the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
The microbiome of the Arctic planktonic foraminifera Neogloboquadrina pachyderma is composed of fermenting and carbohydrate-degrading bacteria and an intracellular diatom chloroplast store
Kate Darling
Rabecca Thiessen
Anna J. Pieńkowski
Neogloboquadrina pachyderma is the only true polar species of planktonic foraminifera. As a key component of the calcite flux, it plays a crucial role in the reconstruction and modelling of seasonality and environmental change within the high latitudes. The rapidly changing environment of the polar regions of the North Atlantic and Arctic oceans poses challenging conditions for this (sub)polar species in terms of temperature, sea-ice decline, calcite saturation, ocean pH, and the progressive contraction of the polar ecosystem. To model the potential future for this important high-latitude species, it is vital to investigate the modern ocean community structure throughout the annual cycle of the Arctic to understand the inter-dependencies of N. pachyderma. This study focusses on the summer ice-free populations in Baffin Bay. We use 16S rDNA metabarcoding and transmission electron microscopy (TEM) to identify the microbial interactions of N. pachyderma and PICRUSt2 to predict the metabolic pathways represented by the amplicon sequencing variants (ASVs) in the foraminiferal microbiome. We demonstrate that the N. pachyderma diet consists of both diatoms and bacteria. The core microbiome, defined as the 16S rDNA ASVs found in 80 % of the individuals investigated, consists of six bacterial ASVs and two diatom chloroplast ASVs. On average, it accounts for nearly 50 % of the total ASVs in any individual. The metabolic pathway predictions based on bacterial ASVs suggest that the foraminiferal microbiome is composed of monosaccharide fermenting and polysaccharide degrading bacterial species in line with those found routinely in the diatom phycosphere. On average, the two chloroplast ASVs constitute 40 % of the core microbiome, and, significantly, an average of 53.3 % of all ASVs in any individual are of chloroplast origin. TEM highlights the importance of diatoms to this species by revealing that intact chloroplasts remain in the foraminiferal cytoplasm in numbers strikingly comparable to the substantial quantities observed in kleptoplastic benthic foraminifera. Diatoms are the major source of kleptoplasts in benthic foraminifera and other kleptoplastic groups, but this adaptation has never been observed in a planktonic foraminifer. Further work is required to understand the association between N. pachyderma, diatoms, and their chloroplasts in the pelagic Arctic realm, but such a strategy may confer an advantage to this species for survival in this extreme habitat.
- Article
(8301 KB) - Full-text XML
- BibTeX
- EndNote
The non-spinose planktonic foraminifera Neogloboquadrina pachyderma is found throughout the global ocean (Morard et al., 2024) but occurs in greatest numbers in polar, sub-polar, and transitional upwelling waters. These different biogeographies and niches are reflected by the distinct N. pachyderma genotypes associated with the divergent ecosystems (Darling et al., 2004, 2007, 2017). For example, while N. pachyderma is the predominant planktonic foraminiferal morphospecies of the polar oceans in both hemispheres (e.g. Bé and Tolderlund, 1971; Bé, 1977), N. pachyderma Type I is the only genotype found in the (sub)polar North Atlantic/Arctic Ocean (Darling et al., 2004; Darling et al., 2007) and is the major marine calcifier (Kohfeld et al., 1996) in the largest ocean carbon sink in the Northern Hemisphere (Gruber et al., 2002). Present-day temperature conditions confine N. pachyderma Type I to the North Atlantic/Arctic Ocean water masses, where summer sea surface temperatures (SSTs) remain below 10 °C (Tolderlund and Bé, 1971; Duplessy et al., 1991). Here, N. pachyderma exhibits strong seasonal productivity in a highly predictable pattern. Winter mixing re-supplies nutrients to surface waters, triggering the seasonal succession of maximal phytoplankton blooms and zooplankton abundance, which is followed by more nutrient-depleted summer conditions (Jonkers and Kučera, 2015).
As the primary component of both the modern and Quaternary fossil (sub)polar assemblage, the calcite tests of N. pachyderma constitute the major contribution to seasonal and environmental change reconstructions within the North Atlantic and Arctic Ocean (e.g. Simstich et al., 2003; Kretschmer et al., 2016; Altuna et al., 2018; Brummer et al., 2020; Livsey et al., 2020). However, the Arctic is now an unremittingly warming ecosystem, with seasonal sea-ice cover constantly reducing and ice-free conditions projected to appear between 2030 and 2055 (Kim et al., 2023; Jahn et al., 2024). The North Atlantic/Arctic N. pachyderma is already considered to be particularly sensitive to the forecasted changes in seawater carbonate chemistry (Manno et al., 2012), with consequent implications for the calcite flux and the biological pump. Under ocean acidification conditions, Arctic N. pachyderma show reduced carbonate production moderated by ocean warming, making it difficult to predict future climate change impacts as the polar habitat of N. pachyderma shrinks. Although the North Atlantic/Arctic N. pachyderma population has clearly survived the extremes of Quaternary climate cyclicity in the past (Brummer et al., 2020), it is unknown whether N. pachyderma will find itself spatially displaced from its adaptive ecological range in the Arctic ecosystem (e.g. Jonkers et al., 2019; Greco et al., 2022), as we transition into the unknown territory of anthropogenically driven extreme global warming.
The warming ocean is affecting all marine organisms at multiple trophic levels (e.g. Poloczanska et al., 2016; Meredith et al., 2019; Deutsch et al., 2015). It has already been demonstrated that some planktonic protist species cannot track their optimal temperatures as their environment changes and may undergo extirpation once local thresholds are exceeded (Trubovitz et al., 2020). Such thresholds are unknown for N. pachyderma, and there may soon be no true polar refugia into which to retreat. At the beginning of the 21st century within our Baffin Bay study area, the Pikialasorsuaq (the former “North Water Polynya”) was considered a region of high biological productivity (Tremblay et al., 2002, 2006). However, increasing oligotrophic conditions have been reported in the last decade driven by meltwater from the Greenland Ice Sheet and nearby glaciers. The increased stratification and reduced mixing/upwelling result in a reduction in diatom-mediated net community production (Bergeron and Tremblay, 2014). Because diatoms are considered a major food source for N. pachyderma (e.g. Schiebel and Hemleben, 2017; Greco et al., 2021), this reduction in diatom primary productivity poses an additional challenge to N. pachyderma populations. To model the impending environmental consequences for this important high-latitude species, going forward, it will be vital to investigate the modern ocean community structure throughout the annual cycle of the Arctic to understand the inter-dependencies of N. pachyderma.
Although our understanding of Arctic (sub)polar N. pachyderma annual/seasonal population structure and ecological behaviour is increasing (e.g. Carstens and Wefer, 1992; Kohfeld et al., 1996; Jonkers et al., 2010, 2013; Greco et al., 2019; Meilland et al., 2022), it is far from complete. The presence of only a single genotype (N. pachyderma Type I) is good news for all the Arctic ecological investigations based on this taxon (e.g. Altuna et al., 2018; Greco et al., 2019; Meilland et al., 2022), as it simplifies analyses. The microbiome, defined as the combined taxa (including food, endobionts/symbionts, and parasites) within a foraminiferal specimen, has already been investigated using 16S metabarcoding of sister Neogloboquadrina species, highlighting the diversity and complexity of the ecological community networks and symbiont/predator/prey interactions that exist between prokaryotes and protists within the water column (Bird et al., 2018). Using the 16S rDNA metabarcoding approach together with fluorescence microscopy or transmission electron microscopy (TEM), Bird et al. (2018) determined the taxonomic character, trophic interactions, food source, and putative symbiotic associations of N. incompta and N. dutertrei in the California Current system. The results highlight their similar feeding strategy of forming feeding cysts of particulate organic matter (POM) in the water column, but such behaviour provides no clues to their choice of prey or potential symbiotic associations. Evidently, ecological concepts of individual planktonic foraminifera must be systematically revised, as each morphospecies and potentially each genotype has most likely evolved individually distinct interactions with the marine microbial assemblage. A recent study used single-cell metabarcoding targeting 18S rDNA to characterise the interactions of N. pachyderma with the local eukaryote community (Greco et al., 2021), as the majority of data on feeding behaviour in planktonic foraminifera suggest that biotic interactions are likely to be mainly with herbivorous eukaryotes (Kohfeld et al., 1996; Manno and Pavlov, 2014; Pados and Spielhagen, 2014; Schiebel and Hemleben, 2017). However, because no direct investigations have been carried out on the feeding behaviour or diet of Arctic N. pachyderma, this remains in question. The data shown in Greco et al. (2021) indicate that the N. pachyderma microbiome is dominated by diatoms, with Crustacea and Syndiniales (a potential parasite) also present. Here, we complement this study by examining the single-cell 16S rDNA metabarcodes of the N. pachyderma microbiome to investigate bacterial and archaeal biotic and trophic interactions and their potential symbiotic associations. In addition, we further investigate the cellular structures within N. pachyderma individual specimens using TEM to examine the cellular position of the bacterial/chloroplast sources of DNA within the N. pachyderma cell. Our results have direct implications for understanding trophic interactions within this at-risk habitat, for modelling future N. pachyderma population dynamics under climate change, and for understanding the evolutionary pressures experienced by this morphospecies.
2.1 Sampling locality and collection methods
Sampling was undertaken in Baffin Bay in July/August 2017 on board CCGS Amundsen as part of ArcticNet Expedition 2017 (Leg 2b; https://amundsenscience.com/expeditions/2017-expedition/, last access: 27 August 2025) and August/September 2018 on board the CCGS Hudson 2018042 expedition (Fig. 1). In both cases, the samples were taken in open water with no sea-ice cover. Details of the sampling stations and collections are listed in Table 1. Sample provenance was either individual foraminifera (Fm) or the water column (WC). Samples were analysed by 16S metabarcoding of the foraminiferal microbiome or the water column bacterial assemblages from stations along the Amundsen cruise track.
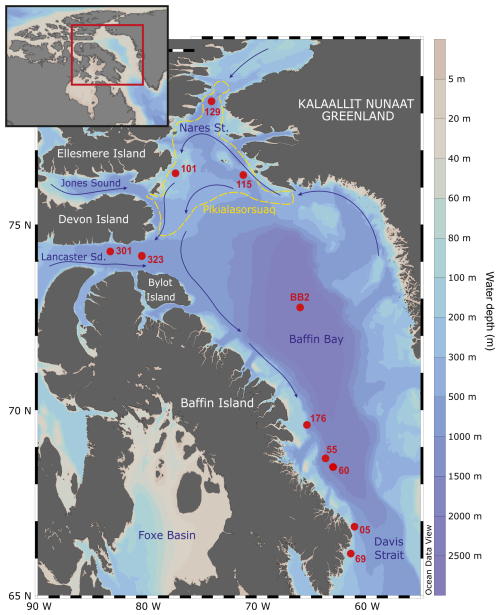
Figure 1Map of sampling stations (numbered red spots) and the site of Pikialasorsuaq (the former “North Water Polynya” (dashed yellow line); Eegeesiak et al., 2017). Stations BB2, 101, 115, 129, 301, and 323 were part of the CCGS Amundsen ArcticNet Expedition 2017, and Stations 05, 55, 60, and 69 were part of the CCGS Hudson 2018042 Expedition 2018. Inset shows the sampling location within the wider region. The base maps were drawn in OceanData View (Schlitzer, Ocean Data View, http://odv.awi.de, 2022).
Table 1Stations and sample information, including provenance, depth, and analysis type. Specimen IDs are either WC (water column) or Fm (foraminifera). This is followed by station identification (e.g. 101), water depth (e.g. 050 = 50 metres), and replicate ID (a, b, c, etc.).
Foraminifera were collected at seven stations by vertical net tow from a 200 m depth to the surface. Foraminifera for genotyping and microbiome analysis were wet picked on board, rinsed in 0.2 µm filtered surface seawater, and preserved in 100 µL RNAlater® (Ambion™). Samples were stored at 4 °C for 4 h and then transferred to a −20 °C freezer until processing.
CTD data and water samples were collected from three stations (101, 115, and 323) in northern Baffin Bay (Fig. 1; Table 1; Amundsen Science Data Collection, 2018) in 2017. Stations 101 and 115 are both situated within the biologically important Pikialasorsuaq between Greenland and Canada, which remains sea-ice-free in winter (Eegeesiak et al., 2017). Station 101 is located close to southeast Ellesmere Island in relatively shallow water (350 m depth). Station 115 is at a similar latitude to Station 101 but deeper (at 653 m) and closer to Greenland. Station 323 is situated further south and outside the Pikialasorsuaq in Lancaster Sound at the entrance to the Northwest Passage. It is the deepest of the three stations (at 789 m).
Water samples were collected from five depths: surface, 50, 100, 150, and 200 m. From each depth at each station, 2 L of seawater was filtered onto 0.2 µm polycarbonate filters. These filters were then individually placed in a 1.5 mL microfuge tube and covered with RNAlater® (Ambion™). The tubes were stored at 4 °C for 4 h and then transferred to a −20 °C freezer until processing. Foraminifera for TEM analysis were collected on the CCGS Hudson cruise (2018042) in 2018 (Table 1). They were wet picked as described above and placed directly in TEM buffer (4 % glutaraldehyde, 2 % paraformaldehyde in salt-adjusted phosphate buffered saline (PBS with 24.62 g L−1 NaCl added)), stored at room temperature for 12 h, then kept at 4 °C until further processing.
2.2 DNA extractions, foraminiferal 18S rDNA genotyping, and 16S rDNA metabarcoding
Downstream washing of individual cells preserved in RNAlater® for genotyping was carried out to remove the test and test-associated external contaminants according to Bird et al. (2017). DNA was extracted from individual foraminifera in 40 µL DOC buffer (Holzmann and Pawlowski, 1996) to identify the specific genotype. PCR amplification of the foraminiferal 18S rDNA gene was performed with three rounds of PCR using a Phire Hot Start DNA polymerase master mix (Thermo-Scientific), 3 % DMSO, and an annealing temperature of 58 °C with 25 cycles. DNA was diluted 1 in 20 in PCR-grade water. Primer pairs were as follows: primary PCR – C5-sB; secondary PCR – N5-N6; and tertiary PCR – 14F1-N6. PCR products between rounds were diluted 1 in 100 PCR-grade water, and 1 µL was used in the following round of PCR. Cloning to account for intra-individual variation was carried out according to Darling et al. (2016). DNA sequencing was carried out using the BigDye® Terminator v3.1 Cycle Sequencing Kit and an ABI 3730 DNA sequencer (both Applied Biosystems). Filtrate from water samples was extracted for DNA using the DNeasy power water kit (Qiagen). Filters were removed from the RNAlater salt (Ambion™), placed in clean 1.5 mL microfuge tubes, and centrifuged for 1 min at 10 000 ×g. Excess RNAlater® (Ambion™) was removed, and the filter was transferred to the bead beating tubes of the DNeasy power water kit and processed following the manufacturer's protocol. A control DNeasy power water kit extraction was carried out in parallel using a clean filter. In addition to the foraminiferal and water sample processing, four reagent controls were processed. These were composed of two PCR controls containing no DNA template, an extraction control containing 2.5 µL DOC buffer only, and an extraction control containing 1 µL of elute from a Qiagen DNA extraction of a clean 0.2 µm polycarbonate filter.
