the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Extracellular enzyme activity in the coastal upwelling system off Peru: a mesocosm experiment
Kristian Spilling
Jonna Piiparinen
Eric P. Achterberg
Javier Arístegui
Lennart T. Bach
Maria T. Camarena-Gómez
Elisabeth von der Esch
Martin A. Fischer
Markel Gómez-Letona
Nauzet Hernández-Hernández
Judith Meyer
Ruth A. Schmitz
Ulf Riebesell
The Peruvian upwelling system is a highly productive ecosystem with a large oxygen minimum zone (OMZ) close to the surface. In this work, we carried out a mesocosm experiment off Callao, Peru, with the addition of water masses from the regional OMZ collected at two different sites simulating two different upwelling scenarios. Here, we focus on the pelagic remineralization of organic matter by the extracellular enzyme activity of leucine aminopeptidase (LAP) and alkaline phosphatase activity (APA). After the addition of the OMZ water, dissolved inorganic nitrogen (N) was depleted, but the standing stock of phytoplankton was relatively high, even after N depletion (mostly > 4 µg chlorophyll a L−1). During the initial phase of the experiment, APA was 0.6 nmol L−1 h−1 even though the PO concentration was > 0.5 µmol L−1. Initially, the dissolved organic phosphorus (DOP) decreased, coinciding with an increase in the PO concentration that was probably linked to the APA. The LAP activity was very high, with most of the measurements in the range of 200–800 nmol L−1 h−1. This enzyme hydrolyzes terminal amino acids from larger molecules (e.g., peptides or proteins), and these high values are probably linked to the highly productive but N-limited coastal ecosystem. Moreover, the experiment took place during a rare coastal El Niño event with higher than normal surface temperatures, which could have affected enzyme activity. Using a nonparametric multidimensional scaling analysis (NMDS) with a generalized additive model (GAM), we found that biogeochemical variables (e.g., nutrient and chlorophyll-a concentrations) and phytoplankton and bacterial communities explained up to 64 % of the variability in APA. The bacterial community best explained the variability (34 %) in LAP. The high hydrolysis rates for this enzyme suggest that pelagic N remineralization, likely driven by the bacterial community, supported the high standing stock of primary producers in the mesocosms after N depletion.
- Article
(2663 KB) - Full-text XML
-
Supplement
(498 KB) - BibTeX
- EndNote
The Peruvian upwelling system is one of the most productive marine ecosystems in the world (FAO, 2018). Its high productivity is driven by the upwelling of deep, nutrient-rich water that fuels primary production upon reaching the sunlit surface. The primary limiting nutrient is nitrogen (N), but iron (Fe) availability is also an important driver for phytoplankton biomass production in addition to light (Chavez et al., 2008; Messié and Chavez, 2015). Part of the phytoplankton biomass passes to higher trophic levels through grazing and predation. As the upwelled water parcel is transported further offshore by Ekman transport, part of the biomass settles out of the euphotic zone and is decomposed in intermediate water layers, thereby creating an extensive oxygen minimum zone (OMZ; Kalvelage et al., 2013). The fate of the biomass produced is consequently of great importance for higher trophic levels and for biogeochemical cycles.
After inorganic nutrients (primarily N) have been depleted, primary production in the surface layer is driven by recycled production. In this process, dissolved organic matter (DOM) must first be broken down into simpler forms before the DOM elements become biologically available. The decomposition of DOM is not a uniform process, as it is affected by both abiotic and biotic variables. Extracellular enzymes hydrolyze complex dissolved organic molecules, and this is the first step in the remineralization of these DOM elements (Arnosti, 2011). Quantifying the rates of pelagic remineralization is important to understand the recycled production and element fluxes in the uppermost water masses. There are a range of different enzymes that are used for hydrolyzing DOM, and two of the most studied ones are leucine aminopeptidase (LAP) and alkaline phosphatase (AP).
LAP hydrolyzes terminal amino acids from larger molecules (e.g., peptides or proteins) and is used extracellularly in aquatic systems by bacteria, some phytoplankton and fungi (Hoppe et al., 1988; Stoecker and Gustafson, 2003; Gutiérrez et al., 2011). It hydrolyzes a broad spectrum of substrates with a free amino group, but it has preference for N-terminal leucine and related amino acids in peptides and proteins (Burley et al., 1990; Steen et al., 2015).
The AP enzyme is produced by a wide range of different organisms, including aquatic bacteria and phytoplankton. Its main function is related to the hydrolysis of phosphate monoesters that separate orthophosphate (PO4) from an organic compound (Perry, 1972; Hoppe, 2003). AP exists either as ectoenzyme (on the cell wall) or is excreted extracellularly; for phytoplankton, AP has commonly been related to P limitation in aquatic environments (Rose and Axler, 1997; Nausch, 1998). Bacterial AP activity (APA) is more complex, as some (especially particle-attached) bacteria take up and use C and N from the organic molecule after hydrolysis and may, for this reason, produce AP even under P-replete conditions (Benitez-Nelson and Buesseler, 1999; Hoppe, 2003; Labry et al., 2016).
The ongoing warming of surface waters caused by climate change is projected to have several consequences for marine ecosystems. For example, increasing temperatures lead to a reduction in gas solubility, thereby causing a decrease in oxygen concentrations; warming will also increase thermal stratification and reduce the ventilation of the deeper ocean (Keeling et al., 2010). Both of these effects will lead to expanding OMZs with potential consequences for biogeochemical cycling (Oschlies et al., 2018). Biogeochemical cycles of nitrogen (N) and phosphorus (P) are affected by O2 depletion, e.g., through denitrification and sediment P release (Canfield et al., 2005). Hence, expanding OMZs may decrease the inorganic N : P ratio in the upwelled water, potentially affecting the seston (i.e., all suspended particles) stoichiometry and plankton community composition (Hauss et al., 2012; Spilling et al., 2019).
In this study, a mesocosm experiment off the coast of Peru was carried out to study the effect of OMZ water on the surface community of plankton, with several papers covering different aspects in this special issue. Here, we were interested in the dynamics of organic matter breakdown. We measured the extracellular LAP and AP activities and used a statistical model to relate them to biogeochemical variables and to plankton and bacterioplankton communities. Our main aim was to understand how much of the variability in enzyme activities could be explained by biogeochemical variables (e.g., nutrient concentrations) and microbial communities.
A detailed description of the mesocosm setup as well as the collection and addition of OMZ water can be found in Bach et al. (2020) within this special issue. Some of the basic variables, such as the inorganic nutrient concentration, can also be found in Bach et al. (2020). In short, the mesocosm bags were 2 m in diameter and extended from the surface down to 19 m depth; the last 2 m of the bags comprised a conical sediment trap. Eight mesocosm bags were used, and they were moored at 12.0555∘ S, 77.2348∘ W, just north of San Lorenzo Island, where the water depth is ∼ 30 m. The mesocosms were closed by attaching the sediment trap to the bottom and pulling the top above the surface on 25 February 2017. The bags were regularly cleaned from the inside and outside. For a full detailed sampling and cleaning timetable, the reader is referred to Bach et al. (2020).
Water (100 m3) from the oxygen minimum zone (OMZ) was collected from two locations and depths. The first was collected from 12.028323∘ S, 77.223603∘ W at 30 m depth, and the second was collected from 12.044333∘ S, 77.377583∘ W at 70 m depth. The original aim was to collect severe- and moderate-OMZ-signature water (differing with respect to, e.g., nitrate concentrations) from the first and second site, respectively. This assumption was based on long-term monitoring data; however, the chemical properties (e.g., nitrate concentration) were more similar in these water masses than anticipated, rather reflecting low and very low OMZ signatures from site 1 and 2, respectively. This was discovered only after the collection, and it was not technically possible to carry out additional collections of OMZ water. For this reason, the data presented here focus on the temporal trend more than the difference between the two treatments; however, for easier comparisons with the other papers in this special issue, we maintain the same graphical interface.
To have a baseline of measured variables, the mesocosms where closed, and environmental and biological variables were determined over 10 d. After this period, the OMZ water was added to the mesocosms in two steps on day 11 and 12 after the closure of the mesocosms. As the mesocosms contain a specific volume (∼ 54 m3), the process of adding the OMZ water started with first removing water from the mesocosms. The water removed (∼ 20 m3) was pumped out from 11–12 m depth. A similar volume of OMZ water from both collection sites was then pumped into four replicate mesocosms each. The OMZ water was pumped into the mesocosms moving the input hose between 14 and 17 m depth. The (low-OMZ-signature) water collected at 30 m depth was pumped into mesocosms M1, M4, M5 and M8, and the (very low-OMZ-signature) water from 70 m depth was pumped into mesocosms M2, M3, M6 and M7. Due to the halocline at 12 m depth (see below), the added OMZ water was not immediately mixed throughout the mesocosm bag.
