the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Ideas and perspectives: Beyond model evaluation – combining experiments and models to advance terrestrial ecosystem science
Benjamin D. Stocker
Teresa E. Gimeno
Richard Nair
Ecosystem manipulative experiments are a powerful tool to understand terrestrial ecosystem responses to global change because they measure real responses in real ecosystems and yield insights into causal relationships. However, their scope is limited in space and time due to cost and labour intensity. This makes generalising results from such experiments difficult, which creates a conceptual gap between local-scale process understanding and global-scale future predictions. Recent efforts have seen results from such experiments used in combination with dynamic global vegetation models, most commonly to evaluate model predictions under global change drivers. However, there is much more potential in combining models and experiments. Here, we discuss the value and potential of a workflow for using ecosystem experiments together with process-based models to enhance the potential of both. We suggest that models can be used prior to the start of an experiment to generate hypotheses, identify data needs, and in general guide experimental design. Models, when adequately constrained with observations, can also predict variables which are difficult to measure frequently or at all, and together with the data they can provide a more complete picture of ecosystem states. Finally, models can be used to help generalise the experimental results in space and time, by providing a framework in which process understanding derived from site-level experiments can be incorporated. We also discuss the potential for using manipulative experiments together with models in formalised model–data integration frameworks for parameter estimation and model selection, a path made possible by the increasing number of ecosystem experiments and diverse observation streams. The ideas presented here can provide a roadmap to future experiment–model studies.
- Article
(691 KB) - Full-text XML
- BibTeX
- EndNote
In the face of a changing climate, understanding how ecosystems will respond to conditions that are unprecedented in the observational record is one of the biggest challenges in Earth system science. To meet this challenge, we must employ all tools available to us – observations, experiments and meta-analyses, and process- and data-based models. All these different tools have strengths and weaknesses in terms of spatial and temporal scales, generality, and predictive capacity. Therefore, using these tools, not only individually but also in combination, is essential for bridging the gap between process understanding and scalability.
Over the last few decades, there have been great advancements in the types and quantity of available ecosystem data, including large databases of field observations (Kattge et al., 2020); continuous ecosystem monitoring (Baldocchi, 2020); remotely sensed Earth observations (Xiao et al., 2019); and spatial datasets with global coverage for climate (Fick and Hijmans, 2017; Karger et al., 2020; Harris et al., 2020), soil (Food and Agriculture Organization and Food and Agriculture Organization of the United Nations, 2008; Hengl et al., 2017), and other spatially upscaled global ecosystem data products (Jung et al., 2020; Poggio et al., 2021). In and of themselves, such observational datasets provide a comprehensive picture of ecosystem properties and structure, as well as carbon and water fluxes. However, the mounting data richness in ecosystem science has not led to a reduction in projection uncertainties in global biogeochemical cycles in a future climate (Bastos et al., 2020; Piao et al., 2020; Arora et al., 2020). Ecological and physiological process understanding – key for predicting outside the domain of observed conditions – often remains weakly constrained despite the large volume of available ecosystem data collected over recent decades. In this context, ecosystem manipulation experiments (EMEs) have a unique and particularly valuable role to play (Stocker et al., 2016; Wieder et al., 2019).
EMEs are outdoor experimental setups in which one or multiple driving factors (e.g. water availability, temperature, nutrient inputs, atmospheric CO2 concentration, perturbations, species composition), or a combination of factors, are controlled to study their separate or interactive effects on ecosystem process subunits within ecosystems such as plants or microbes. They are a useful tool because they contain ecosystem process responses defined by treatments and hypotheses while minimising confounding factors and scaling biases and experimental artefacts from smaller and more limited lab- or mesocosm-based experiments (Dalling et al., 2013). EMEs provide a unique window into the responses of ecosystems to a potential future environment and can reveal complex responses at the level of plants, communities, and ecosystems, shaped by feedbacks between soil and vegetation. While extremely valuable for understanding ecosystem responses to future conditions, due to their realistic scale and multidimensionality of observations, EMEs generate small data – in contrast to, for example, Earth observation. Therefore, it remains challenging to generalise from a relatively small set of local observations to future global predictions. EMEs are usually operated at spatial scales on the order of 1–100 m and are limited in time to typically 1–10 years due to high operating costs and laborious measurements. Thus, it can be difficult to scale the conclusions from experiments with confidence, both in time and in space. Here, we propose and discuss approaches for aiding generalisations from EME observations in combination with process-based models as a potential solution to overcome this scalability challenge.
Process-based ecosystem models are mathematical representations of how plant traits and soil characteristics determine water, energy, and biogeochemical fluxes, as well as the accumulation of organic matter in a cascade of ecosystem pools, given a set of environmental boundary conditions. Depending on the scale and processes represented, such models are termed terrestrial biosphere models, dynamic vegetation models, mechanistic ecosystem models, or land surface models and are implemented as coupled components in Earth system models (Prentice et al., 2007). Irrespective of what we call them, all such models face similar challenges in that they need to balance realism, robustness, and reliability (Prentice et al., 2015), as well as computational efficiency, to achieve reliable predictions for ecosystem fluxes and pools across the globe and across decades into the future. To achieve this, such models need to be confronted with diverse data to leverage complementary constraints. Observations of traits, fluxes, and biomass, obtained at different scales in space and time and across diverse ecosystem types, provide complementary information for calibrating and testing terrestrial biosphere models (Keenan et al., 2012). Recent efforts have been made in using large observational datasets for validating and benchmarking these models (Collier et al., 2018; Seiler et al., 2022). However, as outlined above, model projections remain divergent (Friedlingstein et al., 2022; Bastos et al., 2020), pointing to remaining gaps in critical process understanding from such datasets, and extrapolating observations from past climate conditions to the future often comes with increased uncertainty.
EMEs have been used to parameterise, validate, and test models, ranging from drought (McDowell et al., 2013; Powell et al., 2013) or warming (Parton et al., 2007; Zaehle et al., 2010; Ma et al., 2022) to fertilisation (Meyerholt and Zaehle, 2015; Thomas et al., 2013) or decomposition (Bonan et al., 2013) experiments. A step change came about with the Free-Air CO2 Enrichment (FACE) Model Data Synthesis (MDS) project (Medlyn et al., 2015; De Kauwe et al., 2014; Zaehle et al., 2014; Walker et al., 2014) which synthesised data from two temperate forest FACE EMEs. The project asked not only if the models match the data but why the models match the data, making full use of manipulative experiments and being able to plot a path to model improvement. This approach led to both the identification of key processes that need to be included in models to correctly capture responses to elevated CO2 (e.g. flexible but realistic tissue stoichiometry, flexible biomass allocation, flexible leaf mass per area, organic N uptake) and the identification of areas where more data are needed to identify realistic and reliable process representation (e.g. water limitation effects on photosynthesis and transpiration, wood turnover, detailed photosynthesis information, N losses). Other studies are now following up on these results to further improve models (Caldararu et al., 2020). Since then the FACE-MDS project, there have been several other MDS studies following the same philosophy, including other FACE experiments (Fleischer et al., 2019; Medlyn et al., 2016) and FACE plus warming experiments (De Kauwe et al., 2017). A few such modelling studies were performed prior to the start of the actual experiment (Medlyn et al., 2016; Fleischer et al., 2019; Parton et al., 2007) with the aim of informing experimental design, and in the case of Parton et al. (2007), their model predictions were later tested against experimental results (Dijkstra et al., 2010), showcasing the power of such a study.
There has been an increase in the number of EMEs globally, as well as in the quantity of open data and efforts to synthesise and bring together existing experiments (Van Sundert et al., 2023; Liang et al., 2020; Cleland et al., 2019). The present special issue includes some examples of new experiments including a large-scale drought and fertilisation experiments (Vargas et al., 2023; Baer et al., 2023; Mendoza-Martinez et al., 2023); a transplant and elevation gradient experiment (Ntirugulirwa et al., 2023); and EME–model integration, including studies looking at new nutrient models (Cornut et al., 2022a, b; Li et al., 2022; Eastman et al., 2023; Raoult et al., 2023), the effects of root distribution of water availability response (Kulmatiski et al., 2023), carbon storage in grasslands (Wilcox et al., 2023), and extreme events (Holm et al., 2023).
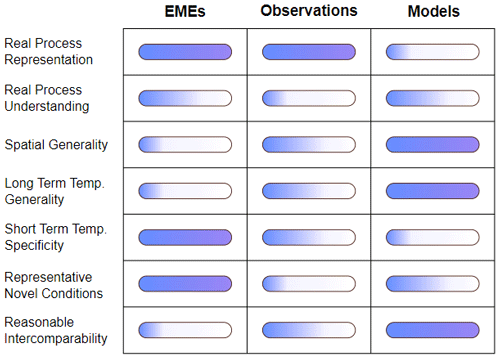
Global observations, ecosystem experiments, and process-based models all have their strengths and weaknesses in terms of scalability and process understanding (Fig. 1). To advance ecosystem science, an integration of complementary sources of observations and models is needed to achieve the full predictive potential of the tools available. The majority of EME–model integration studies have focused on constraining or evaluating models with experimental data; however, there is a large untapped potential to use models and EMEs bidirectionally to bridge the process understanding–scalability gap. While models are capable of making global-scale predictions on long timescales, they also include a detailed process representation which can be a reflection of the insight gained from EMEs. In this paper we set out a roadmap to using models in combination with EMEs not only as a one-way street for validating models but also as an integrated workflow aimed at improving experimental design and generalising experimental conclusions.
In past studies, the focus has been on integrating conclusions from EMEs into models and evaluating such models against experimental data to improve model process representation (Medlyn et al., 2015; Norby et al., 2016). However, this relationship can easily be turned around. Models can be used as digital twins (digital representations of physical objects or environments) or sandboxes to explore different possibilities for experimental design; set realistic and practical treatment levels; and formulate quantitative, defensible hypotheses. Models can also be used to integrate observations gained from one or more experiments for generalising in time and space.