PCR was used to amplify the V4 region of the 16S rDNA gene of bacteria and chloroplasts. PCR reactions using the 515F forward (Parada et al., 2016) and 806R reverse (Apprill et al., 2015) primer pair modified from the original primer pair (Caporaso et al., 2011; Walters et al., 2015) were performed in triplicate. Each reaction contained 1 unit of Phusion DNA polymerase (Thermo-Scientific), 1× Phusion HF buffer, 0.2 mM each dNTP, 0.4 µM of each primer, 0.4 mM MgCl2, and 2.5 µL (foraminifera) or 1 µL (water column) of template DNA in a 50 µL volume made up with PCR grade water (Sigma). All PCR reactions were set up in a UV sterilisation cabinet (GE healthcare). The reaction tubes and PCR mixtures were treated for 15 min with 15 W UV light (wavelength = 254 nm) to destroy contaminating DNA prior to the addition of dNTPs, DNA polymerase primers, and template DNA (Padua et al., 1999). Triplicate PCR reactions were pooled before purification with the Wizard® SV Gel and PCR Clean-Up System (Promega). The purified amplicons were quantified using a Qubit® 2.0 fluorometer (Thermo Fisher Scientific) prior to pooling at equimolar concentrations for DNA sequencing. DNA sequencing was performed at Edinburgh Genomics using an Illumina MiSeq v3 to generate 253 base pair (bp) paired-end reads.
2.3 Quality filtering paired-end reads, rarefaction, taxonomic assignment, and sequence filtering
The Quantitative Insights Into Microbial Ecology 2 pipeline (QIIME2, Bolyen et al., 2019) was used for the initial analyses. Sequences were trimmed and denoising was carried out using the DADA2 plugin (Callahan et al., 2016) for quality filtering, dereplication, the removal of singletons, chimera identification, and the removal and merging of paired-end reads. This method generates amplicon sequence variants (ASVs) and a set of representative sequences. Alpha rarefaction was carried out in QIIME2 (metrics: observed OTUs (operational taxonomic units); Shannon; and Faith PD). Samples were rarefied to the lowest sequencing depth observed across all samples (25 064), and the sampling depth was adequate across all samples (Fig. A1). A total of 60 samples, including 28 foraminiferal samples, 28 water samples, and 4 negative controls, produced a total of 4290 ASVs across 5 802 211 counts. Amplicon sequencing variants with a total frequency count of 50 or less across all 60 samples were removed from the sample set, leaving 1717 ASVs. Taxonomy was assigned using an sklearn classifier pre-trained on the database SILVA-132 99 % OTUs from the V4 515f/806R region of the sequences (Quast et al., 2013). Taxonomic-based filtering was then carried out to remove ASVs assigned to mitochondria, eukaryotes, and those not assigned beyond the kingdom level. Contaminant removal (26 identified ASVs) was carried out in the R package Decontam (Davis et al., 2018) in R v 4.0 (R Core Team, 2017) using the prevalence with batch methods. After filtering, 1548 ASVs remained. For plotting and comparisons, the read counts for each ASV in each sample were converted to percentage reads, i.e. relative abundances. Therefore, all percentages presented are the relative abundances of individual ASVs or groups of ASVs being discussed.
2.4 Statistical analyses
Absolute count data were transformed to centred log-ratios suitable for statistical analysis of compositional data using q2-Gemelli (Martino et al., 2021). Robust Aitchison distances were calculated (Martino et al., 2019), and a PCA was performed in Vegan v2.6-10 (Oksanen et al., 2025) in R (v 4.0 and 4.4.3; R Core Team, 2017). This was visualised with ggplot2 (Wickham, 2009). Statistical analyses were also performed in Vegan v2.6-10. To determine if provenance, sample depth or station significantly affected the assemblages, Adonis2 was used to perform PERMANOVA (default 999 permutations). Pairwise PERMANOVA tests were performed using the pairwise.adonis2 function (pairwiseAdonis v 0.4.1; Martinez Arbizu, 2020) with 999 permutations. To account for multiple testings across pairwise comparisons, raw P-values were corrected using the Benjamini–Hochberg method (implemented via p adjust in base R), and comparisons with adjusted P-values (q-values) < 0.05 were considered statistically significant. Where sample numbers were small and unequal, a dispersion test (implemented with betadisper()) was carried out on the Aitchison dissimilarity matrices to test group dispersion homogeneity, as PERMANOVA is sensitive to unequal sample numbers, particularly when group dispersions differ. To assess differential microbial composition between stations in subsets where PERMANOVA was inappropriate, a Wilcoxon rank-sum test was implemented in ALDEx2 v1.38.0 (Analysis of Differential Abundance Taking Sample and Scale Variation into Account; Fernandes et al., 2014). ALDEx2 models high-throughput compositional data using the centred log-ratio (clr) transformation and Monte Carlo sampling (set to 1000, with denominator = all) from a Dirichlet distribution, which accounts for within-group variance and is robust to small sample sizes and differences in dispersion.
To investigate differential abundance in the taxa associated with the different provenances, the packages Phyloseq v1.50.0 (McMurdie and Holmes, 2013) and DESseq2 v1.46.0 (Love et al., 2014) were used. The package DESeq2 was used rather than ANCOM because ANCOM assumes that < 25 % of the ASVs are changing between provenances, and this assumption does not hold true here (Mandal et al., 2015). Compositional differences and specific taxa that were significantly different between provenances were identified using log2 of fold change analysis in DESeq2 by converting the phyloseq-object, containing the raw frequency counts, to a DESeq2 object. The DESeq2 analysis was run with the size factor type set to “poscounts”, which allows values of 0 in the sample counts and accounts for the data transferal from a phyloseq-object (van den Berge et al., 2018). The significance test was set to “Wald”, and a “local” fit type was selected for the fitting of dispersions.
The core microbiome, here defined as ASVs present across 80 % of the foraminifera, was identified using Microbiome (Lahti and Shetty, 2017).
Functional predictions of the foraminiferal microbiome compared to the wider water column assemblage were made using PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States; Douglas et al., 2020). The inputs to PICRUSt2 were the representative sequences fasta file and the ASV frequency table (converted to BIOM format) generated in QIIME2. The default full pipeline was run with the addition of “–per_sequence_contrib” and “–coverage” to give copy number normalised, community-wide pathway abundances to compare between the provenances. Evolutionary Placement Algorithm-Next Generation (EPA-NG) phylogenetic placement of reads (Barbera et al., 2019) was used with a default cutoff nearest sequence taxon index (NSTI) of 2.0, which removed 8 of 1548 ASVs from downstream analysis, as these could not be satisfactorily placed in the tree. ALDEx2 (Fernandes et al., 2014) was also used to assess the differential abundance of functional pathway predictions between the foraminiferal microbiome and the water column assemblage. PICRUSt2-generated pathway abundance data was rounded to integers and then input to aldex.clr, which generates centred log-ratio transformed values (number of Monte-Carlo instances = 1000, with denominator = iqlr). The Welch's t test (aldex.ttest) and an estimate of the effect size and the within-and-between provenance values (aldex.effect) were calculated from the output of aldex.clr. The dataset was then filtered for pathways that were significantly differentially abundant between provenances (Benjamini–Hochberg-corrected P value < 0.05 and effect size > 1).
2.5 Transmission electron microscopy
TEM was used to observe and document the structural relationships between any endobiotic microorganisms and foraminiferal cells. After fixation in TEM fixative (see methods in Sect. 2.1), specimens were post-fixed in 1 % osmium tetroxide in 0.1 M sodium cacodylate for 45 minutes, followed by a further three 10 min washes in distilled water. Specimens were then set in small cubes of 1 % low melting point agarose and decalcified in 0.1 M EDTA (pH 7.4) for 1 and 48 h at 4 °C. Fixed cells were then dehydrated in 50 %, 70 %, and 90 % ethanol for 2×15 min followed by 100 % ethanol 4×15 min. Two 10 min changes in propylene oxide were carried out prior to being embedded in TAAB 812 resin. Sections, 1 µm thick, were cut on a Leica Ultracut Ultramicrotome, stained with toluidine blue, and then viewed under a light microscope to select suitable specimen areas for investigation. Ultrathin sections, 60 nm thick, were cut from selected areas, stained in uranyl acetate and lead citrate, and then viewed with a JEOL JEM-1400 Plus TEM. Both osmium tetroxide and uranyl acetate used here bind to unsaturated lipids such that they appear dark in TEM imaging.
3.1 Water column
Water samples were taken from three locations: Stations 101, 115, and 323 (Fig. 1, Table 1). At Station 115 to the east of the Pikialasorsuaq, the water is derived from warm Atlantic water (e.g. Melling et al., 2001; Vincent, 2019) with SSTs as high as 4 °C accompanied by a steep thermocline to 40 m (Fig. 2). At Stations 101 (west Pikialasorsuaq) and 323 (southwest of the Pikialasorsuaq), the upper-level water temperatures were colder due to Pacific-derived water entering via the colder Arctic Ocean (Tremblay et al., 2002; Bergeron and Tremblay, 2014). While Station 323 had a surface temperature of 3 °C, it very rapidly dropped to −1 °C by 20 m, and Station 101 had a surface maximum temperature of only 0.75 °C. The chlorophyll maximum was closely associated with the temperature maximum at all stations.
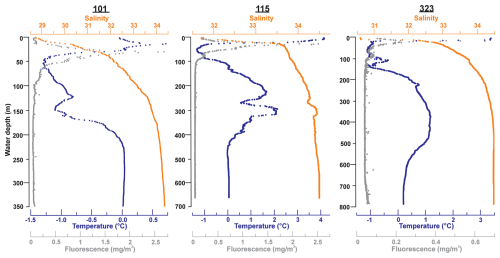
Figure 2CTD plots for temperature, salinity, and fluorescence at Stations 101 (350 m), 115 (653 m), and 323 (789 m) (Amundsen Science Data Collection, 2018), where both water and foraminifera were collected.
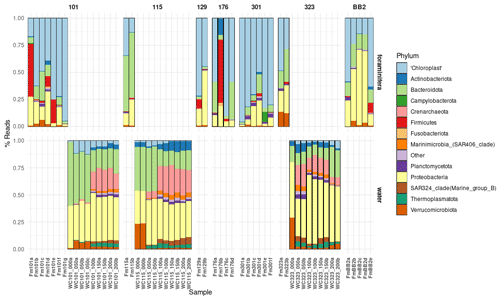
Figure 3The relative abundance of 16S rDNA ASVs generated from the foraminiferal specimens (Fm) and the water column (WC). Note that foraminifera (top row) were successfully processed from seven stations (see Fig. 1) and that water was collected and processed from three stations (bottom row). Taxa are shown at the phylum level except for chloroplast-derived 16S ASVs, which are grouped together. Sample IDs (Table 1) are found on the x axis.
The general microbial assemblages in the water column across the three stations (101, 115, and 323) displayed a similar pattern of assemblage composition with depth (Fig. 3). Surface waters contained either no or extremely low relative abundances of chloroplast or archaeal ASVs. Chloroplast ASV relative abundance increased steeply with depth however, with the highest abundance found in the 50 m water samples before numbers reduced again. This pattern agrees with our CTD data, where the chlorophyll maximum occurred between 20 and 40 m across all stations (Fig. 2). The chloroplast ASVs made up an average of 3.51 % of the ASVs in the water column. Archaeal ASVs (candidate phylum Thermoplasmatota and the Crenarchaeota) are most predominant below 50 m, peaking in the 100–150 m water samples.
3.2 Statistical comparison of water column and foraminiferal ASVs
The combined microbial assemblages of the 28 water column samples and the 28 foraminifera specimens (see Table 1) consisted of 1548 identified ASVs after filtering. By far, the most prevalent ASVs in the foraminifera are those from chloroplasts (averaging 53.3 % relative abundance), and particularly diatom chloroplasts (averaging 44.5%). In the water column, Proteobacteria ASVs are the most abundant (averaging 41.2 %, Fig. 3).
3.2.1 Station and depth as factors influencing microbial assemblages
To assess the influence of environmental factors on microbial assemblages in the water column, the effects of station and depth were tested across the three stations with multiple depth samples. PERMANOVA (Table A1) revealed that depth was a significant driver of community composition, explaining 55 % of the variation (Pr = 0.001). In contrast, station explained only 6.9% and was not significant, either globally (Pr(> F) = 0.458) or in pairwise comparisons (see Pairwise PERMANOVA in the Supplement). PCA ordination (Fig. A2) supported this, showing clearer clustering by depth than by station.
To control for vertical variation, a subset of 50 m water samples was analysed. Station explained 98 % of variation and was significant in the global PERMANOVA (Pr(> F) = 0.012); however, none of the pairwise comparisons were significant, and low sample sizes (n = 2–4) limit interpretation.
In contrast, for foraminiferal samples, station explained 48.3 % of the variation and was significant in the global PERMANOVA (Pr(> F) = 0.003, Table A1). Pairwise comparisons (see Supplement) revealed significant differences in 4 of 21 station pairs (after correction), indicating some degree of spatial structuring, and this is reflected in the PCA ordination of the foraminifera samples (Fig. A3). Specifically, the foraminiferal microbiome at Station 301 differed significantly from those at Stations BB2, 176, and 101. However, samples from Stations 101, 115, and 323, where water column samples were also collected, did not differ significantly, supporting the water column result of minimal microbial differentiation among these three stations. To improve statistical power, the foraminiferal analysis was repeated using only the four best-replicated stations (Stations 101, 176, BB2, and 301; n≥4). In this dataset, station remained significant, explaining 41 % of the variation (Pr(> F) = 0.003), and five of six pairwise comparisons were also significant. These included all previously identified station differences, plus BB2 versus 101.
To enable a direct comparison with the water column dataset, a subset (F3) of foraminiferal samples was created using only those collected at the three stations where water column samples were also available (Stations 101, 115, and 323). This geographically matched subset was used to test further whether the stronger station effect observed in the full foraminiferal dataset simply reflected broader spatial coverage. Due to limited replication (n = 7, 2, and 2) and heterogeneous dispersion (Table A1), PERMANOVA was not appropriate. Instead, ALDEx2 with pairwise Wilcoxon tests identified only one ASV with significantly different abundance between any station pair, suggesting minimal spatial structuring in this subset, consistent with the water column results.
To evaluate whether the observed station effect was maintained across both provenances within a geographically consistent subset, a combined dataset (FW) was created, including all foraminiferal and water column samples from the three co-sampled stations (101, 115, and 323). Testing the effect of station in this FW dataset revealed no significant differences (Pr = 0.423), with station explaining just 5.4 % of the variation. PCA ordination of the FW dataset (Fig. A4) similarly showed stronger separation by provenance than by station, suggesting that sample type has a greater influence on community composition. Together, these results indicate that microbial community composition varies little among these three stations, regardless of sample provenance.
