the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Ice nucleation by viruses and their potential for cloud glaciation
Michael P. Adams
Nina S. Atanasova
Svetlana Sofieva
Janne Ravantti
Aino Heikkinen
Zoé Brasseur
Jonathan Duplissy
Dennis H. Bamford
Benjamin J. Murray
In order to effectively predict the formation of ice in clouds we need to know which subsets of aerosol particles are effective at nucleating ice, how they are distributed and where they are from. A large proportion of ice-nucleating particles (INPs) in many locations are likely of biological origin, and some INPs are extremely small, being just tens of nanometres in size. The identity and sources of such INPs are not well characterized. Here, we show that several different types of virus particles can nucleate ice, with up to about 1 in 20 million virus particles able to nucleate ice at −20 ∘C. In terms of the impact on cloud glaciation, the ice-nucleating ability (the fraction which are ice nucleation active as a function of temperature) taken together with typical virus particle concentrations in the atmosphere leads to the conclusion that virus particles make a minor contribution to the atmospheric ice-nucleating particle population in the terrestrial-influenced atmosphere. However, they cannot be ruled out as being important in the remote marine atmosphere. It is striking that virus particles have an ice-nucleating activity, and further work should be done to explore other types of viruses for both their ice-nucleating potential and to understand the mechanism by which viruses nucleate ice.
- Article
(933 KB) - Full-text XML
-
Supplement
(735 KB) - BibTeX
- EndNote
The formation of ice in clouds is critically important for the planet's radiative balance and our prediction of future changes in climate with increased greenhouse gas concentrations (Vergara-Temprado et al., 2018; Tan et al., 2016). Ice-nucleating particles (INPs) have the potential to cause supercooled liquid cloud droplets, present in mixed-phase clouds, to freeze at temperatures greater than homogenous freezing, which can drastically alter cloud properties such as albedo, composition and lifetime (Murray et al., 2021; Hoose and Möhler 2012; Kanji et al., 2017; Hawker et al., 2021). Despite the potential importance of INPs, there is still a lack of knowledge regarding their characteristics, sources, and ultimately their temporal and spatial distribution around the globe.
Our current knowledge of atmospheric INPs (under mixed-phase cloud conditions) suggests a number of potentially important aerosol types, including mineral dust, marine organics and terrestrial bioaerosols (DeMott et al., 2010; Kanji et al., 2017). The characteristics and source regions for mineral dust are relatively better understood than other potentially important INPs, and mineral dust from both high- (Sanchez-Marroquin et al., 2020; Tobo et al., 2019) and low-latitude sources is thought to be the dominant INP around much of the globe at temperatures ∘C. Marine organics and terrestrial bioaerosols have both been demonstrated to play a major role in the global INP burden, but the nature of these INPs is less well understood than that of mineral dust. Marine organics are of particular importance in remote marine regions where there is little mineral dust (Wilson et al., 2015; Burrows et al., 2013). Terrestrial bioaerosols are thought to outcompete mineral dust in the terrestrial mid-latitudes at temperatures ∘C; however, their source(s) and nature are at present poorly understood (Conen et al., 2016; McCluskey et al., 2018a; O'Sullivan et al., 2018; Vergara-Temprado et al., 2017).
Known INPs of biological origin include bacteria, fungi, pollen and marine organics amongst others (Kanji et al., 2017). Bacteria and fungi exhibit ice nucleation due to the presence of ice-nucleating proteins (Green and Warren, 1985; Pouleur et al., 1992; Lindow et al., 1982), whilst the ice-nucleating ability of pollen has been linked to polysaccharides (Pummer et al., 2012; Dreischmeier et al., 2017). Marine organic INPs, associated with sea spray, are thought to be biogenic and are often smaller than 0.22 µm, but it is currently not clear exactly what these ice-nucleating particles are, and there may be multiple marine INP types (Creamean et al., 2019; DeMott et al., 2016; Irish et al., 2017, 2019; Schnell et al., 1975; Wang et al., 2015; Wilson et al., 2015). Compared to non-biological INPs, some microorganisms such as specific bacteria or fungi nucleate ice at relatively high temperatures; for example, the best-studied ice-nucleating bacterium, Pseudomonas syringae (P. syringae), can nucleate ice at temperatures up to −2 ∘C (Morris et al., 2004, 2013; Lindow et al., 1978). Despite the ice nucleation potential of primary biological aerosol particles, recognized since the 1970s (Schnell et al., 1976), the global distribution and sources of biological INPs remain poorly understood (Murray et al., 2012; Kanji et al., 2017). Hence characterizing the ice-nucleating ability of the various categories of biological aerosol particles is important.
In bacteria, membrane proteins are thought to interact with water and impose order in supercooled water in such a way as to promote nucleation of ice. Pandey et al. (2016) demonstrated that in the case of P. syringae patterned hydrophilic–hydrophobic regions due to the interactions of amino acids belonging to the membrane protein led to the increased ordering of water molecules coupled with efficient removal of thermal energy from the surrounding water molecules into the bacterial cell. This mechanism could potentially protect microorganisms at sub-zero temperatures and preserve their viability and infectivity in the atmosphere (Wilson et al., 2012; Morris et al., 2013). Whether or not a bacterium has the potential to produce ice-nucleating proteins is dependent on the presence of an ice nucleation gene. At present, eight ice-nucleating proteins are known and reviewed in the protein database UniProt, each with an associated gene (protein IDs: O33479, P06620, Q47879, P16239, O30611, P09815, P20469, P18127). It is thought that a single functional ice nucleation protein gene in bacteria is both necessary and sufficient for ice nucleation activity. The ice nucleation activity (INA) of a bacterium that has a gene for the ice-nucleating protein in its genome depends on the expression of the gene (i.e. if the protein coded by the gene is actually produced by the bacterium), the integration of the protein into the outer membrane of the bacterial cell and stabilization of the protein complex by the surrounding membrane constituents.
Viruses are a presently under-studied source with respect to their potential as atmospheric biological INPs. Very little is known about viruses in the atmosphere in general, and even less is known about their potential to influence cloud properties through cloud glaciation. The only studies we are aware of in which the ice-nucleating ability of a virus was examined are those of Junge and Swanson (2008), who studied the polar Colwellia phage 9A, and Cascajo-Castresana et al. (2020), who studied a series of common proteins and a single virus. The former found that these virus particles did not nucleate ice in their experimental system. The latter observed ice nucleation activity in the Tobacco mosaic virus (TMV), a plant virus that infects the family of Solanaceae such us tobacco, tomato or pepper. TMV was shown to be above the baseline of the buffer solution it was suspended in, and it was noted in the study that whilst TMV had a lower onset freezing temperature than other samples in the study (a range of proteins), when normalized to cumulative active site density it was more active.
Compared to bacteria and other micron-sized, single-celled microorganisms, viruses are considerably smaller (from ∼ 25 nm in diameter; except for the nucleocytoplasmic large DNA viruses that are cellular size). The small size of virus particles means that their atmospheric lifetime has the potential to be on the order of many days to weeks in the atmosphere, although this will depend on the size of the particles that they are internally mixed with. This is considerably longer than the lifetime of larger biological particles, especially those larger than ∼ 10 µm, which have lifetimes of only hours (Grythe et al., 2014; Reche et al., 2018) and therefore have atmospheric abundances which decrease rapidly during transport (Hoose et al., 2010).
In addition to supermicron entities such as bacteria, submicron-sized biological particles have also been shown to be effective ice-nucleating particles (O'Sullivan et al., 2015). For example, it has been shown that there are biological INPs belonging to fungal and pollen samples at sizes below 200 nm (Pummer et al., 2012; Fröhlich-Nowoisky et al., 2015). Fertile soil samples when dispersed in water and filtered have also been shown to have a significant number of ice-nucleating particles below 200 nm (O'Sullivan et al., 2015; Hill et al., 2016). O'Sullivan et al. (2015) showed that some ice nucleation persisted in fertile soil samples filtered to 1000 kDa; however, ice nucleation above −10 ∘C was removed by these filtrations. Decayed plant litter was shown to have comparable INP concentrations before and after filtration through 200 nm filter pores and retained a fraction of these INPs when further filtered through 20 nm filter pores (Vali et al., 1976). Ice-nucleating particles below 200 nm were measured in North American Arctic snow samples and in precipitation from North China temperate grassland (Du et al., 2017; Rangel-Alvarado et al., 2015). The snow samples were shown to be of biological origin and subsequently tested for virus-like structures, of which none was observed. Despite this, the authors stated they could not preclude viruses as a potential explanation for the observed ice-nucleating activity, based on the size of the INPs and their likely origin. Measurements of INPs in the Arctic sea surface microlayer showed that most of the observed ice nucleation (in the immersion mode) was caused by particles between 0.02 and 0.2 µm in size and were heat labile; viruses were suggested as a potential explanation (Irish et al., 2017; Wilson et al., 2015). Atmospheric measurements made in the Arctic showed the presence of atmospheric INPs in the size range 150–340 nm (Creamean et al., 2019; Creamean et al., 2018). Size-resolved measurements made in a boreal forest in Hyytiälä, Finland, showed an instance in which INPs in the size range 250–500 nm dominated the atmospheric INP burden at temperatures ∘C, whilst measurements made at near-surface-level locations in the UK showed INPs present at sizes below 250 nm (Porter et al., 2020). There is a growing body of evidence that suggests there is a reservoir of currently unidentified biological particles in the fine mode (<250 nm) present in soil/plant life, the oceans and the atmosphere. In this study, we test the hypothesis that viruses are a potential candidate for the source of these fine-mode INPs.
