotes 0.05 < P < 0.1; * 0.01 < P < 0.05; 0.001 < P < 0.01; P < 0.001).
Table A1Effect of Collembola addition on daily CO2 production over time, repeated-measures ANOVA. P[GG] indicates repeated-measures ANOVA P using Greenhouse–Geisser correction to degrees of freedom to account for violation of the assumption of sphericity (Mauchly's test). P values below 0.05 are denoted in bold; P < 0.001; * 0.01 < P < 0.05. Num. DOFs: numerator degrees of freedom; Den. DOFs: denominator degrees of freedom; GES: generalized Eta-squared measure of effect size; GG ϵ: Greenhouse–Geisser ϵ.
Table A2Effects of Collembola additions on response ratios of cumulative CO2 production at the end of the incubation, excluding (RRsoil) or including (RRgross) estimated Collembola respiration. Holm adjustment for P values of non-orthogonal contrasts. P values below 0.05 are denoted in bold; P < 0.001; 0.001 < P < 0.01; * 0.01 < P < 0.05; dot (⋅) denotes 0.05 < P < 0.1. DOFs: degrees of freedom; Res. DOFs: residual degrees of freedom.
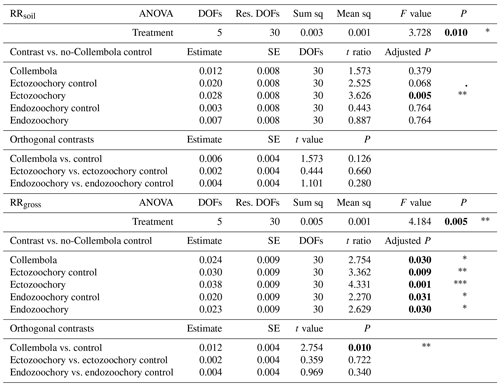
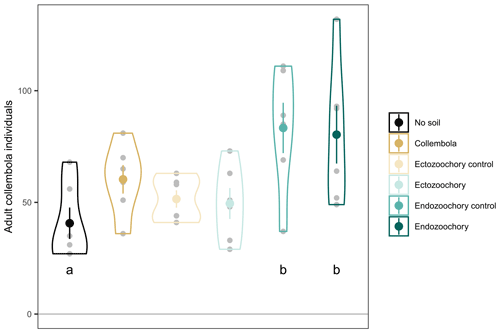
Figure B1Number of adult Collembola individuals per incubation jar. Small grey circles are individual jar values; large symbols are means; error bars are standard errors of the mean (n= 6); violin plots denote the shape of the distributions with maximum width proportional to the number of observations. Different black letters denote statistically significant pairwise differences (estimated marginal means pairwise contrasts with Holm adjustment for multiple comparisons, 0.05 < P < 0.1).
Table C1Effects of Collembola additions on bacterial community composition (permutational multivariate ANOVAs and pairwise contrasts), using distance matrixes based on relative abundances (weighted UniFrac) or presence–absence (non-weighted UniFrac). Holm adjustment for P values of pairwise contrasts. P values below 0.05 are denoted in bold; P < 0.001; 0.001 < P < 0.01; * 0.01 < P < 0.05; dot (⋅) denotes 0.05 < P < 0.1.
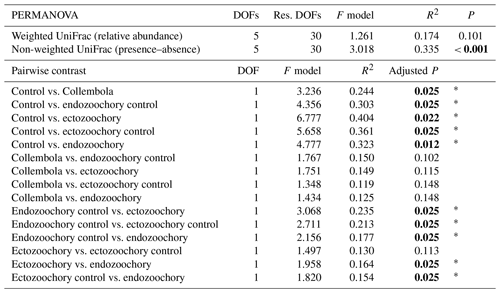
Table C2Effects of Collembola additions on bacterial alpha diversity (ANOVAs and pairwise contrasts), with total number of observed ASVs, abundance-based coverage estimator (ACE), and Shannon's and Fisher's diversity indexes. Holm adjustment for P values of pairwise contrasts. P values below 0.05 are denoted in bold; P < 0.001; 0.001 < P < 0.01; * 0.01 < P < 0.05; dot (⋅) denotes 0.05 < P < 0.1.