3.2.2 Multivariate analyses of foraminiferal versus water column ASV composition
Multivariate analyses were conducted on two datasets to compare the microbial community composition between provenances (foraminifera vs. water column). The first, dataset FW, included all water column and foraminiferal samples from the three co-sampled stations (n = 28 water; n = 11 foraminifera). The second, dataset 101, comprised only samples from Station 101, allowing provenance to be assessed independently of station (n = 9 water; n = 7 foraminifera).
PERMANOVA on the FW dataset revealed a significant difference in ASV composition between provenances, which explained 9.7% of the variation (Pr = 0.013; Table A1). PCA ordination supported this, showing clearer separation by provenance than by station (Fig. A4).
Analysis of dataset 101 reinforced this result where Provenance explained 41.7 % of the variation (Pr = 0.002; Table A1), and PCA ordination showed distinct clustering of foraminiferal samples apart from water column samples (Fig. A5). This indicates that, even within a single water column, microbial communities associated with foraminifera differ significantly from those in surrounding water.
However, although PCA ordinations supported the PERMANOVA results, tests for the homogeneity of the multivariate dispersion indicated significant heterogeneity in dispersion between provenances in both datasets (Table A1), which can violate PERMANOVA assumptions and complicate interpretation of the compositional differences. To address this and identify ASVs potentially driving these compositional differences between provenances, differential abundance analyses were conducted on the ASVs across both datasets.
3.2.3 Differential abundance analysis of ASVs in foraminifera versus the water column
Deseq2 fold change analysis of the FW dataset identified that 572 of 1207 ASVs are driving the significant compositional differences between provenances (P<0.05). All but 13 of those 572 ASVs are significantly more abundant in the water column. These include three Firmicutes (ASVs 1402, 609, 391), three Gammaproteobacteria (ASVs 927, 420, 116), two Bacteroidota (ASVs 743, 46), two Actinobacteriota (ASVs 1403, 509), one chloroplast ASV of unknown origin (ASV 1538), and two chloroplast ASVs from class Bacillariophyceae (ASV355, Fragilariopsis cylindrus, and ASV 956, Chaetoceros gelidus) (Table A2, Fig. 4).
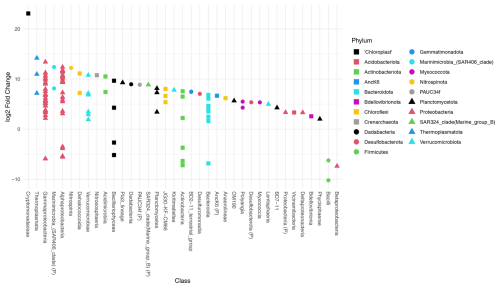
Figure 4Differential abundance testing of FW dataset ASVs between provenances using DESeq2. The log2 fold change in ASVs is the log-ratio of the ASV means in the water column and foraminifera. ASVs with positive log2 fold change are significantly more abundant in the water column assemblage, and ASVs with negative values indicate ASVs that are significantly more abundant in the foraminiferal assemblages. The class, or the highest level of taxonomic assignment available for each ASV, is given on the x axis. (P) = phylum.
Deseq2 fold change analysis of the 101 dataset identified 393 of 1013 ASVs that are driving the significant compositional differences between provenances (P<0.05). All but 14 of those are significantly more abundant in the water column. These include three Firmicutes (ASVs 1402, 609, 391), five Gammaproteobacteria (ASVs 927, 194, 149, 133, 116), two Bacteroidota (ASVs 743, 46), one Actinobacteriota (ASV509), one chloroplast ASV of unknown origin (ASV472), and two chloroplast ASVs from class Bacillariophyceae (ASV355, Fragilariopsis cylindrus, and ASV 956, Chaetoceros gelidus; Table A2, Fig. A6). Importantly, 10 of the ASVs that are significantly more abundant in the foraminifera are supported in both datasets.
3.2.4 Differential abundance analysis of predicted functional pathways in the foraminiferal microbiome versus the water column assemblage
PICRUST2 was used to identify possible functional pathways (using the MetaCyc database; Caspi et al., 2014) and to calculate functional pathway abundances based on estimated abundances of gene families that can be linked to reactions within those pathways. In total, 415 pathways were identified, and their differential abundances were compared between the microbiome of N. pachyderma and the water column assemblage using ALDEx2. Ninety-two pathways were identified as significantly differentially abundant between the two provenances (effect size > 1 and Benjamini-Hochberg adjusted P<0.05). These 92 pathways were grouped according to metabolic types to identify broader metabolic processes that were significantly different within the two provenances (Fig. 5). Of the 92 pathways, 38 were significantly more abundant in the foraminiferal microbiome. They include L-lysine biosynthesis (PWY-2941), peptidoglycan synthesis and recycling (four pathways, 897 ASVs), carbohydrate degradation (nine pathways, 624 ASVs), fermentation (four pathways, 496 ASVs), production of the secondary metabolite palmitate (PWY-1479, 4 ASVs), and butanediol production (two pathways, 49 ASVs, Fig. 5). Table A2 highlights those pathways identified in the significantly differentially abundant ASVs. The remaining 54 pathways were significantly more abundant in the water column assemblage, including 14 pathways involved in aromatic compound degradation, pathways for co-factor carrier and vitamin biosynthesis, inorganic nutrient metabolism, nucleoside and nucleotide biosynthesis, fatty acid and lipid biosynthesis, and C1 compound utilisation.
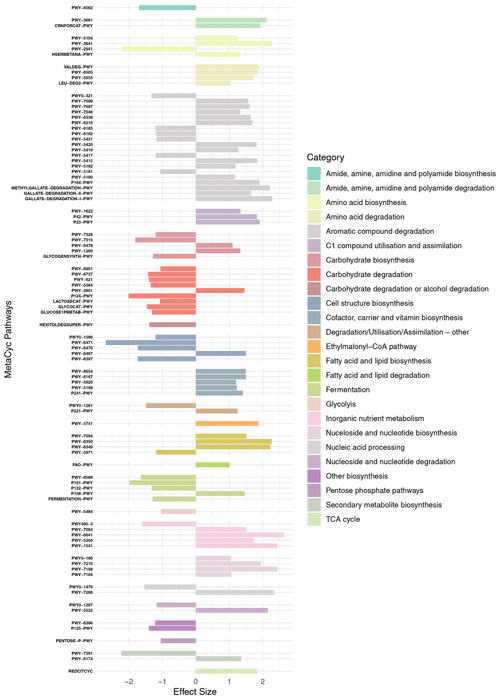
Figure 5MetaCyc pathways identified as being significantly differentially abundant between the two provenances are shown (y axis), with their “Effect Size” indicated by the x axis. Negative values indicate that the pathway is more abundant in the foraminiferal microbiome, and positive values indicate that the pathway is more abundant in the water column assemblage. Pathways are grouped by broader metabolic categories identified in the key.
3.3 Foraminiferal ASV profiles
All the foraminifera were genotyped as N. pachyderma Type I (NCBI GenBank Accession numbers OR137988-OR138014), consistent with it being the only genotype found in the Arctic region to date (Darling et al., 2004, 2007), indicating that variation in ASV composition cannot be driven by genotype differences.
3.3.1 Bacterial ASV profiles in N. pachyderma
Amplicon sequencing variants were assigned to 1367 distinct bacterial and archaeal taxa across all water and foraminifera samples within 29 phyla and 60 classes. The major groups were class Gammaproteobacteria, which contributed on average 18.8 % of ASVs, phylum Bacteroidota, contributing 14.6 %, and class Bacilli (phylum Firmicutes), contributing 5.1 %. The only other groups that contributed > 1 % of ASVs are phylum Actinobacteria, class Alphaproteobacteria, and class Verrucomicrobiae. The other 545 classes all contribute < 1 % each to the ASV total. This distribution reflected the ASVs driving significant compositional differences between the provenances (ASVs from class Gammaproteobacteria, phylum Bacteroidota, phylum Firmicutes, and phylum Actinobacteria; Sect. 3.3, Table A2).
3.3.2 Chloroplast ASV profiles in N. pachyderma
Amplicon sequencing variants are assigned to 181 distinct chloroplast ASVs corresponding to 14 unique chloroplast-containing classes across all water and foraminifera samples, contributing, on average, 53.3 % of all ASVs in the foraminifera and only 3.51 % in the water column (Fig. 3). More specifically, those ASVs assigned to diatom chloroplasts (class Bacillariophyceae) contributed, on average, 44.5 % of all ASVs in the foraminifera and only 2.36 % in the water column, highlighting the major importance of diatoms in the diet of the foraminifera compared to other phytoplankton taxa. Three chloroplast ASVs drive the significant difference between the foraminifera and the water column, due to greater abundance in the foraminifera. These are one ASV (ASV1538) identified only as “Chloroplast” and two diatom ASVs identified as Fragilariopsis cylindrus (ASV355) and Chaetoceros gelidus (ASV956) (Table A3). The relative abundance of chloroplast ASVs in each sample is shown in Fig. 6.
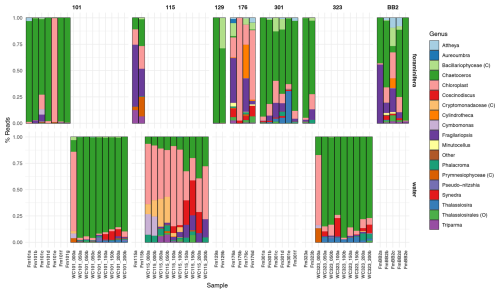
Figure 6The relative abundance of the 13 chloroplast genera identified with a relative abundance above 2 %, along with three classes, and one order identified only for these taxonomic ranks. ASVs with a relative abundance across all samples of less than 2 % were grouped in the “Other” category, and ASVs that could not be taxonomically assigned beyond “Chloroplast” are grouped as such. Bars represent individual samples (foraminiferal specimens = Fm, and the water column = WC). Numbers above indicate the stations (separated by columns), and provenance is separated by row.
ASV956 was the most abundant chloroplast ASV, with a mean relative abundance across all samples of 57.6 % when analysing chloroplast ASVs only. This was identified to the level of class Bacillariophyceae by the SILVA database and further to the diatom species Chaetoceros gelidus (accession NC_063631.1) via a BLAST search of GenBank (100 % identity and coverage) (Fig. 6). ASV956 had a significant log2 fold change of −2.348 (padj = 0.003443). This was not as significant as the other two chloroplast ASVs (Table A3), probably due to its higher relative abundance in the water column (Fig. 6). ASV956 was common within Baffin Bay, with an average relative abundance of 57.2 % (when analysing chloroplasts only) in the water column samples (Fig. 6). ASV956 was therefore relatively common in both the water column and the foraminifera and was clearly a major food source for N. pachyderma in Baffin Bay during the summer months.
At Station 101 (west Pikialasorsuaq polynya; Fig. 1), five out of the seven N. pachyderma specimens contained > 50 % chloroplast ASVs, and only one specimen contained < 20 % chloroplast ASVs (Fm101a; Fig. 3). When analysing only chloroplast ASVs, six of the seven foraminifera specimens contained > 70 % ASV956 (Chaetoceros gelidus). Specimen Fm101e, however, contained over 94 % chloroplast ASVs (472, 548) belonging to two uncharacterised chloroplasts (Fig. 6). Except for specimen Fm101e, the diatom ASVs at the cooler Station 101 can be said to mirror the diatom population profile in the sub-surface water column, where > 85 % of water column chloroplast ASVs were ASV956 (Chaetoceros gelidus) (Fig. 5).
The two foraminifera processed at Station 115 (where Atlantic-derived warmer water is found) contained < 35 % and < 13 % chloroplast ASVs (Fig. 3). Again, this lower chloroplast relative abundance reflected the water column where we found the lowest relative proportion of chloroplast ASVs across the three stations (Fig. 3). Fragilariopsis sp. contributed the greatest proportion of chloroplast ASVs (ASV355) in Station 115 specimens (Fig. 6). The proportion of Fragilariopsis ASVs was higher in the water column at this station relative to other stations, although ASV956 (Chaetoceros gelidus) remained the major diatom ASV present. However, the highest proportion of ASVs were not identified beyond “Chloroplast” at this station. In addition, a non-diatom chloroplast (Triparma laevis ASV471) was also present in both the water column and the foraminiferal specimens. This is a relative of the diatoms and, like most diatom species, forms external siliceous plates. Triparma laevis ASVs were detected only in the upper water column at 50–100 m.
Finally, 48 % and 28 % of the ASVs in the two foraminifera from Station 323 were chloroplast ASVs (Fig. 3), and of those, 94.5 % and 69.8 % were ASV956 (Chaetoceros gelidus; Fig. 6). This reflected the high proportion of ASV956 in the water column at this station (> 73 % across sub-surface samples).
Of the other foraminiferal specimens taken from stations with no comparative water column data, the foraminifera from Station 176 showed the greatest diversity in chloroplast ASVs. Except for Fm176b, which had only three chloroplast ASVs, Fm176a, c, and d contained a much higher relative proportion of Fragilariopsis, Synedra and Cylindrotheca diatoms and other chloroplast ASVs. This may indicate higher comparative diatom and algal diversity at this more southerly station.
3.3.3 The N. pachyderma core microbiome
The core microbiome of N. pachyderma is defined here as ASVs found in 80 % of the foraminiferal specimens across all stations. 16S metabarcoding indicated that there were eight core ASVs: two represented by diatoms, i.e. ASV956, Chaetoceros gelidus ( specimens) and ASV355, identified in BLASTn as Fragilariopsis cylindrus (100 % match to accession NC_045244.1, 24/28 specimens), then six bacterial ASVs, two from the Flavobacteriaceae family (ASV1392, 24/28, and 1447, ), one from the genus Pseudoalteromonas (ASV 1459, 25/28), one from the genus Paraglaciecola (ASV 122, 25/28), one from the family Halieaceae (ASV308, 26/28), and one from the genus Bradyrhizobium (ASV833, ; Fig. 7). Of these eight ASVs, only the two diatom ASVs (355 and 956) were also significantly more abundant in the foraminifera than the water column, driving the significant differences between the provenances. This foraminiferal core microbiome made up, on average, 47.7 % of the ASVs in the N. pachyderma of Baffin Bay, whereas it made up only 9.42 % of ASVs in the water column. Details on the core microbiome, including relative abundances and ASV frequencies in the two provenances, can be seen in Table A3.
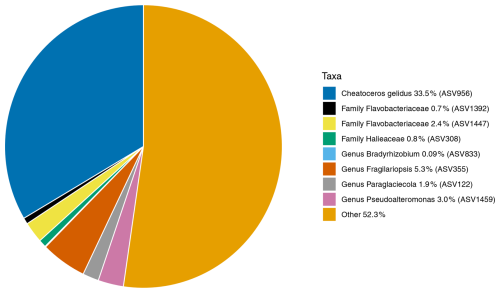
Figure 7The average relative abundances of the 16S rDNA ASVs found across N. pachyderma Type I specimens in Baffin Bay during summer 2017. The average relative abundance of the eight core ASVs (found in ≥ 80 % of specimens) are taxonomically labelled and designated the “core microbiome”. These make up 47.7 % of the ASVs in the microbiome. 52.3 % are ASVs found in fewer than 80 % of specimens, designated as non-core and labelled “Other” above. Colour key starts at 12 O'clock and runs anticlockwise.