It has been estimated that there are ∼ 1031 virus particles in the biosphere (Whitman et al., 1998), with approximately 107 virus particles per millilitre of seawater, 108–109 per millilitre in marine surface sediments (Suttle, 2007, 2005) and 108–109 per gram of soil in different types of terrestrial environments (Srinivasiah et al., 2008). Numerous studies indicate that there are approximately 10–100 times more viruses compared to their host cells in any given environment (Srinivasiah et al., 2008; Cai et al., 2019). With respect to viral abundance in the atmosphere, there is at present a dearth of knowledge. Rastelli et al. (2017) measured the viral abundance in both the seawater microlayer and the aerosol phase directly above using a bubble generator system designed to mimic wave breaking in open seawater. Virus concentrations for seawater and sampled air were 5×1011 and 0.3–3.5 × 105 virus particles m−3, respectively. Virus particles were measured from outdoor air samples using a filter-based technique taken at a university campus, with the atmospheric virus particle concentration being measured as virus particles m−3 (Prussin et al., 2015). The spatial and temporal variability of airborne viruses were investigated in a series of different locations (residential district, forest and an industrial complex), with concentrations of 1.7×106 to 4.0×107 virus particles m−3 being measured (Whon et al., 2012). Overall, the range of outdoor virus concentrations recorded in the literature range between 0.3×105 and 4.0×107 virus particles m−3. It is likely that these numbers do not represent the full variability of virus particle concentration due to the scarcity of measurements.
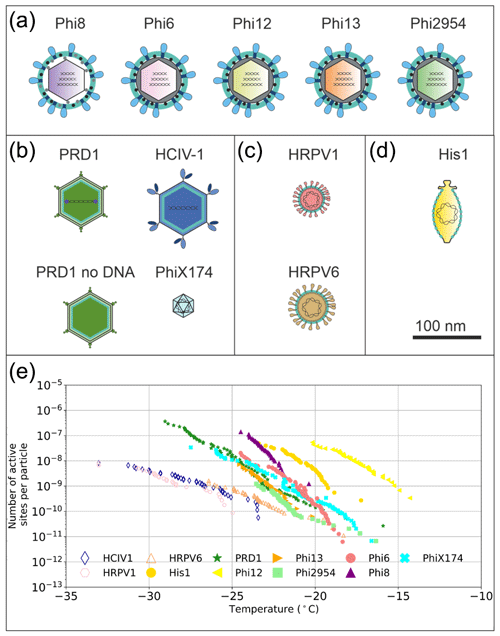
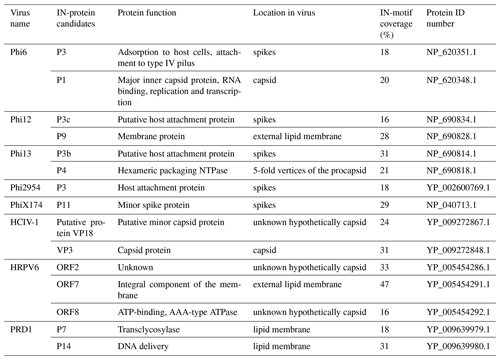
Figure 1Graphical representation of the virus particles used in the ice nucleation study and their ice-nucleating ability. (a) Enveloped icosahedral viruses. (b) Icosahedral viruses. (c) Pleomorphic viruses. (d) Lemon-shaped virus. (e) Ice nucleation activity plots (expressed as nn), where hollow markers indicate limit of detection (LoD) measurements in which the freezing temperatures were consistent with the virus-free saline buffer control. Virus particles are to scale according to the 100 nm scale bar. Temperature values have been corrected for freezing point depression of NaCl.
Despite the large number of virus particles measured in various environments there are a relatively small number of different particle structures a virion (an infective virus particle) can have. This is due to physical constraints of protein fold space that make up the virus particle architecture (Abrescia et al., 2012). Structurally similar viruses can have different host organisms and different geographical source locations (Bamford 2003; Saren et al., 2005; Atanasova et al., 2012). There are several observations of virus isolates with high genome identity originating from spatially distant environments (Atanasova et al., 2015; Pietilä et al., 2012; Saren et al., 2005; Tschitschko et al., 2015). We have chosen virus particles for this study that represent several different symmetric or asymmetric virus architecture types: icosahedral, icosahedral with internal lipid membrane, icosahedral enveloped and lemon-shaped (see Fig. 1a–e). As it would be beyond the realms of feasibility to test even a minute fraction of the 1031 different viruses in the biosphere, we took the approach that we believe allows us to investigate the maximum parameter space and test the hypothesis that virus architecture/structure controls ice-nucleating ability. In this study we present the ice-nucleating ability of viruses with these different architecture types, demonstrate the potential of different structural components in viruses to nucleate ice, and attempt to estimate the potential of viruses as a class of atmospheric ice-nucleating particles.
2.1 Virus growth, purification and production of Phi6 sub-viral particles
Virus particle suspensions were produced under carefully controlled conditions which resulted in suspensions of high purity. The methodology for producing virus particles has been developed over many decades and involves first producing host bacterial cultures which are then infected with a virus. In the case of lytic viruses, the virus causes the bacterial cells to lyse (the cell membranes break down), which releases the contents of the lysed cells (including virus particles) into the growth media. The growth media solution then contains a mixture of components from the cells, cell-wall fragments and virions. This cell debris is removed by centrifugation, and the supernatant (containing the virus) is called the lysate, which is the starting material used for virus purification. Notably, in the case of non-lytic viruses, host cells release high numbers of virions without lysis. In this case, the cells are removed similarly by centrifugation, and the supernatant containing the viruses is used as the virus stock solution. Nevertheless, for simplicity, in this study, the word “virus lysate” is used to refer to the virus stock solution regardless of whether the cells were initially lysed by the virus or not. In this study, all other viruses are lytic, except for Salterprovirus His1 (His1), Alphapleolipovirus HRPV1 (HRPV1) and Alphapleolipovirus HRPV6 (HRPV6), which are non-lytic. In order to examine the ice-nucleating ability of the virus particles without the other components of the lysate, we employ some state-of-the-art purification procedures for each virus, and these purification protocols are described and referred to below. Shortly, the virus particles in the lysate are precipitated using polyethylene glycol and suspended into a small volume of buffer. The precipitate is cleared from impurities by centrifugation and the viruses are purified (1× purification) using rate-zonal centrifugation (in a sucrose gradient) where the separation is based on the velocity of the virus particles. After the purification, the light-scattering virus zone, which now has moved further along the purifying gradient material, is extracted and further purified (2× purification) using equilibrium centrifugation. This type of purification is based on a density gradient, meaning that the virus particles stop moving along the gradient when the density of the viruses equals the density of the gradient medium. The 2× purified virus sample is concentrated by differential ultracentrifugation, and the pellet containing the 2× purified, concentrated virus sample is suspended into a small volume of buffer. For some viruses, only 1× purification is performed, after which the sample is concentrated similarly by differential centrifugation. The concentrated, purified virus sample is used for the ice nucleation activity assay (see below).
Bacterial and archaeal strains and viruses used in this study are listed in Table S1 in the Supplement. Bacterial host strains were aerobically grown in Luria–Bertani broth at 28 ∘C for P. syringae pathovar phaseolicola HB10Y and P. syringae LM2489, and at 37 ∘C for Escherichia (E.) coli HMS174 and E. coli C122 strains. Archaeal host strains were aerobically grown in 23 % modified growth medium (MGM) at 37 ∘C (Nuttall and Smith, 1993).