2.1 Hypothesis generation and experimental design
Ecosystem manipulation experiments are designed around testable research questions and hypotheses, following established theory. Hypotheses are generally of a qualitative and first-order nature; i.e. “if higher A, then B”. For example, nutrient addition leads to increased growth and photosynthesis or drought reduces growth. However, ecosystem responses are affected by multiple feedbacks, operating at diverse timescales. Responses can be non-linear, even threshold-like, and interactive, and they can be affected by plant adaptations, species-specific responses, and plant–soil interactions. Certain processes saturate at higher levels reached over time, such as nutrient addition or elevated CO2, where continued application of the treatment does not lead to a further ecosystem response, as systems become limited by other factors. In the case of edaphic factors, such as nutrient availability, spatial factors, such as the underlying fertility and history of nutrient deposition, interact with future loading. Ecosystem responses have also been shown to vary with the intensity and duration of the treatment (Niu et al., 2022). This means that it is often difficult to (explicitly) generate hypotheses that reflect multiple known individual processes and their (unknown) interactive effects. Using a model, multiple models, or multiple process representations within one modelling framework can aid hypothesis generation.
As many responses to environmental drivers are non-linear, it can be uncertain when designing an experiment what and how many treatment levels are necessary, especially if choosing a response surface rather than a replicated approach (Kreyling et al., 2018). This is particularly important as ecosystem-level experiments are technically challenging and expensive, especially those on the scale of FACE experiments. There are financial, logistical, and practical constraints on how many treatment levels can be considered. Models can easily be run with multiple treatment levels, which would allow for the identification of key non-linearities, and a defined response surface that can be used as a basis for choosing experimental treatments.
Models can also potentially be used to define a sampling strategy. Initial model simulations can help identify which variables are likely to respond to the experimental treatment and at what timescales treatment responses are likely to be detected. If using multiple models or a model with multiple alternative process representations, this can also help identify variables that would help test the proposed hypotheses, by looking at model variables that respond differently for different model formulations. We propose a workflow (Fig. 2) in which one or multiple models are run with local site conditions with multiple treatment levels. This then allows us to identify hypotheses based on predicted responses, ideal treatment levels, and key variables to measure. This can be done multiple times to explore different experimental setups and ideas.
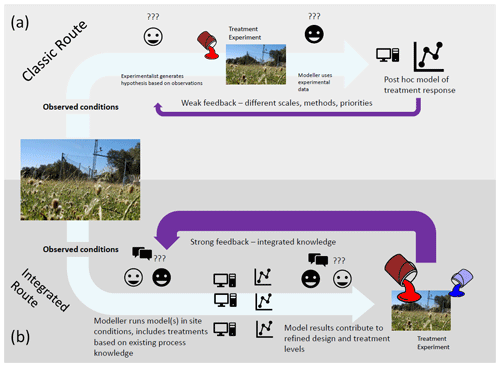
Figure 2Classic workflow (a) contrasted with proposed workflow (b) for using models together with ecosystem manipulative experiments for experimental design and hypothesis generation. Through iterative synthesis of models and experiments in (b) and tuning of experimental parameters based on modelled feedback, stronger predictions are generated.
There is often a gap between the optimum sampling strategy and what is feasible in field experiments – variables often cannot be measured with a sufficient temporal or spatial frequency due to either costs or technical constraints. While we as a community have come a very long way in automating certain measurements – carbon and water fluxes, spectral properties, soil moisture – many remain time- and labour-intensive, such as direct measurements of biomass above- and belowground or of plant and soil chemical composition. Furthermore, other quantities which might be useful to know are not possible to measure; that is, they are latent or conceptual variables such as plant nutrient demand. Such variables are often key in explaining the observed responses but can usually only be guessed at.
In contrast, models estimate variables consistently at regular intervals, including conceptual or latent quantities which can be expressed mathematically and can be used to quantify and explain ecosystem states but cannot be measured. These can therefore be used alongside experimental results to explain observed responses. For example, the issue of nutrient limitation under elevated CO2 involves many plant and soil processes, some of which cannot be monitored easily but are included in nutrient-enabled models (Caldararu et al., 2022). One potential issue with this approach is that to have sufficient confidence in these essentially impossible to measure quantities, models need to be able to accurately represent ecosystem processes at a given particular experimental site. Before being used in such a scenario, models should be evaluated or even calibrated for the site using variables that can commonly be measured to ensure the model performance.
2.2 Generalising in time and space
Most manipulative ecosystem experiments are commonly performed at the plot level and are only maintained for a short time period (1–5 years), with a few exceptional experiments lasting in the range of 10 years or more (Magill et al., 2004; Eastman et al., 2021). While a particular experiment can tell us a lot about the mechanistic process involved, it remains an open question how scalable they are in time and space. While there are increasing efforts to standardise experimental treatments through initiatives such as the Nutrient Network (Borer et al., 2014), only a small and inevitably imperfectly distributed area of the Earth's vegetated surface can ever be covered by experimental treatments. However, models can be run over relatively long periods of time and globally, helping to test the generality of the process understanding informed by EMEs.
The problem of spatial scaling is obvious. Can the particular ecosystem studied represent other ecosystems of the same type, and can plant or soil processes inferred from observed responses be generalised? Upscaling of ecosystem experimental results has been previously performed by accounting for moderating factors of the response, such as climate or soil conditions (Terrer et al., 2021). However, such approaches lack the mechanistic and dynamic underpinning provided by a process-based model and could thus miss important feedbacks.
A particular challenge is posed by the step shape of imposed environmental change in EMEs, which contrasts with the gradual decadal-scale environmental change to which ecosystems are exposed. Several processes make the responses to a continuous change potentially different from those to a step change. Plants acclimate to the new conditions over time (Reich et al., 2018), a process that has been primarily studied for changes in temperature but is likely to occur for most environmental drivers. Species composition is likely to change under different environmental conditions, but it can likely only be observed during the duration of the experiment in systems with high species turnover such as grasslands (Avolio et al., 2014). Although ecosystem responses in observable quantities often cannot be assumed to be time-invariant, particularly if slow plant–soil feedbacks are involved and even if conditions are held constant after a step change, underlying processes often can be. Mechanistic models that simulate responses to step changes and that are constrained by data from step-change experiments are thus a device for translating empirical insights gained from step-change experiments into predictions that respect a priori knowledge of slow processes and their role in shaping decadal-scale ecosystem dynamics.
A challenge for model-enabled temporal upscaling is slow processes that are not represented in models or are insufficiently constrained by relatively short-term observations (1–10 years in EMEs) but can modify ecosystem responses in the longer term. Acclimation of photosynthesis and respiration to changes in temperature and CO2 (Way and Yamori, 2014; Prentice et al., 2014) and plasticity in allocation and effects on plant–soil interactions in response to carbon–nutrient balance changes (Zaehle et al., 2014; Poorter et al., 2012) largely lack reliable representations in models, and they may have a strong influence on decadal to centennial carbon, nitrogen, and phosphorous supply and demand, as well as ecosystem dynamics. On even longer timescales, genetic adaptation through selection under new environmental pressures and changes in species compositions may emerge as strong drivers of ecosystem responses. Some experiments on shorter-lived herbaceous species have shown shifts in species composition in response to elevated CO2 (Reich et al., 2018); similar changes in longer-lived organisms are harder to observe in EMEs. These types of responses and their lack of representation in ecosystem models imply limits for data-constrained and model-based temporal upscaling. In terms of short-term plasticity or acclimation, recent advances in using eco-evolutionary optimality in models (Harrison et al., 2021) can represent plastic plant responses, and they have successfully been used in combination with EMEs (Caldararu et al., 2020; Sabot et al., 2022). However, current ecosystem models are not well suited for dealing with changes in species composition, demography, and competition processes. However, trait-based, individual, or cohort-based vegetation demography models (Fisher et al., 2015) have the potential to resolve this limitation. Particularly for evaluating simulated acclimation and other phenotypic plasticity that operates at timescales of months to years, it will be important to make targeted use of insights gained from EMEs. Since this is a knowledge gap in both EMEs and models, it is an opportunity for both communities to work side by side, rather than sequentially, to advance our knowledge of plant adaptation.
In recent years, there has been a global effort to overcome the limitations of the different approaches and try to solve the need for standardised controlled experiments on wide temporal and spatial scales. Fraser et al. (2013) coined the term “coordinated distributed experiments” (CDEs) to describe a global network of standardised experiments distributed across a wide range of ecosystems and climate zones to account for issues of spatial and temporal scales. While initial CDEs were limited in their spatial coverage to Europe or North America, recent ones are striving to be truly global. Examples of global CDEs include the International Drought Experiment (Knapp et al., 2017), which studies the sensitivity of ecosystems to extreme drought events; the Nutrient Network (Borer et al., 2014), which addresses how grasslands are affected by eutrophication and grazing; or the Tree Diversity Network (Paquette et al., 2018), which includes a global collection of tree biodiversity experiments. Coordinated EME networks with standardised protocols and homogenised compilations of published EME data (Van Sundert et al., 2023) can drastically facilitate model–data integration studies, performed for an extended set of sites and experiments. Multi-experiment modelling may be essential for powerful generalisability tests and uncertainty quantification of model predictions, e.g. by a leave-experiment-out cross-validation (see also Sect. 3). Thus, using experiment networks and compilations together with models would increase confidence in the generality of model predictions and partially deal with the upscaling issues. The use of CDE results could also help to improve the parameterisation of process-based models in ecosystems that are underrepresented, but this would require better coordination between response variables measured in the field and those processes included in the models (Smith et al., 2014).