At the site of the mesocosms, the OMZ is normally close to the surface (< 10 m depth; Graco et al., 2017); consequently, the bottom part of the mesocosm was low in oxygen. To maintain the stratification inside the mesocosm, we added 69 L of concentrated brine on day 13 by carefully inserting it between 12.5 and 17 m depth. The same procedure was repeated on day 33 when 33 L of brine was added. This artificial halocline prevented complete mixing of the mesocosm, and the lower part of the mesocosm had a very different water chemistry compared with the upper 10 m where we did all of our sampling. Right after the experiment, a third addition of brine was carried out to measure the total volume of the mesocosms.
Sampling took place every second day over a period of 50 d; all variables were measured with an integrated water sampler (IWS, Hydro-Bios) preprogrammed to fill from 0 to 10 m depth, and thus all samples were taken from integrated water collected from the upper 10 m. The samples were stored in cool boxes in the dark and were brought back to the laboratory and processed right away. Sampling took place in the morning, and the samples were usually back in the laboratory by around noon.
2.1 Nutrient concentrations
Inorganic nutrients were determined from filtered (0.45 µm filter, Sterivex, Merck) samples immediately after the water arrived at the laboratory. For the measurements, we used a continuous-flow analyzer (QuAAtro AutoAnalyzer, SEAL Analytical) connected to a fluorescence detector (FP-2020, JASCO). Phosphate (PO), nitrate (NO) and nitrite (NO) were determined colorimetrically (Murphy and Riley, 1962; Morris and Riley, 1963) and corrected with the refractive index method reported by Coverly et al. (2012). Ammonium (NH) concentrations were determined fluorometrically (Kérouel and Aminot, 1997). Dissolved inorganic nitrogen (DIN) was calculated by summing NO, NO and NH. Further details on measurement accuracy can be found in Bach et al. (2020), where the individual DIN elements are also presented.
To measure total dissolved nitrogen (TDN) and total dissolved phosphorus (TDP), the samples were first filtered through pre-combusted (5 h at 450 ∘C) Whatman GF/F filters (pore size 0.7 µm). The filtrate was collected in 50 mL acid-cleaned high-density polyethylene (HDPE) bottles and placed directly into a freezer (−20 ∘C). Later, the filtrates were thawed at room temperature over a period of 24 h and divided in two parts. The first half was used to determine inorganic nutrient concentrations, as described above. From the other half, we determined the TDN and TDP concentrations. An oxidizing reagent (Oxisolv, Merck) was added, and the samples were autoclaved for 30 min. TDN and TDP were measured spectrophotometrically (QuAAtro, SEAL Analytical). Dissolved organic nitrogen (DON) concentrations were calculated by subtracting DIN from TDN. Dissolved organic phosphorus (DOP) was calculated as the difference between TDP and PO.
2.2 Fluorescent dissolved organic matter and parallel factor (PARAFAC) analysis
Fluorescent dissolved organic matter (FDOM) was determined by measuring fluorescence in water samples with a Cary Eclipse (Agilent Technologies) spectrofluorometer, using excitation and emission slit widths of 10 nm. Wavelength ranges were set to 230–456 nm for excitation with 2 nm increments and to 290–600 nm for emission with 5 nm increments. The measurements were collected into excitation–emission matrices (EEMs). Blanks were measured with the same settings using ultrapure water.
Raw measurements were processed using the DOMFluor toolbox (v. 1.7; Stedmon and Bro, 2008) for MATLAB (R2017a). The processing consisted of (1) blank subtraction from seawater EEMs; (2) EEM normalization to the Raman area (RA), estimated by applying the trapezoidal rule of integration on the emission scan at the 350 nm excitation wavelength in the blank EEMs; and (3) cropping of the first- and second-order Rayleigh scatter bands. Inner-filter correction was not performed because, for the duration of the experiment, the absorption coefficient at 250 nm (a250) displayed values (mean ± SD = 1.56 ± 0.91 m−1) well below 10 m−1, above which correction is considered necessary (Stedmon and Bro, 2008).
The processed EEMs were analyzed by applying PARAFAC analysis using the DOMFluor toolbox. The PARAFAC model was constructed based on 125 samples (outliers were removed) and validated using split-half validation and random initialization. The resulting model consisted of four components (C1–C4; see Fig. S1 in the Supplement). For each component, the fluorescence maximum (Fmax) was recorded. The identified fluorophores were compared to others found in the literature using the OpenFluor database (http://openfluor.lablicate.com, last access: 5 April 2023; Murphy et al., 2014).
2.3 Phytoplankton community and chlorophyll a
Flow cytometry subsamples were transferred from the IWS into 50 mL beakers and stored cool in the dark until analysis at a maximum of 8 h after sampling. Each sample (650 µL) was analyzed with an Accuri C6 flow cytometer (BD Biosciences) set to a high flow rate (i.e., 66 µL min−1). Phytoplankton groups were differentiated based on the strength of the forward scatter (FSC-A), the side scatter (SSC-A), and the red fluorescence (FL3-A) and orange fluorescence (FL2-A) signal (where “A” refers to the area of the signal integral). Furthermore, we used sequential filtrations with different polycarbonate filters (Whatman, pore sizes of 0.2, 0.4, 0.8, 2, 3, 5 and 8 µm) to distinguish populations in the cytogram based on size. This procedure was helpful to approximate how FSC-A values corresponded to size. We defined the following phytoplankton groups: “Synechococcus-like” cells (Syn; 0.2–2 µm), “Cryptophyte-like” cells (Crypto; ∼ 90 % between 2 and 5 µm), “Picoeukaryotes” (Peuks; 0.2–2 µm), “Nanoeukaryotes” (Nano; 2–20 µm, mostly in the lower range), “Microeukaryotes 1” (Mikro1; ∼ 15–40 µm, occasionally overlapping with Nano), “Microeukaryotes 2” (Mikro2; ∼ > 40 µm, cluster dominated by Akashiwo sanguinea from about day 20 onward), and elongated cells “chains” determined by the ratio of FSC-A to FSC-H, where “H” refers to the height of the forward scatter signal (details about this approach are provided in Paul et al. (2022), this issue, and the goal of this was to detect chain-forming diatoms, which we expected to be an important component of the community).
Samples for chlorophyll-a (Chl-a) determination were filtered onto GF/F filters (Whatman), flash-frozen in liquid nitrogen, and stored at −80∘ (or using dry ice for a brief period during air transfer; ∼ 2 d) until measurement. The Chl a was extracted in acetone, and the concentration was measured using high-performance liquid chromatography calibrated against commercial standards (Barlow et al., 1997). The Chl-a autofluorescence of the phytoplankton community was measured with a handheld fluorometer (AquaPen, Photon Systems Instruments) using 450 nm excitation light. The photochemical efficiency was calculated based on the relationship between the variable and maximal fluorescence ().
2.4 16S-rRNA-gene-based bacterial community determination
The 1 L of surface water obtained from the individual sampling sites was filtered through sterile Millipore Express PLUS membrane filters (polyethersulfone) with a cutoff of 0.22 µm and a diameter of 47 mm (Merck Millipore). After filtration, the filters were flash-frozen in liquid nitrogen and stored at −80 ∘C until nucleic acid extraction. Nucleic acid extraction was performed using the NucleoSpin TriPrep kit (MACHERY-NAGLE) according to manufacturer's instructions but with an additional step at the beginning of the extraction using a pestle to homogenize the sample.
Primers applied for the amplification of the bacterial 16S rRNA gene fragments were annealed to the variable region 1 and 2 and consisted of an initial standardized Illumina adapter (roman text), followed by an eight-nucleotide barcode (X's), a linker region (italic) and a primer sequence (bold). The sequences were for the forward primer Bac27, 5′-AATGATACGGCGACCACCG AGATCTACACXXXXXXXX TATGGTAATTGT AGAGTTTGATCCTGGCTCAG-3′, and the reverse primer Bac338, 5′-CAAGCAGAAGACGGCATACGAGATXXXXXXXX AGTCAGTCAGCC TGCTGCCTCCCGTAGGAGT-3′. The individual PCR reaction contained 100 ng of the extracted DNA. The PCR conditions and purification of the amplification product have been previously described (Fischer et al., 2019a). The final library pool for sequencing was combined from the eluates and contained 100 ng of DNA. Amplicon library sequencing was performed on a MiSeq instrument. Therefore, the library was prepared according to the manufacturer's instructions and was sequenced using v3 chemistry with a 2×300 bp paired end.