Bacteriophages Salmonella virus PRD1 (PRD1) and Pseudomonas virus phi6 (Phi6) were 1× purified as described in Bamford et al. (1995). The 2× purification of PRD1 was performed (Bamford et al., 1995; Lampi et al., 2018). The PRD1 particles devoid of DNA (procapsids) were collected after 1× purification, during which the DNA-containing particles sediment further along the sucrose gradient compared to the empty procapsids. The 1× purified Phi6 was further purified to 2× by equilibrium ultracentrifugation in 20 %–70 % sucrose in 20 mM potassium phosphate buffer pH 7.2 with 1 mM MgCl2 (designated here as potassium phosphate buffer) followed by concentration as described in Bamford et al. (1995). Viruses Pseudomonas virus phi8 (Phi8), Pseudomonas virus phi12 (Phi12), Pseudomonas virus phi13 (Phi13) and Pseudomonas virus phi2954 (Phi2954) were produced and precipitated according to Qiao et al. (2010), and the 1× purification was performed by rate-zonal ultracentrifugation in 5 %–20 % sucrose gradients in potassium phosphate buffer, Sorvall AH629 rotor, 24 000 rpm, 50 min, 15 ∘C, followed by concentration using differential ultracentrifugation, Sorvall T865 rotor, 34 000 rpm, 3 h, 10 ∘C. All other viruses were purified to 1× preparations according to protocols described in Eskelin et al. (2019) (for PhiX174, Escherichia virus phiX174), Pietilä et al. (2009) (for HRPV1), Pietilä et al. (2012) (for HRPV6), Demina et al. (2016) (for HCIV-1, Haloarcula virus HCIV1) and Bath et al. (2006) (for His1).
Phi6 sub-viral particles were prepared according to Bamford et al. (1995), modified by Eskelin and Poranen (2018) (for butylated hydroxytoluene treated particles). Phi6 nucleocapsids (NC) were prepared by adding 1 % final concentration of Triton X-100 to 1× purified Phi6 particles in potassium phosphate buffer and incubating 30 min at 22 ∘C. The treated particles were collected by ultracentrifuge, Ti1270 rotor, 30 000 rpm, 4 h, 15 ∘C. Particles were flushed three times with and resuspended in 0.5 mL potassium phosphate buffer overnight at 5 ∘C.
The protein concentration of viral and sub-viral particles was measured by the Bradford assay using bovine serum albumin as a standard (Bradford, 1976). Virus samples were analysed by sodium dodecyl sulfate 16 % polyacrylamide gel electrophoresis (SDS-PAGE) (Olkkonen and Bamford, 1989) to visualize viral protein profiles.
2.2 Search for ice nucleation motifs
Currently, there are eight referenced ice nucleation proteins identified from bacterial cells according to the public protein database (UniProt, https://www.uniprot.org/, last access: 7 September 2020). The ice nucleation motifs (INMs) predicted based on these genes are short protein sequences conserved in this protein family. They are abundant for the ice nucleation proteins (IN proteins) but scarce in the rest of the bacterial genomes. The group of motifs specific for a protein family can serve as a functional fingerprint indicating similarities in structure and function. It was previously determined that INM3 corresponds to the clathrate structure part of the protein responsible for ice nucleation activity in bacterial IN proteins (Gurian-Sherman and Lindow, 1993; Kajava and Lindow, 1993).
The INMs were acquired from SPRINT, an interface for the PRINTS data bank of protein family fingerprints. SPRINT is a public domain database currently maintained at the University of Manchester (http://130.88.97.239/dbbrowser/sprint/, last access: 28 June 2020). The INMs can be found in SPRINT by the identifier ICENUCLEATN. All known ice nucleation motifs in IUPAC (International Union of Pure and Applied Chemistry) nomenclature are listed in Table S2. Since some of the putative viral proteins are not fully characterized, we used protein INMs from SPRINT to build generalized nucleotide motifs. The annotated viral genomes were acquired from the National Centre for Biotechnology Information (NCBI) genome database (Table S3).
Ice nucleation motifs were searched for in the viral genomes using MEME (Multiple Em for Motif Elicitation) Suite 5.1.0. (Bailey et al., 2009). The search was performed using MCAST (Motif Cluster Alignment Search Tool) and FIMO (Find Individual Motif Occurrences) tools (Charles et al., 2011; Bailey and Noble, 2003). MCAST searches for input motifs in the query sequence for statistically significant clusters of non-overlapping occurrences. FIMO, in turn, searches for individual motif occurrences in the sequences and each motif independently. Each found occurrence was scored with p value. The p score thresholds for significant findings were set to 0.0001.
In order to predict putative IN proteins in the viral genomes, total INM coverage as well as the occurrence of repetitive IN motifs was studied. INM coverage is calculated from the length of the matching INM sequence compared to the total length of the protein. The INMs were annotated to the sequences using Artemis 17.0.1, and the protein alignments were performed using Muscle 3.8.425 and visualized using Geneious Prime 2020.1.1. All the potential IN proteins are listed in Table 1.
2.3 Ice nucleation experiments
Samples for analysis of the ice-nucleating activity of virus particles were prepared by diluting 1× or 2× purified virus particles to specific buffer solutions (Table S1) so that the final concentration of plaque-forming units per microlitre (pfu mL−1) was 1010–1012. One plaque corresponds to the progeny of one virus that initially infected the host cell. Plaque-forming units measure the number of infective virus particles in the sample. Sub-viral particles were used without dilution. Virus host strains were collected by centrifugation (Eppendorf, 13 000 rpm, 5 min, 22 ∘C), diluted into the same buffer as the virus (Table S1), centrifuged (Eppendorf, 13 000 rpm, 5 min, 22 ∘C) and resuspended into buffer according to Table S1. NaCl (500 mM) was added to the buffer for the archaeal cells and viruses due to them being classified as extreme halophiles that require NaCl for optimum growth or infectivity.
Ice nucleation experiments were carried out using the µL-NIPI (nucleation by immersed particles instrument) (Whale et al., 2015). In brief, the µL-NIPI analysis involved pipetting 1 µL droplets of sample suspension onto a hydrophobic-coated glass cover slip that was placed on top of an aluminium cold stage. Then, the cold stage was cooled until the droplets froze. Approximately 50 droplets were used per experiment, with temperatures ranging between 0 to −36 ∘C. The cooling rate was 1 ∘C min−1. Viral samples were agitated on a vortex mixer for 30 s prior to being pipetted to ensure the particles were evenly distributed through the suspension. Droplet freezing was recorded using a camera, with the freezing temperature of each droplet recorded.
The cumulative fraction of droplets frozen on cooling to a temperature, fice(T), is defined by
where nice(T) is the cumulative number of droplets frozen on cooling to T and Ntot is the total number of droplets. The cumulative number of active sites per particle, nn(T), was calculated according to Eq. (2):
where nv is the number of virus particles per 1 µL droplet. The cumulative active sites per unit mass of material, nm(T), is defined by
where mv is the mass of virus particles per droplet.
The freezing point depression of pure water due to NaCl (i.e. in the buffer solutions) was calculated using
where ΔTF is the freezing depression, KF is the cryoscopic constant (1.853 K kg mol−1 for water), b is molality and i is the Van 't Hoff factor (2 for NaCl). Hence, we report the degree of supercooling relative to the melting point of the aqueous saline solution. The correction was typically about 1 ∘C for most virus suspensions (it was around 3 ∘C for an archaeal virus which required a very high salt concentration).
3.1 Ice-nucleating ability of virus particles
We studied virus ice nucleation from a virus structural perspective using the nucleation by immersed particle instrument (µL-NIPI) technique (Whale et al., 2015). We examined the ice nucleation activity (INA) of 11 viruses with different particle architectures, in an effort to probe the hypothesis that virus architecture/structure influences the ice-nucleating ability of virus particles (Fig. 1). These viruses included five enveloped cystoviruses of P. syringae hosts with particle diameters of ∼ 85 nm (Phi6, 8, 12, 13 and 2954; Fig. 1a), two icosahedral viruses with an internal lipid membrane and particle diameters of ∼ 70 nm (Fig. 1b, PRD1 and HCIV-1), one of the icosahedral viruses without the DNA (Fig. 1b, PRD1 no DNA), one 30 nm icosahedral virus without lipids (Fig. 1b, PhiX174), two enveloped pleomorphic viruses with particle diameter of ∼ 50 nm (Fig. 1c), and one lemon-shaped virus (Fig. 1d). Phi6-like viruses are commonly used as models for viruses that cause respiratory illnesses like SARS-CoV-2, the causative agent of COVID-19, due to structure similarity. Of the 11 viruses tested, 9 showed an INA distinct from the INA of the buffer solution they were suspended in (Figs. 1e and S1 in the Supplement).
Phi12, an enveloped virus infecting P. syringae, was found to be the most ice-nucleation-active virus in our study (in terms of the number of INPs per virus particle, nn). Phi12 was observed to trigger freezing from −15 to −21 ∘C, with nn values between and per virus particle. The other structurally similar cystoviruses of P. syringae were all ice nucleation active, although less so compared to Phi12 (Fig. 1e).