Data assimilation (DA) and model–data integration (MDI) are broad umbrella terms for a variety of statistical methods that fit process-based models to observations. The methods are well established and widely used with remote sensing (Smith et al., 2020; Exbrayat et al., 2019) and eddy covariance data (Kuppel et al., 2012; Fox et al., 2009), at both site and global scales. There are now several DA methods available to the community (Fer et al., 2021; Anderson et al., 2009; Huang et al., 2019), which significantly lowers the barrier to entry for those who want to use such tools.
One of the main issues with using DA methods is that the data used to constrain models are observations in present or past conditions, raising questions about the capacity of resulting models to predict ecosystem responses under future conditions. Therefore, using data from manipulative experiments can be extremely valuable in providing information of as yet unobserved conditions. One other common issue with using remote sensing data to parameterise models is that the observations used in DA need to be variables that are represented in models (MacBean et al., 2022), so most remote sensing indices need to be processed further before they can be used. In contrast, experimental observations provide information that can easily be mapped to model variables – biomass, ecosystem fluxes, soil pools, etc.
MDI provides a formalised approach to make best use of naturally sparse EME observations and combine them with a priori understanding embodied in model structures. Integrating diverse ecosystem data can help in estimating the system state, given physical constraints that are built into the model (e.g. mass conservation) (Jiang et al., 2020). MDI can also provide an approach to formalised model selection (Mark et al., 2018) and a systematic treatment for trading off model complexity and fit to the data. EMEs, in contrast to observational data commonly used in MDI studies, often have particularly strong leverage in discriminating between predictions of alternative models, which other data types often lack. Only if an ecosystem's slow biogeochemical cycling is “hit hard”, with altered conditions in an experimental setup, can underlying processes be revealed. EMEs can thus help to resolve equifinality of model parameter combinations that may not be sufficiently constrained by unperturbed field observations (Keenan et al., 2012) and help to avoid cases of getting the “right answer for the wrong reasons”, where different model representations give the same overarching ecosystem-level state variables but can lead to different results under altered environmental conditions. Thus, EMEs provide key information that is required for robust model selection – the discrimination of alternatively formulated model structures that reflect alternative hypotheses of how ecosystem processes work.
However, typical models used for global biogeochemical cycle and climate change impact simulations are often complex and contain a large number of weakly constrained parameters. In view of the sparsity of EME data, this poses a risk of overfitting. Overfitting may be mitigated using a leave-experiment-out cross-validation approach, where one experiment is systematically left out of the model fitting procedure and used as an out-of-sample test. This may be a way to handle the overfitting risk, enable a more robust calibration of model parameters, and provide a more reliable estimate of the spatial generalisation error. However, the environmental space currently covered by EMEs and their available data is limited, and gaps remain particularly in the tropics and for CO2 experiments in all biomes except temperate forests and grasslands (Van Sundert et al., 2023).
Despite the potential for model-informed ecosystem experiments discussed above, data availability still defines model use because, without data, models are impossible to constrain. Advances in measurement techniques and data processing offer an opportunity to increase the types and frequency of measurement that can be gathered via EMEs. The EMEs can be one of the best locations to develop new data streams because EMEs are typically already well parameterised, offering established data in addition to those from novel sources and potentially benefiting from frequent site visits necessary for non-standard instrument development. Thus in addition to the direct feedback with individual EMEs, models can set the agenda for data development through transparent discussion of the uncertainty and parameter sensitivity.
Perhaps the most straightforward EME experiments can be equipped with proximal sensing devices such as phenocams (Brown et al., 2016) or sun-induced fluorescence (SIF) sensors (Yang et al., 2015). These provide continuous regular measurements of (generally) canopy properties. While these types of measurements are available from spaceborne sensors, local measurements provide more spatial detail and often the possibility of measuring each individual plot separately, thus identifying treatment effects in smaller-scale EMEs. Unlike spaceborne instruments, proximal sensors usually require calibrating sensors between different treatments, but on the other hand they provide regular, location-specific information. Such continuous measurements are extremely useful for model evaluation and development as they are often made at a temporal frequency similar to that of a model time step and can provide information on short-term changes in vegetation responses. Additionally, such data can also be used in DA contexts (Sect. 3) much more easily than irregular spot measurements. One application of note of such proximal sensing data on the horizon is capturing patterns of functional diversity, specifically related to morphological, physiological, or phenological traits that can be measured though reflectance. Such data can be validated in the field (Pacheco-Labrador et al., 2022) and through EMEs can be used to explore causative mechanisms behind changes in environment, changes in function, and changes in species or functional diversity. As models move more towards representing species diversity (De Kauwe et al., 2015) and new trait-based demographic models become more widespread (Xu and Trugman, 2021), such measurements in the context of EMEs can provide an invaluable source of process understanding.
Given widespread coverage, data captured from proximal sensing devices or field measurements may also potentially be upscaled via wide networks (which do not necessarily rely on EMEs). Many modern approaches to this problem rely on machine learning (ML) methods to reach from regional or global scales (Lapeyre et al., 2020; Poggio et al., 2021). But in EMEs and similar contexts, ML also offers another complementary tool: data streams which are difficult to either capture or process (Nair et al., 2023) can be gap-filled or interpreted with higher confidence and representativeness. Further development of techniques currently only possible at a laboratory or homogenous agricultural scale may allow dynamic subannual time series of parameters that are difficult to measure such as photosynthetic capacity (Heckmann et al., 2017); nutrient pools both in biomass and available in soil (Tan et al., 2022); or phenological dynamics beyond leaves, especially those belowground (Wang et al., 2022). This is particularly relevant belowground, where data are particularly sparse. However, technical and logistical limitations remain and on-site instrumentation is considerably more challenging than that of laboratory studies. A key challenge here is capturing both spatial and temporal dynamics at the same time. In many cases, many sensor nodes are preferable because ecosystem properties in general and belowground properties in particular are very heterogeneous. However, when such technologies are fully deployable in the field and an EME context, they can provide data streams for models which were not previously available, reducing uncertainty in difficult-to-measure processes.
Eddy covariance (EC) data are one of the most frequently used data streams for model evaluation, parameterisation, and data assimilation, yet pairings of EMEs with such measurements are rare even though they could be potentially extremely useful. The advantage of using EC data is their high temporal frequency and the large number of globally distributed sites where such data are available. However, as detailed above (Fig. 1), they are purely observational measurements and lack the specific advantages of an EME. A small number of EC sites do use a treatment, albeit on an unreplicated treatment scale (Zhao et al., 2022; El-Madany et al., 2021; Gough et al., 2021). EC experiments are laborious to construct and expensive to operate in tall-stature ecosystems, and even in shorter ecosystems the footprint of an EC tower is a challenge for the application of many treatments relevant to global change. Indeed, only a small number of manipulations can be conducted effectively on this scale and thus far are limited to nutrient treatments and mortality disturbance. These designs also necessitate a well-supported baseline of major ecosystem parameters before treatments to partially substitute for replication. This is a necessary compromise considering the scale and unprecedented realism which can be achieved.
A key aspect of several modern designs is the dispensing of replication. While this is controversial in the classic model of the scientific methods, as we highlighted with the EC measurement example, it offers advantages in this case in terms of a level of realism which could not reasonably be captured by other approaches. Similarly, quantitative gradients allow for parameterisation of response surfaces (as opposed to step changes in standard EMEs) but limit possible replication (Kreyling et al., 2018), and they have successfully been used in ecosystem-scale experiments including a multi-factorial grassland experiment (Piepho et al., 2017) and a warming-peatland experiment (Hanson et al., 2020). In a model–EME context, this allows for the models to be evaluated at multiple treatment levels, circumventing the step-change issues. Large, full ecosystem level experiments with sometimes ground-breaking treatments, such as the TEMPEST flooding experiment (Hopple et al., 2023), are also not replicated but provide invaluable insights into ecosystem functioning. An alternative example is the unique Biosphere 2 setup, which allowed for a fully traceable experiment on a “whole ecosystem”, albeit without replication (Werner et al., 2021). This would allow for the intensive tracing of treatment effects in models through a process-by-process validation, rather than the usual validation against end results such as biomass responses.
Ecosystem manipulative experiments and process-based models are both powerful tools for understanding how terrestrial ecosystems are going to respond to future global change. However, on their own, they each suffer from their own particular shortcomings, including a lack of generality in time and space in the case of experiments and lack of process realism in the case of models. We propose that, when used in combination, experiments and models can achieve their full potential. Going beyond a one-sided process where data from experiments are used to inform models, we lay out a roadmap (Fig. 2) for using experiments and models alongside each other, as well as alongside new types of measurements and techniques, as complementary sources of information. This approach will further our understanding of how terrestrial ecosystems work and increase our predictive capability of the future of terrestrial ecosystems.
No data sets were used in this article.
SC, VR, BDS, TEG, and RN have contributed to the conceptualisation and writing of this paper, including original draft preparation, review, and editing.
The contact author has declared that none of the authors has any competing interests.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the special issue “Ecosystem experiments as a window to future carbon, water, and nutrient cycling in terrestrial ecosystems”. It is not associated with a conference.
We would like to acknowledge the help of Sönke Zaehle and Karin Rebel for their comments on early drafts of this opinion piece.
Richard Nair
was supported by ERC ADG “TERRAFORM” (grant ID 101020824), awarded to Jennifer McElwain. Benjamin D. Stocker was funded by the Swiss
National Science Foundation grant PCEFP2_181115. This work is
a contribution to the LEMONTREE (Land Ecosystem Models based On New Theory,
obseRvations and ExperimEnts) project, funded through Eric
and Wendy Schmidt by recommendation of the Schmidt Futures programme (Benjamin D. Stocker).