All code used to process the raw DNA data and produce the figures and tables presented in the text is found at https://doi.org/10.57669/monteux-2022-collembola-1.1.1 (Monteux, 2022).
The code used to generate figures from the processed DNA data is also found at https://doi.org/10.5281/zenodo.7015077 (Monteux et al., 2022).
All 16S sequencing data are found at ENA under project accession number PRJEB51992.
All processed data used to generate the findings presented in the text are found at https://doi.org/10.5281/zenodo.7015077 (Monteux et al., 2022) and https://doi.org/10.57669/monteux-2022-collembola-1.1.1 (Monteux, 2022).
Due to practical constraints, the exact permafrost material and Collembola cultures used throughout the paper are no longer available. Permafrost from the same location may be obtained through contacting the US Army CRREL, while Folsomia candida strains may be obtained by contacting Janine Mariën. Frozen permafrost samples from the end of the incubation period, as well as aliquots of DNA extracts used for this study, may be obtained from the corresponding author upon reasonable request.
SM, JM and EK designed the experiment. JM provided Folsomia candida strains and guidance for culturing. EK provided topsoil from Kärkevagge. SM performed the experiment and data analysis and wrote the manuscript with input from all co-authors.
The contact author has declared that none of the authors has any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the special issue “Global change effects on terrestrial biogeochemistry at the plant–soil interface”. It is not associated with a conference.
This study was funded by a grant from Formas (Dnr 2017-01182) awarded to Eveline J. Krab and by Kempestiftelserna awarded to Sylvain Monteux.
We thank the Department of Forest Mycology and Plant Pathology, SLU, for hosting the molecular work and Thomas H. Douglas from the US Army Cold Regions Research and Engineering Laboratory's permafrost tunnel (Alaska) for assistance and permission to sample. We also thank Frida Keuper from the French National Research Institute for Agriculture, Food and Environment and Ellen Dorrepaal from Umeå University for providing the permafrost sample.
Sequencing was performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Technology Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation.
This research has been supported by the Svenska Forskningsrådet Formas (Dnr 2017-01182 awarded to Eveline J. Krab) and Kempestiftelserna (awarded to Sylvain Monteux).
The article processing charges for this open-access publication were covered by Stockholm University.
This paper was edited by Emily Solly and reviewed by two anonymous referees.
Addison, J. A. and Parkinson, D.: Influence of Collembolan Feeding Activities on Soil Metabolism at a High Arctic Site, Oikos, 30, 529–538, https://doi.org/10.2307/3543348, 1978.
Agamennone, V., Jakupovic, D., Weedon, J. T., Suring, W. J., van Straalen, N. M., Roelofs, D., and Roling, W. F. M.: The microbiome of Folsomia candida: an assessment of bacterial diversity in a Wolbachia-containing animal, FEMS Microbiol. Ecol., 91, fiv128, https://doi.org/10.1093/femsec/fiv128, 2015.
Arbizu, P. M.: pairwiseAdonis: Pairwise multilevel comparison using adonis, R package version 0.4, Github [code], https://github.com/pmartinezarbizu/pairwiseAdonis, last access 22 August 2022.
Bakonyi, G.: Effects of Folsomia candida (Collembola) on the microbial biomass in a grassland soil, Biol. Fert. Soils, 7, 138–141, https://doi.org/10.1007/BF00292572, 1989.
Barbato, R. A., Jones, R. M., Douglas, T. A., Doherty, S. J., Messan, K., Foley, K. L., Perkins, E. J., Thurston, A. K., and Garcia-Reyero, N.: Not all permafrost microbiomes are created equal: Influence of permafrost thaw on the soil microbiome in a laboratory incubation study, Soil Biol. Biochem., 108605, https://doi.org/10.1016/j.soilbio.2022.108605, 2022.
