the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Sedimentary ancient DNA insights into foraminiferal diversity near the grounding line in the western Ross Sea, Antarctica
Ewa Demianiuk
Mateusz Baca
Danijela Popović
Inès Barrenechea Angeles
Ngoc-Loi Nguyen
Jan Pawlowski
John B. Anderson
Foraminifera are important marine environmental indicators widely used in paleoceanography and paleoclimate studies. They are a dominant component of meiobenthic communities around the Antarctic continental shelf, including rarely studied locations below the ice shelves, close to the grounding line. In this study, we use high-throughput sequencing of sedimentary ancient DNA (sedaDNA) targeting foraminifera with two molecular markers, including an ultrashort marker newly designed for this study, in five cores from the western Ross Sea, containing sediments up to 30 000 years old. No foraminiferal DNA is detected in the tills, suggesting a lack of preservation of sedaDNA during glacially induced sediment reworking and transport. We reconstruct diverse foraminiferal communities in the open-marine settings and significantly less diverse communities in sediments from the slopes of the sub-ice-shelf grounding-zone wedges, deposited proximal to the grounding line. Both assemblages are rich in soft-walled monothalamids not preserved in the fossil record and complement the results of earlier micropaleontological studies, allowing for a more complete reconstruction of past biodiversity. The newly designed minibarcode marker provides higher foraminiferal diversity in surface and subsurface samples than the standard barcode and allows for better differentiation between foraminiferal communities in different sediment types. It appears to have great potential for future paleoenvironmental studies, although its taxonomic resolution needs to be evaluated.
- Article
(11151 KB) - Full-text XML
-
Supplement
(530 KB) - BibTeX
- EndNote
The polar regions are the key areas for maintaining the global climate balance (IPCC, 2021) and are major contributors to sea level rise (DeConto and Pollard, 2016). Antarctic ice sheets constitute the largest volume of ice on the planet, totaling almost 60 m of sea level equivalent (IPCC, 2021). Thus, even small changes in the ice sheet volume can significantly contribute to sea level rise. The Ross Sea is a significant drainage outlet for the East Antarctic Ice Sheet and West Antarctic Ice Sheet, which has prompted numerous marine geological investigations, focusing on the behavior of these ice sheets during and following the Last Glacial Maximum (LGM) (e.g., Anderson et al., 2014; Bart et al., 2018; Prothro et al., 2020). These studies have been hampered by the similar composition of sediments deposited in different glacial and glacimarine settings, widespread reworking of mineral grains, mixing of organic sediment fractions and microfossils, and depleted biotic components in subglacial and near-glacial settings (Domack et al., 1999; Prothro et al., 2018). The use of recently developed analytical methods may help to overcome these problems.
This study focuses on foraminifera, protists that are a major component of meiobenthic assemblages in marine ecosystems. They are abundant, are highly diverse, and have short life cycles, making them highly responsive to ecological changes and therefore particularly valuable for environmental monitoring and paleoenvironmental studies (Jorissen et al., 2009). Due to sensitivity to environmental conditions, i.e., temperature, salinity, pH, redox conditions, or food availability, foraminifera are useful indicators of Antarctic paleoenvironments (Melis and Salvi, 2020; Kilfeather et al., 2011; Majewski et al., 2018, 2020). However, foraminiferal assemblages can be decomposed during early diagenesis, reworking, and dissolution due to the strong presence of corrosive bottom waters (Kennett, 1966; Hauck et al., 2012; Seidenstein et al., 2024), blurring the full picture of their diversity and distorting paleoenvironmental interpretations.
In addition to the relatively robust calcareous and multi-chambered agglutinated foraminifera that are the target of routine micropaleontological studies, there is a diverse group of monothalamid (single-chambered) foraminifera, including allogromiids (Gooday et al., 1996; Majewski et al., 2007). Monothalamid foraminifera are particularly well represented in marine restricted environments such as fjords, including environments close to glacier fronts (Majewski, 2010; Korsun et al., 2023), which was also confirmed by metabarcoding analyses (Nguyen et al., 2023a). Due to their fragile, mostly organic-walled shells, they are not preserved in fossil and subfossil archives. Their enormous diversity has been revealed by metabarcoding analyses of sedimentary DNA (Lecroq et al., 2011; Pawlowski et al., 2011; Pawłowska et al., 2014) as well as numerous recent integrative taxonomic studies (e.g., Gooday et al., 2022; Holzmann et al., 2022). Thus, only the metabarcoding analysis of sedimentary ancient DNA (sedaDNA) has the potential to reconstruct their presence in Quaternary deposits.
The sedaDNA analysis examines intracellular and extracellular DNA fragments derived from dead cells or shed by living organisms dispersed in the sediment (Pedersen et al., 2015; Torti et al., 2018). Over the last 4 decades, this method has evolved from analyses of short fragments of mitochondrial or chloroplast DNA (Willerslev et al., 2003; Taberlet et al., 2007) through metabarcoding analyses of amplified DNA fragments using polymerase chain reaction (PCR) or targeted enrichment hybridization (Armbrecht et al., 2021) to broad-based metagenomics analyses covering all groups of living organisms (Slon et al., 2017). The sedaDNA analysis provides new data on past biodiversity or helps to reconstruct paleoenvironmental conditions (Pawłowska et al., 2020a; Armbrecht et al., 2022). Such studies can focus on foraminiferal DNA, improving the reconstruction of their past communities, including rich assemblages of monothalamid foraminifera (Lejzerowicz et al., 2013; Pawłowska et al., 2014, 2020a; Zimmermann et al., 2021; Nguyen et al., 2023b).
This study applies the metabarcoding method to analyze foraminiferal communities in sediments deposited in a unique setting proximal to the paleo-grounding-line of the East Antarctic Ice Sheet in the western Ross Sea. Although attempts have been made to assess the diversity of Antarctic foraminiferal using metabarcoding (Habura et al., 2004; Pawlowski et al., 2011; Li et al., 2023), this is the first time Southern Ocean subsurface sediments have been targeted and one of only a few analyses of subsurface sediments worldwide (Lejzerowicz et al., 2013; Pawłowska et al., 2014, 2020a, b; Szczuciński et al., 2016).
DNA preservation in sediments is strongly related to environmental conditions such as pH, salinity, and temperature; the chemical composition of the sediment; and the biotic activity of living organisms, mainly bacterial communities, which depend on the nature of the nutrient components available in the sediment (Levy-Booth et al., 2007). The factor that significantly increases the possibility of DNA preservation is its ability to bind to mineral and organic grains, protecting DNA from degradation by microbial activity (Blum et al., 1997; Corinaldesi et al., 2008). Strong DNA bonding on clay minerals (Lorenz and Wackernagel, 1987, 1992; Blum et al., 1997; Levy-Booth et al., 2007; Slon et al., 2017), for example, occurs at pH values > 5 (Levy-Booth et al., 2007). pH values in the Ross Sea water column average between 7.9 and 8.3 (Rivaro et al., 2014) with similar values in sediments (Li et al., 2019), which together with low temperatures suggests that the general conditions in the sediments studied are favorable for DNA preservation.
The sediments analyzed in this study are relatively young, at most ca. 20–30 ka, but are highly likely to have experienced repeated mixing and remobilization, especially if deposited subglacially. Considering the delicate structure of DNA and its rapid hydrolysis (Rawlence et al., 2014), significant remobilization of its molecules together with strong sediment mixing and resuspension seems rather unlikely (Willerslev et al., 2004; Armbrecht et al., 2019). However, there is no evidence that sedimentary DNA is affected by weaker mixing processes, such as glacier-induced sediment deformation or redeposition of sediment aggregates, that occur near the grounding line (Prothro et al., 2018; Robinson et al., 2021). Given the potential for highly degraded, short DNA fragments in our samples, we designed a new ultrashort minibarcode marker that may be better suited for analyzing fossil samples.
Based on results from previous studies (Majewski et al., 2020), we combine the micropaleontological data with the newly acquired sedaDNA results. The major goals of this study are (i) to increase our knowledge of foraminiferal communities in glacier-proximal settings of Antarctic shelf seas, including groups not preserved in the fossil record, i.e., monothalamids; (ii) to assess the possibility of preservation of ancient foraminiferal DNA (aDNA) in sediments through glacially induced remobilization and transport and thus the likelihood of preserving the aDNA signal from former interglacials to subglacial tills; and (iii) to test the effectiveness of the shorter, newly designed minibarcode marker compared with a standard, widely used barcode.
2.1 Study area
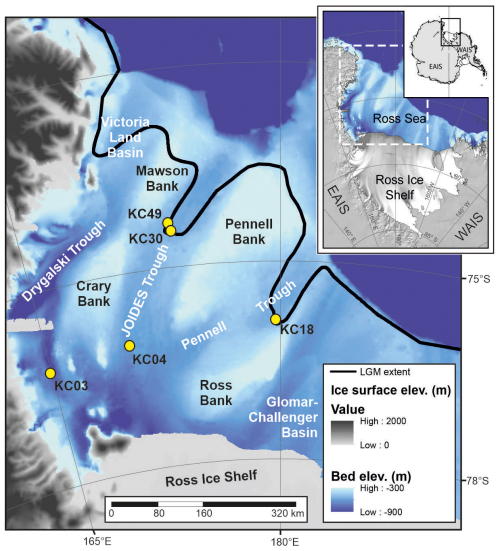
The Ross Sea comprises a vast part of the Antarctic continental shelf. Its drainage area comprises ∼ 25 % of both the East Antarctic Ice Sheet and West Antarctic Ice Sheet (Rignot et al., 2011), which have been retreating since the LGM (Anderson et al., 2014). The seafloor of the Ross Sea was shaped by glacial fluctuations spanning much of the Cenozoic (Barker et al., 2007). Sediment depocenters include large glacial troughs eroded by ice streams during multiple advances of the ice sheet onto the continental shelf (Fig. 1). Importantly, these troughs are largely below the lower limit of iceberg scouring and may therefore provide undisturbed sedimentary archives (Domack et al., 1999). They are separated by several submarine highs or banks that are mostly shallower than 300 m water depth and highly impacted by iceberg scouring (Prothro et al., 2018).
The locations of past ice sheet grounding lines are marked by grounding-zone wedges (GZWs), formed near the grounding line during periods of relative stability. These GZWs are typically up to 100 m thick (Batchelor and Dowdeswell, 2015) and typically overprinted by megascale glacial lineations (MSGLs) formed beneath zones of fast-flowing ice (Spagnolo et al., 2014). These geomorphological features are used to reconstruct ice sheet retreat from the continental shelf that has taken place since the LGM (Halberstadt et al., 2016). In the western Ross Sea, where the East Antarctic Ice Sheet was grounded, subglacial geomorphic features and tills extend to within 30 km of the shelf break (Greenwood et al., 2018; Anderson et al., 2014), where embayments in the northern parts of the JOIDES and Pennell troughs and the Victoria Land Basin (Fig. 1) provided exposure to relatively warm ocean currents.