3.4 TEM analysis
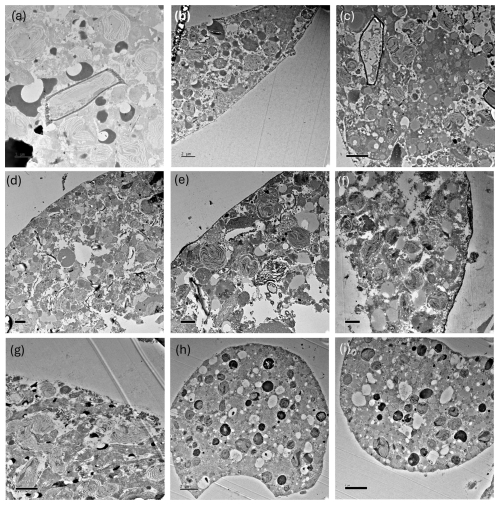
TEM imaging was carried out on samples collected during the 2018 cruise (Fig. 1; Table 1) to further investigate the diet/endobionts in this genotype. Whole diatoms, including frustules, were observed within the foraminiferal cell (Fig. 8a). Empty frustules were also observed both inside and outside the foraminiferal cell. Those outside may have been ejected after the diatom organic material was digested/removed (Fig. 8b) or were part of the diatom-derived POM feeding cyst and were likely caught in the external cytoplasm and rhizopodial network at the time of sampling and fixation, as has been reported previously (Spindler et al., 1984).
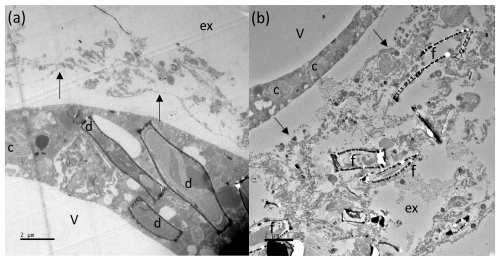
Figure 8TEM images of N. pachyderma (specimen BB2, Table 1) showing internal and external cell and diatom structures. Image (a): intact diatoms (d) observed inside the specimen cell. Black arrows indicate the cross section of pore plugs in the inner organic lining, v = internal cell vacuole, and ex = external to the foraminiferal cell. Image (b): external (ex) to the specimen where debris, including empty diatom frustules (f), is apparent. The organic lining is identified by black arrows, c = chloroplasts inside BB2, and v = internal vacuole. Large vacuoles were observed in several specimens, which may be a result of the fixation process. Scale bars at the bottom left are 2 µm.
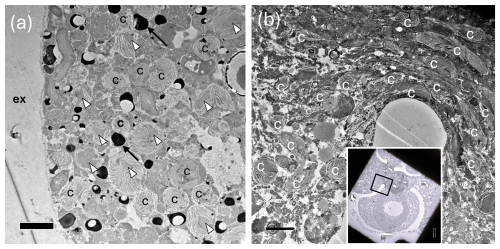
Figure 9TEM images showing abundant chloroplasts throughout the N. pachyderma cell. (a) Clusters of chloroplasts (c) within the specimen BB1 cell (Table 1) close to the cell periphery. Those with obvious pyrenoids are marked with white arrowheads. Due to our staining protocol, black spots could be lipid droplets or electron opaque bodies (LeKieffre et al., 2018; black arrows highlight examples). ex = external to the foraminiferal cell. Scale bar 2 µm. (Inset) Overview of thin section of specimen BB11 (Table 1) showing chambers, and a black square identifying the region of the cell shown in (b). (b) Chloroplasts clustered in numbers at the centre of specimen BB11 also appear stretched where the chambers coalesce. Scale bar 2 µm.
TEM images also show that N. pachyderma contains unexpectedly high numbers of chloroplasts throughout the cell from the cell periphery to the cell centre (Fig. 9a–b, Fig. A7). The level of preservation does not allow us to observe the number of membranes surrounding the chloroplasts or the pyrenoid-dissecting lamellae. Nevertheless, although we cannot unequivocally determine the degradation state of all the chloroplasts present, lenticular pyrenoids and horseshoe-shaped arrays of thylakoid membranes are visible in many chloroplasts (Fig. 9a), as found in Chaetoceros spp. (Bedoshvili et al., 2009). There may also be abundant lipid droplets located among, and immediately adjacent to, the chloroplasts at the cell periphery in some specimens, potentially indicating lipid production by the chloroplasts (Jauffrais et al., 2019a, Figs. 9a, A7a and g).
In this study, our aim was to investigate the microbiome within the polar planktonic foraminifera N. pachyderma Type I from the Arctic Baffin Bay region. We defined the microbiome as the combined taxa identified by taxonomic assignment of 16S ASVs generated by metabarcoding. This included food, any endo(sym)bionts, and chloroplast-containing eukaryotes identified by their chloroplast 16S ASVs. Shedding light on their feeding preferences as well as any microbial associations that form part of the “interactome” in the context of the changing climate may afford some clues as to the ability of N. pachyderma to withstand/adapt to its rapidly changing environment and its contribution to the carbonate cycle and ocean alkalinity. For example, eco-physiological and trait-based models indicate that symbiont-barren foraminifera, which N. pachyderma is understood to be, are predicted to experience reduced numbers and habitat decline (Roy et al., 2015), and the non-spinose species biomass is likely to be reduced by up to 11 % by 2050 (Grigoratou et al., 2022). Sound knowledge of the eco-physiology and traits of foraminifera is required for model accuracy, and to that end the genotype and microbiome of the Arctic polar N. pachyderma was investigated.
4.1 The influence of station and depth on the microbial assemblages
Depth was the dominant factor structuring microbial communities in the water column, likely driven by steep vertical gradients in temperature, salinity, and chlorophyll concentration (Fig. 2). This aligns with previous studies reporting depth-stratified microbial assemblages in marine systems (e.g. Zorz et al., 2019; Reji et al., 2020). Because depth data are unavailable for the foraminiferal samples, its influence on their microbiomes could not be assessed.
In contrast, station exerted a weaker influence on community composition. Although a significant station effect was detected in water samples at 50 m depth, the absence of significant pairwise differences and limited replication reduce confidence in this result. Similarly, analyses of the foraminifera-only dataset from the same three stations (F3), as well as the combined water–foraminifera dataset (FW), revealed minimal spatial structuring, indicating that horizontal variation across these sites was relatively minor.
The distance–decay relationship, which describes how community similarity declines with increasing geographic distance, is a well-documented pattern in microbial biogeography (e.g. Li et al., 2018). To investigate whether this pattern applied to foraminiferal microbiomes, we expanded our analysis to the full foraminiferal dataset, which spans a broader geographic range. This broader scope revealed significant station-level differences in microbiome composition. Although only 4 of 21 pairwise comparisons were significant in the full dataset, these same four comparisons remained significant in a more robustly replicated subset (four stations with ≥ 4 samples each), where five of six comparisons showed significant differences. This consistency suggests that observed differences at certain stations are robust rather than artefacts of low replication.
Notably, Station 301 consistently differed from Stations BB2, 176, and 101, with 176 and BB2 being geographically distant. Located in Lancaster Sound, Station 301 is influenced by an eastward current carrying Beaufort Sea water (Sanderson and LeBlond, 1984), which may shape its distinct microbial assemblages prior to mixing with Atlantic-derived waters in Baffin Bay. Interestingly, Station 301 did not differ significantly from its nearest neighbour Station 323, situated at the mouth of Lancaster Sound. These findings suggest that the stronger station signal observed in the broader foraminiferal dataset is likely attributable to the greater geographic coverage (Fig. 1), consistent with the distance–decay relationship, where community similarity declines with increasing spatial separation.
In addition, selective feeding by N. pachyderma may contribute to the observed patterns. Foraminifera retain a core microbiome that accounts for nearly 48 % of their ASVs. Although individuals from Station 301 exhibited significantly different overall microbiome compositions compared to several other stations, their core microbiome did not significantly diverge from those collected elsewhere (Table A3), suggesting that host-level filtering may buffer against environmental variability and reduce the influence of station. This supports the idea that host selection stabilises key microbial associations despite broader shifts in community composition. The limited number of significant pairwise differences across the full dataset (4 out of 21) may also reflect this buffering effect, alongside subtle or inconsistent geographic influences.
4.2 Divergent feeding strategies in the Neogloboquadrinids
Our findings support previous literature stating that N. pachyderma feeds on diatoms (e.g. Spindler and Dieckmann, 1986; Schiebel and Hemleben, 2017, Greco et al., 2021). This study and that of Greco et al. (2021) indicate that N. pachyderma feeds predominantly on diatoms (class Bacillariophyceae) and occasionally other algae (e.g. Triparmia laevis – this study). Our TEM micrograph observations of intact diatoms (Fig. 8a) also indicate that N. pachyderma feeds on live diatoms and not only detrital (dead) diatoms, as reported by Greco et al. (2021). Our 16S metabarcoding data further suggest that genus Chaetoceros is an important food source for N. pachyderma in Baffin Bay. This agrees with Meilland et al. (2024), who cultured N. pachyderma Type I from several locations, including Baffin Bay, by growing them with a diatom food source. They found that specimens grew faster when fed on the Genera Chaetoceros or Pseudo-nitzschia, compared to Phaeodactylum or Thalassiosira. However, in their cultures, juveniles provided with Pseudo-nitzschia had the longest life span. Despite this, while we recorded Pseudo-nitzschia in the water column and in our foraminiferal specimens (Fig. 6), Chaetoceros was by far the dominant genus identified in our specimens from the natural environment.
Our 16S study further revealed that N. pachyderma also consumed bacteria, and their 16S ASV composition was significantly different from the water column profile (Table A1). This is most likely driven by the feeding behaviour, where particulate organic matter (POM) is gathered around the test to form a feeding cyst. Once formed, the foraminifer remains within the POM microhabitat, becoming isolated from the water column. This behaviour has already been observed in the Neogloboquadrinids N. dutertrei and N. incompta (Bird et al., 2018; Fehrenbacher et al., 2018) and in Globigerinita glutinata (Spindler et al., 1984).
Although the Neogloboquadrinids all feed within the POM microhabitat, we suggest that they are feeding on very different components within the cyst. Work carried out by Bird et al. (2018) demonstrated that of the three Neogloboquadrinids, N. incompta contained the highest proportion of bacterial ASVs (> 99.8 %), indicating that it targets the bacteria within the POM microhabitat. The small proportion of chloroplast ASVs (< 0.2 %) in the N. incompta study indicated that POM was not being passively phagocytosed but that the bacteria, rather than algae, were being specifically selected as food. In contrast, N. dutertrei contained only 2 %–4 % bacterial ASVs and instead maintained a pelagophyte algal endosymbiont population and selectively fed on other protists within the POM. The small proportion of intracellular bacteria in N. dutertrei indicated that it too did not specifically phagocytose POM itself. However, in the case of N. pachyderma, we found a higher proportion of bacterial ASVs than was identified in N. dutertrei and a higher proportion of chloroplast ASVs than found in N. incompta, suggesting that N. pachyderma may feed on the POM directly for food. This was suggested by Greco et al. (2021), who also demonstrated that the N. pachyderma 18S ASV assemblage revealed little difference in intracellular diatom ASVs between the surface-dwelling and deeper-dwelling specimens living in diatom-free waters. This finding led them to suggest that N. pachyderma feeds on dead diatoms contributing to the sinking detritus, which is supportive of a POM-cyst mode of feeding. However, the TEM images (Fig. 8) in our study indicated that they also feed on living diatoms, as intact diatoms were observed in their cells. Further, our evidence indicated that a significant component of the N. pachyderma diet is active or passive consumption of the bacteria living in the diatom “phycosphere” and diatom-derived POM (Bell and Mitchell, 1972; see Sect. 4.3).
4.3 N. pachyderma bacterial ASVs
4.3.1 The core microbiome
The core microbiome (defined as ASVs present across 80 % of the foraminifera) could be made up of organisms that (i) the foraminifera specifically target for food and/or (ii) are routinely passively ingested due to close association with specific food sources and/or (iii) are endo(sym)bionts. However, the bacteria identified in the N. pachyderma core microbiome point to a diatom source and therefore are highly likely to be passively ingested. The core microbiome was composed of just eight ASVs, which accounted for, on average, 47.7 % of the total microbiome. Six bacterial ASVs each contributed small percentages between 0.09 % and 2.96 %, averaging a total of just 8.93 % of the core microbiome between them. The bacterial ASV with the highest average relative abundance in the core microbiome (3 %) was Pseudoalteromonas (ASV1459, Fig. 7, Table A3). Pseudoalteromonas are known hydrocarbon degraders (e.g. Calderon et al., 2018). They are the first bacteria to colonise degrading diatom aggregates (Arandia-Gorostidi et al., 2022; Costanzo et al., 2023), and they are known to be algicidal, releasing diffusible factors (Costanzo et al., 2023) in the diatom phycosphere. There were also two ASVs from the Flavobacteriaceae family (ASV1447 and ASV1392). This is a large family of bacteria that are widely distributed in the marine environment and are often found associated with detritus (as well as algae, fish, and invertebrates; Gavriilidou et al., 2020). Tisserand et al. (2020) isolated 10 species from Baffin Bay, and all were shown to grow on exudates (dissolved organic matter) from two Arctic diatoms (Fragilariopsis cylindrus and Chaetoceros neogracilis). Because members of the Flavobacteriaceae and Pseudoalteromonas are shown to co-occur with diatoms (Amin et al., 2012), it is likely that N. pachyderma passively consumes these bacteria as it feeds on diatom detritus (Greco et al., 2021) and on living diatoms. A BLASTn search (NCBI) identified ASV1392 as 99.6 % identical to a Flavobacteriaceae of the genus Tenacibaculum including T. insulae, T. haliotis, and Tenacibaculum sp. This genus contains many opportunistic fish pathogens, some of which are found to target fish teeth, a high source of calcium shown to promote the bacteria's growth (Hikida et al., 1979; Frisch et al., 2018). Growth promotion by calcium may be a common feature of the Tenacibaculum genus and may be another reason why this ASV is identified with N. pachyderma, and the calcite tests of foraminifera may provide a suitable niche for this genus. Another core ASV was 308, attributed to the OM(NOR) genus of the family Halieaceae, order Cellvibrionales (Spring et al., 2015). The order Cellvibrionales are gram-positive aerobes that are mesophilic and neutrophilic chemoorganotrophs. However, some members of the family Halieaceae may additionally be capable of aerobic photoheterotrophic growth using bacteriochlorophyll a and carotenoids for the harvesting of light. Several strains may also be able to use proteorhodopsin to utilise light as an energy source (Spring et al., 2015). Paraglaciecola (ASV122) was another core ASV (Fig. 7). Numbers of ASVs and the relative abundances were substantial and similar between the provenances (Table A3). Paraglaciecola are a genus of the family Alteromonadaceae. In a BLASTn search, this ASV shows 100 % identity with Paraglaciecola psychrophila, P. arctica, and several other Paraglaciecola sp. sequences. Paraglaciecola psychrophila is a gram-negative, psychrophilic, motile rod-shaped bacteria. Identified from the sea ice of the Canadian Basin and the Greenland Sea, it is aerobic, and optimum growth is at 12 °C (Zhang et al., 2006). Unable to reduce nitrate, it may be associated with POM as an N-source and thus be ingested by N. pachyderma as it feeds on the detritus. The final core ASV Bradyrhizobium (ASV833) constituted on average only 0.09 % of foraminiferal and 0.004 % of water column ASVs. Bradyrhizobium contains mainly nitrogen-fixing species that are part of phylogenetic subcluster IK of nifH (Chien and Zinder, 1994; Gaby and Buckley, 2014; Fernández-Méndez et al., 2016), which encodes the nitrogen-fixing enzyme nitrogenase. Sequences from subcluster IK can make up > 50 % of the nifH sequence abundance in the open waters of the central Arctic Ocean (Fernández-Méndez et al., 2016), supporting our identification of this ASV in the polar waters of Baffin Bay.