Reads generated with amplicon sequencing were trimmed using Trimmomatic software, version 0.33 (Bolger et al., 2014), as described in Fischer et al. (2019b). Briefly, reads were analyzed with a sliding window of 4 bp, and regions were trimmed if the average Phred score (Ewing and Green, 1998; Ewing et al., 1998) within the window was below 30. Trimmed reads were kept within the dataset if the forward and reverse read both survived the quality trimming and were longer than 36 bp. Afterwards, 20 000 reads per sample were kept in the dataset (exceptions were sample M1 on day 10, 5817 reads, and M7 on day 24, 17 660 reads) for further analysis.
Quality-trimmed sequences were analyzed using mothur software, version 1.35.1 (Schloss et al., 2009), as described in Fischer et al. (2019a). The quality-filtered and subsampled reads were concatenated to 1 040 321 contiguous sequences (contigs) using the “make.contig” command. Contigs were filtered for ambiguous bases, homopolymers longer than 8 bases, or sequences longer than 552 bases using the “screen.seqs” command. The resulting 754 310 contigs were checked for redundant sequences using the “unique.seqs” command and clustered to 199 746 unique sequences. The sequences were consecutively aligned to a modified version of the SILVA database, release version 132 (Pruesse et al., 2012), containing only the hypervariable regions V1 and V2 by the “align.seqs” command. Sequences not aligning in the expected region were removed from the dataset using the screen.seqs command. The alignment was further optimized by removing gap-only columns with the “filter.seqs” command. The alignment contained 717 217 sequences (148 760 unique). Rare and closely related sequences were clustered using the unique.seqs and “precluster.seqs” commands. The latter was used to cluster sequences with up to three positional differences, compared with larger sequence clusters, together. Chimeric sequences were removed using the implemented software UCHIME (Edgar et al., 2011) with the “chimera.uchime” command followed by the “remove.seqs” command, leaving 551 142 sequences (29 519 unique) in the dataset. The classification of the sequences was performed against the SILVA database and was done with a bootstrap threshold of 80 %. Operational taxonomic units (OTUs) were formed using the average neighbor clustering method with the “cluster.split” command. A sample-by-OTU table on the 97 % level, containing 10 258 OTUs, was generated using the “ make.shared” command. These OTUs were used for the subsequent analysis. After the removal of mitochondria, chloroplast and singletons, 3225 OTUs were retained. These OTUs were used for downstream analysis.
2.5 Extracellular enzymes
The leucine aminopeptidase (LAP) activity was determined using the method described by Stoecker and Gustafson (2003) in which L-leucine 7-amino-4-methyl-coumarin (Leu-AMC; Sigma Aldrich) is utilized as a substrate. Leu-AMC was added to a final concentration of 500 µmol L−1, which was determined in separate kinetics tests to saturate the enzyme activity. The samples (100–200 µL) were incubated in the dark at in situ surface temperature for 4–6 h. The fluorescence was measured every 30–60 min with a Cary Eclipse (Agilent Technologies) spectrofluorometer using 380 nm excitation and 440 nm emission wavelengths. The results were compared to a standard curve determined using 7-amino-4-methyl-coumarin (AMC; Sigma Aldrich) dissolved in dimethyl sulfoxide (DMSO), and the LAP activity was calculated by linear regression.
Measurements of alkaline phosphatase activity (APA) were conducted with 20 mL subsamples of initial/incubated seawater using 100 nmol L−1 4-methylumbelliferyl phosphate (MUF-P; Sigma-Aldrich) as the organic phosphate substrate (Ammerman, 1993). From this incubation, samples were transferred into a well plate, and fluorescence was measured on a BioTek microplate reader with a Cary Eclipse (Agilent Technologies) spectrofluorometer using 355 nm excitation light and 460 nm emission detection. Following MUF-P addition, fluorescence measurements were performed at 0, 1.5 and 3 h, and APA (h−1) was calculated from the linear increase in fluorescence and calibrated against 4-methylumbelliferone (MUF; Sigma-Aldrich). The assays were performed and incubated in the dark. Ultrapure water (Milli-Q) blanks and paraformaldehyde-killed controls generally yielded fluorescence values similar to t=0 readings.
2.6 Statistical analysis
Before the comparisons of enzyme activity between the two experimental treatments (OMZ water added from two different locations) were conducted, we first constructed a cumulative value where each measured value was summed up for each sampling day. The linear regressions of the cumulative enzyme activity from the two treatments (n=4) were compared using a Student's t test. In addition, the effect of the biogeochemical variables and the phytoplankton and bacterioplankton community composition on the APA and LAP was determined, using the ordination scores of the first and second axis of a nonparametric multidimensional scaling (NMDS) as explanatory variables in generalized additive models (GAMs) with APA or LAP as the dependent variable. NMDS was applied separately to each group of variables: biogeochemical, phytoplankton community and bacterioplankton community. The individual explanatory power of each multidimensional scaling (MDS) score was estimated with a univariate GAM. The visualization of the links was done for each explanatory variable via the prediction of the full model object, setting all other explanatory variables at their mean value. In addition, links to the scores of the biogeochemical variables and phytoplankton community NMDS were estimated with one GAM model. It was not possible to include the bacterioplankton community into this model due to the different sampling regime (lower number of samples); therefore, this community was treated with a second model. NMDS was estimated with the metaMDS function in the vegan package (Oksanen et al., 2022), and GAMs were fitted using the gam function in the mgcv package (Wood, 2017). To explain the deviance, an adjusted coefficient of determination (R2) was used. An adjusted R2 takes the model complexity into account and is more conservative than a non-adjusted R2.
3.1 Nutrients
Inorganic nutrients, dissolved inorganic nitrogen (DIN) and phosphate, were available for the 2 first weeks of the experiment (Fig. 1). The addition of OMZ water increased the phosphate concentrations, whereas the dissolved inorganic nitrogen (DIN) was > 2 µmol L−1 in the mesocosms until the addition of OMZ water (days 11 and 12 of the experiment). After the addition of the OMZ water, the DIN concentration rapidly declined and was depleted at day 15 in most mesocosms except in M3, in which DIN depletion occurred a week later (day 22; Fig. 1). The PO concentration increased after closing the mesocosm and reached ∼ 1.9 µmol L−1 in all mesocosms after OMZ-water addition. There was only a slight reduction to approximately 1.5 µmol PO L−1 over the course of the experiment (Fig. 1).
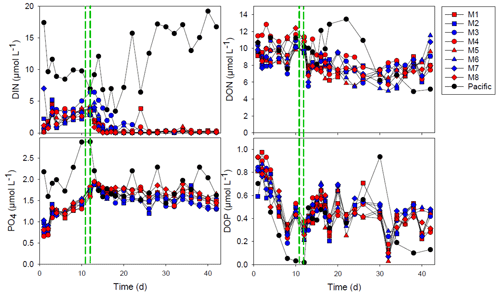
Figure 1The concentration of dissolved inorganic nitrogen (DIN), phosphate (PO), dissolved organic nitrogen (DON) and dissolved organic phosphorus (DOP). The red and blue colors denote the mesocosm bags with the addition of water with a low (closer to shore) and very low (further offshore) oxygen minimum zone (OMZ) signature, respectively. The green dashed lines denote the time of OMZ water addition. Pacific denotes measurements from water collected next to, but outside of, the mesocosms.
The dissolved organic nitrogen (DON) and dissolved organic phosphorus (DOP) concentrations were initially 9–12 and 0.6–1.0 µmol L−1, respectively. There was no drastic change in DON with OMZ-water addition, and there was an overall decrease in DON to 6.0–7.9 µmol L−1 on day 30 after which it increased somewhat again. The DOP concentrations decreased rapidly during the first 8 d to 0.19–0.32 µmol L−1 but increased after OMZ-water addition and remained within the 0.2–0.7 µmol L−1 interval for the rest of the experiment.
The PARAFAC modeling of the EEMs yielded four FDOM components (C1–C4; Figs. 2 and S1). Using the OpenFluor database, we identified multiple fluorophores with strong similarity (TCCex⋅em > 0.95) to our components (Table S1). Components 1 and 3 had characteristics resembling amino-acid-like/protein-like fluorescence, whereas the fluorescence of components 2 and 4 was humic-like (Table S1). All FDOM components increased sharply at day 18. This did not take place in Pacific seawater sampled outside of the mesocosm where the FDOM was relatively stable throughout the experiment. After the increase at day 18, humic-like components (C2 and C4) were relatively stable but decreased slightly after day 28–30. The amino-acid-like components (C1 and C3) exhibited higher variability among mesocosms, and C3 had overall higher variability throughout the experiment. Both humic-like and amino-acid-like components maintained fluorescence values above the initial values until the end of the experiment, but there were no clear differences between the treatments. However, towards the end of the experiment, M1 and M2 had highest fluorescence values of C1. M1 also had highest values of C2 and C3, whereas M3 had the highest values of C4 at the end of the experiment.