The Ross Sea continental shelf is the most productive region in the Southern Ocean (Smith, 2022), responsible for > 25 % of its total CO2 uptake (Arrigo et al., 2008). The productivity in the Ross Sea is highly seasonal due to variability in solar radiation and sea ice cover, and it is only occasionally limited by nutrient depletion (Smith et al., 2014). Diatoms account for about half of this productivity and are a significant component of Ross Sea surface sediments (Domack et al., 1999).
The western Ross Sea is also home to the largest coastal polynya in Antarctica (Park et al., 2018). In such environments, sea ice formation is accompanied by the production of high-salinity shelf water (HSSW). This cold, dense water mass is corrosive to calcium carbonate and dominates the deep basins of the western Ross Sea (Kennett, 1966; Jacobs et al., 1985). It ranges in thickness from 300–400 m on the outer continental shelf to nearly 1000 m in the southern Drygalski and JOIDES troughs. Warm modified Circumpolar Deep Water impinges onto the outer continental shelf and upper continental slope (Picco et al., 1999), resulting in relatively strong bottom currents that are capable of winnowing the seafloor and transporting fine sediments onto the inner shelf of the Ross Sea (Prothro et al., 2018). Above these two water masses, Antarctic surface water is present. Its thickness ranges from just a few tens of meters on the inner shelf to ∼ 100 m on the outer shelf (Orsi and Wiederwohl, 2009).
2.2 Sedimentological and micropaleontological framework
During the NBP1502A cruise on board Nathaniel B. Palmer (RVIB, ice-breaking research vessel) (23 January–20 March 2015), following a high-resolution multibeam bathymetric survey (Halberstadt et al., 2016; Simkins et al., 2018), various geomorphological features were cored in different locations with respect to paleo-grounding-lines to better constrain the deglaciation history of the western Ross Sea (Prothro et al., 2020). In this study, we rely on the results of this investigation, including the sedimentary facies model and chronological framework of Prothro et al. (2018, 2020), along with hard-shelled foraminiferal assemblage data from Majewski et al. (2020).
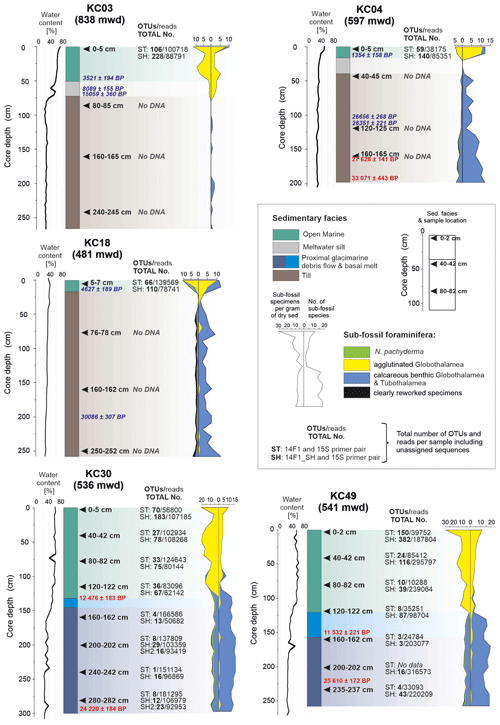
Figure 2Number of reads and operational taxonomic units (OTUs) for standard (ST) and short (SH) primer pairs plotted against environmental conditions (Prothro et al., 2020) and foraminiferal microfossil results (Majewski et al., 2020) in cores collected from the western Ross Sea (Fig. 1). Sample positions in the cores are indicated by black arrows. Radiocarbon-calibrated ages according to Prothro et al. (2020), with those in red measured with foraminiferal tests. Note the presence of the sedaDNA signal only in the top samples taken from open-marine sediments overlying the tills and throughout the marine records in cores KC30 and KC49. mwd – water depth in meters.
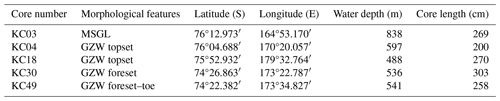
Based on data collected in previous studies (Domack et al., 1999), more recently, Prothro et al. (2018) used sedimentological and micropaleontological characteristics to identify sediment facies associated with different environments, including subglacial, grounding-line-proximal glacimarine, and open marine. Five cores investigated in this study (Fig. 2) recovered sediments deposited in all of these environments, but the entire suite of facies was not sampled for sedaDNA. Facies 1, acquired in the lower parts of cores KC03, KC04, and KC18, sampled MSGL and GZW topsets and is composed of massive diamicton interpreted as till. Facies 2, acquired in cores KC30 and KC49 from the GZW foreset/slope and GZW bottomset/toe, is composed of diamicton with variable sorting and interpreted as debris flows initiated from GZW crests close to the grounding line. Facies 3 is composed of diamicton with abundant granule- to pebble-sized soft sediment clasts. It occurs in thin intervals in the middle of cores KC30 and KC49 and is interpreted as the most ice-proximal deposit formed by basal meltout of debris-laden ice (Prothro et al., 2018; Simkins et al., 2018). Facies 4, interpreted as meltwater plume deposits, occurs in thin intervals in cores KC03 and KC04. Facies 3 and 4 were not sampled for sedaDNA. Finally, facies 5, consisting of olive-grey diatomaceous sandy silt, occurs in the uppermost sections of all cores, and records open-marine conditions. This study focused on facies 1, 2, and 5.
Micropaleontological foraminiferal results (Majewski et al., 2020) show that the tills contain predominantly calcareous foraminifera, including some planktonic forms, at least some of which are clearly reworked. When well-preserved and radiocarbon-dated, as with core KC04, foraminifera from tills yielded ages older than the LGM. The open-marine sediments are dominated by agglutinated foraminifera, mainly Miliammina arenacea and Portatrochammina spp. These are associated with the presence of HSSW and significant primary production in the sediments. Finally, the glacier-proximal sediments, including facies formed below ice shelves, are dominated by calcareous foraminifera, mostly Globocassidulina subglobosa, and radiocarbon-dated between ca. 11 500 and 23 500 cal yr BP (Majewski et al., 2020).
2.3 Sampling
During the NBP1502A cruise, five kasten cores were collected. Cores were taken from locations in different morphological features: KC03 – MSGL, KC04 and KC18 – GZW topsets, KC30 – GZW foreset, and KC49 – GZW foreset–toe, with penetrating sediments deposited in subglacial, proximal–grounding line, and open-marine settings (Table 1 and Fig. 2). The cores were opened immediately after recovery, and undisturbed sediment was collected for sedaDNA, mostly at a regular depth interval of 40 cm. Two replicates were collected at each sampling depth using disposable laboratory gloves and sterile spoons to avoid contamination between samples. Sediment samples were frozen at −20 °C and transported on dry ice to the Laboratory of Paleogenetics and Conservation Genetics, Centre of New Technologies, University of Warsaw, Poland. The cores were then logged and sampled for grain size, subfossil foraminifera, radiocarbon dating, and water content (Prothro et al., 2018, 2020; Majewski et al., 2020).
2.4 Sample preparation and sequencing
Extraction and concentration of total DNA from 34 sediment samples of up to 10 g were conducted with a DNeasy PowerMax Soil Kit (QIAGEN), following the producer protocol in a laboratory dedicated to ancient DNA with no prior history of foraminiferal studies. A hypervariable region 37F of SSU (small subunit) rRNA was amplified using two primer pairs specific to foraminifera and amplifying fragments of different lengths. Firstly, we used the forward primer s14F1 (5'-AAG GGC ACC ACA AGA ACG C-3') (Pawlowski, 2002) paired with the reverse primer s15 (5'-CCA CCT ATC ACA YAA TCA TG-3') (Esling et al., 2015). To enhance detection of strongly degraded DNA from the reworked material, the new forward primer s14F1_SH (5'-GTC CGG ACA CAC TGA GGA TT-3') was designed and paired with the reverse primer s15, resulting in shorter amplicons, i.e., 19 to 132 base pairs (bp) without primers sequences, with the mean around 64 bp compared to the first pair of primers that amplify fragments of ca. 130 bp (89 to 194 bp). The two fragments are referred in this study as short (SH) and standard (ST), respectively. The new primer was designed with Primer3 (https://primer3.org/, last access: 29 May 2025) and confirmed with the NCBI (National Center for Biotechnology Information) Primer-BLAST tool.
The PCR reaction contained 25 µL of AmpliTaq Gold™ 360 Master Mix (Applied Biosystems™), 5 µL of bovine serum albumin, 2 µL of 5 µM primer mix, 15 µL of H2O, and 3 µL of extracted DNA. After denaturation at 94 °C for 5 or 12 min, 60 cycles were applied as follows: 94 °C for 20 s, 52 °C for 20 s, and 72 for 20 s, with final elongation at 72 °C for 2 min. The amplified PCR products were purified on magnetic beads following the Agencourt AMPure PCR purification protocol. Each sample was amplified by both primer pairs in at least five replicates, alongside PCR negative controls. PCR products were examined by electrophoresis on agarose gels. Samples with the PCR product in four or more replicates were transformed into double-indexed Illumina sequencing libraries (Meyer and Kircher, 2010) and sequenced on the MiSeq Illumina platform using MiSeq Reagent Kit v2 (2 × 150 bp). Two replicates were analyzed for SH in samples 200 and 280 cm from KC30.
2.5 Bioinformatics
The paired-end raw reads were first quality-checked using the FastQC program (Andrews, 2010). Then, primers and Illumina tags were removed by Cutadapt (Martin, 2011). Paired-end reads were merged using the fastq_mergepairs module and removed putative chimeric sequences using the uchime_denovo algorithm in VSEARCH v.2.2.2 (Rognes et al., 2016; Edgar et al., 2011), as implemented in SLIM (Dufresne et al., 2019). Subsequently, the remaining reads were dereplicated and clustered at a 97 % similarity threshold into operational taxonomic units (hereafter OTUs), and the abundance of OTUs was calculated with the otu-vsearch module. The possible contaminant OTUs were identified and removed from the dataset using the microDecon package with default settings (McKnight et al., 2019) and negative control samples as a reference. The non-foraminiferal OTUs (without a signature, GACAG), as well as OTUs with less than 10 reads in the total dataset were removed for further analysis for both the ST and SH fragments. OTUs were aligned in BLAST (Altschul et al., 1990) using the BLAST best-hit search against a curated foraminiferal local database based on minimum similarity (−perc_identity 90 %, seven mismatches or gaps accepted) and minimum coverage (−qcov_hsp 90 %) for the taxonomic assignment. The OTUs below 93 % identity were classified at the genus level, if possible, or as unassigned foraminifera. For strict taxonomic analysis, to avoid possible biases, we filtered out the OTUs with less than 10 reads per sample. We were further interested in comparing the OTU composition between two datasets based on the same region (37F). Thus, we first trimmed the ST datasets and identified the shared OTUs using Biopython, and for visualization, we prepared Venn diagrams with shared OTUs and reads.