At this point we address the question of if other components of the bacterial cell lysate might account for the ice-nucleating activity reported in this paper. We consider this possibility very unlikely for the following reasons. (i) We have shown that the non-lysed host cells mostly have no measurable INA (a few have a weak INA) (see Figs. S4 to S7). Several ice-nucleation-active P. syringae strains have been described in previous studies (de Araujo et al., 2019). None of the strains contain functional genes that code ice-nucleating proteins – they only contain partial pseudogenes. (ii) Studies of ice nucleation by bacterial cells which contain INA proteins and cells which are lysed to some degree show that there are no measurable ice nucleation sites that become active below about −12 ∘C (Wex et al., 2015). Hence, the available evidence suggests that bacterial cell lysate does not possess INA in the temperature ranges where we report activity in this study. (iii) The purification steps described in the Methods section remove the vast majority of the cell lysate material as demonstrated by the protein profiles in Fig. S2. (iv) We show that a second purification (2×), which would further remove any cell lysate material, has no effect on the INA of the Phi6 sample. This is consistent with the INA being related to the virus rather than the cell lysates. Hence, we conclude that the activity we observe in our droplet freezing assays of purified virus particle suspensions is most likely related to the virus particles.
The source of the INA was further studied using two of the best-characterized model viruses, Phi6 of P. syringae and PRD1 of E. coli. Regarding Phi6, we used biochemical dissociation to disassemble the virus particles into sub-viral particles (Fig. 2). First, the virus spike proteins were removed using butylated hydroxy toluene (BHT), with the resulting particle referred to as Phi6 BHT and the separate spike proteins referred to as Phi6 P3. Secondly, the lipid envelope and the associated proteins were removed using the anionic detergent Triton X-100, exposing the nucleocapsid (NC) structure of the Phi6 virion (Fig. 2a). Each of the sub-viral particles was shown to have an INA distinguishable from the potassium phosphate buffer (Fig. 2b). Each of the sub-viral components, along with Phi6, was normalized to the mass of particles per volume of sample (nm). When normalized in this manner, each component spanned approximately the same range in nm space, 102–104 (mg−1) across a range of temperatures. The freezing spectrum of each component was similar, with the Phi6 BHT sub-viral components having slightly warmer freezing temperatures than Phi6 at equivalent nm values, whereas spike proteins (P3 in Fig. 2a) had a slightly lower freezing temperature than Phi6. NC was found to be the most IN-active sub-viral particle of Phi6 (Fig. 2c), freezing approximately 4 ∘C warmer across the measured nm range when compared to Phi6. INA is in part related to size (Pummer et al., 2015). Since the spike proteins are only ∼ 20 nm in diameter, whereas the BHT and NC particles are close to 80 nm, the difference in activity may be related to size. It is not clear how virus particles behave in the atmosphere, but several environmental stressors can disrupt virus particles exposing their internal parts. The other Phi6 sub-viral particles were also IN active (Fig. 2), indicating that the virus has broad IN potential, either being active as a whole or in a disrupted form. PRD1 was measured for its INA both with and without DNA. The nn values for PRD1 with and without DNA are shown in Fig. 3 and are similar to one another. This result suggests that the presence of PRD1's DNA is not related to the INA of the particles, indicating that also non-infective viruses (e.g. viruses inactivated by different types of environmental stress) can have INA.
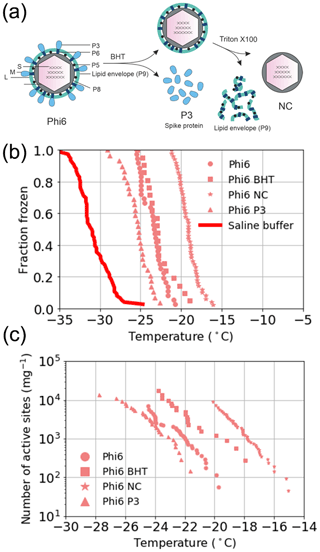
Figure 2Ice nucleation activity of the sub-viral particles of the Phi6 virus. (a) Biochemical dissociation of the Phi6 virion. Small genome fragment is marked as S, medium genome fragment as M and large genome fragment as L; P3 is the spike protein, P5 is the lytic enzyme, P6 is the membrane fusion protein, P8 is the outer capsid lattice protein and P9 is the major envelope protein; BHT means butylated hydroxyl toluene; NC is the nucleocapsid. (b) Fraction frozen curves for Phi6 and its sub-viral components. These values have not been correct for freezing point depression due to NaCl. (c) The INA (expressed as active sites per unit mass, nm) of Phi6 and its sub-viral components normalized to the mass of particle per volume of suspension. Phi6 BHT in panels (b) and (c) refers to spikeless enveloped icosahedral structure. Temperature values have been corrected for freezing point depression of NaCl.
Figure 3The number of active sites per particle (nn) for PRD1 with and without DNA. Temperature values have been corrected for freezing point depression of NaCl.
To further our understanding of the influence of virus structure on IN activity, we tested six other viruses – four archaeal and two bacterial. Of these six viruses, two archaeal viruses (HRPV1 and HRPV6) were enveloped like Phi6 but lack particle symmetry and an NC structure (Fig. 1c). HRPV1 (Fig. S1b) was not distinguishable from the saline buffer (Table S1) it was suspended in, whilst HRPV6 (Fig. S1b) was distinguishable from the buffer but was not distinguishable from its host, and as such they are shown as limiting values (Fig. S5). Viruses with icosahedral symmetry that contain an internal lipid membrane (PRD1 and HCIV-1, Fig. 1b) were also tested to further probe the dependency of viral INA on structure. PRD1, a well-known model virus (Bamford et al., 1995), was shown to be INA with a signal distinguishable from both the potassium phosphate buffer and its host (Fig. S6) and nn values comparable to that of the majority of the P. syringae viruses (excluding Phi12) (Fig. 1e). HCIV-1 did not have an INA distinguishable from the saline buffer it was suspended in and is thus shown as a limiting value (Fig. 1b). Another icosahedral virus, PhiX174, this time without a lipid membrane (Fig. 1b), was tested and had nn values similar to that of PRD1 and the majority of the P. syringae viruses. We further studied the INA dependency on virus architecture by studying an asymmetrical lemon-shaped archaeal virus, His1 (Bath et al., 2006). Interestingly, the virus had a higher INA than all the tested viruses, except for Phi12 (Fig. 1d–e), indicating that structurally different viruses, symmetric or asymmetric, can be IN active. His1 was shown to be distinguishable from the saline buffer solution (Fig. S1b) and its host (Fig. S7).
3.2 Genetic analysis of ice-nucleation-active virus particles
The genomes of the 11 viruses included in this study were explored by bioinformatic analysis to further examine the source of IN activity. The ice nucleation activity observed in bacteria is due to protein structures which mimic ice crystal clathrate structure on the cell surface, thus facilitating ice crystal formation around the cell (Kajava and Lindow, 1993). In viruses, the source of INA might also be of proteinaceous origin. The possibility of the capsid or membrane proteins in virus particles possessing similar structure and function to known bacterial ice nucleation proteins as explanation for their ice nucleation capacity was explored in this study. Proteins with potential ice nucleation activity were screened by searching for conserved short sequences called ice nucleation motifs in the amino acid sequence.
Viral proteins with significant INM coverage and presence of INM3 in their sequence were predicted in eight of the viruses (Table 1). According to the results, only Phi13 and PhiX174 did not have potential IN proteins, with His1 having coverage below 15 % and so is not shown in the table. Other viruses contained at least one potential protein with INM coverage of 15 %–50 % and obligatory INM3 presence. However, the INA of Phi13 and PhiX174 is similar to the majority of the tested viruses such as Phi6. Similarly, HRPV1 and HCIV-1 contain potential IN proteins, but these viruses had the weakest INA of the tested virus particles. Therefore, the presence of INMs in the sequence does not correlate to the capacity to nucleate ice.
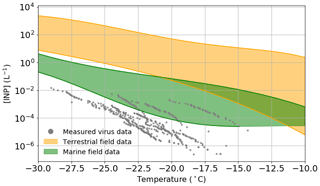
Figure 4Estimated viral INP concentration based on measured ice-nucleating ability of virus particles and upper limit literature values of viral particle concentrations in the atmosphere compared to measured INP concentrations in both terrestrial (orange) and marine/polar (green) environments. Table S4 shows a list of the studies from which the data to create the field measurement envelopes were obtained. Temperature values have been corrected for freezing point depression of NaCl.
In order to estimate the INP concentrations associated with virus particles in the atmosphere, we have combined the nn values shown in Fig. 1e and the upper limit of the concentration of viruses in the atmosphere from literature data (taken as 4×107 particles m−3; see discussion in the introduction). It is important to note that we based these virus INP concentrations on the INA of the specific samples which we studied, and it may be possible that other virus particles have greater INA. However, since there are a limited number of virus architectures and we test a range of these architectures, we tentatively suggest that we capture the typical range of INA of virus particles. Also shown on Fig. 4 are envelopes showing the range of data from field campaigns in terrestrial (orange) and remote marine/polar environments (green) (see Table S4 for a list of representative measurements included in these envelopes).