3.2 Chlorophyll, photochemical efficiency and phytoplankton community
After OMZ-water addition, the Chl-a concentration increased from 2–4 to 4–8 µg L−1 except for mesocosms M3 and M4 where the increase was not as pronounced (Fig. 3). The Chl-a concentration in M3 increased after day 22 to ∼ 4 µg Chl a L−1, whereas the Chl-a concentration remained low (< 2 µg L−1) in M4 throughout most of the experiment (Fig. 3). The photochemical efficiency () was approximately 0.7 throughout the whole experiment with no major difference between mesocosms, except for M4 where it was consistently lower (< 0.6) during the last week of experiment (Fig. 3).
The initial community was dominated by diatoms in terms of biomass, but this group gradually reduced in number after the enclosure of the mesocosms and the mixotrophic dinoflagellate Akashiwo sanguinea instead appeared (Fig. 4). The cell counts done with the flow cytometer were checked with a microscope, and this was the primary species in terms of biomass in the Microeukaryotes 2 group (Fig. 4). The exceptions were mesocosms M3 and M4, where this dinoflagellate was not abundant (M4) or bloomed later (M3) and where there were more chrysophytes. In M4, there was also a bloom of picoeukaryotes starting after day 20 (Fig. 4). The parallels of the same treatment did not develop in the same way in all of the mesocosms, and this was particularly evident from the phytoplankton community composition (Fig. 4).
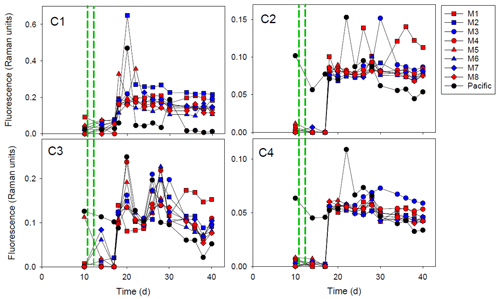
Figure 2The fluorescent dissolved organic matter (FDOM) components (C1–C4) during the experiment. The red and blue colors denote the mesocosm bags with the addition of water with a low (closer to shore) and very low (further offshore) oxygen minimum zone (OMZ) signature, respectively. The green dashed lines denote the time of OMZ-water addition. Pacific denotes measurements from water collected next to, but outside of, the mesocosms.
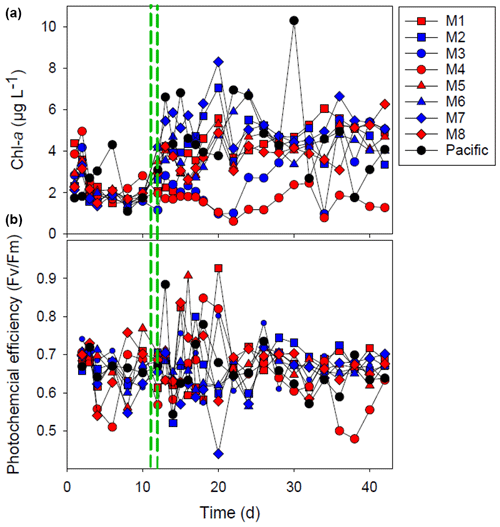
Figure 3The chlorophyll-a (Chl-a) concentration (a) and the photochemical efficiency (b). The red and blue colors denote the mesocosm bags with the addition of water with a low (closer to shore) and very low (further offshore) oxygen minimum zone (OMZ) signature, respectively. The green dashed lines denote the time of OMZ-water addition. Pacific denotes measurements from water collected next to, but outside of, the mesocosms.
3.3 Bacterial community
The bacterial community was dominated by the class Alphaproteobacteria throughout the whole experiment and in all of the mesocosms units, reaching values between 60 % and 88 % of the total sequences at day 16 (Fig. 5). Within Alphaproteobacteria, the Roseobacter lineage (genera HIMB11, Ascidiaceihabitans, Amylibacter and Planktomarina in M1) of the order Rhodobacterales contributed most to the bacterial community in all of the mesocosms (10 %–55 %), in particular on day 16, except in M8 where the SAR11 Ia clade dominated the community (55 % of the total sequence at day 16). The order Parvibaculales had high relative abundances (12 %–20 % of the total sequences) in M4, M5, M6 and M7 before OMZ-water addition (day 10), decreasing in the following week. The relative abundance of order Rickettsiales peaked at day 16 in all of the mesocosms except in M8, decreasing after 1 week. The class Gammaproteobacteria comprised between 20 % and 45 % of the total relative abundance. Within Gammaproteobacteria, the order Thiomicrospirales had a high relative abundance (8 %–17 % total sequences) at day 10 in most of the mesocosms, whereas the order Cellvibrionales and order Oceanospirillales (genus Pseudohongiella) increased from day 24 and by the end of the experiment, respectively. In M8, the abundances of orders Thiomicrospirales and Pseudomonadales (14 % of total sequences) increased at day 24. Other groups that increased in abundance in the second half of the experiment were the deltaproteobacterial orders Desulfobacterales (7 %–20 % in M2, M3, M4 and M5) and Bdellovibrionales (5 %–8 % in M2, M3 and M4). The order Flavobacteriales dominated within Bacteroidetes, and the relative abundance ranged from 1 % to 25 % throughout the experiment, being generally high (10 %–20 %) at day 10 . The flavobacterial genus Aurantivirga contributed > 7 % in M1, M2 and M3.
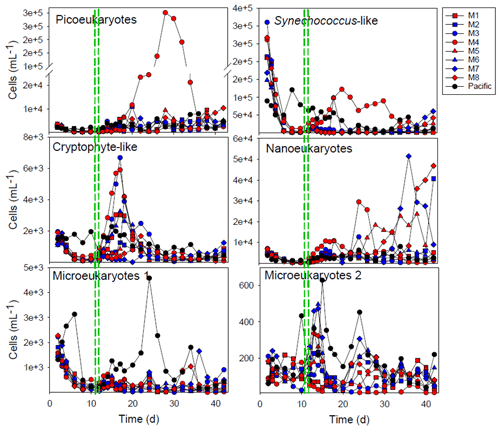
Figure 4Development of the main groups of phytoplankton enumerated by flow cytometry. The red and blue colors denote the mesocosm bags with the addition of water with a low (closer to shore) and very low (further offshore) oxygen minimum zone (OMZ) signature, respectively. The green dashed lines denote the time of OMZ-water addition. Pacific denotes measurements from water collected next to, but outside of, the mesocosms.
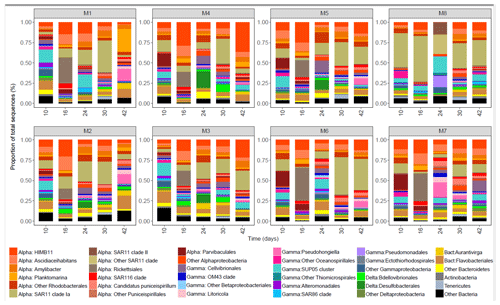
Figure 5The bacterial community composition in the eight mesocosms taken at different time points. The upper row shows mesocosms with water from a low OMZ signature (30 m depth), and the second row shows those with a very low OMZ signature (70 m depth). The y axis indicates the relative abundance of the bacterial taxa. Only the groups that contributed more than 0.5 % of the total sequences are included, and the rest are grouped as “Other Bacteria”. The classification was performed mainly at the class, order and genus levels. The abbreviations indicate the main class levels: Alphaproteobacteria (orange shades), Gammaproteobacteria (blue–pink shades), Deltaproteobacteria (green shades) and Bacteroidia (yellow shades).
3.4 Enzyme activity
The initial LAP activity before OMZ-water addition was relatively low (mean ± SD of 359 ± 81 nmol L−1 h−1), but it increased after the addition of OMZ water in some of the mesocosms (Fig. 6). In M3, the LAP activity was high, reaching 1600 nmol L−1 h−1 directly after OMZ-water addition, but it decreased following OMZ-water addition. The highest overall LAP activity throughout the experiment was in M7: the LAP activity was 716 nmol L−1 h−1 after OMZ-water addition, and the average after day 16 was 657 ± 142 nmol L−1 h−1. There was a slight difference between the treatments with respect to the LAP activity after the addition of the OMZ water until day 16, with the very low-OMZ-signature (lowest NO3 concentration) water producing the highest LAP activity (Student's t test, p=0.047), but this difference disappeared after day 16 (p=0.44).
The alkaline phosphatase activity (APA) was 0.5–0.6 nmol L−1 h−1 at the beginning of the experiment, but it decreased to undetectable levels after day 30 (Fig. 7). There was a noticeable drop in APA after the addition of the OMZ water, and the decrease continued gradually until day 28, after which the APA was very low (< 0.1 nmol L−1 d−1). The APA was similar in all of the mesocosms, and there was no treatment effect (p=0.81). The exception to this was M3: the APA was lower in M3 than in all of the other mesocosms for most of the experiment (Fig. 7).