2.6 Statistics
Before the diversity estimates and statistical analysis, the singleton OTUs (occurring in only one sample) were removed. Statistical analyses were run in R, version 4.1.0 (R Core Team, 2013). All formal hypothesis tests were conducted at the 5 % significance level (α = 0.05), and plots were created using the ggplot2 package (Wickham, 2016). The OTU tables were rarefied using the lowest read depth corresponding to the sample with the least reads (10 336 for ST and 50 654 for SH). Based on the normalized data, alpha diversity, defined as within-sample diversity, was calculated as richness indices (ACE and Chao1) and evenness indices (Shannon, Simpson) for each sample using the estimateR and diversity functions in the vegan package (Oksanen et al., 2019). Significant differences between environmental settings in each sample diversity index in each dataset (ST and SH) were detected using the Wilcoxon test and stat_compare_means function of the ggpubr package (Kassambara, 2023). Beta diversity, defined as the variation in community composition between samples, was estimated as the Bray–Curtis distance and analyzed using nonmetric multidimensional scaling (NMDS) with the metaMDS function of the vegan package.
3.1 The sedaDNA metabarcoding data
Of the 34 samples subjected to sedaDNA extraction, PCR products were obtained for 18 samples in at least 4 replicates each. These included all surface samples from all cores, representing open-marine facies, as well as subsurface samples of open-marine facies and glacimarine facies proximal to the grounding line for cores KC30 and KC49. No PCR products were observed in till samples (in KC03, KC04, and KC18) (Fig. 2). After quality filtering; merging; and removal of chimeras, non-foraminiferal sequences, and control samples, we obtained 4 253 649 reads, including 1 515 729 reads for the ST and 2 737 920 reads for the SH datasets. As shown in the Venn diagram comparing the same fragments that could be amplified by the SH and ST markers (Fig. S1 in the Supplement), 230 OTUs (corresponding to 1 528 719 reads) were shared between the two datasets, with the majority, i.e., 55 % and 81 %, of the OTUs and reads for ST, respectively. The SH dataset had 852 unique OTUs representing 79 % and 44 % of the OTUs and reads, respectively.
Table 2Ranges of OTUs and reads and average values for samples from different types of sediments. Results are shown for standard (ST) and short (SH) primer pairs. n – number of samples. The ST sample from 200 cm depth in KC49 ST was not used for calculating the averages.
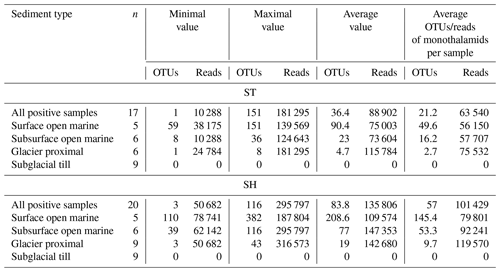
After further removal of rare OTUs (< 10 reads in a single sample, representing < 1 % of the number of subsampled reads), 1383 OTUs (397 of ST and 986 of SH) representing 4 227 450 (1 511 339 of ST and 2 716 111 of SH; Table S1 in the Supplement) reads were used for taxonomic analysis. On average, the number of OTUs and reads per sample reached 36 and 88 902 for 17 ST samples with positive results and 84 and 13 580 for 20 SH positive samples, respectively (Table 2). The ST sample from 200 cm depth in KC49 was not included as no OTUs remained after removing OTUs with < 10 reads.
There are clear differences in the DNA results from different sediment types. No PCR products were obtained from the tills (cores KC03, KC04, and KC18). The number of OTUs strongly graded from the highest in the surface open-marine sediments, i.e., 90 and 209 OTUs for the ST and SH primers, respectively, on average, through subsurface open-marine sediments (23 and 77 OTUs) to the lowest (5 and 19 OTUs) in the glacier-proximal sediments below 120 cm in cores KC30 and KC49 (Table 2).
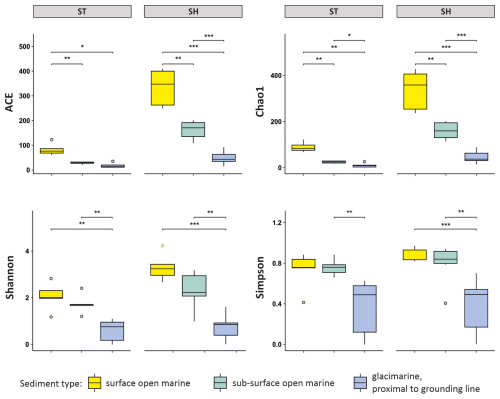
3.2 Alpha diversity
Normalized foraminiferal alpha diversity expressed by the ACE and Chao1 indices are both more than twice as high for SH than for ST and tend to decrease from surface through subsurface open-marine to glacier-proximal samples (Fig. 3). The Shannon and Simpson indices follow a similar pattern. The Shannon index is higher for SH than for ST and shows significantly higher values for open-marine than for glacier-proximal environments for both datasets (Wilcoxon tests, p < 0.01), but the difference between surface and subsurface open-marine values is reduced in comparison with the ACE and Chao1 indices (Fig. 3). For the ST dataset, the Shannon index averages 2.2 for open-marine surface samples and 1.8 for open-marine subsurface samples. For glacier-proximal samples, the Shannon index is lower, reaching only 0.8 on average. For the SH dataset, the Shannon values are higher, averaging 3.2 for open-marine surface samples and 2.4 for open-marine subsurface samples. Shannon values are again lowest in the glacier-proximal environment, averaging only 0.8. The Simpson index shows roughly similar values for ST and SH for surface and subsurface open-marine samples, at ca. 0.8 for both ST and SH, and significantly lower values for glacier-proximal samples, at ca. 0.5 (Wilcoxon tests, p < 0.01, Fig. 3).
3.3 Beta diversity
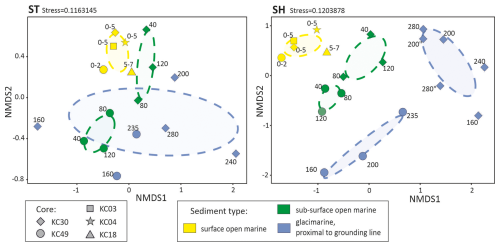
The difference in community composition between the two datasets is reflected in the NMDS plots (Fig. 4). In general, SH and ST datasets produced similar patterns; however, community composition is more scattered in the SH datasets. For the ST dataset, the foraminiferal communities in open-marine samples form clusters for surface and subsurface samples that do not overlap, while those in glacier-proximal samples are largely scattered and overlap with some subsurface open-marine samples (Fig. 4a). For the SH dataset, communities in surface open-marine, subsurface open-marine, and glacier-proximal sediments form distinct clusters (Fig. 4b). Those from cores KC30 and KC49 cluster separately in the case of open-marine subsurface for both datasets and glacier-proximal samples for the SH dataset only. Surface open-marine samples from different cores form a single tight cluster.
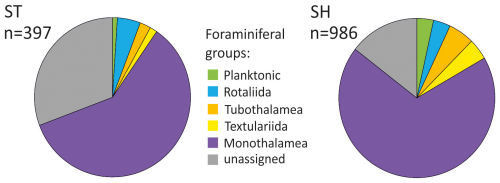
Figure 5Proportion of OTUs of different foraminiferal taxonomic groups detected using standard (ST) and short (SH) primer pairs. n – number of OTUs in the ST/SH dataset. OTUs with less than 10 reads in a single sample excluded.
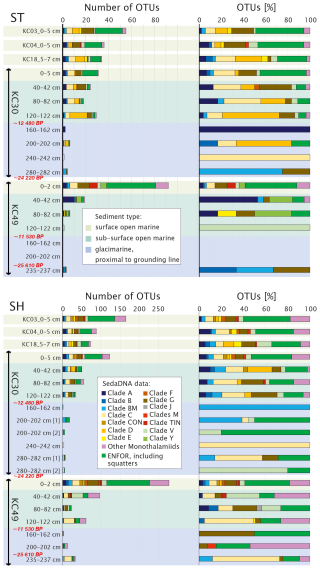
Figure 6Taxonomic composition of monothalamid foraminifera sequenced for standard (ST) and short (SH) primer pairs arranged by sediment type (Prothro et al., 2020). Samples from cores KC30 and KC49 are marked by bars; radiocarbon ages (Prothro et al., 2020) are marked in red. OTU numbers and percentages (only OTUs with ≥ 10 reads in a single sample are shown) are grouped according to the clades identified within the monothalamids (Pawlowski et al., 2011). Environmental foraminifera (ENFOR) correspond to clades known only from environmental sequencing (Pawlowski et al., 2014). Sample depth intervals are indicated.
3.4 Taxonomic composition
The OTUs assigned to the reference sequences represent all major foraminiferal groups (Fig. 5). They are predominant, representing 69.3 % and 85.5 % of the total ST and SH datasets, respectively. When calculated on average per sample, the assigned OTUs accounted for 78.8 % of the ST dataset and as much as 88.3 % of the SH dataset (Table S2 in the Supplement). The assigned OTUs represent 67 genera and 53 named species (Table S3 in the Supplement), representing 33 genera for the ST primers and 59 genera for the SH primers, and they do not always overlap (Table S3). In addition, OTUs assigned to monothalamids represented 17 different clades (Fig. 6), including 12 clades for the ST primers and 17 for the SH primers, together with 10 other divisions based strictly on environmental sequences called ENFOR (environmental foraminifera) clades, 9 detected with ST and 10 detected with SH primers, as well as numerous unclassified environmental sequences (see Table S1).
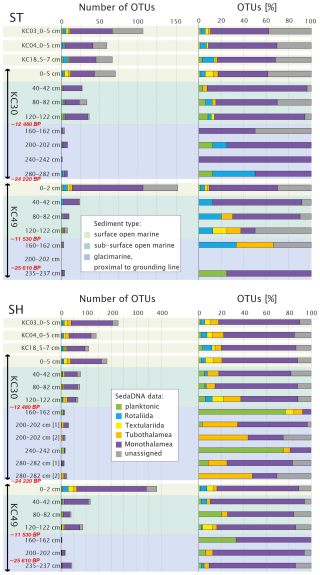
Figure 7Number of OTUs, reads (only OTUs with ≥ 10 reads in a single sample are shown), and percentages of OTUs of different foraminiferal groups for the datasets based on standard (ST) and short (SH) primer pairs plotted against sediment type according to Prothro et al. (2020). Samples from cores KC30 and KC49 are marked by the bars; radiocarbon ages (Prothro et al., 2020) are marked in red. Sample intervals are indicated.
In both datasets, OTUs of Monothalamea dominate overall, reaching an average of 58.5 % for the ST dataset and 58.4 % for the SH dataset. The number of monothalamid OTUs is highest in the surface sample of KC49, with up to 92 OTUs in the ST dataset and 278 OTUs in the SH dataset (Figs. 6 and 7). The rotaliids are the second most abundant group in the ST dataset (9.7 %). In SH, the most abundant groups after Monothalamea are planktonic foraminifera (12.9 %) and Tubothalamea (12.1 %). The other taxonomic groups (e.g., Textulariida) are also recorded but in small numbers, i.e., < 3 % (Table S2). Unassigned OTUs average 11.7 % in the SH dataset and 21.2 % in the ST dataset.