Berman, D. I., Meshcheryakova, E. N., and Mikhaljova, E. V.: Cold Hardiness and Range of the Myriapod Angarozonium Amurense (Polyzoniidae, Diplopoda, Arthropoda) in Permafrost Environments, Cryoletters, 36, 237–242, 2015.
Blume-Werry, G., Krab, E. J., Olofsson, J., Sundqvist, M. K., Väisänen, M., and Klaminder, J.: Invasive earthworms unlock arctic plant nitrogen limitation, Nat. Commun., 11, 1766, https://doi.org/10.1038/s41467-020-15568-3, 2020.
Bottos, E. M., Kennedy, D. W., Romero, E. B., Fansler, S. J., Brown, J. M., Bramer, L. M., Chu, R. K., Tfaily, M. M., Jansson, J. K., and Stegen, J. C.: Dispersal limitation and thermodynamic constraints govern spatial structure of permafrost microbial communities, FEMS Microbiol. Ecol., 94, fiy110, https://doi.org/10.1093/femsec/fiy110, 2018.
Buse, T., Ruess, L., and Filser, J.: Collembola gut passage shapes microbial communities in faecal pellets but not viability of dietary algal cells, Chemoecology, 24, 79–84, https://doi.org/10.1007/s00049-013-0145-y, 2014.
Calderón, K., Spor, A., Breuil, M.-C., Bru, D., Bizouard, F., Violle, C., Barnard, R. L., and Philippot, L.: Effectiveness of ecological rescue for altered soil microbial communities and functions, ISME J., 11, 272–283, https://doi.org/10.1038/ismej.2016.86, 2017.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P.: DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 13, 581–583, https://doi.org/10.1038/nmeth.3869, 2016.
Coulibaly, S. F. M., Winck, B. R., Akpa-Vinceslas, M., Mignot, L., Legras, M., Forey, E., and Chauvat, M.: Functional Assemblages of Collembola Determine Soil Microbial Communities and Associated Functions, Front. Environ. Sci., 7, 52, https://doi.org/10.3389/fenvs.2019.00052, 2019.
Crowther, T. W., Boddy, L., and Hefin Jones, T.: Functional and ecological consequences of saprotrophic fungus–grazer interactions, ISME J., 6, 1992–2001, https://doi.org/10.1038/ismej.2012.53, 2012.
D'Amico, S., Collins, T., Marx, J.-C., Feller, G., Gerday, C., and Gerday, C.: Psychrophilic microorganisms: challenges for life, EMBO Rep., 7, 385–389, https://doi.org/10.1038/sj.embor.7400662, 2006.
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A., and Callahan, B. J.: Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data, Microbiome, 6, 226, https://doi.org/10.1186/s40168-018-0605-2, 2018.
Frouz, J., Novotná, K., Čermáková, L., and Pivokonský, M.: Soil fauna reduce soil respiration by supporting N leaching from litter, Appl. Soil Ecol., 153, 103585, https://doi.org/10.1016/j.apsoil.2020.103585, 2020.
García-Palacios, P., Maestre, F. T., Kattge, J., and Wall, D. H.: Climate and litter quality differently modulate the effects of soil fauna on litter decomposition across biomes, Ecol. Lett., 16, 1045–1053, https://doi.org/10.1111/ele.12137, 2013.
Golovatch, S. and Kime, R.: Millipede (Diplopoda) distributions: a review, Soil Organisms, 81, 565–597, 2009.
Griffiths, H. M., Ashton, L. A., Parr, C. L., and Eggleton, P.: The impact of invertebrate decomposers on plants and soil, New Phytol., 231, 2142–2149, https://doi.org/10.1111/nph.17553, 2021.
Hanlon, R. D. G. and Anderson, J. M.: The effects of collembola grazing on microbial activity in decomposing leaf litter, Oecologia, 38, 93–99, https://doi.org/10.1007/BF00347827, 1979.