As discussed in the introduction, in terrestrial environments, mineral dust is thought to be a very important INP type, with marine organics playing a secondary role (Vergara-Temprado et al., 2017). In addition, there is evidence that biological INPs play an important role in the terrestrial-influenced atmosphere in some locations (O'Sullivan et al., 2018; Hill et al., 2016; Šantl-Temkiv et al., 2019; Conen et al., 2016; Pratt et al., 2009; Gong et al., 2020). Figure 4 shows that across the entire temperature spectra relevant for mixed-phase clouds, the concentrations of virus INPs are lower than the lowest typical INP concentrations in terrestrial-influenced areas. The closest the terrestrial envelope and the virus data points come to overlapping is between −18 and −22 ∘C, at which temperatures the difference in INP concentration is approximately 1 order of magnitude, i.e. at most they might contribute about 10 % of the INP population at around −20 ∘C. Furthermore, one might expect that in environments where there is a strong source of virus particles there is also a strong source of other biological materials; hence the influence of virus INP may be overestimated in our simple analysis. Overall, these results suggest that virus INPs generally play a minor role in regions influenced by terrestrial INPs.
Remote marine locations are less influenced by active terrestrial sources, and thus the INP populations there are different from those of the terrestrial atmosphere (Creamean et al., 2019; DeMott et al., 2016; McCluskey et al., 2018a, b). Marine organics and sea spray aerosol have been shown to be INP sources of first-order importance in such environments (DeMott et al., 2016; Vergara-Temprado et al., 2017; Wilson et al., 2015). Tests indicate that these INPs are sensitive to heat and can pass through 200 nm filters (Wilson et al., 2015; Schnell and Vali, 1975), and while it is unclear exactly which component of the seawater nucleate ice it has been pointed out that virus particles are of the right size (Wilson et al., 2015). Field measurements made in remote marine environments have reported remarkably low INP concentrations. McCluskey et al. (2018a) measured INP concentrations in a pristine marine environment at the Mace Head research station in 2015, with INP concentrations as low as 10−3 L−1 at −20 ∘C. In a separate field campaign, measurements were made in the Southern Ocean, and INP concentrations range between and at −20 ∘C (McCluskey et al., 2018b). Figure 4 shows overlap between the virus INP data points and the marine envelope in the temperature range from −15 to −27 ∘C, with the most active of the virus INPs being approximately 15× higher than the lower limit of the marine envelope at −20 ∘C. Whilst this by no means proves that virus INPs are important in remote marine environment, it indicates they may contribute to the atmospheric INP burden in such regions. However, the lowest INP concentrations in the remote marine environment are most likely associated with periods when the aerosol concentrations were lowest, as a result of the combined effect of precipitation scavenging and weak sources. Under these conditions, virus particles would also presumably be depleted.
In this study we show that a range of viruses can nucleate ice heterogeneously when immersed in supercooled solution droplets. A selection of virus types with diverse architectures are shown to have ice-nucleating abilities spanning 3 orders of magnitude at −20 ∘C, when normalized to particle number. We probed the virus ice-nucleating ability dependence on virus particle structure/architecture, showing that for our selected viruses there was not a dependency on certain virus architecture. Bioinformatic analysis shows that our current knowledge of ice nucleation related to ice nucleation protein genes in bacterial ice nucleators is likely insufficient to understand why viruses nucleate ice, which can be due to e.g. the overall arrangement of structural proteins making up the virion.
Our results are based on a small subsample of virus types but include several of the most prominent viral architectures. Nine out of 11 tested viruses were ice nucleation active, indicating that several structurally different viruses can have IN potential. In addition, it has been shown previously that a helical virus, Tobacco mosaic virus (TMV), can also nucleate ice (Cascajo-Castresana et al., 2020). While we have selected virus particles with a range of architectures that are relatively common in nature, it is possible that other virus particles nucleate ice more or less effectively. In particular, the specific virus types we have studied here are from the terrestrial and freshwater aquatic environment; the isolation and testing of a range of marine viruses presents an important next step in quantifying the importance of viral ice nucleators. More work needs to be done to understand what drives viral ice nucleation and whether it would be dependent on virus structure/morphology, host or some other factor.
This study shows the potential role viruses play as atmospheric INPs in certain environments. The ubiquity of viruses in the atmosphere implies they could serve as a baseline of INPs in situations where other, better-known atmospheric INPs are absent in any meaningful quantity. However, our estimates for the upper limit of virus INPs suggest they do not play a meaningful role in terrestrial environment but may contribute to the INP population in marine environments. More work needs to be done to understand both why viruses nucleate ice and what role they play in regional and global atmospheric ice nucleation.
The data associated with this paper are openly available from the University of Leeds data repository (https://doi.org/10.5518/1019, Adams et al., 2021).
The supplement related to this article is available online at: https://doi.org/10.5194/bg-18-4431-2021-supplement.
MPA, NSA, SS, AH and ZB performed the experiments. SS designed and performed bioinformatic analyses. DHB developed the NC purification protocol. MPA, NSA, BM and DHB designed the experiments. All authors participated in data analysis and interpretation of results. NSA, JD, JR, BJM and DHB supervised and supported the project. The manuscript was written by MPA, NSA, SS, BJM and DHB. All authors reviewed and approved the manuscript.
The authors declare that they have no conflict of interest.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Helin Veskiväli and Emeline Vidal are thanked for technical assistance. We thank Leonard Mindich for providing bacteriophages Phi12, Phi13, Phi2954 and the P.syringae bacterial strain. Ben Fane is thanked for providing the PhiX174 bacteriophage. The use of the facilities and expertise of the Instruct-HiLIFE Biocomplex unit, member of Biocenter Finland and Instruct-FI, is gratefully acknowledged.
This research has been supported by the European Research Council (CryoProtect (grant no. 713664) and MarineIce (grant no. 648661)), the Natural Environment Research Council (grant no. NE/T00648X/1), and the Academy of Finland (grant no. 309570). Nina S. Atanasova was supported by the Academy of Finland postdoctoral grant 309570 and the Scientific Advisory Board for Defence grant VN/627/2020-PLM-9.
This paper was edited by Paul Stoy and reviewed by two anonymous referees.
Abrescia, N. G. A., Bamford, D. H., Grimes, J. M., and Stuart, D. I.: Structure Unifies the Viral Universe, Annu. Rev. Biochem., 81, 795–822, https://doi.org/10.1146/annurev-biochem-060910-095130, 2012.
Adams, M. P., Atanasova, N. S., Sofieva, S., Ravantti, J., Heikkinen, A., Brasseur, Z., Duplissy, J., Bamford, D. H., and Murray, B. J.: Data for “Ice nucleation by viruses and their potential for cloud glaciation”, University of Leeds, https://doi.org/10.5518/1019, 2021.
Atanasova, N. S., Roine, E., Oren, A., Bamford, D. H., and Oksanen, H. M.: Global network of specific virus-host interactions in hypersaline environments, Environ. Microbiol., 14, 426–440, https://doi.org/10.1111/j.1462-2920.2011.02603.x, 2012.
Atanasova, N. S., Demina, T. A., Buivydas, A., Bamford, D. H., and Oksanen, H. M.: Archaeal viruses multiply: Temporal screening in a solar saltern, Viruses, 7, 1902–1926, https://doi.org/10.3390/v7041902, 2015.
Bailey, T. L. and Noble, W. S.: Searching for statistically significant regulatory modules, Bioinformatics, 19 19, 16–25, https://doi.org/10.1093/bioinformatics/btg1054, 2003.
Bailey, T. L., Bodén, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., Ren, J., Li, W. W., and Noble, W. S.: MEME SUITE: tools for motif discovery and searching, Nucleic Acids Res., https://doi.org/10.1093/nar/gkp335, 2009.
Bamford, D. H.: Do viruses form lineages across different domains of life?, 154, 231–236, https://doi.org/10.1016/S0923-2508(03)00065-2, 2003.
Bamford, D. H., Ojala, P. M., Frilander, M., Walin, L., and Bamford, J. K. H.: Isolation, purification, and function of assembly intermediates and subviral particles of bacteriophages PRD1 and σ6, Methods Mol. Genet., 6, 455–474, https://doi.org/10.1016/S1067-2389(06)80028-2, 1995.
Bath, C., Cukalac, T., Porter, K., and Dyall-Smith, M. L.: His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus, Virology, 350, 228–239, https://doi.org/10.1016/j.virol.2006.02.005, 2006.
Bradford, M. M.: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem., 72, 248–254, https://doi.org/10.1016/0003-2697(76)90527-3, 1976.
Burrows, S. M., Hoose, C., Pöschl, U., and Lawrence, M. G.: Ice nuclei in marine air: biogenic particles or dust?, Atmos. Chem. Phys., 13, 245–267, https://doi.org/10.5194/acp-13-245-2013, 2013.
Cai, L., Jørgensen, B. B., Suttle, C. A., He, M., Cragg, B. A., Jiao, N., and Zhang, R.: Active and diverse viruses persist in the deep sub-seafloor sediments over thousands of years, ISME J., 13, 1857–1864, https://doi.org/10.1038/s41396-019-0397-9, 2019.