In the ST dataset, the most commonly detected foraminifera, i.e., taxa identified in the largest number of samples, are the planktonic Neogloboquadrina pachyderma in 11 out of 17 samples, the monothalamid genera Micrometula and Hippocrepinella in 9 samples, the rotaliids Nonionella auris and Cibicidoides wuellerstorfi in 6 samples, and the monothalamid genus Psammophaga in 5 samples (Table S1). All of these foraminifera are found in several surface open-marine sites and at least one glacier-proximal site. In the SH dataset, the most common are N. pachyderma, found in 18 of 20 samples; the miliolids Cornuspira antarctica in 16 samples and Cornuspiramia sp. in 11 samples; the monothalamids Gloiogullmia sp. and Micrometula sp. in 11 samples; Hippocrepinella sp. and Saccammina sp. in 7 samples; and the rotaliids N. auris and G. subglobosa, found in 7 and 6 samples, respectively, along with a few clades of monothalamids (Table S1). Of these most commonly identified taxa, only G. subglobosa is not found in glacier-proximal samples.
3.5 Downcore assemblage variability
The proportions of the major taxonomic groups change with core depth. From the depth of 40 to 120 cm, corresponding to the subsurface open-marine system in KC30 and KC49, the number of OTUs is lower than in surface samples but the assemblage is relatively diverse, with a strong dominance of monothalamids and unassigned OTUs. From a depth of 160 cm, corresponding to the glacier-proximal marine samples in those cores, the number of OTUs decreases sharply. For the glacier-proximal samples, in the ST dataset, there is a strong dominance of Monothalamea and an absence of Tubothalamea, Textulariida, and unassigned OTUs (Fig. 7); benthic and planktonic Rotaliida may also be missing in some samples. In the SH dataset of KC30 samples, Monothalamea is less dominant in glacier-proximal samples below 120 cm than in overlying open-marine samples and planktonic Rotaliida and Tubothalamea may be more abundant. Textulariida and benthic Rotaliida are absent below 120 cm. Unassigned OTUs vary from 0 % to ca. 30 %. In KC49 samples, on the other hand, Monothalamea are more dominant, especially in the two bottom samples, but Textulariida are consistently absent below 120 cm and benthic Rotaliida are present only in the bottom sample.
The occurrence of particular OTUs/species is difficult to follow downcore as it is highly irregular with depth. The variability in the overall assemblage, including the discontinuity below core depths of 120 cm, is shown by different clusters in the NMDS plot for SH (Fig. 4) and presence/absence in open-marine vs. glacier-proximal facies in Table S3. Except for two monothalamid species, Bathysiphon flexilis in KC30 at 200 cm depth and Conqueria laevis in KC49 at 235 cm depth, both recognized by SH, there are no assigned foraminiferal taxa specific to glacier-proximal sediments that were not detected in open-marine facies (Table S3). Among Globothalamea and Tubothalamea, only a few are genetically detected in the glacier-proximal facies (Table S3), all of which are also present in open-marine samples. In the glacier-proximal facies, with the ST primers, it was possible to amplify G. subglobosa and C. wuellerstorfi from single samples in KC30 and N. auris and Cornuspiramia sp. (Tubothalamea) in KC49. The SH primers revealed the presence of N. auris in the deepest sample of KC49, within the family of Trochamminidae (Textulariida) in KC30, and abundant C. antarctica (Tubothalamea) and planktonic N. pachyderma in cores KC30 and KC49 (Table S1).
Compared to globothalamids and tubothalamids, monothalamids are more abundant in the glacier-proximal samples. In core KC30, OTUs belonging to the monothalamid clades A, B (Bowseria sp.), BM (B. flexilis and Micrometula sp.), C (Gloiogullmia sp. and Hippocrepinella sp.), D, G, V, and the environmental clades (ENFOR2 and ENFOR3) are recorded for the ST (13 OTUs) and SH (29 OTUs) datasets (Tables S1 and S3). In the KC49 samples from the same facies, we observed only three monothalamid OTUs for the ST dataset (Psammosphaera sp., Micrometula sp., and an OTU belonging to clade G), while for the SH dataset, we obtained 43 OTUs. Most of them, 33 OTUs, are from the deepest sample at 235 cm depth, including numerous OTUs representing the genera Micrometula, Saccamina and Gloiogullmia and a few OTUs assigned to C. laevis and Bathyallogromia sp., as well as clades G and M3 and ENFORs 2, 3, and 4 (Table S1).
4.1 Absence of the ancient DNA signal in glacially redeposited sediments
Numerous studies have focused on the marine record of Antarctic deglaciation, particularly in the Ross Sea, but the details of ice sheet behavior remain uncertain (Anderson et al., 2014; Prothro et al., 2020). This is largely due to the widespread reworking of sediments during multiple ice sheet retreats and expansions and difficulties in distinguishing between subglacial and glacimarine sediments (Domack et al., 1999; Prothro et al., 2018). The reworking process also affects biogenic carbonates and organic matter (Domack et al., 1999; Prothro et al., 2020), raising the question of whether the DNA signal representing past glacial cycles can be transferred to post-LGM sediments.
In our study, we were unable to detect foraminiferal DNA in subsurface samples from sites located in areas with MSGL (KC03) or GZW topsets (KC04 and KC18) for either the ST or ultrashort SH primer pairs. In addition, spectrometric measurement of DNA content after extraction and concentration failed to detect its presence. Organic carbon and foraminiferal tests from these deposits yielded radiocarbon ages of < 30 000 years (Fig. 2), so these samples are young enough to allow for the preservation of sedaDNA (Lejzerowicz et al., 2013; Pawłowska et al., 2020b; Armbrecht et al., 2022). Consequently, the lack of measurable DNA could be attributed to sediment mixing and dilution, which may cause DNA degradation. This is based on the interpretation that the MSGL and GZW topsets are products of bed deformation and sediment reworking during long-distance transport beneath the ice sheet (Domack et al., 1999; Spagnolo et al., 2014; Halberstadt et al., 2018; Robinson et al., 2021).
Ross Sea tills are typically overcompacted, stiff diamictons with low porewater content (Domack et al., 1999; Prothro et al., 2018), indicating that these sediments must have been under considerable pressure during ice sheet expansion (Tulaczyk et al., 2001; Robinson et al., 2021). Perhaps this was another factor affecting the preservation of sedaDNA. Tills in our samples have < 30 % water content, i.e., from 25.4 % in KC03 and 27.5 % in KC04 to 29.3 % in KC18 on average (Fig. 2), whereas in overlying open-marine sediments water content reaches up to 70 % (Prothro et al., 2018). This is a significant difference, but values similar to those recorded in the tills have also been measured in glacier-proximal sediments in the lower parts of KC30 (30.8 % on average) and KC49 (28.6 %), which revealed the presence of sedaDNA, suggesting that compaction was not a critical factor in preserving DNA.
Overall, it is not clear whether the paucity of DNA in the tills observed in KC03, KC04, and KC18 (Fig. 2) applies to all types of subglacial sediments and all types of DNA material. This needs to be confirmed in further studies of relatively young tills with well-preserved microfossils. The absence of DNA in glacial sediments can be further confirmed, for example, by shotgun sequencing or the target enrichment technique by hybridization method based on a single-strain library, i.e., 30 bp long, which is able to capture very short sedaDNA fragments of a dozen to several tens of base pairs (Wales et al., 2015), performed not only on foraminifera but also on other marine organisms.
4.2 Low foraminiferal diversity in sedaDNA samples near the grounding line
Although foraminifera are the key microfossils for reconstructing past paleoenvironments, their distribution in some important Antarctic habitats, especially under ice shelves, remains unclear. Recent assemblages have been documented from only two sites below the Ross Ice Shelf; i.e., the testate forms from about 400 km south of the calving front (Lipps et al., 1979; Dameron et al., 2024), and the monothalamid foraminifera form from 10 km south of it (Pawlowski et al., 2005). Similar studies have been carried out beneath the Amery Ice Shelf in East Antarctica, revealing abundant planktonic foraminifera (Hemer et al., 2007), which, together with other microplankton, increased just prior to the colonization of sub-ice-shelf habitats by benthic infauna and filter feeders (Post et al., 2007). These direct observations are rare and cannot provide the baseline data needed for reliable environmental reconstructions.
An estimate of sub-ice-shelf communities can also be based on studies in restricted, low-productivity Antarctic settings, such as Explorers Cove in McMurdo Sound, which is characterized by cold and nutrient-poor waters derived from beneath the Ross Ice Shelf (Barry and Dayton, 1988) and semi-permanent sea ice (Gooday et al., 1996). The fauna of Explorers Cove is heterogeneous (Bernhard, 1987) and includes a variety of hard- and soft-shelled forms representing widespread and endemic species (Gooday et al., 1996). Subfossil foraminifera from sub-ice-shelf habitats have also been studied in sediment cores, where the multi-proxy approach has allowed for robust interpretation of past conditions (Kilfeather et al., 2011; Majewski et al., 2018, 2020; Bombard et al., 2024; Seidenstein et al., 2024). These micropaleontological data seem to be more complete than our metabarcoding results in the case of Rotaliida and Textulariida (Table S3), but they do not include monothalamids, which carry important ecological information (e.g., Habura et al., 2004; Lecroq et al., 2011; Pawlowski et al., 2002a, b). Our study of the small subunit of the rRNA region now allows us to extend our knowledge of foraminifera that are not preserved in the fossil record in poorly studied glacier-proximal habitats.
The most striking feature of the KC30 and KC49 sedaDNA records is the significantly lower foraminiferal diversity in the glacier-proximal sediments than in the subsurface open-marine sediments represented by ST and SH (Figs. 6 and 7). Except for two monothalamid species, B. flexilis and C. laevis, there are no assigned foraminiferal taxa specific to the glacier-proximal environment. Furthermore, these two are also known elsewhere from more open-marine environments (Höglund, 1947; Gooday and Pawlowski, 2004). All other taxa identified in glacier-proximal sediments also occur in open-marine settings, so the glacier-proximal assemblage appears to represent a subset of foraminifera present in open-marine settings. Nevertheless, representatives of 16 genera are present in the glacier-proximal samples from KC30 and KC49 (Table S3), including 9 monothalamid genera, together with 7 and 19 OTUs of unnamed monothalamids belonging to various clades for ST and SH, respectively, and 8 OTUs representing ENFOR clades (Pawlowski et al., 2011; see Table S1). Their presence in the highly restricted environment below the ice shelf and in relative proximity to the grounding line is consistent with the findings of Habura et al. (2004), which revealed unexpectedly high foraminiferal diversity, with ca. 90 % of environmental DNA reads belonging to Monothalamea in McMurdo Sound (Gooday et al., 1996).