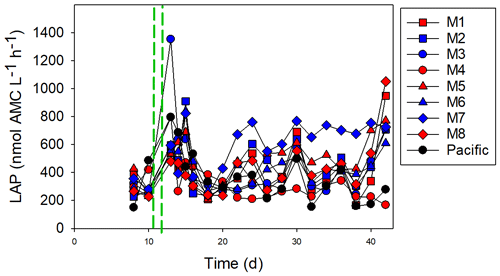
Figure 6The leucine aminopeptidase (LAP) activity. The red and blue colors denote the mesocosm bags with the addition of water with a low (closer to shore) and very low (further offshore) oxygen minimum zone (OMZ) signature, respectively. The green dashed lines denote the time of OMZ-water addition. Pacific denotes measurements from water collected next to, but outside of, the mesocosms.
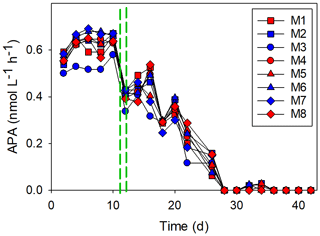
Figure 7The alkaline phosphatase activity (APA). The red and blue colors denote the mesocosm bags with the addition of water with a low (closer to shore) and very low (further offshore) oxygen minimum zone (OMZ) signature, respectively. The green dashed lines denote the time of OMZ-water addition.
The variability in the APA was better explained by the measured variables than LAP (Fig. 8). The biogeochemical variables and bacterioplankton community separately explained 62 % of the variability in the APA, whereas the phytoplankton community alone explained 57 % of the variability. Combining both the biogeochemical variables and the phytoplankton community increased the explanatory power to 74 % (the bacterioplankton community was not included, as the number of sample points was lower). The variability in LAP activity was best explained by the bacterioplankton community (38 %), followed by biogeochemical variables (20 %) and the phytoplankton community (18 %). The combined biochemical variables and phytoplankton community explained 28 % of the LAP variability.
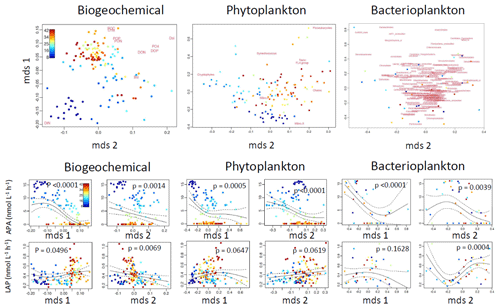
Figure 8Nonparametric multidimensional scaling (NMDS) plots for the biochemical variables and phytoplankton and bacterioplankton communities (upper row). From the NMDS scores, generalized additive models (GAMs) were made (lower two rows); for these models, we used alkaline phosphatase activity (APA) and leucine aminopeptidase (LAP) as the dependent variables. The output scores (mds1 and mds2) of the NMDS are depicted in the lower two rows.
After mesocosm closure and the addition of OMZ water, there was rapid phytoplankton growth in the upper 5 m of the mesocosms, with low-light conditions limiting primary production deeper down (Bach et al., 2020). The DIN concentrations were depleted by around day 18, coinciding with an increase in several of the FDOM components (both amino-acid-like and humic-like components), also matching the end of the phytoplankton bloom. There was, however, relatively constant and low export of carbon out of the mesocosms (Bach et al., 2020) and, at the same time, a relatively high Chl-a concentration (mostly > 4 µg Chl a L−1) under conditions with depleted DIN (Fig. 3). In addition, the photochemical efficiency was relatively high overall (> 0.5) throughout the experiment, suggesting regenerated primary production driven by recycling of nutrients. The measured hydrolysis rates, particularly LAP, indicated that extracellular enzyme activity plays an important role in this recycled production.
The main aim of this study was to relate the biogeochemical variables and microbial community to the extracellular enzyme activity. A more detailed description of the temporal development and biomass comparison of microbial groups will be presented elsewhere in this special issue (e.g., Bach et al., 2020; Schulz et al., 2021; Chen et al., 2022; Paul et al., 2022). Among phytoplankton, diatoms typically dominate following upwelling events (Anabalón et al., 2016), whereas dinoflagellates tend to become more dominant after the establishment of stratification (Margalef et al., 1979). This was also seen in our mesocosm, as the dinoflagellate Akashiwo sanguinea, a mixotrophic species that may form red tides (Jeong et al., 2005; Badylak et al., 2014), quickly appeared in some mesocosms with some exceptions. In M3, it appeared a little later than in most mesocosms, whereas it did not bloom at all in M4. Interestingly, these two mesocosms had a higher concentration of cryophytes, and M4 also had a bloom event of picoeukaryotes. Being mixotrophic, A. sanguinea is known to prey on smaller species (Jeong et al., 2005), and lower grazing pressure could be the reason for the bloom of picoeukaryotes in M4.
The bacterial community composition changed during the experiment but without clear treatment effects. The dominant bacterial groups were the class Alphaproteobacteria (Parvibaculales, SAR11 subclade Ia, Roseobacter clade and Rickettsiales), class Gammaproteobacteria (SAR116 clade, Cellvibrionales, Oceanospirillales and SUP05 clade), and (to lesser extent) the class Deltaproteobacteria (Desulfobacterales) and class Bacteroidia (order Flavobacteriales). SAR11 subclade Ia, Roseobacter clade, SAR116 clade, SUP05 clade and Desulfobacterales are known to utilize inorganic and organic sulfur components, such as hydrogen sulfide (H2S), sulfate (SO4) and dimethylsulfoniopropionate (DMSP) metabolites, for their metabolic requirements (Nowinski et al., 2019) and are coupled with the nitrogen cycle (Schunck et al., 2013). Specifically, the sulfur-oxidizing SUP05 oxidizes H2S coupled with nitrate reduction and potentially produces nitrite (Shah et al., 2017), whereas Desulfobacterales play an important role in N2 fixation (Gier et al., 2016). These bacterial taxa associated with the sulfur cycle are typically found in the OMZ regions (Pajares et al., 2020). We observed a temporal shift in the bacterial community through the experiment between sulfur-oxidizing (SUP05) and sulfate-reducing (Desulfobacterales) bacteria, probably linked to the nitrate availability, i.e., more DIN at closure of the mesocosms and, thus, a higher relative abundance of SUP05. We also observed a shift within the phytoplankton-associated bacteria (Roseobacter lineage, Gammaproteobacteria and Flavobacteriales) that likely responded to the availability of DOM during the experiment (Buchan et al., 2014; Chafee et al., 2017). The high relative abundance of Flavobacteriales and genera from the Roseobacter lineage on days 10 and 16, respectively, coincided with the increase in Chl a and high LAP activity until day 16. Positive correlations have been observed between Chl a, Bacteroides and Deltaproteobacteria, and LAP during phytoplankton blooms (Shi et al., 2019). However, we do not have gene expression data and cannot make any firm conclusion about the connection between these groups and the production of LAP.
The temporal shift in the bacterial community indicates niche partitioning between bacterial taxa that assimilate different organic substrates or inorganic sulfur components, produced during phytoplankton bloom events or from sulfidic events (Schunck et al., 2013; Callbeck et al., 2018; Nowinski et al., 2019). Our results support previous studies that have demonstrated the important role of the sulfur cycle in shaping the bacterial community composition in poorly oxygenated waters (Schunck et al., 2013; Aldunate et al., 2018). It is worth noting that the conditions at the bottom of the mesocosms were suboxic, and there might have been a clear depth gradient in the bacterial community that was not picked up by our integrated 0–10 m sampling method.
Overall, there was a treatment effect of the different OMZ waters on the LAP activity, with higher LAP in the very low-OMZ-signature addition, but this effect was only observed right after the addition of the OMZ water. There were also slightly higher NO3 concentrations in this water (Bach et al., 2020). However, this difference in both DIN and LAP was relatively small and disappeared a week after the OMZ-water addition, most likely because the collected OMZ water was more similar between the two locations than anticipated, with relatively similar concentrations of DIN. Although there were differences between individual mesocosms in terms of the plankton community structure, there were no clear differences between treatments, and we can conclude that the availability of nutrients alone can shift LAP production.