Harding, T., Jungblut, A. D., Lovejoy, C., and Vincent, W. F.: Microbes in High Arctic Snow and Implications for the Cold Biosphere, Appl. Environ. Microb., 77, 3234–3243, https://doi.org/10.1128/AEM.02611-10, 2011.
Hensel, R., Helbig, R., Aland, S., Voigt, A., Neinhuis, C., and Werner, C.: Tunable nano-replication to explore the omniphobic characteristics of springtail skin, NPG Asia Mater., 5, e37, https://doi.org/10.1038/am.2012.66, 2013a.
Hensel, R., Helbig, R., Aland, S., Braun, H.-G., Voigt, A., Neinhuis, C., and Werner, C.: Wetting Resistance at Its Topographical Limit: The Benefit of Mushroom and Serif T Structures, Langmuir, 29, 1100–1112, https://doi.org/10.1021/la304179b, 2013b.
Hultman, J., Waldrop, M. P., Mackelprang, R., David, M. M., McFarland, J., Blazewicz, S. J., Harden, J., Turetsky, M. R., McGuire, A. D., Shah, M. B., VerBerkmoes, N. C., Lee, L. H., Mavrommatis, K., and Jansson, J. K.: Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes, Nature, 521, 208–212, https://doi.org/10.1038/nature14238, 2015.
Inglese, C. N., Christiansen, C. T., Lamhonwah, D., Moniz, K., Montross, S. N., Lamoureux, S., Lafrenière, M., Grogan, P., and Walker, V. K.: Examination of Soil Microbial Communities After Permafrost Thaw Subsequent to an Active Layer Detachment in the High Arctic, Arct. Antarct. Alp. Res., 49, 455–472, https://doi.org/10.1657/AAAR0016-066, 2017.
Johnston, E. R., Hatt, J. K., He, Z., Wu, L., Guo, X., Luo, Y., Schuur, E. A. G., Tiedje, J. M., Zhou, J., and Konstantinidis, K. T.: Responses of tundra soil microbial communities to half a decade of experimental warming at two critical depths, P. Natl. Acad. Sci. USA, 116, 15096–15105, https://doi.org/10.1073/pnas.1901307116, 2019.
Kaneda, S. and Kaneko, N.: Collembolans feeding on soil affect carbon and nitrogen mineralization by their influence on microbial and nematode activities, Biol. Fert. Soils, 44, 435–442, https://doi.org/10.1007/s00374-007-0222-x, 2008.
Knoblauch, C., Beer, C., Liebner, S., Grigoriev, M. N., and Pfeiffer, E.-M.: Methane production as key to the greenhouse gas budget of thawing permafrost, Nat. Clim. Change, 8, 309–312, https://doi.org/10.1038/s41558-018-0095-z, 2018.
Koltz, A. M., Classen, A. T., and Wright, J. P.: Warming reverses top-down effects of predators on belowground ecosystem function in Arctic tundra, P. Natl. Acad. Sci. USA, 115, E7541–E7549, https://doi.org/10.1073/pnas.1808754115, 2018.
Lenth, R.: Least-Squares Means: The R Package lsmeans, J. Stat. Softw., 69, 1–33, https://doi.org/10.18637/jss.v069.i01, 2016.
Leo, C., Nardi, F., Cucini, C., Frati, F., Convey, P., Weedon, J. T., Roelofs, D., and Carapelli, A.: Evidence for strong environmental control on bacterial microbiomes of Antarctic springtails, Sci. Rep.-UK, 11, 2973, https://doi.org/10.1038/s41598-021-82379-x, 2021.
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J., and Knight, R.: UniFrac: an effective distance metric for microbial community comparison, ISME J., 5, 169–172, https://doi.org/10.1038/ismej.2010.133, 2011.