It is also important to note that among the testate Globothalamea and Tubothalamea, only a few are genetically detected in the glacier-proximal facies (Table S3); G. subglobosa, C. wuellerstorfi, N. auris, and Cornuspiramia sp. were present in individual samples of the ST dataset, while the SH results revealed the presence of N. auris, C. antarctica, and the family Trochamminidae (Table S1). Importantly, sequences assigned to N. pachyderma are more abundant in the glacier-proximal facies than those that belong to benthic Globothalamea, reflecting the high dispersal potential of the planktonic foraminifera. This agrees with an influx of abundant N. pachyderma at least 100 km beneath Amery Ice Shelf reported by Hemer et al. (2007). Furthermore, in the ST dataset, a single but abundant OTU of planktonic N. pachyderma was detected in the deepest samples in KC30 (280 cm) and in KC49 (235 cm), dated to ca. 25 ka (Fig. 7). Their presence may indicate a rich influx of microplankton with open-ocean water close to the grounding line near the time of the LGM. In addition, the very strong presence of N. pachyderma in the KC30 record (Fig. 7), shown by the SH data just prior to ice shelf retreat and establishment of open-marine conditions, corresponds to the microfossil record of abundant N. pachyderma prior to ice shelf collapse in Pine Island Bay (Kirshner et al., 2012; Totten et al., 2017). In reality, we do not observe the same signal in KC49, nor in the ST and micropaleontological data (Majewski et al., 2020), so this intriguing interpretation remains problematic.
In summary, our data do not support the presence of index species indicative of a glacier-proximal environment. This is due to the low diversity of the glacier-proximal assemblage, which appears to represent a subset of foraminifera found in open-marine facies. However, the sedaDNA records of KC30 and KC49 from JOIDES Trough are very different (Fig. 4). At the site of KC30, the SH marker revealed a stronger presence of N. pachyderma and Tubothalamea (Fig. 7), which may reflect less restricted conditions. If not due to incomplete records, this suggests considerable faunal variability in glacier-proximal benthic foraminiferal communities, which was not detected by fairly consistent micropaleontological results (Majewski et al., 2018, 2020).
4.3 Does the length of the marker matter?
Several physicochemical factors present in natural environments, i.e., UV radiation and the hydrolysis process, as well as the biological activity of bacterial deoxyribonuclease, contribute to the degradation of DNA structure, causing its fragments to become shorter with time (Blum et al., 1997; Levy-Booth et al., 2007) and core depth (Armbrecht et al., 2021). In order to analyze more degraded material and to improve our reconstruction of past foraminiferal assemblages, we designed a new primer, s14F1_SH, which allows for the amplification of shorter DNA fragments than the routinely used ST marker.
Our results confirm that the amplicon length has a direct impact on the quantitative analysis of metabarcoding data. Overall, after filtering, we detected almost 2.5 times more OTUs with the newly designed SH than with the ST marker, i.e., 986 vs. 397 (Fig. 5), which do not always overlap (Fig. S1). This increased effectiveness of the ultrashort SH marker is manifested by significantly higher values of the alpha diversity indices (Fig. 3). Importantly, except the Simpson index, all indices show significantly higher values for the SH marker not only in subsurface but also in surface samples, suggesting its higher effectiveness in analyzing not only fossil, i.e., degraded, but also modern DNA. The SH marker also allows for better differentiation between OTUs from different sediment types and from different cores but mainly from subsurface samples (Fig. 4).
The higher performance of the ultrashort SH marker could be explained by several factors. First, the newly designed forward primer can amplify a wider range of foraminiferal taxa, especially in the case of monothalamids. Indeed, only up to 15 monothalamid OTUs are detected by the ST marker in glacier-proximal samples, whereas up to 69 OTUs are detected by the SH marker (Table S1). The SH marker also appears to perform better than ST at detecting planktonic foraminifera and Tubothalamea (Fig. 5). Some species present in the SH dataset are absent from the ST dataset (e.g., Astrononion echolsi; Table S3). However, each of these species could be amplified using the ST primers, suggesting that primer specificity is not the real cause of the increased number of OTUs.
The most plausible reason for the quantitative difference between ST and SH markers is the ability of the latter to amplify very short fragments of highly degraded DNA. The usefulness of short barcodes, even < 100 bp, has been demonstrated on several occasions, e.g., for plant metabarcoding using the ultrashort trnL marker (Taberlet et al., 2007; Mallott et al., 2018). It has also been shown that some foraminiferal species can only be detected in sediment samples when targeted using species-specific fragments (Lejzerowicz et al., 2013). However, our study does not clearly show that the proportion of SH and ST metabarcodes changes with sediment age.
Furthermore, the decreasing length of DNA barcodes may reduce their taxonomic resolution and lead to misidentifications, as illustrated in our data by the assignment of some SH OTUs to tropical genera such as Borelis or Planoperculina (Table S3). The choice of marker used for metabarcoding should therefore be a compromise between the ability to amplify degraded DNA and taxonomic resolution. In light of our results, the SH marker has the potential to become a new standard for foraminiferal paleogenomics. However, its taxonomic resolution needs to be evaluated and its performance needs to be tested in other environments.
4.4 Complementarity of sedaDNA and micropaleontological records
When comparing the metabarcoding results with the paleontological record (Table S3), the low degree of overlap is striking. The discrepancy is due to the abundance of testate forms, i.e., Textulariida, Rotaliida, and Tubothalamea in the fossil data, as reported by Majewski et al. (2020), and the dominance of soft-walled Monothalamea in the sedaDNA data.
The paucity of fragile monothalamids in the fossil record is well known and established (Gooday et al., 1996). More intriguing is the low abundance of Textulariida, Rotaliida, and Tubothalamea (Table S3) in the metabarcoding data. Excluding surface samples, there is a limited sedaDNA record of Textulariida in open-marine samples, which dominate the microfossil record, and sparse sedaDNA record of Rotaliida in glacier-proximal samples, despite abundant tests of calcareous foraminifera present in the same samples (Majewski et al., 2020). In fact, only 1 and 11 (ST and SH) OTUs, respectively, representing agglutinated Globothalamea (Reophax subfusiformis and Arenoparrella mexicana) were recognized in subsurface open-marine samples (Table S1) and only in samples from 120 cm depth in KC30 and KC49, which were directly adjacent to the layer representing ice-proximal sediments. The OTUs represented by the rotaliids (C. wuellerstorfi, Epistominella sp., N. auris, Bolivinellina pseudopunctata, G. subglobosa, and Stainforthia sp.) and Tubothalamea (Cornuspiramia sp., C. antarctica, Cyclogyra sp., and Spirophtalmidium sp.) were more abundant in subsurface samples, but similar to Textulariida they were mostly in single samples, while for many species their subfossils were present throughout large parts of cores KC30 and KC49 (Fig. 2). The Tubothalamea species detected by sedaDNA are actually not recognized in the fossil record (Table S3). Only the planktonic N. pachyderma is widespread in the sedaDNA record, but a second planktonic species (Globorotalia scitula) is also noted in the SH results, the presence of which was not identified in the fossil record.
A general inconsistency between the fossil and molecular record could be due to the random nature of the PCR (Vosberg, 1989) and different amplification efficiencies due to different strengths of DNA binding depending on the lithology of the sediment. It is also possible that other natural factors, such as significant genetic polymorphism (Weber and Pawlowski, 2014) and highly variable numbers of rRNA copies at different life stages and between different species (Weber and Pawlowski, 2013), may bias the sedaDNA results. However, the striking inconsistency between fossil and molecular records found in this study (Table S3) seems more likely to be due to more specific causes. It could be due to natural sedimentary and diagenetic processes resulting in increased microfossil diversity due to reworking and selective preservation of subfossil tests. However, this possibility is unlikely as the fossil assemblages are consistent between sites and the reworking of foraminiferal tests from older strata does not appear to have occurred at the sites of KC30 and KC49 (Majewski et al., 2020). In addition, radiocarbon dating of foraminiferal tests confirms that the calcareous assemblage is in situ in glacier-proximal sediments (Prothro et al., 2020; Majewski et al., 2020). The selective preservation of subfossil assemblages can be also an issue, but we observe an underrepresentation rather than an overrepresentation of genetically identified Textulariida and Rotaliida.
Alternatively, the strong underrepresentation of rotaliids in the sedaDNA results from the glacier-proximal samples could be explained by the strong binding of DNA fragments to carbonate grains, to the point where they are difficult to extract or the DNA is highly degraded (Barton et al., 2006; Levy-Booth et al., 2007; Freeman et al., 2023). Good preservation of resilient calcareous specimens, as observed in the fossil record, may further enhance these processes. However, it is still unclear why DNA from fragile monothalamids, which dominate the sedaDNA results (Fig. 7), is better represented than agglutinated Textulariida in the open-marine facies. It is possible that monothalamid DNA, being released quickly due to the delicate nature of their tests, binds much more rapidly to sediment grains before bacterial deoxyribonuclease intensifies its destructive activity (Blum et al., 1997; Levy-Booth et al., 2007) than is in the case for robustly testate Rotaliida and Textulariida.
To sum up, although metabarcoding is increasingly used to study modern (Li et al., 2023; Nguyen et al., 2023a) and past (Pawłowska et al., 2014; Nguyen et al., 2023b) foraminiferal diversity, it is important to keep in mind that sedaDNA analysis and micropaleontological results can be highly divergent (Lejzerowicz et al., 2013; Pawłowska et al., 2014) but at the same time highly complementary. By combining these two approaches, it is possible to reconstruct a more complete and ecologically meaningful picture of the diversity of foraminiferal assemblages. The advantage of the sedaDNA metabarcoding method is particularly important in marginal marine environments, where fragile Monothalamea are dominant (Gooday et al., 1996; Nguyen et al., 2023a).
In this study, high-throughput sequencing of sedaDNA is used to improve the understanding of foraminiferal communities inhabiting open-marine and glacier-proximal environments of the western Ross Sea, mainly by adding the record of abundant and diverse monothalamids not preserved in the fossil record. By using the newly designed forward primer s14F1_SH, which allows for the amplification of DNA fragments that are ca. 50 bp shorter, we were able to detect higher diversity in surface and subsurface samples and discriminate between foraminiferal assemblages from different sediment types and different cores better than with the standard approach. Thus, the newly designed ultrashort marker appears to be potentially more useful for paleoecological studies.
Our results, showing a consistent absence of a foraminiferal DNA signal in tills, suggest an absence of their DNA in sediments overridden and reworked by advancing ice sheets during the last glaciation. Foraminiferal assemblages from the open-marine environment show significantly greater alpha diversity than sediments deposited on the slopes of a grounding-zone wedge proximal to the grounding line. The metabarcoding method appears to be particularly useful in restricted marine environments, such as those proximal to the grounding line, where fragile monothalamids predominate. Foraminifera surviving in such an environment represent a subset of the species present in open-marine facies.
At the same time, the sedaDNA records from sites KC30 and KC49, which are located along the Last Glacial Maximum grounding-zone wedge foreset in the JOIDES Trough, are significantly different. If not due to undersampling, this observation suggests considerable variability in glacier-proximal foraminiferal communities. Interestingly, this variability is not reflected in the micropaleontological data, which diverge strongly from the sedaDNA results. These two approaches are highly complementary and, when combined, provide enriched information on past biodiversity.