Cascajo-Castresana, M., David, R. O., Iriarte-Alonso, M. A., Bittner, A. M., and Marcolli, C.: Protein aggregates nucleate ice: the example of apoferritin, Atmos. Chem. Phys., 20, 3291–3315, https://doi.org/10.5194/acp-20-3291-2020, 2020.
Charles, E. G., Bailey, T. L., and Stafford, W.: FIMO: scanning for occurrences of a given motif, Bioinformatics, 27, 1017–1018, https://doi.org/10.1093/bioinformatics/btr064, 2011.
Conen, F., Stopelli, E., and Zimmermann, L.: Clues that decaying leaves enrich Arctic air with ice nucleating particles, Atmos. Environ., 129, 91–94, https://doi.org/10.1016/j.atmosenv.2016.01.027, 2016.
Creamean, J. M., Kirpes, R. M., Pratt, K. A., Spada, N. J., Maahn, M., de Boer, G., Schnell, R. C., and China, S.: Marine and terrestrial influences on ice nucleating particles during continuous springtime measurements in an Arctic oilfield location, Atmos. Chem. Phys., 18, 18023–18042, https://doi.org/10.5194/acp-18-18023-2018, 2018.
Creamean, J. M., Cross, J. N., Pickart, R., McRaven, L., Lin, P., Pacini, A., Hanlon, R., Schmale, D. G., Ceniceros, J., Aydell, T., Colombi, N., Bolger, E., and DeMott, P. J.: Ice Nucleating Particles Carried From Below a Phytoplankton Bloom to the Arctic Atmosphere, Geophys. Res. Lett., 46, 8572–8581, https://doi.org/10.1029/2019GL083039, 2019.
de Araujo, G. G., Rodrigues, F., Gonçalves, F. L. T., and Galante, D.: Survival and ice nucleation activity of Pseudomonas syringae strains exposed to simulated high-altitude atmospheric conditions, Sci. Rep.-UK, 9, 1–11, https://doi.org/10.1038/s41598-019-44283-3, 2019.
Demina, T. A., Pietilä, M. K., Svirskaitė, J., Ravantti, J. J., Atanasova, N. S., Bamford, D. H., and Oksanen, H. M.: Archaeal haloarcula californiae icosahedral virus 1 highlights conserved elements in icosahedral membrane-containing DNA viruses from extreme environments, MBio, 7, e00699-16, https://doi.org/10.1128/mBio.00699-16, 2016.
DeMott, P. J., Prenni, A. J., Liu, X., Kreidenweis, S. M., Petters, M. D., Twohy, C. H., Richardson, M. S., Eidhammer, T., and Rogers, D. C.: Predicting global atmospheric ice nuclei distributions and their impacts on climate, P. Natl. Acad. Sci. USA, 107, 11217–11222, https://doi.org/10.1073/pnas.0910818107, 2010.
DeMott, P. J., Hill, T. C. J., McCluskey, C. S., Prather, K. A., Collins, D. B., Sullivan, R. C., Ruppel, M. J., Mason, R. H., Irish, V. E., Lee, T., Hwang, C. Y., Rhee, T. S., Snider, J. R., McMeeking, G. R., Dhaniyala, S., Lewis, E. R., Wentzell, J. J. B., Abbatt, J., Lee, C., Sultana, C. M., Ault, A. P., Axson, J. L., Diaz Martinez, M., Venero, I., Santos-Figueroa, G., Stokes, M. D., Deane, G. B., Mayol-Bracero, O. L., Grassian, V. H., Bertram, T. H., Bertram, A. K., Moffett, B. F., and Franc, G. D.: Sea spray aerosol as a unique source of ice nucleating particles, P. Natl. Acad. Sci. USA, 113, 5797–803, https://doi.org/10.1073/pnas.1514034112, 2016.
Dreischmeier, K., Budke, C., Wiehemeier, L., Kottke, T., and Koop, T.: Boreal pollen contain ice-nucleating as well as ice-binding “antifreeze” polysaccharides, Sci. Rep.-UK, 7, 1–13, https://doi.org/10.1038/srep41890, 2017.
Du, R., Du, P., Lu, Z., Ren, W., Liang, Z., Qin, S., Li, Z., Wang, Y., and Fu, P.: Evidence for a missing source of efficient ice nuclei, Sci. Rep.-UK, 7, 1–8, https://doi.org/10.1038/srep39673, 2017.
Hawker, R. E., Miltenberger, A. K., Wilkinson, J. M., Hill, A. A., Shipway, B. J., Cui, Z., Cotton, R. J., Carslaw, K. S., Field, P. R., and Murray, B. J.: The temperature dependence of ice-nucleating particle concentrations affects the radiative properties of tropical convective cloud systems, Atmos. Chem. Phys., 21, 5439–5461, https://doi.org/10.5194/acp-21-5439-2021, 2021.
Eskelin, K. and Poranen, M. M.: Controlled disassembly and purification of functional viral subassemblies using asymmetrical flow field-flow fractionation (AF4), Viruses, 10, 579, https://doi.org/10.3390/v10110579, 2018.
Eskelin, K., Poranen, M. M., and Oksanen, H. M.: Asymmetrical flow field-flow fractionation on virus and virus-like particle applications, Microorganisms, 7, 555, https://doi.org/10.3390/microorganisms7110555, 2019.
Fröhlich-Nowoisky, J., Hill, T. C. J., Pummer, B. G., Yordanova, P., Franc, G. D., and Pöschl, U.: Ice nucleation activity in the widespread soil fungus Mortierella alpina, Biogeosciences, 12, 1057–1071, https://doi.org/10.5194/bg-12-1057-2015, 2015.
Gong, X., Wex, H., van Pinxteren, M., Triesch, N., Fomba, K. W., Lubitz, J., Stolle, C., Robinson, T.-B., Müller, T., Herrmann, H., and Stratmann, F.: Characterization of aerosol particles at Cabo Verde close to sea level and at the cloud level – Part 2: Ice-nucleating particles in air, cloud and seawater, Atmos. Chem. Phys., 20, 1451–1468, https://doi.org/10.5194/acp-20-1451-2020, 2020.
Green, R. L. and Warren, G. J.: Physical and functional repetition in a bacterial ice nucleation gene, Nature, 317, 645–648, https://doi.org/10.1038/317645a0, 1985.
Grythe, H., Ström, J., Krejci, R., Quinn, P., and Stohl, A.: A review of sea-spray aerosol source functions using a large global set of sea salt aerosol concentration measurements, Atmos. Chem. Phys., 14, 1277–1297, https://doi.org/10.5194/acp-14-1277-2014, 2014.
Gurian-Sherman, D. and Lindow, S. E.: Bacterial ice nucleation: significance and molecular basis, FASEB J., 7, 1338–1343, https://doi.org/10.1096/fasebj.7.14.8224607, 1993.
Hill, T. C. J., DeMott, P. J., Tobo, Y., Fröhlich-Nowoisky, J., Moffett, B. F., Franc, G. D., and Kreidenweis, S. M.: Sources of organic ice nucleating particles in soils, Atmos. Chem. Phys., 16, 7195–7211, https://doi.org/10.5194/acp-16-7195-2016, 2016.
Hoose, C. and Möhler, O.: Heterogeneous ice nucleation on atmospheric aerosols: a review of results from laboratory experiments, Atmos. Chem. Phys., 12, 9817–9854, https://doi.org/10.5194/acp-12-9817-2012, 2012.
Hoose, C., Kristjánsson, J. E., and Burrows, S. M.: How important is biological ice nucleation in clouds on a global scale?, Environ. Res. Lett., 5, 024009, https://doi.org/10.1088/1748-9326/5/2/024009, 2010.
Irish, V. E., Elizondo, P., Chen, J., Chou, C., Charette, J., Lizotte, M., Ladino, L. A., Wilson, T. W., Gosselin, M., Murray, B. J., Polishchuk, E., Abbatt, J. P. D., Miller, L. A., and Bertram, A. K.: Ice-nucleating particles in Canadian Arctic sea-surface microlayer and bulk seawater, Atmos. Chem. Phys., 17, 10583–10595, https://doi.org/10.5194/acp-17-10583-2017, 2017.
Irish, V. E., Hanna, S. J., Xi, Y., Boyer, M., Polishchuk, E., Ahmed, M., Chen, J., Abbatt, J. P. D., Gosselin, M., Chang, R., Miller, L. A., and Bertram, A. K.: Revisiting properties and concentrations of ice-nucleating particles in the sea surface microlayer and bulk seawater in the Canadian Arctic during summer, Atmos. Chem. Phys., 19, 7775–7787, https://doi.org/10.5194/acp-19-7775-2019, 2019.
Junge, K. and Swanson, B. D.: High-resolution ice nucleation spectra of sea-ice bacteria: implications for cloud formation and life in frozen environments, Biogeosciences, 5, 865–873, https://doi.org/10.5194/bg-5-865-2008, 2008.