4.3.2 The differentially abundant ASVs and PICRUSt2 pathways
Metabolic pathway abundances were predicted using PICRUST2. It is important to note that these are predictions indicating potential functional capacity and are not indicative of active processes. Nevertheless, many of the predicted pathways are consistent with the hypothesis that the bacteria in the foraminiferal microbiome are derived from foraminiferal feeding on diatoms and diatom-derived POM.
A POM feeding cyst will contain an oxygen gradient with oxygen concentrations in the centre significantly below ambient seawater (Alldredge and Cohen, 1987). Interestingly, facultatively anaerobic fermentation pathways are present in higher abundance in the foraminiferal microbiome, which may be a result of preferential ingestion of fermenting bacteria due to their presence in the centre of a low-oxygen POM-feeding cyst. There are two pathways categorised as “Other biosynthesis” pathways (P125-PWY and PWY-7391, Fig. 5) that are involved in the biosynthesis of the antifreeze butanediol, via the fermentation of pyruvate (Caspi et al., 2014). These pathways are attributed to 49 ASVs predominantly from class Gamma- and Alphaproteobacteria, phylum Firmicutes, and phylum Actinobacteriota and include two of the differentially abundant ASVs in the foraminiferal microbiome (Fig. 4; Table A2). Four other identified fermentation pathways utilise monosaccharides to produce ATP and reducing power (NADH). These monosaccharides are abundant in the diatom exopolysaccharide (EPS) exudates in the phycosphere (Daly et al., 2023), and therefore, again, the phycosphere or diatom-derived POM is likely to be the foraminiferal microbiome source of these fermenting bacteria. A total of 496 ASVs contribute to this pathway, with three of the differentially abundant ASVs doing so (Table A2).
Peptidoglycan synthesis pathways are also more abundant in the foraminiferal microbiome; this is driven by 897 ASVs, of which four are also significantly differentially abundant in the foraminiferal microbiome (Table A2). A pathway for synthesis of a single amino acid L-lysine (PWY-2941) is more prevalent here, compared to higher abundance of three different amino acid biosynthesis pathways in the water column assemblage. L-lysine is essential for cell wall biosynthesis (Gillner et al., 2013), and this pathway also produces meso-diaminopimelate, which is another component of the peptidoglycan cell wall (Weinberger and Gilvarg, 1970). Supporting this, there are four further peptidoglycan synthesis and recycling pathways (grouped in the category “Cell structure biosynthesis”, Fig. 5). Pathway PWY-6471 is specific to gram-positive bacteria, and the greater abundance of these cell wall synthesis pathways in general might indicate a greater relative abundance of gram-positive bacteria in the foraminiferal microbiome compared to the water column, although at present it is unclear why.
The degradation of certain carbohydrates was also key in the foraminiferal microbiome, with 624 ASVs associated with these pathways. For example, pathways for the degradation of the polysaccharides glycogen and starch and the sugars sucrose, glucose, and xylose were differentially abundant, and given these are abundant sugars in the diatom phycosphere (Daly et al., 2023), this is further supporting evidence that the majority of bacteria in the foraminiferal microbiome are derived from the phycosphere or diatom-derived POM.
Of interest were two additional pathways. The first was norspermidine biosynthesis (PWY-6562), which plays a central role in biofilm formation in Vibrio spp. (Wotanis et al., 2017) while inhibiting biofilm formation by other species and, in particular, other gram-negative bacteria (Qu et al., 2016). Norspermidine biosynthesis was identified across seven Gammaproteobacterial ASVs, including three Alteromonadales and four Vibrionales, one of which (ASV116, genus Vibrio) was differentially abundant in the foraminiferal microbiome.
Finally, interestingly, the palmitate biosynthesis II pathway (PWY-5971) is more abundant in the foraminiferal microbiome. Palmitate is a long-chain saturated fatty acid. It is produced both by algae such as diatoms and by bacteria (Allan et al., 2023), but in our dataset, the ASVs responsible for this pathway are one ASV of the genus Alteromonas and three ASVs from the genus Cellvibrio. Palmitate, or palmitic acid, is a very abundant saturated fatty acid and key precursor for phospholipids and lipopolysaccharides essential for the bacterial plasma membrane (Cronan and Thomas, 2009). Given the universal nature of this requirement, it is surprising that genes encoding enzymes in this pathway are more abundant in the foraminiferal microbiome. Interestingly, however, palmitic acid is used by pathogenic bacteria to modify their proteins and glycoproteins to avoid detection by host immune system TLR4 receptors (Toll-like-receptor family) (Sobocińska et al., 2018), thereby increasing infectivity and the production of biofilms. However, TLR4 evolved 500 million years ago near the beginning of vertebrate evolution (Beutler and Rehli, 2002) and is not known to be present in protists. Therefore, the reason for increased abundance of the palmitate biosynthesis II pathway in the foraminiferal microbiome compared to the water column is currently unknown.
4.4 N. pachyderma chloroplast ASVs
Two diatom chloroplasts contributed 5.3 % (ASV355, Fragilariopsis cylindrus) and 33.46 % (ASV956 Chaetoceros gelidus) of the core microbiome (Fig. 7). Both also contributed to the significant difference in assemblage composition between the foraminifera and the water column (Fig. Table A2). Chloroplasts were exceptionally abundant in our TEM images (Figs. 9 and A7), which appear very similar to observations made in kleptoplastic benthic species (e.g. Bernhard and Bowser, 1999; Jauffrais et al., 2018; Jesus et al., 2022). This raises important questions about the nature of the relationship between the chloroplasts and N. pachyderma Type I and is discussed below. Empty diatom frustules were also observed in the TEM images, which is highly consistent with previous reports that diatoms are a significant part of the N. pachyderma diet (e.g. Hemleben et al., 1989; Schiebel and Hemleben, 2017; Greco et al., 2021).
On average, 53.3 % of all 16S rDNA ASVs in the foraminifera belong to chloroplast-containing taxa (Fig. 2; Sect. 3.3.2). This contrasts with the 3.51 % average proportion found in the water column. Most of the foraminiferal intracellular chloroplast ASVs are dominated by ASV956, Chaetoceros gelidus (BLASTn), from the diatom class Bacillariophyceae. The compositional dominance of ASV956 in the foraminifera reflects the chloroplast ASV composition of the water column (Fig. 6), although found in much higher proportions in the foraminifera (Fig. 3). The presence of ASV956, Chaetoceros gelidus, in all other specimens indicates its importance to N. pachyderma in this location and season. It is characteristic of northern temperate and polar waters (Chamnansinp et al., 2013), and it is a known important biomass fraction in Baffin Bay (Crawford et al., 2018). In fact, eight strains were isolated from Baffin Bay only during bloom development or bloom peak (Ribeiro et al., 2020), and Chaetoceros's reputation for bloom forming (Booth et al., 2002) is reflected here in its high abundances compared to other species.
The diatom chloroplast 16S ASVs identified in this study (Fig. 6) are also consistent with the diatoms found by Greco et al. (2021), who identified Chaetoceros and Fragilariopsis as major components of the N. pachyderma 18S ASVs from Baffin Bay. Both Chaetoceros (ASV1413, ASV956) and Fragilariopsis (ASV355) 16S ASVs were among those ASVs driving the significant difference between the foraminifera and the water column, and both are major constituents of the core microbiome. Intact Fragilariopsis were also identified in the foraminiferal TEM images (Fig. 8a), hinting that, like Ammonia sp. and the miliolid Hauerina diversa, N. pachyderma may phagocytose the entire diatom before digesting the cell and then extruding the silicate frustules (Jauffrais et al., 2018; Pinko et al., 2023).
4.5 Observation of abundant chloroplasts throughout the cytoplasm of N. pachyderma Type I
To our knowledge, this is the first report of large numbers of chloroplasts observed by TEM imaging and recorded via metabarcoding in any planktonic foraminiferal species. These observations cover two summers in different regions of Baffin Bay. The high numbers observed, and the relative abundance of diatom chloroplasts recorded, could indicate a kleptoplastic behaviour in N. pachyderma Type I, a strategy that is well known in several protist lineages such as benthic foraminifera (e.g. Jesus et al., 2022), dinoflagellates (e.g. Takano et al., 2014; Yamada et al., 2023), and ciliates (e.g. Johnson et al., 2007). Kleptoplasty refers to the phenomenon where an organism sequesters chloroplasts from its microalgal prey. The original definition does not include the requirement for temporary photosynthesis to continue in the host (Clark et al., 1990; Jauffrais et al., 2018) and is appropriate because chloroplasts are known to perform many functions in addition to photosynthesis. These include amino acid, nucleotide, and fatty acid synthesis as well as N and S assimilation (e.g. Cedhagen, 1991; Bobik and Burch-Smith, 2015). Further, the benthic foraminiferal species Nonionellina labradorica retains chloroplasts despite living in sediments below the photic zone. The photosynthetic pathway of their retained chloroplasts is therefore not functional (Cedhagen, 1991; Jauffrais et al., 2019b), and the reason for chloroplast retention in this and other species that live in the aphotic zone is unknown (Bernhard and Bowser 1999). However, its importance is reflected by the discovery that the kleptoplast genome in Nonionella stella, another benthic species that lives below the photic zone, is transcribed in the host (Gomaa et al., 2021; Powers et al., 2022). Where kleptoplasts do continue to photosynthesise in the new host, evidence suggests that this has been important in supporting major evolutionary innovations crucial to the current ecological roles of such protists in the marine environment (Stoecker et al., 2009). Therefore, the role of the retained chloroplasts remains a fascinating question in many species of foraminifera, including N. pachyderma Type I. It is important to assess and further investigate potential kleptoplast roles to understand any contribution they make to N. pachyderma evolution and their ability to inhabit the true polar habitat, as well as to evaluate their potential resilience to future climate change.
4.5.1 Evidence for potential kleptoplasty in N. pachyderma Type I
Foraminifera such as N. pachyderma that eat diatoms (and other algae) would be expected to contain some chloroplasts in their cytoplasm as a byproduct of their grazing. For example, 18S metabarcoding demonstrates that the non-kleptoplastic benthic foraminifer Ammonia sp. (Jauffrais et al., 2016) grazes on diatoms in a comparable way to the kleptoplastic Elphidium sp. and Haynesina germanica (Chronopoulou et al., 2019), and chloroplasts are indeed observed within the cytoplasm of Ammonia sp. Yet, the relative plastid abundance in Ammonia sp. is reported as “rare” compared to “abundant” in Elphidium sp. and Haynesina sp. (Goldstein et al., 2004; Cesbron et al., 2017; Jauffrais et al., 2018), with a high proportion of chloroplasts in Ammonia sp. undergoing degradation (Jauffrais et al., 2018; LeKieffre et al., 2018). In contrast, our TEM images show high numbers of chloroplasts in N. pachyderma, congruent with or greater than the abundance observed in the TEM images of kleptoplastic foraminifera such as Elphidium sp. and H. germanica (Jauffrais et al., 2018; Figs. 9, A7).
Kleptoplasty is common among benthic foraminifera (e.g. Lopez, 1979; Lee et al., 1988; Cedhagen 1991; Tsuchiya et al., 2018; Jauffrais et al., 2018; Pinko et al., 2023). The molecular studies identifying the source of kleptoplasts in benthic foraminifera to date would suggest a diatom source from the family Thalassiosiraceae, but, potentially, kleptoplasts from more than one diatom species can be present (e.g. Pillet et al., 2011; Lechliter, 2014; Jauffrais et al., 2019a; Tsuchiya et al., 2020; Pinko et al., 2023). More than 20 diatom species have been identified in benthic foraminifera that host intact diatom symbionts (Lee et al., 1995; Schmidt et al., 2018), with potentially up to three different symbionts within a single foraminifer at the same time (Lee, 2011). In addition, diatom symbiont shuffling appears to be an adaptation to changing environmental conditions such as heat stress (Schmidt et al., 2018). These studies indicate that host–symbiont or host–kleptoplast relationships are not strictly species-specific, supporting our findings of multiple-diatom ASVs.
The chloroplasts in N. pachyderma are distributed throughout the foraminiferal cytoplasm (Fig. 9). In the benthic kleptoplastic species, chloroplast location is specific to the foraminiferal host species, where some kleptoplasts may be associated with the cellular periphery, while others, as observed here, may be distributed throughout the cell cytosol (e.g. Jauffrais et al., 2018; Pinko et al., 2023). Chloroplast placement therefore cannot provide a clear-cut indicator of kleptoplasty.
The degradation state of the N. pachyderma chloroplasts in this study is uncertain due to poor fixation of the samples and therefore, unfortunately, cannot provide information on how intact they are. However, in benthic foraminifera, actively photosynthesising kleptoplasts can remain active from just a few days to a few months before being digested (e.g. Grzymski et al., 2002; Jauffrais et al., 2018). Given that turnover rates are extremely variable, the number of degrading versus intact kleptoplasts must also be highly variable from species to species.
It is important to note that no photosynthetic potential was found in six N. pachyderma Type VII individuals from the North Pacific using fast repetition rate (FRR) fluorometry (Takagi et al., 2019). There was also no evidence of non-functional chlorophyll using this technique (Takagi et al., 2019). This is extremely surprising given the herbivorous nature of N. pachyderma (e.g. Spindler and Dieckmann, 1986; Schiebel and Hemleben, 2017; Greco et al., 2021; this study) and may reflect different feeding strategies in the two genotypes, whereby Type I retains chloroplasts but Type VII does not, or Type VII has a broader-ranging diet and so there is no resultant build-up of chloroplasts in the cytoplasm. Neogloboquadrina pachyderma Type I should now be tested using FRR fluorometry to identify whether retained chloroplasts have photosynthetic potential and therefore behave as traditional kleptoplasts or not. This potential difference could represent a divergent evolutionary adaptation in N. pachyderma Type I to survive and flourish in the extreme Arctic environment. Neogloboquadrina pachyderma has genetically diversified to inhabit a wide range of extreme environments from the Arctic and Antarctic polar waters to the frontal and upwelling systems of the transitional to tropical zones (e.g. Darling and Wade, 2008). Type I N. pachyderma diverged from its Southern Ocean counterparts during the early Quaternary (Darling et al., 2004), allowing substantial time for distinct adaptations to develop in its North Atlantic and Arctic habitat.