Lubbers, I. M., Berg, M. P., De Deyn, G. B., van der Putten, W. H., and van Groenigen, J. W.: Soil fauna diversity increases CO2 but suppresses N2O emissions from soil, Glob. Change Biol., 26, 1886–1898, https://doi.org/10.1111/gcb.14860, 2020.
Lucas, J. M., McBride, S. G., and Strickland, M. S.: Trophic level mediates soil microbial community composition and function, Soil Biol. Biochem., 143, 107756, https://doi.org/10.1016/j.soilbio.2020.107756, 2020.
Mackelprang, R., Waldrop, M. P., DeAngelis, K. M., David, M. M., Chavarria, K. L., Blazewicz, S. J., Rubin, E. M., and Jansson, J. K.: Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw, Nature, 480, 368–371, https://doi.org/10.1038/nature10576, 2011.
Mackelprang, R., Burkert, A., Haw, M., Mahendrarajah, T., Conaway, C. H., Douglas, T. A., and Waldrop, M. P.: Microbial survival strategies in ancient permafrost: insights from metagenomics, ISME J., 11, 2305–2318, https://doi.org/10.1038/ismej.2017.93, 2017.
Marshall, V. G. and Kevan, D. K. McE.: Preliminary Observations on the Biology of Folsomia candida Willem, 1902 (Collembola: Isotomidae), Can. Entomol., 94, 575–586, https://doi.org/10.4039/Ent94575-6, 1962.
Martin, M.: Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet.journal, 17, 10–12, https://doi.org/10.14806/ej.17.1.200, 2011.
Marushchak, M. E., Kerttula, J., Diáková, K., Faguet, A., Gil, J., Grosse, G., Knoblauch, C., Lashchinskiy, N., Martikainen, P. J., Morgenstern, A., Nykamb, M., Ronkainen, J. G., Siljanen, H. M. P., van Delden, L., Voigt, C., Zimov, N., Zimov, S., and Biasi, C.: Thawing Yedoma permafrost is a neglected nitrous oxide source, Nat. Commun., 12, 7107, https://doi.org/10.1038/s41467-021-27386-2, 2021.
Masson-Delmotte, V., Zhai, P., Pirani, A., Connors, S. L., Péan, C., Berger, S., Caud, N., Chen, Y., Goldfarb, L., Gomis, M. I., Huang, M., Leitzell, K., Lonnoy, E., Matthews, J. B. R., Maycock, T. K., Waterfield, T., Yelekçi, Ö., Yu, R., and Zhou, B. (Eds.): Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change, Cambridge University Press, 2021.
Mitchell, M. J.: An improved method for microrespirometry using gas chromatography, Soil Biol. Biochem., 5, 271–274, https://doi.org/10.1016/0038-0717(73)90011-4, 1973.
Monteux, S., Weedon, J. T., Blume-Werry, G., Gavazov, K., Jassey, V. E. J., Johansson, M., Keuper, F., Olid, C., and Dorrepaal, E.: Long-term in situ permafrost thaw effects on bacterial communities and potential aerobic respiration, ISME J., 12, 2129–2141, https://doi.org/10.1038/s41396-018-0176-z, 2018.
Monteux, S., Keuper, F., Fontaine, S., Gavazov, K., Hallin, S., Juhanson, J., Krab, E. J., Revaillot, S., Verbruggen, E., Walz, J., Weedon, J. T., and Dorrepaal, E.: Carbon and nitrogen cycling in Yedoma permafrost controlled by microbial functional limitations, Nat. Geosci., 13, 794–798, https://doi.org/10.1038/s41561-020-00662-4, 2020.
Monteux, S.: Data analysis pipeline for bacterial community composition and CO2 fluxes in a permafrost-Collembola incubation, Software version 1.1.1, Bolin Centre Code Repository [code], https://doi.org/10.57669/monteux-2022-collembola-1.1.1, 2022.
Monteux, S., Mariën, J., and Krab, E.: Data supporting “Dispersal of bacteria and stimulation of permafrost decomposition by Collembola”, Zenodo [data set], https://doi.org/10.5281/zenodo.7015077, 2022.