The article processing charges for this open-access publication were covered by the Max Planck Society.
This paper was edited by Ivonne Trebs and reviewed by two anonymous referees.
Anderson, J., Hoar, T., Raeder, K., Liu, H., Collins, N., Torn, R., and Avellano, A.: The Data Assimilation Research Testbed: A Community Facility, Bull. Am. Meteorol. Soc., 90, 1283–1296, https://doi.org/10.1175/2009BAMS2618.1, 2009.
Arora, V. K., Katavouta, A., Williams, R. G., Jones, C. D., Brovkin, V., Friedlingstein, P., Schwinger, J., Bopp, L., Boucher, O., Cadule, P., Chamberlain, M. A., Christian, J. R., Delire, C., Fisher, R. A., Hajima, T., Ilyina, T., Joetzjer, E., Kawamiya, M., Koven, C. D., Krasting, J. P., Law, R. M., Lawrence, D. M., Lenton, A., Lindsay, K., Pongratz, J., Raddatz, T., Séférian, R., Tachiiri, K., Tjiputra, J. F., Wiltshire, A., Wu, T., and Ziehn, T.: Carbon–concentration and carbon–climate feedbacks in CMIP6 models and their comparison to CMIP5 models, Biogeosciences, 17, 4173–4222, https://doi.org/10.5194/bg-17-4173-2020, 2020.
Avolio, M. L., Koerner, S. E., La Pierre, K. J., Wilcox, K. R., Wilson, G. W. T., Smith, M. D., and Collins, S. L.: Changes in plant community composition, not diversity, during a decade of nitrogen and phosphorus additions drive above-ground productivity in a tallgrass prairie, J. Ecol., 102, 1649–1660, 2014.
Baer, T., Furrer, G., Zimmermann, S., and Schleppi, P.: Long-term additions of ammonium nitrate to montane forest ecosystems may cause limited soil acidification, even in presence of soil carbonate, Biogeosciences Discuss. [preprint], https://doi.org/10.5194/bg-2023-38, in review, 2023.
Baldocchi, D. D.: How eddy covariance flux measurements have contributed to our understanding of Global Change Biology, Glob. Change Biol., 26, 242–260, https://doi.org/10.1111/gcb.14807, 2020.
Bastos, A., O'Sullivan, M., and Ciais, P.: Sources of Uncertainty in Regional and Global Terrestrial CO2 Exchange Estimates, Global Biogeochem. Cy., 34, e2019GB006393, https://doi.org/10.1029/2019GB006393, 2020.
Bonan, G. B., Hartman, M. D., Parton, W. J., and Wieder, W. R.: Evaluating litter decomposition in earth system models with long-term litterbag experiments: an example using the Community Land Model version 4 (CLM4), Glob. Change Biol., 19, 957–974, https://doi.org/10.1111/gcb.12031, 2013.
Borer, E. T., Harpole, W. S., Adler, P. B., Lind, E. M., Orrock, J. L., Seabloom, E. W., and Smith, M. D.: Finding generality in ecology: a model for globally distributed experiments, Method. Ecol. Evol., 5, 65–73, https://doi.org/10.1111/2041-210x.12125, 2014.
Brown, T. B., Hultine, K. R., Steltzer, H., Denny, E. G., Denslow, M. W., Granados, J., Henderson, S., Moore, D., Nagai, S., SanClements, M., Sánchez-Azofeifa, A., Sonnentag, O., Tazik, D., and Richardson, A. D.: Using phenocams to monitor our changing Earth: toward a global phenocam network, Front. Ecol. Environ., 14, 84–93, https://doi.org/10.1002/fee.1222, 2016.
Caldararu, S., Thum, T., Yu, L., and Zaehle, S.: Whole-plant optimality predicts changes in leaf nitrogen under variable CO2 and nutrient availability, New Phytol., 225, 2331–2346, 2020.
Caldararu, S., Thum, T., Yu, L., Kern, M., Nair, R., and Zaehle, S.: Long-term ecosystem nitrogen limitation from foliar δ15N data and a land surface model, Glob. Change Biol., 28, 493–508, https://doi.org/10.1111/gcb.15933, 2022.
Cleland, E. E., Lind, E. M., DeCrappeo, N. M., DeLorenze, E., Wilkins, R. A., Adler, P. B., Bakker, J. D., Brown, C. S., Davies, K. F., Esch, E., Firn, J., Gressard, S., Gruner, D. S., Hagenah, N., Stanley Harpole, W., Hautier, Y., Hobbie, S. E., Hofmockel, K. S., Kirkman, K., Knops, J., Kopp, C. W., La Pierre, K. J., MacDougall, A., McCulley, R. L., Melbourne, B. A., Moore, J. L., Prober, S. M., Riggs, C., Risch, A. C., Schuetz, M., Stevens, C., Wragg, P. D., Wright, J., Borer, E. T., and Seabloom, E. W.: Belowground Biomass Response to Nutrient Enrichment Depends on Light Limitation Across Globally Distributed Grasslands, Ecosystems 22, 1466–1477, https://doi.org/10.1007/s10021-019-00350-4, 2019.
Collier, N., Hoffman, F. M., Lawrence, D. M., Keppel-Aleks, G., Koven, C. D., Riley, W. J., Mu, M., and Randerson, J. T.: The international land model benchmarking (ILAMB) system: Design, theory, and implementation, J. Adv. Model. Earth Syst., 10, 2731–2754, https://doi.org/10.1029/2018ms001354, 2018.
Cornut, I., Delpierre, N., Laclau, J.-P., Guillemot, J., Nouvellon, Y., Campoe, O., Stape, J. L., Fernanda Santos, V., and le Maire, G.: Potassium-limitation of forest productivity, part 1: A mechanistic model simulating the effects of potassium availability on canopy carbon and water fluxes in tropical eucalypt stands, EGUsphere [preprint], https://doi.org/10.5194/egusphere-2022-883, 2022a.
Cornut, I., le Maire, G., Laclau, J.-P., Guillemot, J., Nouvellon, Y., and Delpierre, N.: Potassium-limitation of forest productivity, part 2: CASTANEA-MAESPA-K shows a reduction in photosynthesis rather than a stoichiometric limitation of tissue formation, EGUsphere [preprint], https://doi.org/10.5194/egusphere-2022-884, 2022b.
Dalling, J. W., Winter, K., Andersen, K. M., and Turner, B. L.: Artefacts of the pot environment on soil nutrient availability: implications for the interpretation of ecological studies, Plant Ecol., 214, 329–338, https://doi.org/10.1007/s11258-013-0172-3, 2013.
De Kauwe, M. G., Medlyn, B. E., Zaehle, S., Walker, A. P., Dietze, M. C., Wang, Y.-P., Luo, Y., Jain, A. K., El-Masri, B., Hickler, T., Wårlind, D., Weng, E., Parton, W. J., Thornton, P. E., Wang, S., Prentice, I. C., Asao, S., Smith, B., McCarthy, H. R., Iversen, C. M., Hanson, P. J., Warren, J. M., Oren, R., and Norby, R. J.: Where does the carbon go? A model-data intercomparison of vegetation carbon allocation and turnover processes at two temperate forest free-air CO2 enrichment sites, New Phytol., 203, 883–899, https://doi.org/10.1111/nph.12847, 2014.
De Kauwe, M. G., Zhou, S.-X., Medlyn, B. E., Pitman, A. J., Wang, Y.-P., Duursma, R. A., and Prentice, I. C.: Do land surface models need to include differential plant species responses to drought? Examining model predictions across a mesic-xeric gradient in Europe, Biogeosciences, 12, 7503–7518, https://doi.org/10.5194/bg-12-7503-2015, 2015.
De Kauwe, M. G., Medlyn, B. E., Walker, A. P., Zaehle, S., Asao, S., Guenet, B., Harper, A. B., Hickler, T., Jain, A. K., Luo, Y., Lu, X., Luus, K., Parton, W. J., Shu, S., Wang, Y.-P., Werner, C., Xia, J., Pendall, E., Morgan, J. A., Ryan, E. M., Carrillo, Y., Dijkstra, F. A., Zelikova, T. J., and Norby, R. J.: Challenging terrestrial biosphere models with data from the long-term multifactor Prairie Heating and CO2 Enrichment experiment, Glob. Change Biol., 23, 3623–3645, https://doi.org/10.1111/gcb.13643, 2017.
Dijkstra, F. A., Blumenthal, D., Morgan, J. A., Pendall, E., Carrillo, Y., and Follett, R. F.: Contrasting effects of elevated CO2 and warming on nitrogen cycling in a semiarid grassland, New Phytol., 187, 426–437, https://doi.org/10.1111/j.1469-8137.2010.03293.x, 2010.
Eastman, B. A., Adams, M. B., Brzostek, E. R., Burnham, M. B., Carrara, J. E., Kelly, C., McNeil, B. E., Walter, C. A., and Peterjohn, W. T.: Altered plant carbon partitioning enhanced forest ecosystem carbon storage after 25 years of nitrogen additions, New Phytol., 230, 1435–1448, https://doi.org/10.1111/nph.17256, 2021.
Eastman, B. A., Wieder, W. R., Hartman, M. D., Brzostek, E. R., and Peterjohn, W. T.: Can models adequately reflect how long-term nitrogen enrichment alters the forest soil carbon cycle?, Biogeosciences Discuss. [preprint], https://doi.org/10.5194/bg-2023-36, in review, 2023.