The LAP activity in our study was very high (∼ 10 times higher than most literature data). In a comparable study but further offshore in Peru, the LAP activity was 20–65 nmol L−1 h−1 in surface waters (Maßmig et al., 2020). Further to the south, in Chile (30∘30.80′ S), values up to 230 nmol L−1 h−1 have been recorded, with a clear seasonal cycle linked to upwelling events (Gutiérrez et al., 2011). With most of our data ranging between 200 and 800 nmol L−1 h−1, it is clear that these LAP activities are linked to upwelling, which is more intense near the coast and also more constant at the study site compared with sites further south. The enzyme activity in sediments can be up to 3 orders of magnitude higher than what we found (Hoppe et al., 2002), and values an order of magnitude higher have been observed in a eutrophic saltwater lake (Song et al., 2019). The high LAP activities are likely a reflection of the high microbial activity in the Peruvian upwelling system. The experiment also took place during a rare coastal El Niño event (Garreaud, 2018) with anomalous higher surface temperatures (20–22 ∘C), which could be a reason for the high values that we recorded, as LAP activity is known to increase with temperature (Christian and Karl, 1995).
There was also some loss of N due to denitrification, estimated to be 0.2–4.2 nmol N2 L−1 h−1, during the experiment (Schulz et al., 2021). For comparison, the LAP activity suggested an average of 417 nmol L−1 h−1 hydrolyzation of N-containing compounds, but this should be seen as the maximal potential rather than the actual rate. The use of fluorescently labeled substrates for measuring extracellular activity is a proxy method that has some drawbacks. The primary drawback is that the molecular structure of the substrate used is never equivalent to the high-molecular-weight DOM in the water. This means that the measured hydrolysis rates could be an overestimation of the actual hydrolysis rates of DOM (e.g., Arnosti, 2011). The primary benefit of the method is that it is straightforward and has been in widespread use for decades, which means that comparisons with other ecosystems is possible. For our purpose, we use this method to better understand how much of the variability can be explained by the other measured variables.
Considering the APA, the most interesting aspect was that it was measurable at the beginning of the experiment at a high PO concentration. This high APA activity at a high PO concentration has been observed in deep oceans (Hoppe and Ullrich, 1999; Baltar et al., 2016). Baltar et al. (2016) also observed an increase in the APA in experiments amended with organic matter, suggesting that the activity of APA was linked to organic matter supply, independently of the PO concentration. This could be due to bacterial APA, which is more complex than for phytoplankton, in that it can be linked to the hydrolysis and acquisition of C (Hoppe, 2003). In our experiment, the initial decrease in DOP and increase in PO concentrations indicates that P released by AP hydrolysis was added to the PO pool. This suggests that APA was not used for P acquisition.
It is known that APA remains suspended and active for a long time in marine environments, and cell-free APA was reduced by only 25 % over 16 d in the experiment by Thomson et al. (2019). If this enzyme is viable for this long, it suggests that there was no new production of AP after the closure of the mesocosms, which is supported by the dilution effect of adding the OMZ water. In that case, the disappearance of the initial AP took 30 d.
The hydrolysis rates of AP were relatively low compared with most published data, probably linked to the clear surplus of PO. It is worth noting, however, that we were most likely not measuring the maximal potential hydrolysis rates, as substrate addition was relatively low (100 nmol L−1), and values would likely have been higher with more added substrate. This could be the reason for the apparent discrepancy between the measured hydrolysis rates and the change in the PO and DOP pools during the first 10 d of the experiment. During this time, there was a decrease of approximately 0.5 µmol DOP L−1 and an increase of 0.6 µmol PO L−1, suggesting an actual hydrolysis rate of 2.0–2.5 nmol L−1 h−1 (assuming 500–600 nmol over 10 d). This is a factor of 3–4 higher compared with the initially measured APA of ∼ 0.6 nmol L−1 h−1.
The statistical model that we applied was better at explaining the variability in APA compared with the LAP activity. APA gradually decreased during the initial phase of the experiment to undetectable levels after the middle of the experiment. Any correlation does not mean causality, and the higher coefficient of determination is probably rather a reflection of the clear temporal development in APA. If the AP was produced before the closure of the mesocosm and slowly degraded as discussed above, any connection with the biogeochemical variables or plankton community was likely due to unrelated temporal development; for example, the DIN also decreased over time.
For the LAP activity, the overall explanatory power with respect to the biogeochemical variables and plankton community composition was less than for APA; however, interestingly, the bacterioplankton community composition clearly explained the variability better (38 %) than the combined biogeochemical variables and phytoplankton community (28 %). Considering that the bacterial community was not sampled as frequently as the biogeochemical variables and flow cytometer counts, we suspect that the explanatory power would have increased with more frequent sampling. It is likely that bacteria were producing the LAP activity, and some taxa are more reliant on enzyme production for nutrient acquisition than others (Ramin and Allison, 2019). Some dinoflagellates are also known to produce LAP, and most of the mesocosms had high dinoflagellate biomass (except M4). However, the phytoplankton community only explained 18 % of the variability in LAP activity, and these dinoflagellates were likely not producing any substantial amount of this enzyme.
In conclusion, there was measurable APA at the start of the experiment, but this gradually declined to undetectable levels in all of the mesocosms midway through the experiment (∼ 30 d). With high concentrations of PO, low APA is not surprising, and AP is a relatively slowly degrading enzyme that could have been fully dissolved and produced before the closure of the mesocosms. Our statistical model better explained the variability in APA (74 %) compared with LAP activity, probably due to the clear temporal development of APA that was likely independent of some of the other temporal trends, such as decreasing DIN. We found very high levels of LAP activity (mostly in the range of 200–800 nmol L−1 h−1), which is an order of magnitude higher than most literature data. This is probably linked to upwelling supporting high levels of microbial activity in combination with general DIN limitation in the coastal Peruvian upwelling. The bacterioplankton community composition best explained the variability in LAP activity (38 %) compared with the combined biochemical variable and phytoplankton community model (28 %). With more than 50 % of the variability unaccounted for, we are still missing important pieces of the puzzle with respect to understanding the variability in LAP activity. The high hydrolysis rates for LAP suggest that pelagic N remineralization supported the relatively high standing stock of primary producers (mostly > 4 µg Chl a L−1) in the mesocosms after N depletion.
All data are available upon request. The DNA sequencing data will be submitted to NCBI SRA.
The supplement related to this article is available online at: https://doi.org/10.5194/bg-20-1605-2023-supplement.
Samples were taken by KS, JP, JA, LTB, EvdE, MAF, NHH, JM and UR. In addition to the sampling crew, further data analysis was conducted by MTCG and MGL. UR developed the experimental design and sampling strategy as well as coordinating the mesocosm campaign. All co-authors contributed to the data interpretation. KS wrote the manuscript with contributions from all co-authors.
The contact author has declared that none of the authors has any competing interests.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the special issue “Ecological and biogeochemical functioning of the coastal upwelling system off Peru: an in situ mesocosm study”. It is not associated with a conference.
We thank all participants of the KOSMOS 2017 Peru study for assisting with mesocosm sampling and maintenance, in particular Andrea Ludwig, Jana Meyer, Jean-Pierre Bednar, Gabriela Chavez, Susanne Feiersinger, Peter Fritsche, Paul Stange, Anna Schukat and Michael Krudewig. We are particularly thankful to the staff of IMARPE for their support during the planning and to the Marina de Guerra del Perú, the Dirección General de Capitanías y Guardacostas and the Club Náutico Del Centro Naval for their great support. The NMDS plots and GAM models were produced by Riina Klais-Peets at EcoStat Ltd. This study used the SYKE marine research infrastructure as a part of the Finnish FINMARI consortium. The publication of sequencing data was approved by the Peruvian Ministry of Production with respect to the access and benefit sharing regulations of the Nagoya Protocol.
The experiment was funded through the German Research Foundation (DFG) Collaborative Research Centre project “Climate–biogeochemistry interactions in the tropical ocean” (SFB 754). Additional funding came from the Academy of Finland (decision no. 259164; Kristian Spilling and Jonna Piiparinen), the EU project AQUACOSM (Ulf Riebesell) under grant no. 731065 of the European Union's Horizon 2020 Research and Innovation program, the Leibniz Award 2012 of the German Research Foundation (Ulf Riebesell) and the Helmholtz International Fellow Award 2015 (Javier Arístegui).
This paper was edited by Tina Treude and reviewed by two anonymous referees.
Aldunate, M., De la Iglesia, R., Bertagnolli, A. D., and Ulloa, O.: Oxygen modulates bacterial community composition in the coastal upwelling waters off central Chile, Deep-Sea Res. Pt. II, 156, 68–79, 2018.
Ammerman, J.: Microbial cycling of inorganic and organic phosphorus in the water column, Handbook Method. Aquat. Microb. Ecol., 1, 649–660, 1993.
Anabalón, V., Morales, C., González, H., Menschel, E., Schneider, W., Hormazabal, S., Valencia, L., and Escribano, R.: Micro-phytoplankton community structure in the coastal upwelling zone off Concepción (central Chile): Annual and inter-annual fluctuations in a highly dynamic environment, Prog. Oceanogr., 149, 174–188, 2016.