4.5.2 Potential chloroplast storage to facilitate overwintering and reproduction
The cytoplasmic chloroplasts observed in N. pachyderma Type I (Fig. 9) are retained from their diatom food source, in particular from Chaetoceros spp. (Booth et al., 2002; this study). These chloroplasts represent a rich source of amino acids, fatty acids, lipids, vitamin E, pro-vitamin A, lutein, Cu, Fe, Zn, and Mn (Gedi et al., 2017). Therefore, either functioning photosynthetic kleptoplasts and/or chloroplasts themselves could potentially provide N. pachyderma Type I with a substantial additional energy resource in the challenging Arctic environment. If chloroplasts can be retained in the cytoplasm over many months before consumption, they could provide a valuable source of nutrition for the overwintering population. A similar overwintering survival strategy citing the high nutrient levels of stored chloroplasts has been proposed for the benthic foraminifera N. labradorica (Salonen et al., 2021), as it is known that no photosynthesis occurs in these kleptoplasts in the winter months (Cedhagen, 1991).
Ecological processes in the Arctic are largely governed by sea ice and light dynamics. There is a general perception of minimal biological activity in the Arctic marine surface layers during the Arctic winter due to the low light intensity producing minimal photosynthetic activity. However, studies around Svalbard in January 2012–2015 revealed unexpectedly high biological activity in the Arctic winter, with high respiration rates per unit of biomass in the upper 100 m water column (Berge et al., 2015a, b; Falk-Petersen et al., 2015), and an earlier winter Calanus copepod (Arthropoda) presence than previously thought (Espinasse et al., 2022). In Baffin Bay, low but significant phytoplankton growth was also observed during winter under the sea ice at extremely low light levels (Randelhoff et al., 2020). Because N. pachyderma Type I are thought to feed on both POM (including Arthropoda) and live diatoms (Greco et al., 2021, and this study), such wintertime POM-producing biological activity combined with stored chloroplasts (whether photosynthesising or not) could provide significant nutritional resources for an overwintering population of foraminifera.
These factors potentially combine to provide N. pachyderma with a significant nutritional resource to survive over the winter months, but questions remain about its behaviour in the water column and the form in which it may overwinter. Sediment traps in the Irminger Sea indicate a very low-level population of overwintering N. pachyderma, and their isotopic signature profiles imply that a dormant non-calcifying population of N. pachyderma may remain in the water column during winter (Jonkers et al., 2010). However, it is possible that the N. pachyderma population they detected may not fully represent the true winter population size, as sieve sizes of 150 µm would not retain smaller mature/immature N. pachyderma specimens. Potentially, N. pachyderma could also remain buoyant in the water column as non-reproducing immature cells, slowing down their cellular metabolism as largely quiescent cells during the most challenging winter months. In culture, several specimens of N. pachyderma Type I exhibited extended periods of dormancy or inactivity, followed by recovery (Westgård et al., 2023).
4.6 Palaeoenvironments and geochemical signatures
The biological adaptations and interactions of calcifying foraminifera have varying influences on the geochemistry of their test, as photosymbionts are known to influence test geochemistry (e.g. Spero and Lea, 1993; Bemis et al., 1998, 2002; Anand et al., 2003; Russell et al., 2004), and symbiont–host respiration and potentially respiration of endobiont bacteria may increase the use of metabolic (respired) carbon in their tests (e.g. Rink et al., 1998; Wolf-Gladrow et al., 1999; Hönisch et al., 2003; Eggins et al., 2004; Bird et al., 2017). To fully understand variations in the geochemical signatures of Arctic N. pachyderma tests through time in the fossil record, we need to improve our understanding of the ecology and interactions between N. pachyderma and the intracellular microorganisms that it hosts. Interactions may exhibit ontogenetic or strong seasonal differences and may be facultative or obligate. Recent geochemical studies have used high-resolution single-specimen and even single-chamber analyses to investigate both the biological and seasonal influences on test geochemistry throughout the lifetime of calcareous foraminifera (e.g. Spindler and Dieckmann, 1986; Takagi et al., 2015, 2016; Lougheed et al., 2018; Pracht et al., 2019; Metcalfe et al., 2019). Single test analysis of δ18O isotope values has identified two distinct populations of morphologically identical N. pachyderma populations in the North Atlantic during the last deglacial period. Isotope values indicate a temperature difference of about 4 °C, potentially due to a bi-modal seasonal population with peak abundances separated temporally in late spring/early summer and late summer (Brummer et al., 2020). Spatial difference in the assemblage water depth, driven by low salinity meltwater (Brummer et al., 2020), may also contribute towards these seasonal differences. Because potential kleptoplasty (this study) could occur seasonally, obligately or facultatively, in N. pachyderma Type I, it is imperative to understand if the stored chloroplasts photosynthesise or not because photosynthesis is known to influence δ18O values (e.g. Spero and Lea, 1993; Bemis et al., 1998).
The N. pachyderma Type I microbiome consisted of a range of bacterial ASVs that are significantly different from those of the water column. The genera profile and the putative metabolic pathways identified imply that the likely source of the bacterial ASVs in the foraminiferal microbiome derive from the phycosphere of their diatom food source or from the diatom-derived POM. Although N. pachyderma Type I clearly utilises diatom-associated bacteria as a food source, it is most likely passively consumed during ingestion and digestion of the diatom prey and diatom-derived POM. Neogloboquadrina pachyderma Type I also retains large numbers of intact diatom chloroplasts in its cytoplasm. While our TEM images cannot account for the degradation state of the chloroplasts, they appear commensurate with TEM images derived from kleptoplastic benthic foraminifera. 16S metabarcoding data suggest that most of the chloroplasts inside N. pachyderma Type I across Baffin Bay during the summer of 2017 derived from the diatom Chaetoceros gelidus and that these comprised the majority component of the core microbiome at that time. Work still remains to understand the relationship between the chloroplasts and the foraminiferal “host” to determine the chloroplast role in the nutrition of N. pachyderma Type I, be it photosynthesis, a facultative nutrient-rich overwintering store, or simply a build-up of chloroplasts due to gorging on diatoms. Improving our understanding of the biology, ecology, and seasonal microbial interactions of Arctic N. pachyderma is essential to disentangle the palaeoproxies for this species and develop an understanding of its susceptibility/adaptability to climate change in the rapidly contracting Arctic biome.
Table A1Results of PERMANOVA (Adonis2, Vegan) and tests for the homogeneity of multivariate dispersions on robust Aitchison distance matrices of foraminifera and water column samples. Dataset FW includes all foraminifera and all water samples from Stations 101, 115, and 323. Dataset 101 includes all water column and foraminiferal samples from Station 101. Dataset F3 includes all foraminifera from Stations 101, 115, and 323.
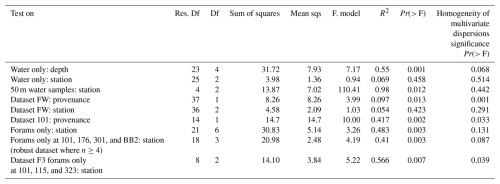
Table A2The ASVs demonstrating significantly higher differential abundance in the foraminiferal samples compared to the water column samples based on DESeq2 analysis of the FW and 101 datasets (see Sect. 3.2.3). The contribution of the bacterial ASVs to differentially abundant pathways identified in PICRUSt2 (Sect. 3.2.4) is also shown. Note that NA represents not available.
Table A3The taxonomic assignment, log2 fold change, and abundance characteristics of the eight ASVs that make up the foraminiferal core microbiome.
Figure A1Alpha rarefaction curves of all foraminiferal and water column samples (Shannon, Observed Features, and Faith PD). Each line represents a single sample. The levelling out of the curves demonstrates that the sequencing depth is adequate for all samples.
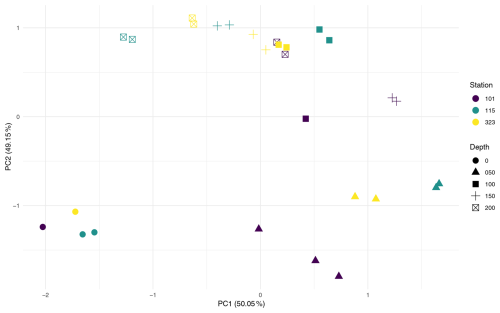
Figure A2Aitchison dissimilarity PCA plot of water samples from different depths at three stations. Colours represent the station, and shapes represent the depth. Depth drives 55 % of the variability (Pr = 0.001) compared to station, which drives 6.9 % of the variability (Pr = 0.458, Adonis2, Vegan).
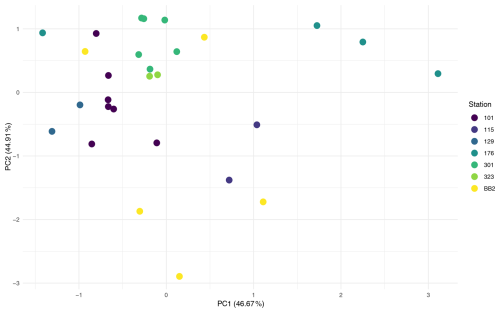
Figure A3Aitchison dissimilarity PCA ordination of foraminiferal samples across all stations. There is a degree of clustering of foraminifera by station, with 48.3 % of the variation in the foraminifera driven by station (Pr = 0.003; Adonis2, Vegan).
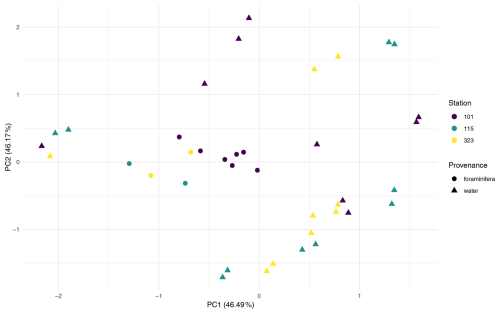
Figure A4Aitchison dissimilarity PCA ordination of the foraminiferal and water column samples (dataset FW). Provenance drives 9.7 % of the variation, and the microbial assemblage between provenances is statistically different (Pr = 0.013; Adonis2, Vegan).
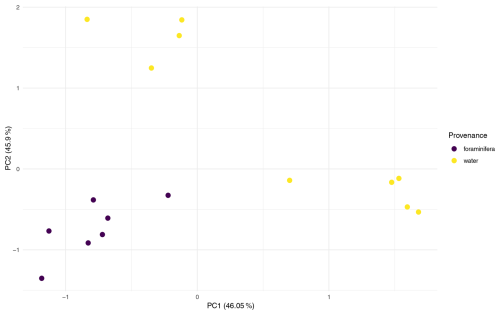
Figure A5Aitchison dissimilarity PCA plot of water column (yellow) and foraminiferal samples (purple) from Station 101. Of the differences in ASV composition, 41.7 % are driven by provenance (Pr = 0.002; Adonis2, Vegan).
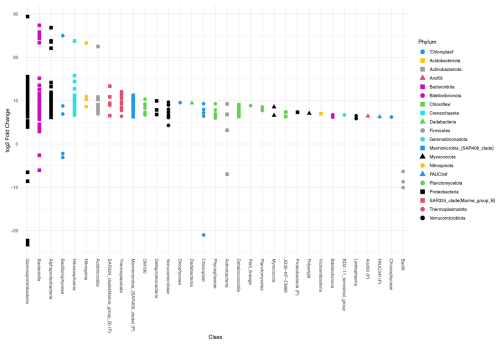
Figure A6Differential abundance testing of ASVs between provenances at Station 101 using DESeq2. The log2 fold change in ASVs is the log-ratio of the ASV means in the water column and foraminifera. ASVs with positive log2 fold change are significantly more abundant in the water column assemblage, and ASVs with negative values indicate ASVs that are significantly more abundant in the foraminiferal assemblages. The class, or the highest level of taxonomic assignment available for each ASV, is given on the x axis. (P) = phylum.
The raw 16S metabarcoding dataset was submitted to the NCBI Sequencing Read Archive (BioProject accession PRJNA984332). N. pachyderma 18S sequences can be found at NCBI, GenBank Accession numbers OR137988–OR138014. The environmental data, pairwise PERMANOVA summaries, ASV table, and sequence files used for this analysis can be found on Figshare (https://doi.org/10.6084/m9.figshare.28915598, Bird, 2025). All code used was standard scripts for each package utilised.
CB contributed to the conception and design of the work, gained funding to carry out the molecular work, and wrote the paper. KD contributed significantly to the writing of the paper and conception of the work. AJP made a substantial contribution to the conception of the work, the acquisition of funding for sample collection, collected samples in 2017, and contributed to the editing of the paper. RT collected samples in 2018 and contributed to the editing of the paper.
The contact author has declared that none of the authors has any competing interests.
The views expressed in this publication do not necessarily represent the views of Amundsen Science or that of its partners.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
The authors wish to acknowledge the work of Steve Mitchell at the University of Edinburgh TEM facility for substantial contributions to sample processing and TEM analysis. Some of the data presented herein were collected by the Canadian research icebreaker CCGS Amundsen and made available by the Amundsen Science programme, which was supported by the Canada Foundation for Innovation and the Natural Sciences and Engineering Research Council of Canada. Transmission electron microscopy costs were covered by the Marine Alliance for Science and Technology for Scotland (MASTS) small grant scheme.
This research has been supported by the Natural Sciences and Engineering Research Council of Canada (grant no. RGPIN-2016-05457) and the Carnegie Trust for the Universities of Scotland (grant no. RIG008299).
This paper was edited by Cindy De Jonge and reviewed by two anonymous referees.
Allan, E., Douglas, P. M. J., Vernal, A. de, Gélinas, Y., and Mucci, A. O.: Palmitic Acid Is Not a Proper Salinity Proxy in Baffin Bay and the Labrador Sea but Reflects the Variability in Organic Matter Sources Modulated by Sea Ice Coverage, Geochem. Geophy. Geosy., 24, e2022GC010837, https://doi.org/10.1029/2022gc010837, 2023.
Alldredge, A. L. and Cohen, Y.: Can Microscale Chemical Patches Persist in the Sea? Microelectrode Study of Marine Snow, Fecal Pellets, Science, 235, 689–691, https://doi.org/10.1126/science.235.4789.689, 1987.
Altuna, N. E. B., Pieńkowski, A. J., Eynaud, F., and Thiessen, R.: The morphotypes of Neogloboquadrina pachyderma: Isotopic signature and distribution patterns in the Canadian Arctic Archipelago and adjacent regions, Mar. Micropaleontol., 142, 13–24, https://doi.org/10.1016/j.marmicro.2018.05.004, 2018.