El-Madany, T. S., Reichstein, M., Carrara, A., Martín, M. P., Moreno, G., Gonzalez-Cascon, R., Peñuelas, J., Ellsworth, D. S., Burchard-Levine, V., Hammer, T. W., Knauer, J., Kolle, O., Luo, Y., Pacheco-Labrador, J., Nelson, J. A., Perez-Priego, O., Rolo, V., Wutzler, T., and Migliavacca, M.: How nitrogen and phosphorus availability change water use efficiency in a Mediterranean Savanna ecosystem, J. Geophys. Res.-Biogeo., 126, e2020JG006005, https://doi.org/10.1029/2020jg006005, 2021.
Exbrayat, J.-F., Bloom, A. A., Carvalhais, N., Fischer, R., Huth, A., MacBean, N., and Williams, M.: Understanding the Land Carbon Cycle with Space Data: Current Status and Prospects, Surv. Geophys., 40, 735–755, https://doi.org/10.1007/s10712-019-09506-2, 2019.
Fer, I., Gardella, A. K., Shiklomanov, A. N., Campbell, E. E., Cowdery, E. M., De Kauwe, M. G., Desai, A., Duveneck, M. J., Fisher, J. B., Haynes, K. D., Hoffman, F. M., Johnston, M. R., Kooper, R., LeBauer, D. S., Mantooth, J., Parton, W. J., Poulter, B., Quaife, T., Raiho, A., Schaefer, K., Serbin, S. P., Simkins, J., Wilcox, K. R., Viskari, T., and Dietze, M. C.: Beyond ecosystem modeling: A roadmap to community cyberinfrastructure for ecological data-model integration, Glob. Change Biol., 27, 13–26, https://doi.org/10.1111/gcb.15409, 2021.
Fick, S. E. and Hijmans, R. J.: WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas, Int. J. Climatol, 37, 4302–4315, https://doi.org/10.1002/joc.5086, 2017.
Fisher, R. A., Muszala, S., Verteinstein, M., Lawrence, P., Xu, C., McDowell, N. G., Knox, R. G., Koven, C., Holm, J., Rogers, B. M., Spessa, A., Lawrence, D., and Bonan, G.: Taking off the training wheels: the properties of a dynamic vegetation model without climate envelopes, CLM4.5(ED), Geosci. Model Dev., 8, 3593–3619, https://doi.org/10.5194/gmd-8-3593-2015, 2015.
Fleischer, K., Rammig, A., De Kauwe, M. G., Walker, A. P., Domingues, T. F., Fuchslueger, L., Garcia, S., Goll, D. S., Grandis, A., Jiang, M., Haverd, V., Hofhansl, F., Holm, J. A., Kruijt, B., Leung, F., Medlyn, B. E., Mercado, L. M., Norby, R. J., Pak, B., von Randow, C., Quesada, C. A., Schaap, K. J., Valverde-Barrantes, O. J., Wang, Y.-P., Yang, X., Zaehle, S., Zhu, Q., and Lapola, D. M.: Amazon forest response to CO2 fertilization dependent on plant phosphorus acquisition, Nat. Geosci., 12, 736–741, https://doi.org/10.1038/s41561-019-0404-9, 2019.
Wieder, W. R., Boehnert, J., Bonan, G. B., and Langseth, M.: Regridded Harmonized World Soil Database v1.2, ORNL DAAC, Oak Ridge, Tennessee, USA, https://doi.org/10.3334/ORNLDAAC/1247, 2014.
Fox, A., Williams, M., Richardson, A. D., Cameron, D., Gove, J. H., Quaife, T., Ricciuto, D., Reichstein, M., Tomelleri, E., Trudinger, C. M., and Wijk, M. T. V.: The REFLEX project: Comparing different algorithms and implementations for the inversion of a terrestrial ecosystem model against eddy covariance data, Agr. Forest Meteorol., 149, 1597–1615, https://doi.org/10.1016/j.agrformet.2009.05.002, 2009.
Fraser, L. H., Henry, H. A. L., Carlyle, C. N., White, S. R., Beierkuhnlein, C., Cahill, J. F., Jr, Casper, B. B., Cleland, E., Collins, S. L., Dukes, J. S., Knapp, A. K., Lind, E., Long, R., Luo, Y., Reich, P. B., Smith, M. D., Sternberg, M., and Turkington, R.: Coordinated distributed experiments: an emerging tool for testing global hypotheses in ecology and environmental science, Front. Ecol. Environ., 11, 147–155, https://doi.org/10.1890/110279, 2013.
Friedlingstein, P., O'Sullivan, M., Jones, M. W., Andrew, R. M., Gregor, L., Hauck, J., Le Quéré, C., Luijkx, I. T., Olsen, A., Peters, G. P., Peters, W., Pongratz, J., Schwingshackl, C., Sitch, S., Canadell, J. G., Ciais, P., Jackson, R. B., Alin, S. R., Alkama, R., Arneth, A., Arora, V. K., Bates, N. R., Becker, M., Bellouin, N., Bittig, H. C., Bopp, L., Chevallier, F., Chini, L. P., Cronin, M., Evans, W., Falk, S., Feely, R. A., Gasser, T., Gehlen, M., Gkritzalis, T., Gloege, L., Grassi, G., Gruber, N., Gürses, Ö., Harris, I., Hefner, M., Houghton, R. A., Hurtt, G. C., Iida, Y., Ilyina, T., Jain, A. K., Jersild, A., Kadono, K., Kato, E., Kennedy, D., Klein Goldewijk, K., Knauer, J., Korsbakken, J. I., Landschützer, P., Lefèvre, N., Lindsay, K., Liu, J., Liu, Z., Marland, G., Mayot, N., McGrath, M. J., Metzl, N., Monacci, N. M., Munro, D. R., Nakaoka, S.-I., Niwa, Y., O'Brien, K., Ono, T., Palmer, P. I., Pan, N., Pierrot, D., Pocock, K., Poulter, B., Resplandy, L., Robertson, E., Rödenbeck, C., Rodriguez, C., Rosan, T. M., Schwinger, J., Séférian, R., Shutler, J. D., Skjelvan, I., Steinhoff, T., Sun, Q., Sutton, A. J., Sweeney, C., Takao, S., Tanhua, T., Tans, P. P., Tian, X., Tian, H., Tilbrook, B., Tsujino, H., Tubiello, F., van der Werf, G. R., Walker, A. P., Wanninkhof, R., Whitehead, C., Willstrand Wranne, A., Wright, R., Yuan, W., Yue, C., Yue, X., Zaehle, S., Zeng, J., and Zheng, B.: Global Carbon Budget 2022, Earth Syst. Sci. Data, 14, 4811–4900, https://doi.org/10.5194/essd-14-4811-2022, 2022.
Gough, C. M., Bohrer, G., Hardiman, B. S., Nave, L. E., Vogel, C. S., Atkins, J. W., Bond-Lamberty, B., Fahey, R. T., Fotis, A. T., Grigri, M. S., Haber, L. T., Ju, Y., Kleinke, C. L., Mathes, K. C., Nadelhoffer, K. J., Stuart-Haëntjens, E., and Curtis, P. S.: Disturbance-accelerated succession increases the production of a temperate forest, Ecol. Appl., 31, e02417, https://doi.org/10.1002/eap.2417, 2021.
Hanson, P. J., Griffiths, N. A., Iversen, C. M., and Norby, R. J.: Rapid net carbon loss from a whole-ecosystem warmed Peatland, AGU Advances, 1, e2020AV000163, https://doi.org/10.1029/2020AV000163, 2020.
Harris, I., Osborn, T. J., Jones, P., and Lister, D.: Version 4 of the CRU TS monthly high-resolution gridded multivariate climate dataset, Sci. Data, 7, 109, https://doi.org/10.1038/s41597-020-0453-3, 2020.
Harrison, S. P., Cramer, W., Franklin, O., Prentice, I. C., Wang, H., Brännström, Å., de Boer, H., Dieckmann, U., Joshi, J., Keenan, T. F., Lavergne, A., Manzoni, S., Mengoli, G., Morfopoulos, C., Peñuelas, J., Pietsch, S., Rebel, K. T., Ryu, Y., Smith, N. G., Stocker, B. D., and Wright, I. J.: Eco-evolutionary optimality as a means to improve vegetation and land-surface models, New Phytol., 231, 2125–2141, https://doi.org/10.1111/nph.17558, 2021.
Heckmann, D., Schlüter, U., and Weber, A. P. M.: Machine Learning Techniques for Predicting Crop Photosynthetic Capacity from Leaf Reflectance Spectra, Mol. Plant, 10, 878–890, https://doi.org/10.1016/j.molp.2017.04.009, 2017.
Hengl, T., Mendes de Jesus, J., Heuvelink, G. B. M., Ruiperez Gonzalez, M., Kilibarda, M., Blagotić, A., Shangguan, W., Wright, M. N., Geng, X., Bauer-Marschallinger, B., Guevara, M. A., Vargas, R., MacMillan, R. A., Batjes, N. H., Leenaars, J. G. B., Ribeiro, E., Wheeler, I., Mantel, S., and Kempen, B.: SoilGrids250m: Global gridded soil information based on machine learning, PLoS One, 12, e0169748, https://doi.org/10.1371/journal.pone.0169748, 2017.
Holm, J. A., Medvigy, D. M., Smith, B., Dukes, J. S., Beier, C., Mishurov, M., Xu, X., Lichstein, J. W., Allen, C. D., Larsen, K. S., Luo, Y., Ficken, C., Pockman, W. T., Anderegg, W. R. L., and Rammig, A.: Exploring the impacts of unprecedented climate extremes on forest ecosystems: hypotheses to guide modeling and experimental studies, Biogeosciences, 20, 2117–2142, https://doi.org/10.5194/bg-20-2117-2023, 2023.