Nickerl, J., Helbig, R., Schulz, H.-J., Werner, C., and Neinhuis, C.: Diversity and potential correlations to the function of Collembola cuticle structures, Zoomorphology, 132, 183–195, https://doi.org/10.1007/s00435-012-0181-0, 2013.
Oksanen, J., Simpson, G. L., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., Solymos, P., Stevens, M. H. H., Szoecs, E., Wagner, H., Barbour, M., Bedward, M., Bolker, B., Borcard, D., Carvalho, G., Chirico, M., Caceres, M. D., Durand, S., Evangelista, H. B. A., FitzJohn, R., Friendly, M., Furneaux, B., Hannigan, G., Hill, M. O., Lahti, L., McGlinn, D., Ouellette, M.-H., Cunha, E. R., Smith, T., Stier, A., Braak, C. J. F. T., and Weedon, J.: vegan: Community Ecology Package, R package [code], https://CRAN.R-project.org/package=vegan, last access: 22 August 2022.
Potapov, A., Bellini, B. C., Chown, S. L., Deharveng, L., Janssens, F., Kováč, Ĺ., Kuznetsova, N., Ponge, J.-F., Potapov, M., Querner, P., Russell, D., Sun, X., Zhang, F., and Berg, M. P.: Towards a global synthesis of Collembola knowledge: challenges and potential solutions, soil organisms, 92, 161–188, https://doi.org/10.25674/so92iss3pp161, 2020.
Potapov, A. M., Beaulieu, F., Birkhofer, K., Bluhm, S. L., Degtyarev, M. I., Devetter, M., Goncharov, A. A., Gongalsky, K. B., Klarner, B., Korobushkin, D. I., Liebke, D. F., Maraun, M., Mc Donnell, R. J., Pollierer, M. M., Schaefer, I., Shrubovych, J., Semenyuk, I. I., Sendra, A., Tuma, J., Tůmová, M., Vassilieva, A. B., Chen, T.-W., Geisen, S., Schmidt, O., Tiunov, A. V., and Scheu, S.: Feeding habits and multifunctional classification of soil-associated consumers from protists to vertebrates, Biol. Rev., 97, 1057–1117, https://doi.org/10.1111/brv.12832, 2022.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glöckner, F. O.: The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucl. Acids Res., 41, D590–D596, https://doi.org/10.1093/nar/gks1219, 2013.
R Core Team: R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/, last access: 22 August 2022.
Seastedt, T. R. and Crossley Jr., D. A.: The Influence of Arthropods on Ecosystems, BioScience, 34, 157–161, https://doi.org/10.2307/1309750, 1984.
Sfenthourakis, S. and Hornung, E.: Isopod distribution and climate change, Zookeys, 801, 25–61, https://doi.org/10.3897/zookeys.801.23533, 2018.
Shatilovich, A. V., Tchesunov, A. V., Neretina, T. V., Grabarnik, I. P., Gubin, S. V., Vishnivetskaya, T. A., Onstott, T. C., and Rivkina, E. M.: Viable Nematodes from Late Pleistocene Permafrost of the Kolyma River Lowland, Dokl. Biol. Sci., 480, 100–102, https://doi.org/10.1134/S0012496618030079, 2018.
Shmakova, L., Malavin, S., Iakovenko, N., Vishnivetskaya, T., Shain, D., Plewka, M., and Rivkina, E.: A living bdelloid rotifer from 24,000-year-old Arctic permafrost, Curr. Biol., 31, R712–R713, https://doi.org/10.1016/j.cub.2021.04.077, 2021.
Shur, Y., French, H. M., Bray, M. T., and Anderson, D. A.: Syngenetic permafrost growth: cryostratigraphic observations from the CRREL tunnel near Fairbanks, Alaska, Permafrost Periglac., 15, 339–347, https://doi.org/10.1002/ppp.486, 2004.