Arnosti, C.: Microbial extracellular enzymes and the marine carbon cycle, Ann. Rev. Mar. Sci., 3, 401–425, 2011.
Bach, L. T., Paul, A. J., Boxhammer, T., von der Esch, E., Graco, M., Schulz, K. G., Achterberg, E., Aguayo, P., Arístegui, J., Ayón, P., Baños, I., Bernales, A., Boegeholz, A. S., Chavez, F., Chavez, G., Chen, S.-M., Doering, K., Filella, A., Fischer, M., Grasse, P., Haunost, M., Hennke, J., Hernández-Hernández, N., Hopwood, M., Igarza, M., Kalter, V., Kittu, L., Kohnert, P., Ledesma, J., Lieberum, C., Lischka, S., Löscher, C., Ludwig, A., Mendoza, U., Meyer, J., Meyer, J., Minutolo, F., Ortiz Cortes, J., Piiparinen, J., Sforna, C., Spilling, K., Sanchez, S., Spisla, C., Sswat, M., Zavala Moreira, M., and Riebesell, U.: Factors controlling plankton community production, export flux, and particulate matter stoichiometry in the coastal upwelling system off Peru, Biogeosciences, 17, 4831–4852, https://doi.org/10.5194/bg-17-4831-2020, 2020.
Badylak, S., Phlips, E. J., and Mathews, A. L.: Akashiwo sanguinea (Dinophyceae) blooms in a sub-tropical estuary: an alga for all seasons, Plankt. Benthos. Res., 9, 147–155, 2014.
Baltar, F., Lundin, D., Palovaara, J., Lekunberri, I., Reinthaler, T., Herndl, G. J., and Pinhassi, J.: Prokaryotic responses to ammonium and organic carbon reveal alternative CO2 fixation pathways and importance of alkaline phosphatase in the mesopelagic North Atlantic, Front. Microbiol., 7, 1670, https://doi.org/10.3389/fmicb.2016.01670, 2016.
Barlow, R., Cummings, D., and Gibb, S.: Improved resolution of mono-and divinyl chlorophylls a and b and zeaxanthin and lutein in phytoplankton extracts using reverse phase C-8 HPLC, Mar. Ecol. Prog. Ser., 161, 303–307, 1997.
Benitez-Nelson, C. R. and Buesseler, K. O.: Variability of inorganic and organic phosphorus turnover rates in the coastal ocean, Nature, 398, 502–505, 1999.
Bolger, A. M., Lohse, M., and Usadel, B.: Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 30, 2114–2120, 2014.
Buchan, A., LeCleir, G. R., Gulvik, C. A., and González, J. M.: Master recyclers: Features and functions of bacteria associated with phytoplankton blooms, Nat. Rev. Microbiol., 12, 686–698, https://doi.org/10.1038/nrmicro3326, 2014.
Burley, S. K., David, P. R., Taylor, A., and Lipscomb, W. N.: Molecular structure of leucine aminopeptidase at 2.7-A resolution, P. Natl. Acad. Sci. USA, 87, 6878–6882, 1990.
Canfield, D., Kristensen, E., and Thamdrup, B.: Aquatic geomicrobiology, Elsevier, ISBN: 978-0-12-026147-5, 2005.
Callbeck, C. M., Lavik, G., Ferdelman, T. G., Fuchs, B., Gruber-Vodicka, H. R., Hach, P. F., Littmann, S., Schoffelen, N. J., Kalvelage, T., and Thomsen, S.: Oxygen minimum zone cryptic sulfur cycling sustained by offshore transport of key sulfur oxidizing bacteria, Nat. Commun., 9, 1–11, 2018.
Chafee, M., Fernàndez-Guerra, A., Buttigieg, P. L., Gerdts, G., Eren, A. M., Teeling, H., and Amann, R. I.: Recurrent patterns of microdiversity in a temperate coastal marine environment, ISME J., 12, 237–252, https://doi.org/10.1038/ismej.2017.165, 2017.
Chavez, F. P., Bertrand, A., Guevara-Carrasco, R., Soler, P., and Csirke, J.: The northern Humboldt Current System: Brief history, present status and a view towards the future, Prog. Oceanogr., 79, 95–105, 2008.
Chen, S.-M., Riebesell, U., Schulz, K. G., von der Esch, E., Achterberg, E. P., and Bach, L. T.: Temporal dynamics of surface ocean carbonate chemistry in response to natural and simulated upwelling events during the 2017 coastal El Niño near Callao, Peru, Biogeosciences, 19, 295–312, https://doi.org/10.5194/bg-19-295-2022, 2022.
Christian J. R. and Karl D. M.: Bacterial ectoenzymes in marine waters: activity ratios and temperature responses in three oceanographic provinces, Limnol. Oceanogr., 40, 1042–1049, 1995.
Coverly, S., Kérouel, R., and Aminot, A.: A re-examination of matrix effects in the segmented-flow analysis of nutrients in sea and estuarine water, Anal. Chim. Ac., 712, 94–100, 2012.
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R.: UCHIME improves sensitivity and speed of chimera detection, Bioinformatics, 27, 2194–2200, 2011.
Ewing, B. and Green, P.: Base-calling of automated sequencer traces using phred, II. Error probabilities, Genome Res., 8, 186–194, 1998.
Ewing, B., Hillier, L., Wendl, M. C., and Green, P.: Base-calling of automated sequencer traces usingPhred, I. Accuracy assessment, Genome Res., 8, 175–185, 1998.
FAO: The state of world fisheries and aquaculture, Food and Agriculture Organization of the United Nations, FAO, Rome, 223 pp., ISBN 978-92-5-130562-1, 2018.
Fischer, M. A., Güllert, S., Refai, S., Künzel, S., Deppenmeier, U., Streit, W. R., and Schmitz, R. A.: Long-term investigation of microbial community composition and transcription patterns in a biogas plant undergoing ammonia crisis, Microb. Biotechnol., 12, 305–323, 2019a.
Fischer, M. A., Ulbricht, A., Neulinger, S. C., Refai, S., Waßmann, K., Künzel, S., and Schmitz-Streit, R. A.: Immediate effects of ammonia shock on transcription and composition of a biogas reactor microbiome, Front. Microbiol., 10, 2064, https://doi.org/10.3389/fmicb.2019.02064, 2019b.
Garreaud, R. D.: A plausible atmospheric trigger for the 2017 coastal El Niño, Int. J. Climatol., 38, e1296–e1302, https://doi.org/10.1002/joc.5426, 2018.
Gier, J., Sommer, S., Löscher, C. R., Dale, A. W., Schmitz, R. A., and Treude, T.: Nitrogen fixation in sediments along a depth transect through the Peruvian oxygen minimum zone, Biogeosciences, 13, 4065–4080, https://doi.org/10.5194/bg-13-4065-2016, 2016.
Graco, M. I., Purca, S., Dewitte, B., Castro, C. G., Morón, O., Ledesma, J., Flores, G., and Gutiérrez, D.: The OMZ and nutrient features as a signature of interannual and low-frequency variability in the Peruvian upwelling system, Biogeosciences, 14, 4601–4617, https://doi.org/10.5194/bg-14-4601-2017, 2017.
Gutiérrez, M., Pantoja, S., Tejos, E., and Quiñones, R.: The role of fungi in processing marine organic matter in the upwelling ecosystem off Chile, Mar. Biol., 158, 205–219, 2011.
Hauss, H., Franz, J. M., and Sommer, U.: Changes in N: P stoichiometry influence taxonomic composition and nutritional quality of phytoplankton in the Peruvian upwelling, J. Sea Res., 73, 74–85, 2012.
Hoppe, H.-G.: Phosphatase activity in the sea, Hydrobiology, 493, 187–200, 2003.
Hoppe, H.-G., Kim, S.-J., and Gocke, K.: Microbial decomposition in aquatic environments: combined process of extracellular enzyme activity and substrate uptake, Appl. Environ. Microbiol., 54, 784–790, 1988.
Hoppe H.-G. and Ullrich S.: Profiles of ectoenzymes in the Indian Ocean: phenomena of phosphatase activity in the mesopelagic zone, Aquat. Microb. Ecol., 19, 139–148, 1999.
Hoppe, H.-G., Arnosti, C., and Herndl, G.: Ecological significance of bacterial enzymes in the marine environment, in: Enzymes in the Environment: Activity, Ecology, and Applications, edited by: Burns R. G and Dick R. P., CRC Press, Boca Raton, 73–107, https://doi.org/10.1201/9780203904039, 2002.
Jeong, H. J., Du Yoo, Y., Park, J. Y., Song, J. Y., Kim, S. T., Lee, S. H., Kim, K. Y., and Yih, W. H.: Feeding by phototrophic red-tide dinoflagellates: five species newly revealed and six species previously known to be mixotrophic, Aquat. Microb. Ecol., 40, 133–150, 2005.