Hopple, A. M., Doro, K. O., Bailey, V. L., Bond-Lamberty, B., McDowell, N., Morris, K. A., Myers-Pigg, A., Pennington, S. C., Regier, P., Rich, R., Sengupta, A., Smith, R., Stegen, J., Ward, N. D., Woodard, S. C., and Megonigal, J. P.: Attaining freshwater and estuarine-water soil saturation in an ecosystem-scale coastal flooding experiment, Environ. Monit. Assess., 195, 425, https://doi.org/10.1007/s10661-022-10807-0, 2023.
Huang, Y., Stacy, M., Jiang, J., Sundi, N., Ma, S., Saruta, V., Jung, C. G., Shi, Z., Xia, J., Hanson, P. J., Ricciuto, D., and Luo, Y.: Realized ecological forecast through an interactive Ecological Platform for Assimilating Data (EcoPAD, v1.0) into models, Geosci. Model Dev., 12, 1119–1137, https://doi.org/10.5194/gmd-12-1119-2019, 2019.
Jiang, M., Medlyn, B. E., Drake, J. E., Duursma, R. A., Anderson, I. C., Barton, C. V. M., Boer, M. M., Carrillo, Y., Castañeda-Gómez, L., Collins, L., Crous, K. Y., De Kauwe, M. G., Dos Santos, B. M., Emmerson, K. M., Facey, S. L., Gherlenda, A. N., Gimeno, T. E., Hasegawa, S., Johnson, S. N., Kännaste, A., Macdonald, C. A., Mahmud, K., Moore, B. D., Nazaries, L., Neilson, E. H. J., Nielsen, U. N., Niinemets, Ü., Noh, N. J., Ochoa-Hueso, R., Pathare, V. S., Pendall, E., Pihlblad, J., Piñeiro, J., Powell, J. R., Power, S. A., Reich, P. B., Renchon, A. A., Riegler, M., Rinnan, R., Rymer, P. D., Salomón, R. L., Singh, B. K., Smith, B., Tjoelker, M. G., Walker, J. K. M., Wujeska-Klause, A., Yang, J., Zaehle, S., and Ellsworth, D. S.: The fate of carbon in a mature forest under carbon dioxide enrichment, Nature, 580, 227–231, https://doi.org/10.1038/s41586-020-2128-9, 2020.
Jung, M., Schwalm, C., Migliavacca, M., Walther, S., Camps-Valls, G., Koirala, S., Anthoni, P., Besnard, S., Bodesheim, P., Carvalhais, N., Chevallier, F., Gans, F., Goll, D. S., Haverd, V., Köhler, P., Ichii, K., Jain, A. K., Liu, J., Lombardozzi, D., Nabel, J. E. M. S., Nelson, J. A., O'Sullivan, M., Pallandt, M., Papale, D., Peters, W., Pongratz, J., Rödenbeck, C., Sitch, S., Tramontana, G., Walker, A., Weber, U., and Reichstein, M.: Scaling carbon fluxes from eddy covariance sites to globe: synthesis and evaluation of the FLUXCOM approach, Biogeosciences, 17, 1343–1365, https://doi.org/10.5194/bg-17-1343-2020, 2020.
Karger, D. N., Schmatz, D. R., Dettling, G., and Zimmermann, N. E.: High-resolution monthly precipitation and temperature time series from 2006 to 2100, Sci. Data, 7, 248, https://doi.org/10.1038/s41597-020-00587-y, 2020.
Kattge, J., Bönisch, G., Díaz, S., Lavorel, S., Prentice, I. C., Leadley, P., Tautenhahn, S., Werner, G. D. A., Aakala, T., Abedi, M., Acosta, A. T. R., Adamidis, G. C., Adamson, K., Aiba, M., Albert, C. H., Alcántara, J. M., Alcázar C, C., Aleixo, I., Ali, H., Amiaud, B., Ammer, C., Amoroso, M. M., Anand, M., Anderson, C., Anten, N., Antos, J., Apgaua, D. M. G., Ashman, T.-L., Asmara, D. H., Asner, G. P., Aspinwall, M., Atkin, O., Aubin, I., Baastrup-Spohr, L., Bahalkeh, K., Bahn, M., Baker, T., Baker, W. J., Bakker, J. P., Baldocchi, D., Baltzer, J., Banerjee, A., Baranger, A., Barlow, J., Barneche, D. R., Baruch, Z., Bastianelli, D., Battles, J., Bauerle, W., Bauters, M., Bazzato, E., Beckmann, M., Beeckman, H., Beierkuhnlein, C., Bekker, R., Belfry, G., Belluau, M., Beloiu, M., Benavides, R., Benomar, L., Berdugo-Lattke, M. L., Berenguer, E., Bergamin, R., Bergmann, J., Bergmann Carlucci, M., Berner, L., Bernhardt-Römermann, M., Bigler, C., Bjorkman, A. D., Blackman, C., Blanco, C., Blonder, B., Blumenthal, D., Bocanegra-González, K. T., Boeckx, P., Bohlman, S., Böhning-Gaese, K., Boisvert-Marsh, L., Bond, W., Bond-Lamberty, B., Boom, A., Boonman, C. C. F., Bordin, K., Boughton, E. H., Boukili, V., Bowman, D. M. J. S., Bravo, S., Brendel, M. R., Broadley, M. R., Brown, K. A., Bruelheide, H., Brumnich, F., Bruun, H. H., Bruy, D., Buchanan, S. W., Bucher, S. F., Buchmann, N., Buitenwerf, R., Bunker, D. E., et al.: TRY plant trait database – enhanced coverage and open access, Glob. Change Biol., 26, 119–188, https://doi.org/10.1111/gcb.14904, 2020.
Keenan, T. F., Davidson, E. A., Munger, J. W., and Richardson, A. D.: Rate my data: quantifying the value of ecological data for the development of models of the terrestrial carbon cycle, Ecol. Appl., 23, 273–286, https://doi.org/10.1890/12-0747.1, 2012.
Knapp, A. K., Avolio, M. L., Beier, C., Carroll, C. J. W., Collins, S. L., Dukes, J. S., Fraser, L. H., Griffin-Nolan, R. J., Hoover, D. L., Jentsch, A., Loik, M. E., Phillips, R. P., Post, A. K., Sala, O. E., Slette, I. J., Yahdjian, L., and Smith, M. D.: Pushing precipitation to the extremes in distributed experiments: recommendations for simulating wet and dry years, Glob. Change Biol., 23, 1774–1782, https://doi.org/10.1111/gcb.13504, 2017.
Kreyling, J., Schweiger, A. H., Bahn, M., Ineson, P., Migliavacca, M., Morel-Journel, T., Christiansen, J. R., Schtickzelle, N., and Larsen, K. S.: To replicate, or not to replicate – that is the question: how to tackle nonlinear responses in ecological experiments, Ecol. Lett., 21, 1629–1638, https://doi.org/10.1111/ele.13134, 2018.
Kulmatiski, A., Holdrege, M. C., Chirvasa, C., and Beard, K. H.: Root distributions predict shrub-steppe responses to precipitation intensity, Biogeosciences Discuss. [preprint], https://doi.org/10.5194/bg-2023-13, in review, 2023.
Kuppel, S., Peylin, P., Chevallier, F., Bacour, C., Maignan, F., and Richardson, A. D.: Constraining a global ecosystem model with multi-site eddy-covariance data, Biogeosciences, 9, 3757–3776, https://doi.org/10.5194/bg-9-3757-2012, 2012.
Lapeyre, C. J., Cazard, N., Roy, P. T., Ricci, S., and Zaoui, F.: Reconstruction of Hydraulic Data by Machine Learning, Advances in Hydroinformatics, Springer Water. Springer, Singapore, 701–715, https://doi.org/10.1007/978-981-15-5436-0_54, 2020.
Li, S., Waring, B. G., Powers, J. S., and Medvigy, D.: Tropical Dry Forest Response to Nutrient Fertilization: A Model Validation and Sensitivity Analysis, Biogeosciences Discuss. [preprint], https://doi.org/10.5194/bg-2022-243, in review, 2022.
Liang, X., Zhang, T., Lu, X., Ellsworth, D. S., BassiriRad, H., You, C., Wang, D., He, P., Deng, Q., Liu, H., Mo, J., and Ye, Q.: Global response patterns of plant photosynthesis to nitrogen addition: A meta-analysis, Glob. Change Biol., 26, 3585–3600, https://doi.org/10.1111/gcb.15071, 2020.
Ma, S., Jiang, L., Wilson, R. M., Chanton, J. P., Bridgham, S., Niu, S., Iversen, C. M., Malhotra, A., Jiang, J., Lu, X., Huang, Y., Keller, J., Xu, X., Ricciuto, D. M., Hanson, P. J., and Luo, Y.: Evaluating alternative ebullition models for predicting peatland methane emission and its pathways via data–model fusion, Biogeosciences, 19, 2245–2262, https://doi.org/10.5194/bg-19-2245-2022, 2022.
MacBean, N., Liddy, H., Quaife, T., Kolassa, J., and Fox, A.: Building a Land Data Assimilation Community to Tackle Technical Challenges in Quantifying and Reducing Uncertainty in Land Model Predictions, Bull. Am. Meteorol. Soc., 103, E733–E740, https://doi.org/10.1175/BAMS-D-21-0228.1, 2022.
Magill, A. H., Aber, J. D., Currie, W. S., Nadelhoffer, K. J., Martin, M. E., McDowell, W. H., Melillo, J. M., and Steudler, P.: Ecosystem response to 15 years of chronic nitrogen additions at the Harvard Forest LTER, Massachusetts, USA, Forest Ecol. Manag., 196, 7–28, https://doi.org/10.1016/j.foreco.2004.03.033, 2004.
Mark, C., Metzner, C., Lautscham, L., Strissel, P. L., Strick, R., and Fabry, B.: Bayesian model selection for complex dynamic systems, Nat. Commun., 9, 1803, https://doi.org/10.1038/s41467-018-04241-5, 2018.