Amin, S. A., Parker, M. S., and Armbrust, V. E.: Interactions between Diatoms and Bacteria, Microbiol. Mol. Biol. R., 76, 667–684, https://doi.org/10.1128/mmbr.00007-12, 2012.
Amundsen Science Data Collection: CTD data collected by the CCGS Amundsen in the Canadian Arctic. ArcticNet Inc., Quebec, Canada, Processed data, Version 1, Canadian Cryospheric Information Network (CCIN), Waterloo, Canada, https://doi.org/10.5884/12713, 2018.
Anand, P., Elderfield, H., and Conte, M. H.: Calibration of Mg Ca thermometry in planktonic foraminifera from a sediment trap time series, Paleoceanography, 18, 1050, https://doi.org/10.1029/2002pa000846, 2003.
Apprill, A., McNally, S., Parsons, R., and Weber, L.: Minor revision to V4 region SSU rDNA 806R gene primer greatly increases detection of SAR11 bacterioplankton, Aquat. Microb. Ecol., 75, 129–137, https://doi.org/10.3354/ame01753, 2015.
Arandia-Gorostidi, N., Berthelot, H., Calabrese, F., Stryhanyuk, H., Klawonn, I., Iversen, M., Nahar, N., Grossart, H.-P., Ploug, H., and Musat, N.: Efficient carbon and nitrogen transfer from marine diatom aggregates to colonizing bacterial groups, Sci. Rep., 12, 14949, https://doi.org/10.1038/s41598-022-18915-0, 2022.
Barbera, P., Kozlov, A. M., Czech, L., Morel, B., Darriba, D., Flouri, T., and Stamatakis, A.: EPA-ng: Massively Parallel Evolutionary Placement of Genetic Sequences, Syst. Biol., 68, 365–369, https://doi.org/10.1093/sysbio/syy054, 2019.
Bé, A. W. H.: An ecological, zoogeographic and taxonomic review of recent planktonic foraminifera., in: Oceanographic Micropaleontology, vol. 1, edited by: Ramsay, A. T. S., Academic Press, London, 1–100, ISBN 0125773013, 1977.
Bé, A. W. H. and Tolderlund, D.: Distribution and ecology of living planktonic foraminifera in surface waters of the Atlantic and Indian Oceans, in: The Micropalaeontology of Oceans, edited by: Funnell, B. M. and Riedel, W. R., Cambridge University Press, New York, 105–149, https://doi.org/10.1017/S0016756800042321, 1971.
Bedoshvili, Ye. D., Popkova, T. P., and Likhoshway, Ye. V.: Chloroplast structure of diatoms of different classes, Cell Tissue Biol., 3, 297–310, https://doi.org/10.1134/s1990519x09030122, 2009.
Bell, W. and Mitchell, R.: Chemotactic and growth responses of marine bacteria to algal extracellular products, Biol. Bull., 134, 265–277, https://doi.org/10.2307/1540052, 1972.
Bemis, B. E., Spero, H. J., Bijma, J., and Lea, D. W.: Re-evaluation of the oxygen isotopic composition of planktonic foraminifera: Experimental results and revised paleotemperature equations, Paleoceanography, 13, 150–160, https://doi.org/10.1029/98pa00070, 1998.
Bemis, B., Spero, H., and Thunell, R.: Using species-specific paleotemperature equations with foraminifera: a case study in the Southern California Bight, Mar. Micropaleontol., 46, 405–430, https://doi.org/10.1016/S0377-8398(02)00083-X, 2002.
Bernhard, J. M. and Bowser, S. S.: Benthic foraminifera of dysoxic sediments: chloroplast sequestration and functional morphology, Earth-Sci. Rev., 46, 149–165, https://doi.org/10.1016/s0012-8252(99)00017-3, 1999.
Berge, J., Daase, M., Renaud, P. E., Ambrose, W. G., Darnis, G., Last, K. S., Leu, E., Cohen, J. H., Johnsen, G., Moline, M. A., Cottier, F., Varpe, Ø., Shunatova, N., Bałazy, P., Morata, N., Massabuau, J.-C., Falk-Petersen, S., Kosobokova, K., Hoppe, C. J. M., Węsławski, J. M., Kukliński, P., Legeżyńska, J., Nikishina, D., Cusa, M., Kędra, M., Włodarska-Kowalczuk, M., Vogedes, D., Camus, L., Tran, D., Michaud, E., Gabrielsen, T. M., Granovitch, A., Gonchar, A., Krapp, R., and Callesen, T. A.: Unexpected Levels of Biological Activity during the Polar Night Offer New Perspectives on a Warming Arctic, Curr. Biol., 25, 2555–2561, https://doi.org/10.1016/j.cub.2015.08.024, 2015a.
Berge, J., Renaud, P. E., Darnis, G., Cottier, F., Last, K., Gabrielsen, T. M., Johnsen, G., Seuthe, L., Weslawski, J. M., Leu, E., Moline, M., Nahrgang, J., Søreide, J. E., Varpe, Ø., Lønne, O. J., Daase, M., and Falk-Petersen, S.: In the dark: A review of ecosystem processes during the Arctic polar night, Prog. Oceanogr., 139, 258–271, https://doi.org/10.1016/j.pocean.2015.08.005, 2015b.
Bergeron, M. and Tremblay, J.: Shifts in biological productivity inferred from nutrient drawdown in the southern Beaufort Sea (2003–2011) and northern Baffin Bay (1997–2011), Canadian Arctic, Geophys. Res. Lett., 41, 3979–3987, https://doi.org/10.1002/2014gl059649, 2014.
Beutler, B. and Rehli, M.: Toll-Like Receptor Family Members and Their Ligands, Curr. Top Microbiol., 270, 1–21, https://doi.org/10.1007/978-3-642-59430-4_1, 2002.
Bird, C.: Baffin Bay foraminifera 16S metabarcoding ASV table, sequences and metadata, figshare [data set], https://doi.org/10.6084/m9.figshare.28915598, 2025.
Bird, C., Darling, K. F., Russell, A. D., Davis, C. V., Fehrenbacher, J., Free, A., Wyman, M., and Ngwenya, B. T.: Cyanobacterial endobionts within a major marine planktonic calcifier (Globigerina bulloides, Foraminifera) revealed by 16S rRNA metabarcoding, Biogeosciences, 14, 901–920, https://doi.org/10.5194/bg-14-901-2017, 2017.
Bird, C., Darling, K. F., Russell, A. D., Fehrenbacher, J. S., Davis, C. V., Free, A., and Ngwenya, B. T.: 16S rRNA gene metabarcoding and TEM reveals different ecological strategies within the genus Neogloboquadrina (planktonic foraminifer), PLOS ONE, 13, e0191653, https://doi.org/10.1371/journal.pone.0191653, 2018.
Bobik, K. and Burch-Smith, T. M.: Chloroplast signaling within, between and beyond cells, Front Plant Sci., 6, 781, https://doi.org/10.3389/fpls.2015.00781, 2015.
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., Alexander, H., Alm, E. J., Arumugam, M., Asnicar, F., Bai, Y., Bisanz, J. E., Bittinger, K., Brejnrod, A., Brislawn, C. J., Brown, C. T., Callahan, B. J., Caraballo-Rodríguez, A. M., Chase, J., Cope, E. K., Silva, R. D., Diener, C., Dorrestein, P. C., Douglas, G. M., Durall, D. M., Duvallet, C., Edwardson, C. F., Ernst, M., Estaki, M., Fouquier, J., Gauglitz, J. M., Gibbons, S. M., Gibson, D. L., Gonzalez, A., Gorlick, K., Guo, J., Hillmann, B., Holmes, S., Holste, H., Huttenhower, C., Huttley, G. A., Janssen, S., Jarmusch, A. K., Jiang, L., Kaehler, B. D., Kang, K. B., Keefe, C. R., Keim, P., Kelley, S. T., Knights, D., Koester, I., Kosciolek, T., Kreps, J., Langille, M. G. I., Lee, J., Ley, R., Liu, Y.-X., Loftfield, E., Lozupone, C., Maher, M., Marotz, C., Martin, B. D., McDonald, D., McIver, L. J., Melnik, A. V., Metcalf, J. L., Morgan, S. C., Morton, J. T., Naimey, A. T., Navas-Molina, J. A., Nothias, L. F., Orchanian, S. B., Pearson, T., Peoples, S. L., Petras, D., Preuss, M. L., Pruesse, E., Rasmussen, L. B., Rivers, A., Robeson, M. S., Rosenthal, P., Segata, N., Shaffer, M., Shiffer, A., Sinha, R., Song, S. J., Spear, J. R., Swafford, A. D., Thompson, L. R., Torres, P. J., Trinh, P., Tripathi, A., Turnbaugh, P. J., Ul-Hasan, S., van der Hooft, J. J. J., Vargas, F., Vázquez-Baeza, Y., Vogtmann, E., von Hippel, M., Walters, W., Wan, Y., Wang, M., Warren, J., Weber, K. C., Williamson, C. H. D., Willis, A. D., Xu, Z. Z., Zaneveld, J. R., Zhang, Y., Zhu, Q., Knight, R., Caporaso, J. G.: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 37, 852–857, https://doi.org/10.1038/s41587-019-0209-9, 2019.
Booth, B. C., Larouche, P., Bélanger, S., Klein, B., Amiel, D., and Mei, Z.-P.: Dynamics of Chaetoceros socialis blooms in the North Water, Deep-Sea Res Pt. II, 49, 5003–5025, https://doi.org/10.1016/s0967-0645(02)00175-3, 2002.
Brummer, G.-J. A., Metcalfe, B., Feldmeijer, W., Prins, M. A., van 't Hoff, J., and Ganssen, G. M.: Modal shift in North Atlantic seasonality during the last deglaciation, Clim. Past, 16, 265–282, https://doi.org/10.5194/cp-16-265-2020, 2020.
Calderon, L. J. P., Potts, L. D., Gontikaki, E., Gubry-Rangin, C., Cornulier, T., Gallego, A., Anderson, J. A., and Witte, U.: Bacterial Community Response in Deep Faroe-Shetland Channel Sediments Following Hydrocarbon Entrainment With and Without Dispersant Addition, Frontiers Mar. Sci., 5, 159, https://doi.org/10.3389/fmars.2018.00159, 2018.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P.: DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 13, 581–583, https://doi.org/10.1038/nmeth.3869, 2016.
Carstens, J. and Wefer, G.: Recent distribution of planktonic foraminifera in the Nansen Basin, Arctic Ocean, Deep Sea Res, 39, S507–S524, https://doi.org/10.1016/s0198-0149(06)80018-x, 1992.
Caporaso, G. J., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., Fierer, N., and Knight, R.: Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample, P. Natl. Acad. Sci. USA, 108, 4516–4522, https://doi.org/10.1073/pnas.1000080107, 2011.
Caspi, R., Altman, T., Billington, R., Dreher, K., Foerster, H., Fulcher, C. A., Holland, T. A., Keseler, I. M., Kothari, A., Kubo, A., Krummenacker, M., Latendresse, M., Mueller, L. A., Ong, Q., Paley, S., Subhraveti, P., Weaver, D. S., Weerasinghe, D., Zhang, P., and Karp, P. D.: The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases, Nucleic Acids Res., 42, D459–D471, https://doi.org/10.1093/nar/gkt1103, 2014.
Cedhagen, T.: Retention of chloroplasts and bathymetric distribution in the Sublittoral Foraminiferan Nonionellina Labradorica, Ophelia, 33, 17–30, https://doi.org/10.1080/00785326.1991.10429739, 1991.
Cesbron, F., Geslin, E., Kieffre, C. L., Jauffrais, T., Nardelli, M. P., Langlet, D., Mabilleau, G., Jorissen, F. J., Jézéquel, D., and Metzger, E.: sequestered chloroplasts in the benthic foraminifer Haynesina germanica: cellular organization, oxygen fluxes and potential ecological implications, J. Foramin. Res., 47, 268–278, https://doi.org/10.2113/gsjfr.47.3.268, 2017.
Chamnansinp, A., Li, Y., Lundholm, N., and Moestrup, Ø.: Global diversity of two widespread, colony-forming diatoms of the marine plankton, Chaetoceros socialis (syn. C. radians) and Chaetoceros gelidus sp. nov., J. Phycol., 49, 1128–1141, https://doi.org/10.1111/jpy.12121, 2013.
Chien, Y. T. and Zinder, S. H.: Cloning, DNA sequencing, and characterization of a nifD-homologous gene from the archaeon Methanosarcina barkeri 227 which resembles nifD1 from the eubacterium Clostridium pasteurianum, J. Bacteriol., 176, 6590–6598, https://doi.org/10.1128/jb.176.21.6590-6598.1994, 1994.
Chronopoulou, P.-M., Salonen, I., Bird, C., Reichart, G.-J., and Koho, K. A.: Metabarcoding Insights into the Trophic Behavior and Identity of Intertidal Benthic Foraminifera, Front. Microbiol., 10, 1169, https://doi.org/10.3389/fmicb.2019.01169, 2019.
Clark, K. B., Jensen, K. E., and Stirts, H. M.: Survey for Functional Kleptoplasty Among West Atlantic Ascoglossa (= Sarcoglossa) (Mollusca: Opisthobranchia), Veliger, 33, 339–345, 1990.
Costanzo, F. D., Dato, V. D., and Romano, G.: Diatom–Bacteria Interactions in the Marine Environment: Complexity, Heterogeneity, and Potential for Biotechnological Applications, Microorganisms, 11, 2967, https://doi.org/10.3390/microorganisms11122967, 2023.
Crawford, D. W., Cefarelli, A. O., Wrohan, I. A., Wyatt, S. N., and Varela, D. E.: Spatial patterns in abundance, taxonomic composition and carbon biomass of nano- and microphytoplankton in Subarctic and Arctic Seas, Prog. Oceanogr., 162, 132–159, https://doi.org/10.1016/j.pocean.2018.01.006, 2018.
Cronan, J. E. and Thomas, J.: Chapter 17 Bacterial Fatty Acid Synthesis and its Relationships with Polyketide Synthetic Pathways, Methods Enzymol., 459, 395–433, https://doi.org/10.1016/s0076-6879(09)04617-5, 2009.
Daly, G., Decorosi, F., Viti, C., and Adessi, A.: Shaping the phycosphere: Analysis of the EPS in diatom-bacterial co-cultures, J. Phycol., 59, 791–797, https://doi.org/10.1111/jpy.13361, 2023.
Darling, K. F., Kucera, M., Pudsey, C. J., and Wade, C. M.: Molecular evidence links cryptic diversification in polar planktonic protists to Quaternary climate dynamics, P. Natl. Acad. Sci. USA, 101, 7657–7662, https://doi.org/10.1073/pnas.0402401101, 2004.
Darling, K. F., Kucera, M., and Wade, C. M.: Global molecular phylogeography reveals persistent Arctic circumpolar isolation in a marine planktonic protist, P. Natl. Acad. Sci. USA, 104, 5002–5007, https://doi.org/10.1073/pnas.0700520104, 2007.