Kajava, A. V. and Lindow, S. E.: A model of the three-dimensional structure of ice nucleation proteins, 232, 709–717, https://doi.org/10.1006/jmbi.1993.1424, 5 August 1993.
Kanji, Z. A., Ladino, L. A., Wex, H., Boose, Y., Burkert-Kohn, M., Cziczo, D. J., Krämer, M., Kanji, Z. A., Ladino, L. A., Wex, H., Boose, Y., Burkert-Kohn, M., Cziczo, D. J., and Krämer, M.: Overview of Ice Nucleating Particles, Meteorol. Monogr., 58, 1.1–1.33, https://doi.org/10.1175/amsmonographs-d-16-0006.1, 2017.
Lampi, M., Oksanen, H. M., Meier, F., Moldenhauer, E., Poranen, M. M., Bamford, D. H., and Eskelin, K.: Asymmetrical flow field-flow fractionation in purification of an enveloped bacteriophage φ6, J. Chromatogr. B, 1095, 251–257, https://doi.org/10.1016/j.jchromb.2018.07.008, 2018.
Lindow, S. E., Arny, D. C., and Upper, C. D.: The role of bacterial ice nuclei in frost injury to sensitive plants, New York Acad. Press., 249–263, 1978.
Lindow, S. E., Arny, D. C., and Upper, C. D.: Bacterial Ice Nucleation: A Factor in Frost Injury to Plants', Plant Physiol., 70, 1084–1089, 1982.
McCluskey, C. S., Ovadnevaite, J., Rinaldi, M., Atkinson, J., Belosi, F., Ceburnis, D., Marullo, S., Hill, T. C. J., Lohmann, U., Kanji, Z. A., O'Dowd, C., Kreidenweis, S. M., and DeMott, P. J.: Marine and Terrestrial Organic Ice-Nucleating Particles in Pristine Marine to Continentally Influenced Northeast Atlantic Air Masses, J. Geophys. Res.-Atmos., 123, 6196–6212, https://doi.org/10.1029/2017JD028033, 2018a.
McCluskey, C. S., Hill, T. C. J., Humphries, R. S., Rauker, A. M., Moreau, S., Strutton, P. G., Chambers, S. D., Williams, A. G., McRobert, I., Ward, J., Keywood, M. D., Harnwell, J., Ponsonby, W., Loh, Z. M., Krummel, P. B., Protat, A., Kreidenweis, S. M., and DeMott, P. J.: Observations of Ice Nucleating Particles Over Southern Ocean Waters, Geophys. Res. Lett., 45, 11989–11997, https://doi.org/10.1029/2018GL079981, 2018b.
Morris, C. E., Georgakopoulos, D. G., and Sands, D. C.: Ice nucleation active bacteria and their potential role in precipitation, J. Phys. IV, 121, 87–103, https://doi.org/10.1051/jp4:2004121004, 2004.
Morris, C. E., Monteil, C. L., and Berge, O.: The Life History of Pseudomonas syringae: Linking Agriculture to Earth System Processes, Annu. Rev. Phytopathol., 51, 85–104, https://doi.org/10.1146/annurev-phyto-082712-102402, 2013.
Murray, B. J., O'Sullivan, D., Atkinson, J. D., and Webb, M. E.: Ice nucleation by particles immersed in supercooled cloud droplets, Chem. Soc. Rev., 41, 6519, https://doi.org/10.1039/c2cs35200a, 2012.
Murray, B. J., Carslaw, K. S., and Field, P. R.: Opinion: Cloud-phase climate feedback and the importance of ice-nucleating particles, Atmos. Chem. Phys., 21, 665–679, https://doi.org/10.5194/acp-21-665-2021, 2021.
Nuttall, S. D. and Smith, M. L. D.: HF1 and HF2: Novel bacteriophages of halophilic archaea, Virology, 197, 678–684, https://doi.org/10.1006/viro.1993.1643, 1993.
O'Sullivan, D., Murray, B. J., Ross, J. F., Whale, T. F., Price, H. C., Atkinson, J. D., Umo, N. S., and Webb, M. E.: The relevance of nanoscale biological fragments for ice nucleation in clouds, Sci. Rep.-UK, 5, 8082, https://doi.org/10.1038/srep08082, 2015.
O'Sullivan, D., Adams, M. P., Tarn, M. D., Harrison, A. D., Vergara-Temprado, J., Porter, G. C. E., Holden, M. A., Sanchez-Marroquin, A., Carotenuto, F., Whale, T. F., McQuaid, J. B., Walshaw, R., Hedges, D. H. P., Burke, I. T., Cui, Z., and Murray, B. J.: Contributions of biogenic material to the atmospheric ice-nucleating particle population in North Western Europe, Sci. Rep., 8, 13821, https://doi.org/10.1038/s41598-018-31981-7, 2018.
Olkkonen, V. M. and Bamford, D. H.: Quantitation of the adsorption and penetration stages of bacteriophage φ6 infection, Virology, 171, 229–238, https://doi.org/10.1016/0042-6822(89)90530-8, 1989.
Pandey, R., Usui, K., Livingstone, R. A., Fischer, S. A., Pfaendtner, J., Backus, E. H. G., Nagata, Y., Fröhlich-Nowoisky, J., Schmüser, L., Mauri, S., Scheel, J. F., Knopf, D. A., Pöschl, U., Bonn, M., and Weidner, T.: Ice-nucleating bacteria control the order and dynamics of interfacial water, Sci. Adv., 2, e1501630, https://doi.org/10.1126/sciadv.1501630, 2016.
Pietilä, M. K., Roine, E., Paulin, L., Kalkkinen, N., and Bamford, D. H.: An ssDNA virus infecting archaea: A new lineage of viruses with a membrane envelope, Mol. Microbiol., 72, 307–319, https://doi.org/10.1111/j.1365-2958.2009.06642.x, 2009.
Pietilä, M. K., Atanasova, N. S., Manole, V., Liljeroos, L., Butcher, S. J., Oksanen, H. M., and Bamford, D. H.: Virion Architecture Unifies Globally Distributed Pleolipoviruses Infecting Halophilic Archaea, J. Virol., 86, 5067–5079, https://doi.org/10.1128/jvi.06915-11, 2012.
Porter, G. C. E., Sikora, S. N. F., Adams, M. P., Proske, U., Harrison, A. D., Tarn, M. D., Brooks, I. M., and Murray, B. J.: Resolving the size of ice-nucleating particles with a balloon deployable aerosol sampler: the SHARK, Atmos. Meas. Tech., 13, 2905–2921, https://doi.org/10.5194/amt-13-2905-2020, 2020.
Pouleur, S., Richard, C., Martin, J., and Antoun, H.: Ice Nucleation Activity in Fusarium acuminatum and Fusarium avenaceumt, ASM Journals, 58, 2960–2964, https://doi.org/10.1128/aem.58.9.2960-2964.1992, 1992.
Pratt, K. A., Demott, P. J., French, J. R., Wang, Z., Westphal, D. L., Heymsfield, A. J., Twohy, C. H., Prenni, A. J., and Prather, K. A.: In situ detection of biological particles in cloud ice-crystals, Nat. Geosci., 2, 398–401, https://doi.org/10.1038/NGEO521, 2009.
Prussin, A. J., Garcia, E. B., and Marr, L. C.: Total concentrations of virus and bacteria in indoor and outdoor air, Environ. Sci. Technol. Lett., 2, 84–88, https://doi.org/10.1021/acs.estlett.5b00050, 2015.
Pummer, B. G., Bauer, H., Bernardi, J., Bleicher, S., and Grothe, H.: Suspendable macromolecules are responsible for ice nucleation activity of birch and conifer pollen, Atmos. Chem. Phys., 12, 2541–2550, https://doi.org/10.5194/acp-12-2541-2012, 2012.
Pummer, B. G., Budke, C., Augustin-Bauditz, S., Niedermeier, D., Felgitsch, L., Kampf, C. J., Huber, R. G., Liedl, K. R., Loerting, T., Moschen, T., Schauperl, M., Tollinger, M., Morris, C. E., Wex, H., Grothe, H., Pöschl, U., Koop, T., and Fröhlich-Nowoisky, J.: Ice nucleation by water-soluble macromolecules, Atmos. Chem. Phys., 15, 4077–4091, https://doi.org/10.5194/acp-15-4077-2015, 2015.
Qiao, X., Sun, Y., Qiao, J., Di Sanzo, F., and Mindich, L.: Characterization of Φ2954, a newly isolated bacteriophage containing three dsRNA genomic segments, BMC Microbiol., 10, 55, https://doi.org/10.1186/1471-2180-10-55, 2010.
Rangel-Alvarado, R. B., Nazarenko, Y., and Ariya, P. A.: Snow-borne nanosized particles: Abundance, distribution, composition, and significance in ice nucleation processes, J. Geophys. Res.-Atmos., 120, 11760–11774, https://doi.org/10.1002/2015JD023773, 2015.