Raw sequencing reads generated in this study were deposited in the European Nucleotide Archive under project PRJEB85670. Table S1 is available at https://doi.org/10.18150/9ABUGS (Demianiuk, 2025), and the remaining data used for this study can be found in the Supplement.
The supplement related to this article is available online at https://doi.org/10.5194/bg-22-2601-2025-supplement.
ED and WM designed the study and participated in the fieldwork. JBA organized the fieldwork. ED performed the laboratory work under the supervision of MB, and DP analyzed the data. IBA and NLN performed bioinformatic and statistical analyses. JP helped to interpret the results. ED and WM drafted the paper, and ED prepared the figures. All authors participated in the revision of the first draft.
The contact author has declared that none of the authors has any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
Inès Barrenechea Angeles is funded by the Swiss National Science Foundation (project number: 221959). We would like to thank R. Mark Leckie and an anonymous reviewer for their comments that helped to improve this paper.
This research has been supported by the National Science Foundation (grant no. ANT-1246353) and the Narodowe Centrum Nauki (grant no. NCN-2015/17/B/ST10/03346).
This paper was edited by Chiara Borrelli and reviewed by R. Mark Leckie and one anonymous referee.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J.: Basic local alignment search tool, J. Mol. Biol., 215, 403–410, https://doi.org/10.1016/S0022-2836(05)80360-2, 1990.
Anderson, J. B., Conway, H., Bart, P. J., Witus, A. E., Greenwood, S. L., McKay, R. M., Hall, B. L., Ackert, R. P., Licht, K., Jakobsson, M., and Stone, J. O.: Ross Sea paleo-ice sheet drainage and deglacial history during and since the LGM, Quaternary Sci. Rev., 100, 31–54, https://doi.org/10.1016/j.quascirev.2013.08.020, 2014.
Andrews, S.: FASTQC. A quality control tool for high throughput sequence data, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (last access: 29 May 2025), 2010.
Armbrecht, L. H., Coolen, M. J., Lejzerowicz, F., George, S. C., Negandhi, K., Suzuki, Y., Young, J., Foster, N. R., Armand, L. K., Cooper, A., Ostrowski, M., Focardi, A., Stat, M., Moreau, J. W., and Weyrich, L. S.: Ancient DNA from marine sediments: precautions and considerations for seafloor coring, sample handling and data generation, Earth Sci. Rev., 196, 102887, https://doi.org/10.1016/j.earscirev.2019.102887, 2019.
Armbrecht, L., Hallegraeff, G., Bolch, C. J. S., Woodward, C., and Cooper, A.: Hybridisation capture allows DNA damage analysis of ancient marine eukaryotes, Sci. Rep.-UK, 11, 3220, https://doi.org/10.1038/s41598-021-82578-6, 2021.
Armbrecht, L., Weber, M. E., Raymo, M. E., Peck, V. L., Williams, T., Warnock, J., Kato, Y., Hernández-Almeida, I., Hoem, F., Reilly, B., Hemming, S., Bailey, I., Martos, Y. M., Gutjahr, M., Percuoco, V., Allen, C., Brachfeld, S., Cardillo, F. G., Du, Z., Fauth, G., Fogwill, C., Garcia, M., Glüder, A., Guitard, M., Hwang, J. H., Iizuka, M., Kenlee, B., O'Connell, S., Pérez, L. F., Ronge, T. A., Seki, O., Tauxe, L., Tripathi, S., and Zheng, X.: Ancient marine sediment DNA reveals diatom transition in Antarctica, Nat. Commun., 13, 5787, https://doi.org/10.1038/s41467-022-33494-4, 2022.
Arrigo, K. R., van Dijken, G., and Long, M.: Coastal Southern Ocean: A strong anthropogenic CO2 sink, Geophys. Res. Lett., 35, L21602, https://doi.org/10.1029/2008GL035624, 2008.
Barker, B. F., Diekmann, B., and Escutia, C.: Onset of Cenozoic Antarctic glaciation, Deep-Sea Res. Pt. II, 54, 2293–2307, https://doi.org/10.1016/j.dsr2.2007.07.027, 2007.
Barry, J. P. and Dayton, P. K.: Current patterns in McMurdo Sound, Antarctica and their relationship to local biotic communities, Polar Biol., 8, 367–376, https://doi.org/10.1007/BF00442028, 1988.
Bart, P. J., DeCesare, M., Rosenheim, B. E., Majewski, W., and McGlannan, A.: A centuries-long delay between a paleo-ice-shelf collapse and grounding-line retreat in the Whales Deep Basin, eastern Ross Sea, Antarctica, Sci. Rep.-UK, 8, 12392, https://doi.org/10.1038/s41598-018-29911-8, 2018.
Barton, H. A., Taylor, N. M., Lubbers, B. R., and Pemberton, A. C.: DNA extraction from low-biomass carbonate rock: an improved method with reduced contamination and the low-biomass contaminant database, J. Microbiol. Meth., 66, 21–31, https://doi.org/10.1016/j.mimet.2005.10.005, 2006.
Batchelor, C. L. and Dowdeswell, J. A.: Ice-sheet grounding-zone wedges (GZWs) on high-latitude continental margins, Mar. Geol., 363, 65–92, https://doi.org/10.1016/j.margeo.2015.02.001, 2015.
Bernhard, J. M.: Foraminiferal biotopes in Explorers Cove, McMurdo Sound, Antarctica, J. Foramin. Res., 17, 286–297, https://doi.org/10.2113/gsjfr.17.4.286, 1987.
Blum, S. A., Lorenz, M. G., and Wackernagel, W.: Mechanism of retarded DNA degradation and prokaryotic origin of DNases in nonsterile soils, Syst. Appl. Microbiol., 20, 513–521, https://doi.org/10.1016/S0723-2020(97)80021-5, 1997.
Bombard, S. E., Leckie, R. M., Browne, I. M., Shevenell, A. E., McKay, R. M., Harwood, D. M., and the IODP Expedition 374 Scientists: Miocene Climatic Optimum and Middle Miocene Climate Transition: a foraminiferal record from the central Ross Sea, Antarctica, J. Micropalaeontol., 43, 383–421, https://doi.org/10.5194/jm-43-383-2024, 2024.
Corinaldesi, C., Beolchini, F., and Dell'Anno, A.: Damage and degradation rates of extracellular DNA in marine sediments: implications for the preservation of gene sequences, Mol. Ecol., 17, 3939–3951, https://doi.org/10.1111/j.1365-294X.2008.03880.x, 2008.
Dameron, S. N., Leckie, R. M., Harwood, D., Scherer, R., and Webb, P.-N.: Return to the Ross Ice Shelf Project (RISP), Site J-9 (1977–1979): perspectives of West Antarctic Ice Sheet history from Miocene and Holocene benthic foraminifera, J. Micropalaeontol., 43, 187–209, https://doi.org/10.5194/jm-43-187-2024, 2024.
DeConto, R. M. and Pollard, D.: Contribution of Antarctica to past and future sea-level rise, Nature, 531, 591–597, https://doi.org/10.1038/nature17145, 2016.
Demianiuk, E.: Sedimentary ancient DNA insights into foraminiferal diversity near the grounding line in the western Ross Sea, Antarctica, RepOD [data set], https://doi.org/10.18150/9ABUGS, 2025.
Domack, E. W., Jacobson, E. A., Shipp, S., and Anderson, J. B.: Late Pleistocene–Holocene retreat of the West Antarctic Ice-Sheet system in the Ross Sea: Part 2—sedimentologic and stratigraphic signature, Geol. Soc. Am. Bull., 111, 1517–1536, https://doi.org/10.1130/0016-7606(1999)111<1517:LPHROT>2.3.CO;2, 1999.
Dufresne, Y., Lejzerowicz, F., Perret-Gentil, L. A., Pawlowski, J., and Cordier, T.: SLIM: a flexible web application for the reproducible processing of environmental DNA metabarcoding data, BMC Bioinformatics, 20, 1–6, https://doi.org/10.1186/s12859-019-2663-2, 2019.
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R.: UCHIME improves sensitivity and speed of chimera detection, Bioinformatics, 27, 2194–2200, https://doi.org/10.1093/bioinformatics/btr381, 2011.
Esling, P., Lejzerowicz, F., and Pawlowski, J.: Accurate multiplexing and filtering for high-throughput amplicon-sequencing, Nucleic Acids Res., 43, 2513–2524, https://doi.org/10.1093/nar/gkv107, 2015.
Freeman, C. L., Dieudonné, L., Agbaje, O. B. A., Žure, M., Sanz, J. Q., Collins, M., and Sand, K. K.: Survival of environmental DNA in sediments: Mineralogic control on DNA taphonomy, Environ. DNA, 5, 1691–1705, https://doi.org/10.1002/edn3.482, 2023.
Gooday, A. J. and Pawlowski, J.: Conqueria laevis gen. and sp. nov., a new soft-walled, monothalamous foraminiferan from the deep Weddell Sea, J. Mar. Biol. Assoc. UK, 84, 919–924, https://doi.org/10.1017/S0025315404010197h, 2004.
Gooday, A. J., Bowser, S. S., and Bernhard, J. M.: Benthic foraminiferal assemblages in Explorers Cove, Antarctica: A shallow-water site with deep-sea characteristics, Prog. Oceanogr., 37, 117–166, https://doi.org/10.1016/S0079-6611(96)00007-9, 1996.
Gooday, A. J., Holzmann, M., Majewski, W., and Pawlowski, J.: New species of Gromia (Protista, Rhizaria) from South Georgia and the Falkland Islands, Polar Biol., 45, 647–666, https://doi.org/10.1007/s00300-022-03017-4, 2022.
Greenwood, S. L., Simkins, L. M., Halberstadt, A. R. W., Prothro, L. O., and Anderson, J. B.: Holocene reconfiguration and readvance of the East Antarctic Ice Sheet, Nat. Commun., 9, 3176, https://doi.org/10.1038/s41467-018-05625-3, 2018.
Habura, A., Pawlowski, J., Hanes, S. D., and Bowser, S. S.: Unexpected foraminiferal diversity revealed by small-subunit rDNA analysis of Antarctic sediment, J. Eukaryot. Microbiol., 51, 173–179, https://doi.org/10.1111/j.1550-7408.2004.tb00542.x, 2004.
Halberstadt, A. R. W., Simkins, L. M., Greenwood, S. L., and Anderson, J. B.: Past ice-sheet behaviour: retreat scenarios and changing controls in the Ross Sea, Antarctica, The Cryosphere, 10, 1003–1020, https://doi.org/10.5194/tc-10-1003-2016, 2016.
Halberstadt, A. R. W., Simkins, L. M., Anderson, J. B., Prothro, L. O., and Bart, P. J.: Characteristics of the deforming bed: till properties on the deglaciated Antarctic continental shelf, J. Glaciol., 64, 1014–1027, https://doi.org/10.1017/jog.2018.92, 2018.