Kalvelage, T., Lavik, G., Lam, P., Contreras, S., Arteaga, L., Löscher, C. R., Oschlies, A., Paulmier, A., Stramma, L., and Kuypers, M. M.: Nitrogen cycling driven by organic matter export in the South Pacific oxygen minimum zone, Nat. Geosci., 6, 228–234, 2013.
Keeling, R. F., Körtzinger, A., and Gruber, N.: Ocean deoxygenation in a warming world, Ann. Rev. Mar. Sci., 2, 199–229, 2010.
Kérouel, R. and Aminot, A.: Fluorometric determination of ammonia in sea and estuarine waters by direct segmented flow analysis, Mar. Chem., 57, 265–275, 1997.
Labry, C., Delmas, D., Youenou, A., Quere, J., Leynaert, A., Fraisse, S., Raimonet, M., and Ragueneau, O.: High alkaline phosphatase activity in phosphate replete waters: The case of two macrotidal estuaries, Limnol. Oceanogr., 61, 1513–1529, 2016.
Margalef, R., Estrada, M., and Blasco, D.: Functional morphology of organisms involved in red tides, as adapted to decaying turbulence, in: Toxic dinoflagellate blooms, edited by: Taylor, D. L. and Seliger, H. H., Elsevier-North Holland, Amsterdam, 89–94, 1979.
Maßmig, M., Lüdke, J., Krahmann, G., and Engel, A.: Bacterial degradation activity in the eastern tropical South Pacific oxygen minimum zone, Biogeosciences, 17, 215–230, https://doi.org/10.5194/bg-17-215-2020, 2020.
Messié, M. and Chavez, F. P.: Seasonal regulation of primary production in eastern boundary upwelling systems, Prog. Oceanogr., 134, 1–18, 2015.
Morris, A. and Riley, J.: The determination of nitrate in sea water, Anal. Chim. Ac., 29, 272–279, 1963.
Murphy, J. and Riley, J. P.: A modified single solution method for the determination of phosphate in natural waters, Anal. Chim. Ac., 27, 31–36, 1962.
Murphy, K. R., Stedmon, C. A., Wenig, P., and Bro, R.: OpenFluor–an online spectral library of auto-fluorescence by organic compounds in the environment, Anal. Meth., 6, 658–661, 2014.
Nausch, M.: Alkaline phosphatase activities and the relationship to inorganic phosphate in the Pomeranian Bight (southern Baltic Sea), Aquat. Microb. Ecol., 16, 87–94, 1998.
Nowinski, B., Motard-Côté, J., Landa, M., Preston, C. M., Scholin, C. A., Birch, J. M., Kiene, R. P., and Moran, M. A.: Microdiversity and temporal dynamics of marine bacterial dimethylsulfoniopropionate genes, Environ. Microbiol., 21, 1687–1701, 2019.
Oksanen, J., Simpson, G. L., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O'Hara, R. B., Solymos, P., Stevens, M. H. H., Szoecs, E., Wagner, H., Barbour, M., Bedward, M., Bolker, B., Borcard, D., Carvalho, G., Chirico, M., De Caceres, M., Durand, S., Evangelista, H. B. A., FitzJohn, R., Friendly, M., Furneaux, B., Hannigan, G., Hill, M. O., Lahti, L., McGlinn, D., Ouellette, M.-H., Cunha, E. R., Smith, T., Stier, A., Ter Braak, C. J. F., and Weedon, J.: Ordination methods, diversity analysis and other functions for community and vegetation ecologists. Vegan: Community Ecology Package, https://CRAN.R-project.org/package=vegan, last access: 6 April 2023, 2022.
Oschlies, A., Brandt, P., Stramma, L., and Schmidtko, S.: Drivers and mechanisms of ocean deoxygenation, Nat. Geosci., 11, 467–473, 2018.
Pajares, S., Varona-Cordero, F., and Hernández-Becerril, D. U.: Spatial Distribution Patterns of Bacterioplankton in the Oxygen Minimum Zone of the Tropical Mexican Pacific, Microb. Ecol., 80, 519–536, https://doi.org/10.1007/s00248-020-01508-7, 2020.
Paul, A. J., Bach, L. T., Arístegui, J., von der Esch, E., Hernández-Hernández, N., Piiparinen, J., Ramajo, L., Spilling, K., and Riebesell, U.: Upwelled plankton community modulates surface bloom succession and nutrient availability in a natural plankton assemblage, Biogeosciences, 19, 5911–5926, https://doi.org/10.5194/bg-19-5911-2022, 2022.
Perry, M.: Alkaline phosphatase activity in subtropical Central North Pacific waters using a sensitive fluorometric method, Mar. Biol., 15, 113–119, 1972.
Pruesse, E., Peplies, J., and Glöckner, F. O.: SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes, Bioinformatics, 28, 1823–1829, 2012.
Rose, C. and Axler, R. P.: Uses of alkaline phosphatase activity in evaluating phytoplankton community phosphorus deficiency, Hydrobiology, 361, 145–156, 1997.
Ramin K. I. and Allison S. D.: Bacterial tradeoffs in growth rate and extracellular enzymes, Front. Microbiol., 10, 2956, https://doi.org/10.3389/fmicb.2019.02956, 2019.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., Lesniewski, R. A., Oakley, B. B., Parks, D. H., and Robinson, C. J.: Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities, Appl. Environ. Microbiol., 75, 7537–7541, 2009.
Schulz, K. G., Achterberg, E. P., Arístegui, J., Bach, L. T., Baños, I., Boxhammer, T., Erler, D., Igarza, M., Kalter, V., Ludwig, A., Löscher, C., Meyer, J., Meyer, J., Minutolo, F., von der Esch, E., Ward, B. B., and Riebesell, U.: Nitrogen loss processes in response to upwelling in a Peruvian coastal setting dominated by denitrification – a mesocosm approach, Biogeosciences, 18, 4305–4320, https://doi.org/10.5194/bg-18-4305-2021, 2021.
Schunck, H., Lavik, G., Desai, D. K., Großkopf, T., Kalvelage, T., Löscher, C. R., Paulmier, A., Contreras, S., Siegel, H., and Holtappels, M.: Giant hydrogen sulfide plume in the oxygen minimum zone off Peru supports chemolithoautotrophy, PloS One, 8, e68661, https://doi.org/10.1371/journal.pone.0068661, 2013.
Shah, V., Chang, B. X., and Morris, R. M.: Cultivation of a chemoautotroph from the SUP05 clade of marine bacteria that produces nitrite and consumes ammonium, ISME J., 11, 263–271, 2017.
Shi, Z., Xu, J., Li, X., Li, R., and Li, Q.: Links of extracellular enzyme activities, microbial metabolism, and community composition in the river‐impacted coastal waters, J. Geophys. Res.-Biogeo., 124, 3507–3520, 2019.
Song, C., Cao, X., Zhou, Y., Azzaro, M., Monticelli, L. S., Maimone, G., Azzaro, F., La Ferla, R., and Caruso, G.: Nutrient regeneration mediated by extracellular enzymes in water column and interstitial water through a microcosm experiment, Sci. Total Environ., 670, 982–992, 2019.
Spilling, K., Camarena-Gómez, M.-T., Lipsewers, T., Martinez-Varela, A., Díaz-Rosas, F., Eronen-Rasimus, E., Silva, N., von Dassow, P., and Montecino, V.: Impacts of reduced inorganic N : P ratio on three distinct plankton communities in the Humboldt upwelling system, Mar. Biol., 166, 114, https://doi.org/10.1007/s00227-019-3561-x, 2019.
Steen, A. D., Vazin, J. P., Hagen, S. M., Mulligan, K. H., and Wilhelm, S. W.: Substrate specificity of aquatic extracellular peptidases assessed by competitive inhibition assays using synthetic substrates, Aquat. Microb. Ecol., 75, 271–281, 2015.
Stedmon, C. A. and Bro, R.: Characterizing dissolved organic matter fluorescence with parallel factor analysis: a tutorial, Limnol. Oceanogr. Method., 6, 572–579, 2008.
Stoecker, D. K. and Gustafson, D. E.: Cell-surface proteolytic activity of photosynthetic dinoflagellates, Aquat. Microb. Ecol., 30, 175–183, 2003.
Thomson, B., Wenley, J., Currie, K., Hepburn, C., Herndl, G. J., and Baltar, F.: Resolving the paradox: continuous cell-free alkaline phosphatase activity despite high phosphate concentrations, Mar. Chem., 214, 103671, https://doi.org/10.1016/j.marchem.2019.103671, 2019.
Wood, S. N.: Generalized additive models: an introduction with R, CRC press, New York, 2nd Edn., https://doi.org/10.1201/9781315370279, 2017.