Rastelli, E., Corinaldesi, C., Dell'Anno, A., Lo Martire, M., Greco, S., Cristina Facchini, M., Rinaldi, M., O'Dowd, C., Ceburnis, D., and Danovaro, R.: Transfer of labile organic matter and microbes from the ocean surface to the marine aerosol: an experimental approach, Sci. Rep.-UK, 7, 11475, https://doi.org/10.1038/s41598-017-10563-z, 2017.
Reche, I., D'Orta, G., Mladenov, N., Winget, D. M., and Suttle, C. A.: Deposition rates of viruses and bacteria above the atmospheric boundary layer, ISME J., 12, 1154–1162, https://doi.org/10.1038/s41396-017-0042-4, 2018.
Sanchez-Marroquin, A., Arnalds, O., Baustian-Dorsi, K. J., Browse, J., Dagsson-Waldhauserova, P., Harrison, A. D., Maters, E. C., Pringle, K. J., Vergara-Temprado, J., Burke, I. T., McQuaid, J. B., Carslaw, K. S., and Murray, B. J.: Iceland is an episodic source of atmospheric ice-nucleating particles relevant for mixed-phase clouds, Sci. Adv., 6, eaba8137, https://doi.org/10.1126/sciadv.aba8137, 2020.
Šantl-Temkiv, T., Sikoparija, B., Maki, T., Carotenuto, F., Amato, P., Yao, M., Morris, C. E., Schnell, R., Jaenicke, R., Pöhlker, C., DeMott, P. J., Hill, T. C. J., and Huffman, J. A.: Bioaerosol field measurements: Challenges and perspectives in outdoor studies, 54, 520–546, https://doi.org/10.1080/02786826.2019.1676395, 2019.
Saren, A. M., Ravantti, J. J., Benson, S. D., Burnett, R. M., Paulin, L., Bamford, D. H., and Bamford, J. K. H.: A snapshot of viral evolution from genome analysis of the Tectiviridae family, J. Mol. Biol., 350, 427–440, https://doi.org/10.1016/j.jmb.2005.04.059, 2005.
Schnell, R. C., Vali, G., and Schnell', R. C.: Freezing nuclei in marine waters, Tellus, 27, 321–323, https://doi.org/10.3402/tellusa.v27i3.9911, 1975.
Schnell, R. C., Vali, G., Schnell, R. C., and Vali, G.: Biogenic Ice Nuclei: Part I. Terrestrial and Marine Sources, J. Atmos. Sci., 33, 1554–1564, https://doi.org/10.1175/1520-0469(1976)033<1554:BINPIT>2.0.CO;2, 1976.
Srinivasiah, S., Bhavsar, J., Thapar, K., Liles, M., Schoenfeld, T., and Wommack, K. E.: Phages across the biosphere: contrasts of viruses in soil and aquatic environments, Res. Microbiol., 159, 349–357, https://doi.org/10.1016/j.resmic.2008.04.010, 2008.
Suttle, C. A.: Viruses in the sea, Nature, 437, 356–361, https://doi.org/10.1038/nature04160, 2005.
Suttle, C. A.: Marine viruses – Major players in the global ecosystem, Nat. Rev. Microbiol., 5, 801–812, https://doi.org/10.1038/nrmicro1750, 2007.
Tan, I., Storelvmo, T., and Zelinka, M. D.: Observational constraints on mixed-phase clouds imply higher climate sensitivity, Science, 352, 224–227, https://doi.org/10.1126/science.aad5300, 2016.
Tobo, Y., Adachi, K., DeMott, P. J., Hill, T. C. J., Hamilton, D. S., Mahowald, N. M., Nagatsuka, N., Ohata, S., Uetake, J., Kondo, Y., and Koike, M.: Glacially sourced dust as a potentially significant source of ice nucleating particles, Nat. Geosci., 12, 253–258, https://doi.org/10.1038/s41561-019-0314-x, 2019.
Tschitschko, B., Williams, T. J., Allen, M. A., Páez-Espino, D., Kyrpides, N., Zhong, L., Raftery, M. J., and Cavicchioli, R.: Antarctic archaea-virus interactions: Metaproteome-led analysis of invasion, evasion and adaptation, ISME J., 9, 2094–2107, https://doi.org/10.1038/ismej.2015.110, 2015.
Vali, G., Christensen, M., Fresh, R. W., Galyan, E. L., Maki, L. R., Schnell, R. C., Vali, G., Christensen, M., Fresh, R. W., Galyan, E. L., Maki, L. R., and Schnell, R. C.: Biogenic Ice Nuclei. Part II: Bacterial Sources, J. Atmos. Sci., 33, 1565–1570, https://doi.org/10.1175/1520-0469(1976)033<1565:BINPIB>2.0.CO;2, 1976.
Vergara-Temprado, J., Murray, B. J., Wilson, T. W., O'Sullivan, D., Browse, J., Pringle, K. J., Ardon-Dryer, K., Bertram, A. K., Burrows, S. M., Ceburnis, D., DeMott, P. J., Mason, R. H., O'Dowd, C. D., Rinaldi, M., and Carslaw, K. S.: Contribution of feldspar and marine organic aerosols to global ice nucleating particle concentrations, Atmos. Chem. Phys., 17, 3637–3658, https://doi.org/10.5194/acp-17-3637-2017, 2017.
Vergara-Temprado, J., Miltenberger, A. K., Furtado, K., Grosvenor, D. P., Shipway, B. J., Hill, A. A., Wilkinson, J. M., Field, P. R., Murray, B. J., and Carslaw, K. S.: Strong control of Southern Ocean cloud reflectivity by ice-nucleating particles, Proc. Natl. Acad. Sci. USA, 115, 2687–2692, https://doi.org/10.1073/pnas.1721627115, 2018.
Wang, X., Sultana, C. M., Trueblood, J., Hill, T. C. J., Malfatti, F., Lee, C., Laskina, O., Moore, K. A., Beall, C. M., McCluskey, C. S., Cornwell, G. C., Zhou, Y., Cox, J. L., Pendergraft, M. A., Santander, M. V., Bertram, T. H., Cappa, C. D., Azam, F., DeMott, P. J., Grassian, V. H., and Prather, K. A.: Microbial control of sea spray aerosol composition: A tale of two blooms, ACS Cent. Sci., 1, 124–131, https://doi.org/10.1021/acscentsci.5b00148, 2015.
Wex, H., Augustin-Bauditz, S., Boose, Y., Budke, C., Curtius, J., Diehl, K., Dreyer, A., Frank, F., Hartmann, S., Hiranuma, N., Jantsch, E., Kanji, Z. A., Kiselev, A., Koop, T., Möhler, O., Niedermeier, D., Nillius, B., Rösch, M., Rose, D., Schmidt, C., Steinke, I., and Stratmann, F.: Intercomparing different devices for the investigation of ice nucleating particles using Snomax® as test substance, Atmos. Chem. Phys., 15, 1463–1485, https://doi.org/10.5194/acp-15-1463-2015, 2015.
Whale, T. F., Murray, B. J., O'Sullivan, D., Wilson, T. W., Umo, N. S., Baustian, K. J., Atkinson, J. D., Workneh, D. A., and Morris, G. J.: A technique for quantifying heterogeneous ice nucleation in microlitre supercooled water droplets, Atmos. Meas. Tech., 8, 2437–2447, https://doi.org/10.5194/amt-8-2437-2015, 2015.
Whitman, W. B., Coleman, D. C., and Wiebe, W. J.: Prokaryotes: The unseen majority, P. Natl. Acad. Sci. USA, 95, 6578–6583, https://doi.org/10.1073/pnas.95.12.6578, 1998.
Whon, T. W., Kim, M.-S., Roh, S. W., Shin, N.-R., Lee, H.-W., and Bae, J.-W.: Metagenomic characterization of airborne viral DNA diversity in the near-surface atmosphere, J. Virol., 86, 8221–8231, https://doi.org/10.1128/JVI.00293-12, 2012.
Wilson, S. L., Grogan, P., and Walker, V. K.: Prospecting for ice association: Characterization of freeze-thaw selected enrichment cultures from latitudinally distant soils, Can. J. Microbiol., 58, 402–412, https://doi.org/10.1139/W2012-010, 2012.
Wilson, T. W., Ladino, L. A., Alpert, P. A., Breckels, M. N., Brooks, I. M., Browse, J., Burrows, S. M., Carslaw, K. S., Huffman, J. A., Judd, C., Kilthau, W. P., Mason, R. H., McFiggans, G., Miller, L. A., Nájera, J. J., Polishchuk, E., Rae, S., Schiller, C. L., Si, M., Temprado, J. V., Whale, T. F., Wong, J. P. S., Wurl, O., Yakobi-Hancock, J. D., Abbatt, J. P. D., Aller, J. Y., Bertram, A. K., Knopf, D. A., and Murray, B. J.: A marine biogenic source of atmospheric ice-nucleating particles, Nature, 525, 234–238, https://doi.org/10.1038/nature14986, 2015.