Hauck, J., Gerdes, D., Hillenbrand, C. D., Hoppema, M., Kuhn, G., Nehrke, G., Völker, C. G., and Wolf-Gladrow, D. A.: Distribution and mineralogy of carbonate sediments on Antarctic shelves, J. Marine Syst., 90, 77–87, https://doi.org/10.1016/j.jmarsys.2011.09.005, 2012.
Hemer, M. A., Post, A. L., O'Brien, P. E., Craven, M., Truswell, E. M., Roberts, D., and Harris, P. T.: Sedimentological signatures of the sub-Amery Ice Shelf circulation, Antarct. Sci., 19, 497–506, https://doi.org/10.1017/S0954102007000697, 2007.
Höglund, H.: Foraminifera in the Gullmar Fjord and the Skagerak, Zool. Bidr. Upps., 26, 1–328, 1947.
Holzmann, M., Gooday, A. J., Majewski, W., and Pawlowski, J.: Molecular and morphological diversity of monothalamous foraminifera from South Georgia and the Falkland Islands: description of four new species, Eur. J. Protistol., 85, 125909, https://doi.org/10.1016/j.ejop.2022.125909, 2022.
Jacobs, S. S., Fairbanks, R. G., and Horibe, Y.: Origin and evolution of water masses near the Antarctic continental margin: Evidence form H218O/H216O ratios in seawater, Antarct. Res. Ser., 43, 59–85, 1985.
Jorissen, F. J., Bicchi, E., Duchemin, G., Durrieu, J., Galgani, F., Cazes, L., Gaultier M., and Camps, R.: Impact of oil-based drill mud disposal on benthic foraminiferal assemblages on the continental margin off Angola, Deep-Sea Res. Pt. II, 56, 2270–2291, https://doi.org/10.1016/j.dsr2.2009.04.009, 2009.
Kassambara, A.: ggpubr: 'ggplot2' Based Publication Ready Plots, R package version 0.6.0, https://rpkgs.datanovia.com/ggpubr/ (last access: 29 May 2025), 2023.
Kennett, J. P.: Foraminiferal evidence for a shallow calcium carbonate solution boundary, Ross Sea, Antarctica, Science, 153, 191–193, https://doi.org/10.1126/science.153.3732.191, 1966.
Kilfeather, A. A., O'Cofaigh, C., Lloyd, J. M., Dowdeswell, J. A., Xu, S., and Moreton, S. G.: Ice-stream retreat and ice-shelf history in Marguerite Trough, Antarctic Peninsula: Sedimentological and foraminiferal signatures, Geol. Soc. Am. Bull., 123, 997–1015, https://doi.org/10.1130/B30282.1, 2011.
Kirshner, A. E., Anderson, J. B., Jakobsson, M., O'Regan, M., Majewski, W., and Nitsche, F. O.: Post-LGM deglaciation in Pine Island Bay, west Antarctica, Quaternary Sci. Rev., 38, 11–26, https://doi.org/10.1016/j.quascirev.2012.01.017, 2012.
Korsun, S., Kniazeva, O., Majewski, W., Godoi, M. A., Hromic, T., Varfolomeeva, M., and Pawlowski, J.: Foraminifera in temperate fjords strongly affected by glacial meltwater, Tierra del Fuego, South America, Mar. Micropaleontol., 181, 102248, https://doi.org/10.1016/j.marmicro.2023.102248, 2023.
IPCC: Summary for Policymakers, in: Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change, edited by: Masson-Delmotte, V., Zhai, P., Pirani, A., Connors, S.L., Péan, C., Berger, S., Caud, N., Chen, Y., Goldfarb, L., Gomis, M.I., Huang, M., Leitzell, K., Lonnoy, E., Matthews, J. B. R., Maycock, T. K., Waterfield, T., Yelekçi, O., Yu, R., and Zhou, B., Cambridge University Press, Cambridge, https://doi.org/10.1017/9781009157896, 2021.
Lecroq, B., Lejzerowicz, F., Bachar, D., Christen, R., Esling, P., Baerlocher, L., Østerås M., Farinelli, L., and Pawlowski, J.: Ultra-deep sequencing of foraminiferal microbarcodes unveils hidden richness of early monothalamous lineages in deep-sea sediments, P. Natl. Acad. Sci. USA, 108, 13177–13182, https://doi.org/10.1073/pnas.1018426108, 2011.
Lejzerowicz, F., Esling, P., Majewski, W., Szczuciński, W., Decelle, J., Obadia, C., Arbizu, P. M., and Pawlowski, J.: Ancient DNA complements microfossil record in deep-sea subsurface sediments, Biol. Lett.-UK, 9, 20130283, https://doi.org/10.1098/rsbl.2013.0283, 2013.
Levy-Booth, D. J., Campbell, R. G., Gulden, R. H., Hart, M. M., Powell, J. R., Klironomos, J. N., Pauls, K. P., Swanton C. J., Trevors, J. T., and Dunfield, K. E.: Cycling of extracellular DNA in the soil environment, Soil Biol. Biochem., 39, 2977–2991, https://doi.org/10.1016/j.soilbio.2007.06.020, 2007.
Li, A. Z., Han, X. B., Zhang, M. X., Zhou, Y., Chen, M., Yao, Q., and Zhu, H. H.: Culture-dependent and-independent analyses reveal the diversity, structure, and assembly mechanism of benthic bacterial community in the Ross Sea, Antarctica, Front. Microbiol., 10, 2523, https://doi.org/10.3389/fmicb.2019.02523, 2019.
Li, Q., Lei, Y., Li, H., and Li, T.: Distinct responses of abundant and rare foraminifera to environmental variables in the Antarctic region revealed by DNA metabarcoding, Front. Mar. Sci., 10, 1089482, https://doi.org/10.3389/fmars.2023.1089482, 2023.
Lipps, J. H., Ronan, T. E., and Delaca, T. E.: Life below the Ross Ice Shelf, Antarctica, Science, 203, 447–449, https://doi.org/10.1126/science.203.4379.447, 1979.
Lorenz, M. G. and Wackernagel, W.: Adsorption of DNA to sand and variable degradation rates of adsorbed DNA, Appl. Environ. Microb., 53, 2948–2952, https://doi.org/10.1128/aem.53.12.2948-2952.1987, 1987.
Lorenz, M. G. and Wackernagel, W.: DNA binding to various clay minerals and retarded enzymatic degradation of DNA in a sand/clay microcosm, in: Gene Transfers and Environment: Proceedings of the Third European Meeting on Bacterial Genetics and Ecology (BAGECO-3), 20–22 November 1991, Villefranche-sur-Mer, France, 103–113, 1992.
Majewski, W.: Benthic foraminifera from West Antarctic fiord environments: An overview, Pol. Polar Res., 31, 61–82, https://doi.org/10.4202/ppres.2010.05, 2010.
Majewski, W., Lecroq, B., Sinniger, F., and Pawlowski, J.: Monothalamous foraminifera from Admiralty Bay, King George Island, West Antarctica, Pol. Polar Res., 28, 187–210, 2007.
Majewski, W., Bart, P. J., and McGlannan, A. J.: Foraminiferal assemblages from ice-proximal paleo-settings in the Whales Deep Basin, eastern Ross Sea, Antarctica, Palaeogeogr. Palaeocl., 493, 64–81, https://doi.org/10.1016/j.palaeo.2017.12.041, 2018.
Majewski, W., Prothro, L. O., Simkins, L. M., Demianiuk, E. J., and Anderson, J. B.: Foraminiferal patterns in deglacial sediment in the Western Ross Sea, Antarctica: Life near grounding lines, Paleoceanogr. Paleoclimatol., 35, e2019PA003716, https://doi.org/10.1029/2019PA003716, 2020.
Mallott, E. K., Garber, P. A., and Malhi, R. S.: trnL outperforms rbcL as a DNA metabarcoding marker when compared with the observed plant component of the diet of wild white-faced capuchins (Cebus capucinus, Primates), PLOS ONE, 13, e0199556, https://doi.org/10.1371/journal.pone.0199556, 2018.
Martin, M.: Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet. J., 17, 10–12, https://doi.org/10.14806/ej.17.1.200, 2011.
McKnight, D. T., Huerlimann, R., Bower, D. S., Schwarzkopf, L., Alford, R. A., and Zenger, K. R.: microDecon: A highly accurate read-subtraction tool for the post-sequencing removal of contamination in metabarcoding studies, Environ. DNA, 1, 14–25, https://doi.org/10.1002/edn3.11, 2019.
Melis, R. and Salvi, G.: Foraminifer and Ostracod Occurrence in a Cool-Water Carbonate Factory of the Cape Adare (Ross Sea, Antarctica): A Key Lecture for the Climatic and Oceanographic Variations in the Last 30,000 Years, Geosciences, 10, 413, https://doi.org/10.3390/geosciences10100413, 2020.
Meyer, M. and Kircher, M.: Illumina Sequencing Library Preparation for Highly Multiplexed Target Capture and Sequencing, Cold Spring Harbor Protocols, 2010, pdb.prot5448, https://doi.org/10.1101/pdb.prot5448, 2010.
Nguyen, N.-L., Pawłowska, J., Angeles, I. B., Zajaczkowski, M., and Pawłowski, J.: Metabarcoding reveals high diversity of benthic foraminifera linked to water masses circulation at coastal Svalbard, Geobiology 21, 133–150, https://doi.org/10.1111/gbi.12530, 2023a.
Nguyen, N. L., Devendra, D., Szymańska, N., Greco, M., Angeles, I. B., Weiner, A. K., Ray, J. L., Cordier, T., De Schepper, S., Pawłowska, J.: Sedimentary ancient DNA: a new paleogenomic tool for reconstructing the history of marine ecosystems, Front. Mar. Sci., 10, 1185435, https://doi.org/10.3389/fmars.2023.1185435, 2023b.
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., Minchin, R. O'Hara B., Simpson, G. L., Solymos, P., Stevens, H. H., Szoecs, E., and Wagner, E.: Package `vegan', Community ecology package, https://vegan.r-forge.r-project.org/ (lasst access: 29 May 2025), 2019.
Orsi, A. H. and Wiederwohl, C. L.: A recount of Ross Sea waters, Deep-Sea Res. Pt. II, 56, 778–795, https://doi.org/10.1016/j.dsr2.2008.10.033, 2009.
Park, J., Kim, H. C., Jo, Y. H., Kidwell, A., and Hwang, J.: Multi-temporal variation of the Ross Sea Polynya in response to climate forcings, Polar Res., 37, 1444891, 91, https://doi.org/10.1080/17518369.2018.1444891, 2018.
Pawłowska, J., Lejzerowicz, F., Esling, P., Szczuciński, W., Zajączkowski, M., and Pawlowski, J.: Ancient DNA sheds new light on the Svalbard foraminiferal fossil record of the last millennium, Geobiology, 12, 277–288, https://doi.org/10.1111/gbi.12087, 2014.
Pawłowska, J., Ła̧cka, M., Kucharska, M., Pawlowski, J., and Zaja̧czkowski, M.: Multiproxy evidence of the Neoglacial expansion of Atlantic Water to eastern Svalbard, Clim. Past, 16, 487–501, https://doi.org/10.5194/cp-16-487-2020, 2020a.