McDowell, N. G., Fisher, R. A., Xu, C., Domec, J. C., Hölttä, T., Mackay, D. S., Sperry, J. S., Boutz, A., Dickman, L., Gehres, N., Limousin, J. M., Macalady, A., Martínez-Vilalta, J., Mencuccini, M., Plaut, J. A., Ogée, J., Pangle, R. E., Rasse, D. P., Ryan, M. G., Sevanto, S., Waring, R. H., Williams, A. P., Yepez, E. A., and Pockman, W. T.: Evaluating theories of drought-induced vegetation mortality using a multimodel-experiment framework, New Phytol., 200, 304–321, https://doi.org/10.1111/nph.12465, 2013.
Medlyn, B. E., Zaehle, S., De Kauwe, M. G., Walker, A. P., Dietze, M. C., Hanson, P. J., Hickler, T., Jain, A. K., Luo, Y., Parton, W., Prentice, I. C., Thornton, P. E., Wang, S., Wang, Y.-P., Weng, E., Iversen, C. M., McCarthy, H. R., Warren, J. M., Oren, R., and Norby, R. J.: Using ecosystem experiments to improve vegetation models, Nat. Clim. Change, 5, 528–534, https://doi.org/10.1038/nclimate2621, 2015.
Medlyn, B. E., De Kauwe, M. G., Zaehle, S., Walker, A. P., Duursma, R. A., Luus, K., Mishurov, M., Pak, B., Smith, B., Wang, Y.-P., Yang, X., Crous, K. Y., Drake, J. E., Gimeno, T. E., Macdonald, C. A., Norby, R. J., Power, S. A., Tjoelker, M. G., and Ellsworth, D. S.: Using models to guide field experiments: a priori predictions for the CO2 response of a nutrient- and water-limited native Eucalypt woodland, Glob. Change Biol., 22, 2834–2851, https://doi.org/10.1111/gcb.13268, 2016.
Mendoza-Martinez, V., Collins, S. L., and McLaren, J. R.: Long-term fertilization increases soil but not plant or microbial N in a Chihuahuan Desert Grassland, Biogeosciences Discuss. [preprint], https://doi.org/10.5194/bg-2023-41, in review, 2023.
Meyerholt, J. and Zaehle, S.: The role of stoichiometric flexibility in modelling forest ecosystem responses to nitrogen fertilization, New Phytol., 208, 1042–1055, https://doi.org/10.1111/nph.13547, 2015.
Nair, R., Strube, M., Hertel, M., Kolle, O., Rolo, V., and Migliavacca, M.: High frequency root dynamics: sampling and interpretation using replicated robotic minirhizotrons, J. Exp. Bot., 74, 769–786, https://doi.org/10.1093/jxb/erac427, 2023.
Niu, G., Wang, Y., Wang, R., Ning, Q., Guan, H., Yang, J., Lu, X., Han, X., and Huang, J.: Intensity and duration of nitrogen addition jointly alter soil nutrient availability in a temperate grassland, J. Geophys. Res.-Biogeo., 127, e2021JG006698, https://doi.org/10.1029/2021jg006698, 2022.
Norby, R. J., De Kauwe, M. G., Domingues, T. F., Duursma, R. A., Ellsworth, D. S., Goll, D. S., Lapola, D. M., Luus, K. A., MacKenzie, A. R., Medlyn, B. E., Pavlick, R., Rammig, A., Smith, B., Thomas, R., Thonicke, K., Walker, A. P., Yang, X., and Zaehle, S.: Model-data synthesis for the next generation of forest free-air CO2 enrichment (FACE) experiments, New Phytol., 209, 17–28, https://doi.org/10.1111/nph.13593, 2016.
Ntirugulirwa, B., Zibera, E., Epaphrodite, N., Manishimwe, A., Nsabimana, D., Uddling, J., and Wallin, G.: Contrasting growth and mortality responses of different species lead to shifts in tropical montane tree community composition in a warmer climate, Biogeosciences Discuss. [preprint], https://doi.org/10.5194/bg-2023-42, in review, 2023.
Pacheco-Labrador, J., Migliavacca, M., Ma, X., Mahecha, M. D., Carvalhais, N., Weber, U., Benavides, R., Bouriaud, O., Barnoaiea, I., Coomes, D. A., Bohn, F. J., Kraemer, G., Heiden, U., Huth, A., and Wirth, C.: Challenging the link between functional and spectral diversity with radiative transfer modeling and data, Remote Sens. Environ., 280, 113170, https://doi.org/10.1016/j.rse.2022.113170, 2022.
Paquette, A., Hector, A., Castagneyrol, B., Vanhellemont, M., Koricheva, J., Scherer-Lorenzen, M., Verheyen, K., and TreeDivNet: A million and more trees for science, Nat. Ecol. Evol., 2, 763–766, https://doi.org/10.1038/s41559-018-0544-0, 2018.
Parton, W. J., Morgan, J. A., Wang, G., and Del Grosso, S.: Projected ecosystem impact of the Prairie Heating and CO2 Enrichment experiment, New Phytol., 174, 823–834, https://doi.org/10.1111/j.1469-8137.2007.02052.x, 2007.
Piao, S., Wang, X., Wang, K., Li, X., Bastos, A., Canadell, J. G., Ciais, P., Friedlingstein, P., and Sitch, S.: Interannual variation of terrestrial carbon cycle: Issues and perspectives, Glob. Change Biol., 26, 300–318, https://doi.org/10.1111/gcb.14884, 2020.
Piepho, H.-P., Herndl, M., Pötsch, E. M., and Bahn, M.: Designing an experiment with quantitative treatment factors to study the effects of climate change, J. Agron. Crop Sci., 203, 584–592, https://doi.org/10.1111/jac.12225, 2017.
Poggio, L., de Sousa, L. M., Batjes, N. H., Heuvelink, G. B. M., Kempen, B., Ribeiro, E., and Rossiter, D.: SoilGrids 2.0: producing soil information for the globe with quantified spatial uncertainty, SOIL, 7, 217–240, https://doi.org/10.5194/soil-7-217-2021, 2021.
Poorter, H., Niklas, K. J., Reich, P. B., Oleksyn, J., Poot, P., and Mommer, L.: Biomass allocation to leaves, stems and roots: meta-analyses of interspecific variation and environmental control, New Phytol., 193, 30–50, https://doi.org/10.1111/j.1469-8137.2011.03952.x, 2012.
Powell, T. L., Galbraith, D. R., Christoffersen, B. O., Harper, A., Imbuzeiro, H. M. A., Rowland, L., Almeida, S., Brando, P. M., da Costa, A. C. L., Costa, M. H., Levine, N. M., Malhi, Y., Saleska, S. R., Sotta, E., Williams, M., Meir, P., and Moorcroft, P. R.: Confronting model predictions of carbon fluxes with measurements of Amazon forests subjected to experimental drought, New Phytol., 200, 350–365, https://doi.org/10.1111/nph.12390, 2013.
Prentice, I. C., Colin Prentice, I., Bondeau, A., Cramer, W., Harrison, S. P., Hickler, T., Lucht, W., Sitch, S., Smith, B., and Sykes, M. T.: Dynamic Global Vegetation Modeling: Quantifying Terrestrial Ecosystem Responses to Large-Scale Environmental Change, in: Terrestrial Ecosystems in a Changing World, edited by: Canadell, J. G., Pataki, D. E., and Pitelka, L. F., Spinger, 175–192, https://doi.org/10.1007/978-3-540-32730-1_15, 2007.
Prentice, I. C., Dong, N., Gleason, S. M., Maire, V., and Wright, I. J.: Balancing the costs of carbon gain and water transport: testing a new theoretical framework for plant functional ecology, Ecol. Lett., 17, 82–91, https://doi.org/10.1111/ele.12211, 2014.
Prentice, I. C., Liang, X., Medlyn, B. E., and Wang, Y.-P.: Reliable, robust and realistic: the three R's of next-generation land-surface modelling, Atmos. Chem. Phys., 15, 5987–6005, https://doi.org/10.5194/acp-15-5987-2015, 2015.
Raoult, N., Edouard-Rambaut, L.-A., Vuichard, N., Bastrikov, V., Lansø, A. S., Guenet, B., and Peylin, P.: Using free air CO2 enrichment data to constrain land surface model projections of the terrestrial carbon cycle, EGUsphere [preprint], https://doi.org/10.5194/egusphere-2023-360, 2023.
Reich, P. B., Hobbie, S. E., Lee, T. D., and Pastore, M. A.: Unexpected reversal of C3 versus C4 grass response to elevated CO2 during a 20-year field experiment, Science, 360, 317–320, https://doi.org/10.1126/science.aas9313, 2018.
Sabot, M. E. B., De Kauwe, M. G., Pitman, A. J., Ellsworth, D. S., Medlyn, B. E., Caldararu, S., Zaehle, S., Crous, K. Y., Gimeno, T. E., Wujeska-Klause, A., Mu, M., and Yang, J.: Predicting resilience through the lens of competing adjustments to vegetation function, Plant Cell Environ., 45, 2744–2761, https://doi.org/10.1111/pce.14376, 2022.
Seiler, C., Melton, J. R., Arora, V. K., Sitch, S., Friedlingstein, P., Anthoni, P., Goll, D., Jain, A. K., Joetzjer, E., Lienert, S., Lombardozzi, D., Luyssaert, S., Nabel, J. E. M. S., Tian, H., Vuichard, N., Walker, A. P., Yuan, W., and Zaehle, S.: Are Terrestrial Biosphere Models Fit for Simulating the Global Land Carbon Sink?, J. Adv. Model. Earth Syst., 14, e2021MS002946, https://doi.org/10.1029/2021MS002946, 2022.