Sørensen, L. I., Holmstrup, M., Maraldo, K., Christensen, S., and Christensen, B.: Soil fauna communities and microbial respiration in high Arctic tundra soils at Zackenberg, Northeast Greenland, Polar Biol., 29, 189–195, https://doi.org/10.1007/s00300-005-0038-9, 2006.
Strauss, J., Schirrmeister, L., Grosse, G., Fortier, D., Hugelius, G., Knoblauch, C., Romanovsky, V., Schädel, C., Schneider von Deimling, T., Schuur, E. A. G., Shmelev, D., Ulrich, M., and Veremeeva, A.: Deep Yedoma permafrost: A synthesis of depositional characteristics and carbon vulnerability, Earth-Sci. Rev., 172, 75–86, https://doi.org/10.1016/j.earscirev.2017.07.007, 2017.
Theenhaus, A., Scheu, S., and Schaefer, M.: Contramensal interactions between two collembolan species: effects on population development and on soil processes, Funct. Ecol., 13, 238–246, https://doi.org/10.1046/j.1365-2435.1999.00303.x, 1999.
Väisänen, M., Krab, E. J., Monteux, S., Teuber, L. M., Gavazov, K., Weedon, J. T., Keuper, F., and Dorrepaal, E.: Meshes in mesocosms control solute and biota exchange in soils: A step towards disentangling (a)biotic impacts on the fate of thawing permafrost, Appl. Soil Ecol., 151, 103537, https://doi.org/10.1016/j.apsoil.2020.103537, 2020.
Wang, M., Zhang, W., Xia, H., Huang, J., Wu, Z., and Xu, G.: Effect of Collembola on mineralization of litter and soil organic matter, Biol. Fert. Soils, 53, 563–571, https://doi.org/10.1007/s00374-017-1200-6, 2017.
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R.: Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy, Appl. Environ. Microb., 73, 5261–5267, https://doi.org/10.1128/AEM.00062-07, 2007.
Wang, Y., Naumann, U., Wright, S. T., and Warton, D. I.: mvabund – an R package for model-based analysis of multivariate abundance data, Methods Ecol. Evol., 3, 471–474, https://doi.org/10.1111/j.2041-210X.2012.00190.x, 2012.
Warton, D. I., Wright, S. T., and Wang, Y.: Distance-based multivariate analyses confound location and dispersion effects, Methods Ecol. Evol., 3, 89–101, https://doi.org/10.1111/j.2041-210X.2011.00127.x, 2012.
Xiang, Q., Zhu, D., Chen, Q.-L., Delgado-Baquerizo, M., Su, J.-Q., Qiao, M., Yang, X.-R., and Zhu, Y.-G.: Effects of diet on gut microbiota of soil collembolans, Sci. Total Environ., 676, 197–205, https://doi.org/10.1016/j.scitotenv.2019.04.104, 2019.
Yashina, S., Gubin, S., Maksimovich, S., Yashina, A., Gakhova, E., and Gilichinsky, D.: Regeneration of whole fertile plants from 30,000-y-old fruit tissue buried in Siberian permafrost, P. Natl. Acad. Sci. USA, 109, 4008–4013, https://doi.org/10.1073/pnas.1118386109, 2012.
- Abstract
- Introduction
- Materials and methods
- Results
- Discussion
- Conclusions
- Appendix A
- Appendix B
- Appendix C
- Code availability
- Data availability
- Sample availability
- Author contributions
- Competing interests
- Disclaimer
- Special issue statement
- Acknowledgements
- Financial support
- Review statement
- References
- Abstract
- Introduction
- Materials and methods
- Results
- Discussion
- Conclusions
- Appendix A
- Appendix B
- Appendix C
- Code availability
- Data availability
- Sample availability
- Author contributions
- Competing interests
- Disclaimer
- Special issue statement
- Acknowledgements
- Financial support
- Review statement
- References