Pawłowska, J., Wollenburg, J. E., Zajączkowski, M., and Pawlowski, J.: Planktonic foraminifera genomic variations reflect paleoceanographic changes in the Arctic: evidence from sedimentary ancient DNA, Sci. Rep.-UK, 10, 1–10, https://doi.org/10.1038/s41598-020-72146-9, 2020b.
Pawlowski, J.: Phylogeny of allorgomiid foraminifera inferred from SSU rRNA gene sequences, J. Foramin. Res., 32, 334–343, https://doi.org/10.2113/0320334, 2002.
Pawlowski, J., Fahrni, J., and Bowser, S. S.: Phylogenetic analysis and genetic diversity of Notodendrodes hyalinosphaira, J. Foramin. Res., 32: 173–176, https://doi.org/10.2113/0320173, 2002a.
Pawlowski, J., Fahrni, J., Brykczynska, U., Habura, A., and Bowser, S. S.: Molecular data reveal high taxonomic diversity of allogromiid Foraminifera in Explorers Cove (McMurdo Sound, Antarctica), Polar Biol., 25, 96–105, https://doi.org/10.1007/s003000100317, 2002b.
Pawlowski, J., Fahrni, J. F., Guiard, J., Conlan, K., Hardecker, J., Habura, A., and Bowser, S. S.: Allogromiid foraminifera and gromiids from under the Ross Ice Shelf: morphological and molecular diversity, Polar Biol., 28, 514–522, https://doi.org/10.1007/s00300-005-0717-6, 2005.
Pawlowski, J., Fontaine, D., da Silva, A. A., and Guiard, J.: Novel lineages of Southern Ocean deep-sea foraminifera revealed by environmental DNA sequencing, Deep-Sea Res. Pt. II, 58, 1996–2003, https://doi.org/10.1016/j.dsr2.2011.01.009, 2011.
Pedersen, M. W., Overballe-Petersen, S., Ermini, L., Sarkissian, C. D., Haile, J., Hellstrom, M., Spens, J., Thomsen, P. F., Bohmann, K., Cappellini, E., Schnell, I. B., Wales, N. A., Carøe, C., Campos, P. F., Schmidt, A. M., Gilbert, T. P., Hansen, A. J., Orlando, L., and Willerslev, E.: Ancient and modern environmental DNA, Philos. T. Roy. Soc. B, 370, 20130383, https://doi.org/10.1098/rstb.2013.0383, 2015.
Picco, P., Amici, L., Meloni, R., Langone, L., and Ravaioli, M.: Temporal variability of currents in the Ross Sea (Antarctica), in: Oceanography of the Ross Sea, Antarctica, edited by: Spezie, G. and Manzella, G. M. R., Springer, https://doi.org/10.1007/978-88-470-2250-8_7, pp. 103–117, 1999.
Post, A. L., Hemer, M. A., Philip, E. O., Roberts, D., and Craven, M.: History of benthic colonisation beneath the Amery ice shelf, East Antarctica, Mar. Ecol. Prog. Ser., 344, 29–37, https://doi.org/10.3354/meps06966, 2007.
Prothro, L. O., Simkins, L. M., Majewski, W., and Anderson, J. B.: Glacial retreat patterns and processes determined from integrated sedimentology and geomorphology records, Mar. Geol., 395, 104–119, https://doi.org/10.1016/j.margeo.2017.09.012, 2018.
Prothro, L. O., Majewski, W., Yokoyama, Y., Simkins, L. M., Anderson, J. B., Yamane, M., Miyairi, Y., and Ohkouchi, N.: Timing and pathways of east Antarctic ice sheet retreat, Quaternary Sci. Rev., 230, 106166, https://doi.org/10.1016/j.quascirev.2020.106166, 2020.
R Core Team: R: A language and environment for statistical computing, Vienna, Austria, https://www.R-project.org/ (lass access: 29 May 2025), 2013.
Rawlence, N. J., Lowe, D. J., Wood, J. R., Young, J. M., Churchman, G. J., Huang, Y. T., and Cooper, A.: Using palaeoenvironmental DNA to reconstruct past environments: progress and prospects, J. Quaternary Sci., 29, 610–626, https://doi.org/10.1002/jqs.2740, 2014.
Rignot, E., Mouginot, J., and Scheuchl, B.: Ice flow of the Antarctic ice sheet, Science, 333, 1427–1430, https://doi.org/10.1126/science.1208336, 2011.
Rivaro, P., Messa, R., Ianni, C., Magi, E., and Budillon, G.: Distribution of total alkalinity and pH in the Ross Sea (Antarctica) waters during austral summer 2008, Polar Res., 33, 20403, https://doi.org/10.3402/polar.v33.20403, 2014.
Robinson, D. E., Menzies, J., Wellner, J. S., and Clark, R. W.: Subglacial sediment deformation in the Ross Sea, Antarctica, Quat. Sci. Adv., 4, 100029, https://doi.org/10.1016/j.qsa.2021.100029, 2021.
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F.: VSEARCH: a versatile open source tool for metagenomics, PeerJ, 4, e2584, https://doi.org/10.7717/peerj.2584, 2016.
Seidenstein, J. L., Leckie, R. M., McKay, R., De Santis, L., Harwood, D., and IODP Expedition 374 Scientists: Pliocene–Pleistocene warm-water incursions and water mass changes on the Ross Sea continental shelf (Antarctica) based on foraminifera from IODP Expedition 374, J. Micropalaeontol., 43, 211–238, https://doi.org/10.5194/jm-43-211-2024, 2024.
Simkins, L. M., Greenwood, S. L., and Anderson, J. B.: Diagnosing ice sheet grounding line stability from landform morphology, The Cryosphere, 12, 2707–2726, https://doi.org/10.5194/tc-12-2707-2018, 2018.
Slon, V., Hopfe, C., Weiß, C., Mafessoni, F., De La Rasilla, M., Lalueza-Fox, C., and Meyer, M.: Neandertal and Denisovan DNA from Pleistocene sediments, Science, 356, 605–608, https://doi.org/10.1126/science.aam9695, 2017.
Smith Jr., W. O.: Primary productivity measurements in the Ross Sea, Antarctica: a regional synthesis, Earth Syst. Sci. Data, 14, 2737–2747, https://doi.org/10.5194/essd-14-2737-2022, 2022.
Smith Jr, W. O., Ainley, D. G., Arrigo, K. R., and Dinniman, M. S.: The oceanography and ecology of the Ross Sea, Annu. Rev. Mar. Sci., 6, 469–487, https://doi.org/10.1146/annurev-marine-010213-135114, 2014.
Spagnolo, M., Clark, C. D., Ely, J. C., Stokes, C. R., Anderson, J. B., Andreassen, K., Graham, A. G. C., and King, E. C.: Size, shape and spatial arrangement of mega-scale glacial lineations from a large and diverse dataset, Earth Surf. Proc. Land., 39, 1432–1448, https://doi.org/10.1002/esp.3532, 2014.
Szczuciński, W., Pawłowska, J., Lejzerowicz, F., Nishimura, Y., Kokociński, M., Majewski, W., Nakamura, Y., and Pawlowski, J.: Ancient sedimentary DNA reveals past tsunami deposits, Mar. Geol., 381, 29–33, https://doi.org/10.1016/j.margeo.2016.08.006, 2016.
Taberlet, P., Coissac, E., Pompanon, F., Gielly, L., Miquel, C., Valentini, A., Vermat, T., Corthier, G., Brochmann, C., and Willerslev, E.: Power and limitations of the chloroplast trn L (UAA) intron for plant DNA barcoding, Nucleic Acids Res., 35, e14–e14, https://doi.org/10.1093/nar/gkl938, 2007.
Torti, A., Jørgensen, B. B., and Lever, M. A.: Preservation of microbial DNA in marine sediments: insights from extracellular DNA pools, Environ. Microbiol., 20, 4526–4542, https://doi.org/10.1111/1462-2920.14401, 2018.
Totten, R. L., Majewski, W., Anderson, J. B., Yokoyama, Y., Fernandez, R., and Jakobsson, M.: Oceanographic influences on the stability of the Cosgrove Ice Shelf, Antarctica, Holocene, 27, 1645–1658, https://doi.org/10.1177/0959683617702226, 2017.
Tulaczyk, S., Kamb, B., and Engelhardt, H. F.: Estimates of effective stress beneath a modern West Antarctic ice stream from till preconsolidation and void ratio, Boreas, 30, 101–114, https://doi.org/10.1111/j.1502-3885.2001.tb01216.x, 2001.
Vosberg, H. P.: The polymerase chain reaction: an improved method for the analysis of nucleic acids, Hum. Genet., 83, 1–15, https://doi.org/10.1007/BF00274139, 1989.
Wales, N., Carøe, C., Sandoval-Velasco, M., Gamba, C., Barnett, R., Samaniego, J. A., Madrigal, J. R., Orland, L., and Gilbert, M. T. P.: New insights on single-stranded versus double-stranded DNA library preparation for ancient DNA, Biotechniques, 59, 368–371, https://doi.org/10.2144/000114364, 2015.
Weber, A. A. and Pawlowski, J.: Can abundance of protists be inferred from sequence data: a case study of Foraminifera, PLOS ONE, 8, e56739, https://doi.org/10.1371/journal.pone.0056739, 2013.
Weber, A. A. and Pawlowski, J.: Wide occurrence of SSU rDNA intragenomic polymorphism in foraminifera and its implications for molecular species identification, Protist, 165, 645–661, https://doi.org/10.1016/j.protis.2014.07.006, 2014.
Wickham, H.: ggplot2: Elegant Graphics for Data Analysis, Springer-Verlag, New York, ISBN: 978-3-319-24277-4, https://ggplot2.tidyverse.org (last access: 29 May 2025), 2016.
Willerslev, E., Hansen, A. J., Binladen, J., Brand, T. B., Gilbert, M. T. P., Shapiro, B., Bunce, M., Wiuf, C., Gilichinsky, D. A., and Cooper, A.: Diverse plant and animal genetic records from Holocene and Pleistocene sediments, Science, 300, 791–795, https://doi.org/10.1126/science.1084114, 2003.
Willerslev, E., Hansen, A. J., Rønn, R., Brand, T. B., Barnes, I., Wiuf, C., Gilichinsky, D., Mitchell, D., and Cooper, A.: Long-term persistence of bacterial DNA, Curr. Biol., 14, R9–R10, https://doi.org/10.1016/j.cub.2003.12.012, 2004.
Zimmermann, H. H., Stoof-Leichsenring, K. R., Kruse, S., Nürnberg, D., Tiedemann, R., and Herzschuh, U.: Sedimentary ancient DNA from the subarctic North Pacific: How sea ice, salinity, and insolation dynamics have shaped diatom composition and richness over the past 20,000 years, Paleoceanogr. Paleoclimatol., 36, e2020PA004091, https://doi.org/10.1029/2020PA004091, 2021.