Smith, N. G., Rodgers, V. L., Brzostek, E. R., Kulmatiski, A., Avolio, M. L., Hoover, D. L., Koerner, S. E., Grant, K., Jentsch, A., Fatichi, S., and Niyogi, D.: Toward a better integration of biological data from precipitation manipulation experiments into Earth system models, Rev. Geophys., 52, 412–434, https://doi.org/10.1002/2014RG000458, 2014.
Smith, W. K., Fox, A. M., MacBean, N., Moore, D. J. P., and Parazoo, N. C.: Constraining estimates of terrestrial carbon uptake: new opportunities using long-term satellite observations and data assimilation, New Phytol., 225, 105–112, https://doi.org/10.1111/nph.16055, 2020.
Stocker, B. D., Prentice, I. C., Cornell, S. E., Davies-Barnard, T., Finzi, A. C., Franklin, O., Janssens, I., Larmola, T., Manzoni, S., Näsholm, T., Raven, J. A., Rebel, K. T., Reed, S., Vicca, S., Wiltshire, A., and Zaehle, S.: Terrestrial nitrogen cycling in Earth system models revisited, New Phytol., 210, 1165–1168, https://doi.org/10.1111/nph.13997, 2016.
Tan, B., You, W., Tian, S., Xiao, T., Wang, M., Zheng, B., and Luo, L.: Soil Nitrogen Content Detection Based on Near-Infrared Spectroscopy, Sensors, 22, 8013, https://doi.org/10.3390/s22208013, 2022.
Terrer, C., Phillips, R. P., Hungate, B. A., Rosende, J., Pett-Ridge, J., Craig, M. E., van Groenigen, K. J., Keenan, T. F., Sulman, B. N., Stocker, B. D., Reich, P. B., Pellegrini, A. F. A., Pendall, E., Zhang, H., Evans, R. D., Carrillo, Y., Fisher, J. B., Van Sundert, K., Vicca, S., and Jackson, R. B.: A trade-off between plant and soil carbon storage under elevated CO2, Nature, 591, 599–603, https://doi.org/10.1038/s41586-021-03306-8, 2021.
Thomas, R. Q., Zaehle, S., Templer, P. H., and Goodale, C. L.: Global patterns of nitrogen limitation: confronting two global biogeochemical models with observations, Glob. Change Biol., 19, 2986–2998, https://doi.org/10.1111/gcb.12281, 2013.
Van Sundert, K., Van Sundert, K., Leuzinger, S.-F. Bader, M. K., Chang, S. X., De Kauwe, M. G., Dukes, J. S., Adam Langley, J., Ma, Z., Mariën, B., Reynaert, S., Ru, J., Song, J., Stocker, B., Terrer, C., Thoresen, J., Vanuytrecht, E., Wan, S., Yue, K., and Vicca, S.: When things get MESI: The Manipulation Experiments Synthesis Initiative – A coordinated effort to synthesize terrestrial global change experiments, Glob. Change Biol., 29, 1922–1938, https://doi.org/10.1111/gcb.16585, 2023.
Vargas Gutiérrez, G., Pérez-Aviles, D., Raczka, N., Pereira-Arias, D., Tijerín-Triviño, J., Pereira-Arias, L. D., Medvigy, D., Waring, B. G., Morrisey, E., Brzostek, E., and Powers, J. S.: Throughfall exclusion and fertilization effects on tropical dry forest tree plantations, a large-scale experiment, Biogeosciences, 20, 2143–2160, https://doi.org/10.5194/bg-20-2143-2023, 2023.
Walker, A. P., Hanson, P. J., De Kauwe, M. G., Medlyn, B. E., Zaehle, S., Asao, S., Dietze, M., Hickler, T., Huntingford, C., Iversen, C. M., Jain, A., Lomas, M., Luo, Y., McCarthy, H., Parton, W. J., Prentice, I. C., Thornton, P. E., Wang, S., Wang, Y.-P., Warlind, D., Weng, E., Warren, J. M., Woodward, F. I., Oren, R., and Norby, R. J.: Comprehensive ecosystem model-data synthesis using multiple data sets at two temperate forest free-air CO2 enrichment experiments: Model performance at ambient CO2 concentration : FACE MODEL-DATA SYNTHESIS, J. Geophys. Res.-Biogeo., 119, 937–964, https://doi.org/10.1002/2013JG002553, 2014.
Wang, B., Luke McCormack, M., Ricciuto, D. M., Yang, X., and Iversen, C. M.: Embracing fine-root system complexity to improve the predictive understanding of ecosystem functioning, bioRxiv, 2022.10.07.511037, https://doi.org/10.1101/2022.10.07.511037, 2022.
Way, D. A. and Yamori, W.: Thermal acclimation of photosynthesis: on the importance of adjusting our definitions and accounting for thermal acclimation of respiration, Photosynth. Res., 119, 89–100, https://doi.org/10.1007/s11120-013-9873-7, 2014.
Werner, C., Meredith, L. K., Ladd, S. N., Ingrisch, J., Kübert, A., van Haren, J., Bahn, M., Bailey, K., Bamberger, I., Beyer, M., Blomdahl, D., Byron, J., Daber, E., Deleeuw, J., Dippold, M. A., Fudyma, J., Gil-Loaiza, J., Honeker, L. K., Hu, J., Huang, J., Klüpfel, T., Krechmer, J., Kreuzwieser, J., Kühnhammer, K., Lehmann, M. M., Meeran, K., Misztal, P. K., Ng, W.-R., Pfannerstill, E., Pugliese, G., Purser, G., Roscioli, J., Shi, L., Tfaily, M., and Williams, J.: Ecosystem fluxes during drought and recovery in an experimental forest, Science, 374, 1514–1518, https://doi.org/10.1126/science.abj6789, 2021.
Wieder, W. R., Lawrence, D. M., Fisher, R. A., Bonan, G. B., Cheng, S. J., Goodale, C. L., Grandy, A. S., Koven, C. D., Lombardozzi, D. L., Oleson, K. W., and Thomas, R. Q.: Beyond Static Benchmarking: Using Experimental Manipulations to Evaluate Land Model Assumptions, Global Biogeochem. Cy., 33, 1289–1309, https://doi.org/10.1029/2018GB006141, 2019.
Wilcox, K. R., Collins, S. L., Knapp, A. K., Pockman, W., Shi, Z., Smith, M. D., and Luo, Y.: Assessing carbon storage capacity and saturation across six central US grasslands using data–model integration, Biogeosciences, 20, 2707–2725, https://doi.org/10.5194/bg-20-2707-2023, 2023.
Xiao, J., Chevallier, F., Gomez, C., Guanter, L., Hicke, J. A., Huete, A. R., Ichii, K., Ni, W., Pang, Y., Rahman, A. F., Sun, G., Yuan, W., Zhang, L., and Zhang, X.: Remote sensing of the terrestrial carbon cycle: A review of advances over 50 years, Remote Sens. Environ., 233, 111383, https://doi.org/10.1016/j.rse.2019.111383, 2019.
Xu, X. and Trugman, A. T.: Trait-Based Modeling of Terrestrial Ecosystems: Advances and Challenges Under Global Change, Curr. Clim. Change Rep., 7, 1–13, https://doi.org/10.1007/s40641-020-00168-6, 2021.
Yang, X., Tang, J., Mustard, J. F., Lee, J.-E., Rossini, M., Joiner, J., Munger, J. W., Kornfeld, A., and Richardson, A. D.: Solar-induced chlorophyll fluorescence that correlates with canopy photosynthesis on diurnal and seasonal scales in a temperate deciduous forest, Geophys. Res. Lett., 42, 2977–2987, https://doi.org/10.1002/2015gl063201, 2015.
Zaehle, S., Friedlingstein, P., and Friend, A. D.: Terrestrial nitrogen feedbacks may accelerate future climate change, Geophys. Res. Lett., 37, L01401, https://doi.org/10.1029/2009gl041345, 2010.
Zaehle, S., Medlyn, B. E., De Kauwe, M. G., Walker, A. P., Dietze, M. C., Hickler, T., Luo, Y., Wang, Y.-P., El-Masri, B., Thornton, P., Jain, A., Wang, S., Warlind, D., Weng, E., Parton, W., Iversen, C. M., Gallet-Budynek, A., McCarthy, H., Finzi, A., Hanson, P. J., Prentice, I. C., Oren, R., and Norby, R. J.: Evaluation of 11 terrestrial carbon–nitrogen cycle models against observations from two temperate Free-Air CO2 Enrichment studies, New Phytol., 202, 803–822, https://doi.org/10.1111/nph.12697, 2014.
Zhao, P., Chi, J., Nilsson, M. B., Löfvenius, M. O., Högberg, P., Jocher, G., Lim, H., Mäkelä, A., Marshall, J., Ratcliffe, J., Tian, X., Näsholm, T., Lundmark, T., Linder, S., and Peichl, M.: Long-term nitrogen addition raises the annual carbon sink of a boreal forest to a new steady-state, Agr. Forest Meteorol., 324, 109112, https://doi.org/10.1016/j.agrformet.2022.109112, 2022.
- Abstract
- Introduction
- Not a one-sided relationship: what can models do for experiments?
- Model–data integration
- New data for EME–model synthesis
- Conclusions and outlook
- Data availability
- Author contributions
- Competing interests
- Disclaimer
- Special issue statement
- Acknowledgements
- Financial support
- Review statement
- References
- Abstract
- Introduction
- Not a one-sided relationship: what can models do for experiments?
- Model–data integration
- New data for EME–model synthesis
- Conclusions and outlook
- Data availability
- Author contributions
- Competing interests
- Disclaimer
- Special issue statement
- Acknowledgements
- Financial support
- Review statement
- References