Darling, K. F., Schweizer, M., Knudsen, K. L., Evans, K. M., Bird, C., Roberts, A., Filipsson, H. L., Kim, J.-H., Gudmundsson, G., Wade, C. M., Sayer, M. D. J., and Austin, W. E. N.: The genetic diversity, phylogeography and morphology of Elphidiidae (Foraminifera) in the Northeast Atlantic, Mar. Micropaleontol., 129, 1–23, https://doi.org/10.1016/j.marmicro.2016.09.001, 2016.
Darling, K. F. and Wade, C. M.: The genetic diversity of planktic foraminifera and the global distribution of ribosomal RNA genotypes, Mar. Micropaleontol., 67, 216–238, https://doi.org/10.1016/j.marmicro.2008.01.009, 2008.
Darling, K. F., Wade, C. M., Siccha, M., Trommer, G., Schulz, H., Abdolalipour, S., and Kurasawa, A.: Genetic diversity and ecology of the planktonic foraminifers Globigerina bulloides, Turborotalita quinqueloba and Neogloboquadrina pachyderma off the Oman margin during the late SW Monsoon, Mar. Micropaleontol., 137, 64–77, https://doi.org/10.1016/j.marmicro.2017.10.006, 2017.
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A., and Callahan, B. J.: Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data, Microbiome, 6, 226, https://doi.org/10.1186/s40168-018-0605-2, 2018.
Deutsch, C., Ferrel, A., Seibel, B., Pörtner, H.-O., and Huey, R. B.: Climate change tightens a metabolic constraint on marine habitats, Science, 348, 1132–1135, https://doi.org/10.1126/science.aaa1605, 2015.
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., Huttenhower, C., and Langille, M. G. I.: PICRUSt2 for prediction of metagenome functions, Nat. Biotechnol., 38, 685–688, https://doi.org/10.1038/s41587-020-0548-6, 2020.
Duplessy, J.-C., Labeyrie, L., Juillet-Leclerc, A., Maitre, F., Duprat, J., and Sarnthein, M.: Surface salinity reconstruction of the North Atlantic Ocean during the Last glacial maximum, Oceanol. Ac., 14, 311–324, 1991.
Eegeesiak, O., Aariak, E., and Kleist, K. V.: People of the Ice Bridge: The future of the Pikialasorsuaq, Report of the Pikialasorsuaq Commission, Inuit Circumpolar Council Canada, Ottawa, Canada, http://pikialasorsuaq.org/en/Resources/Reports (last access: 27 August 2025), 2017.
Eggins, S., Sadekov, A., and Deckker, D. P.: Modulation and daily banding of Mg/Ca in Orbulina universa tests by symbiont photosynthesis and respiration: a complication for seawater thermometry?, Earth Planet Sc. Lett., 225, 411–419, 2004.
Espinasse, B., Daase, M., Halvorsen, E., Reigstad, M., Berge, J., and Basedow, S. L.: Surface aggregations of Calanus finmarchicus during the polar night, Ices J. Mar. Sci., 79, 803–814, https://doi.org/10.1093/icesjms/fsac030, 2022.
Falk-Petersen, S., Pavlov, V., Berge, J., Cottier, F., Kovacs, K. M., and Lydersen, C.: At the rainbow's end: high productivity fuelled by winter upwelling along an Arctic shelf, Polar Biol., 38, 5–11, https://doi.org/10.1007/s00300-014-1482-1, 2015.
Fehrenbacher, J. S., Russell, A. D., Davis, C. V., Spero, H. J., Chu, E., and Hönisch, B.: Ba Ca ratios in the non-spinose planktic foraminifer Neogloboquadrina dutertrei: Evidence for an organic aggregate microhabitat, Geochim. Cosmochim. Ac., 236, 361–372, https://doi.org/10.1016/j.gca.2018.03.008, 2018.
Fernandes, A. D., Reid, J. N., Macklaim, J. M., McMurrough, T. A., Edgell, D. R., and Gloor, G. B.: Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis, Microbiome, 2, 15, https://doi.org/10.1186/2049-2618-2-15, 2014.
Fernández-Méndez, M., Turk-Kubo, K. A., Buttigieg, P. L., Rapp, J. Z., Krumpen, T., Zehr, J. P., and Boetius, A.: Diazotroph Diversity in the Sea Ice, Melt Ponds, and Surface Waters of the Eurasian Basin of the Central Arctic Ocean, Front. Microbiol., 7, 1884, https://doi.org/10.3389/fmicb.2016.01884, 2016.
Frisch, K., Småge, S. B., Johansen, R., Duesund, H., Brevik, Ø. J., and Nylund, A.: Pathology of experimentally induced mouthrot caused by Tenacibaculum maritimum in Atlantic salmon smolts, PLOS ONE, 13, e0206951, https://doi.org/10.1371/journal.pone.0206951, 2018.
Gaby, J. and Buckley, D. H.: A comprehensive aligned nifH gene database: a multipurpose tool for studies of nitrogen-fixing bacteria, Database, 2014, bau001, https://doi.org/10.1093/database/bau001, 2014.
Gavriilidou, A., Gutleben, J., Versluis, D., Forgiarini, F., Passel, M. W. J. van, Ingham, C. J., Smidt, H., and Sipkema, D.: Comparative genomic analysis of Flavobacteriaceae: insights into carbohydrate metabolism, gliding motility and secondary metabolite biosynthesis, BMC Genomics, 21, 569, https://doi.org/10.1186/s12864-020-06971-7, 2020.
Gedi, M. A., Briars, R., Yuseli, F., Zainol, N., Darwish, R., Salter, A. M., and Gray, D. A.: Component analysis of nutritionally rich chloroplasts: recovery from conventional and unconventional green plant species, J. Food Sci. Tech. Mys., 54, 2746–2757, https://doi.org/10.1007/s13197-017-2711-8, 2017.
Gillner, D. M., Becker, D. P., and Holz, R. C.: Lysine biosynthesis in bacteria: a metallodesuccinylase as a potential antimicrobial target, JBIC J. Biol. Inorg. Chem., 18, 155–163, https://doi.org/10.1007/s00775-012-0965-1, 2013.
Goldstein, S. T., Bernhard, J. M., and Richardson, E. A.: Chloroplast Sequestration in the Foraminifer Haynesina germanica: Application of High Pressure Freezing and Freeze Substitution, Microsc. Microanal., 10, 1458–1459, https://doi.org/10.1017/s1431927604885891, 2004.
Gomaa, F., Utter, D. R., Powers, C., Beaudoin, D. J., Edgcomb, V. P., Filipsson, H. L., Hansel, C. M., Wankel, S. D., Zhang, Y., and Bernhard, J. M.: Multiple integrated metabolic strategies allow foraminiferan protists to thrive in anoxic marine sediments, Sci. Adv., 7, eabf1586, https://doi.org/10.1126/sciadv.abf1586, 2021.
Greco, M., Jonkers, L., Kretschmer, K., Bijma, J., and Kucera, M.: Depth habitat of the planktonic foraminifera Neogloboquadrina pachyderma in the northern high latitudes explained by sea-ice and chlorophyll concentrations, Biogeosciences, 16, 3425–3437, https://doi.org/10.5194/bg-16-3425-2019, 2019.
Greco, M., Morard, R., and Kucera, M.: Single-cell metabarcoding reveals biotic interactions of the Arctic calcifier Neogloboquadrina pachyderma with the eukaryotic pelagic community, J. Plankton Res., 43, 113–125, https://doi.org/10.1093/plankt/fbab015, 2021.
Greco, M., Werner, K., Zamelczyk, K., Rasmussen, T. L., and Kucera, M.: Decadal trend of plankton community change and habitat shoaling in the Arctic gateway recorded by planktonic foraminifera, Glob. Change Biol., 28, 1798–1808, https://doi.org/10.1111/gcb.16037, 2022.
Grigoratou, M., Monteiro, F. M., Wilson, J. D., Ridgwell, A., and Schmidt, D. N.: Exploring the impact of climate change on the global distribution of non-spinose planktonic foraminifera using a trait-based ecosystem model, Glob. Change Biol., 28, 1063–1076, https://doi.org/10.1111/gcb.15964, 2022.
Gruber, N., Keeling, C. D., and Bates, N. R.: Interannual Variability in the North Atlantic Ocean Carbon Sink, Science, 298, 2374–2378, https://doi.org/10.1126/science.1077077, 2002.
Grzymski, J., Schofield, O. M., Falkowski, P. G., and Bernhard, J. M.: The function of plastids in the deep-sea benthic foraminifer, Nonionella stella, Limnol. Oceanogr., 47, 1569–1580, https://doi.org/10.4319/lo.2002.47.6.1569, 2002.
Hemleben, C., Spindler, M., and Anderson, O. R.: Modern Planktonic Foraminifera, Springer-Verlag, New York, https://doi.org/10.1007/978-1-4612-3544-6, 1989.
Hikida, M., Wakabayashi, H., Egusa, S., and Masumura, K.: Flexibacter sp., a Gliding Bacterium Pathogenic to Some Marine Fishes in Japan, B. Jpn. Soc. Sci. Fish., 45, 421–428, https://doi.org/10.2331/suisan.45.421, 1979.
Holzmann, M. and Pawlowski, J.: Preservation of Foraminifera for DNA extraction and PCR amplification, J. Foramin. Res., 26, 264–267, https://doi.org/10.2113/gsjfr.26.3.264, 1996.
Hönisch, B., Bijma, J., Russell, A. D., Spero, H. J., Palmer, M. R., Zeebe, R. E., and Eisenhauer, A.: The influence of symbiont photosynthesis on the boron isotopic composition of foraminifera shells, Mar. Micropaleontol., 49, 8796, https://doi.org/10.1016/s0377-8398(03)00030-6, 2003.
Jahn, A., Holland, M. M., and Kay, J. E.: Projections of an ice-free Arctic Ocean, Nat. Rev. Earth Environ., 5, 164–176, https://doi.org/10.1038/s43017-023-00515-9, 2024.
Jauffrais, T., Jesus, B., Metzger, E., Mouget, J.-L., Jorissen, F., and Geslin, E.: Effect of light on photosynthetic efficiency of sequestered chloroplasts in intertidal benthic foraminifera (Haynesina germanica and Ammonia tepida), Biogeosciences, 13, 2715–2726, https://doi.org/10.5194/bg-13-2715-2016, 2016.
Jauffrais, T., LeKieffre, C., Koho, K. A., Tsuchiya, M., Schweizer, M., Bernhard, J. M., Meibom, A., and Geslin, E.: Ultrastructure and distribution of kleptoplasts in benthic foraminifera from shallow-water (photic) habitats, Mar. Micropaleontol., 138, 46–62, https://doi.org/10.1016/j.marmicro.2017.10.003, 2018.
Jauffrais, T., LeKieffre, C., Schweizer, M., Jesus, B., Metzger, E., and Geslin, E.: Response of a kleptoplastidic foraminifer to heterotrophic starvation: photosynthesis and lipid droplet biogenesis, Fems Microbiol. Ecol., 95, fiz046, https://doi.org/10.1093/femsec/fiz046, 2019a.
Jauffrais, T., LeKieffre, C., Schweizer, M., Geslin, E., Metzger, E., Bernhard, J. M., Jesus, B., Filipsson, H. L., Maire, O., and Meibom, A.: Kleptoplastidic benthic foraminifera from aphotic habitats: insights into assimilation of inorganic C, N and S studied with sub-cellular resolution, Environ. Microbiol., 21, 125–141, https://doi.org/10.1111/1462-2920.14433, 2019b.
Jesus, B., Jauffrais, T., Trampe, E. C. L., Goessling, J. W., LeKieffre, C., Meibom, A., Kühl, M., and Geslin, E.: Kleptoplast distribution, photosynthetic efficiency and sequestration mechanisms in intertidal benthic foraminifera, ISME J., 16, 822–832, https://doi.org/10.1038/s41396-021-01128-0, 2022.
Johnson, M. D., Oldach, D., Delwiche, C. F., and Stoecker, D. K.: Retention of transcriptionally active cryptophyte nuclei by the ciliate Myrionecta rubra, Nature, 445, 426–428, https://doi.org/10.1038/nature05496, 2007.
Jonkers, L., Brummer, G. A., Peeters, F. J., Aken, H. M., and Jong, F. M.: Seasonal stratification, shell flux, and oxygen isotope dynamics of left coiling N. pachyderma and T. quinqueloba in the western subpolar North Atlantic, Paleoceanography, 25, PA2204, https://doi.org/10.1029/2009pa001849, 2010.
Jonkers, L., Heuven, S., Zahn, R., and Peeters, F. J. C.: Seasonal patterns of shell flux, δ18O and δ13C of small and large N. pachyderma (s) and G. bulloides in the subpolar North Atlantic, Paleoceanography, 28, 164–174, https://doi.org/10.1002/palo.20018, 2013.
Jonkers, L., Hillebrand, H., and Kucera, M.: Global change drives modern plankton communities away from the pre-industrial state, Nature, 570, 372–375, https://doi.org/10.1038/s41586-019-1230-3, 2019.
Jonkers, L. and Kučera, M.: Global analysis of seasonality in the shell flux of extant planktonic Foraminifera, Biogeosciences, 12, 2207–2226, https://doi.org/10.5194/bg-12-2207-2015, 2015.
Kim, Y.-H., Min, S.-K., Gillett, N. P., Notz, D., and Malinina, E.: Observationally constrained projections of an ice-free Arctic even under a low emission scenario, Nat. Commun., 14, 3139, https://doi.org/10.1038/s41467-023-38511-8, 2023.
Kohfeld, K. E., Fairbanks, R. G., Smith, S. L., and Walsh, I. D.: Neogloboquadrina pachyderma (sinistral coiling) as paleoceanographic tracers in polar oceans: Evidence from northeast water polynya plankton tows, sediment traps, and surface sediments, Paleoceanography, 11, 679–699, https://doi.org/10.1029/96pa02617, 1996.
Kretschmer, K., Kucera, M., and Schulz, M.: Modelling the distribution and seasonality of Neogloboquadrina pachyderma in the North Atlantic Ocean during Heinrich Stadial 1, Paleoceanography, 31, 986–1010, https://doi.org/10.1002/2015pa002819, 2016.
Lahti, L. and Shetty, S.: Tools for microbiome analysis in R, Microbiome package version 1.7.21 R/Bioconductor, https://bioconductor.org/packages/release/bioc/html/microbiome.html (last access: 12 October 2024), 2017.
Lechliter, S.: Preliminary Study of Kleptoplasty in Foraminifera of South Carolina, Bridges, 8, 44–54, https://digitalcommons.coastal.edu/bridges/vol8/iss8/4 (last access: 26 August 2024), 2014.
Lee, J. J.: The Diatom World -Diatoms as endosymbionts, Cell Origin Life Ext., 19, 437–464, https://doi.org/10.1007/978-94-007-1327-7_20, 2011.
Lee, J. J., Lanners, E., and Kuile, B. T.: The retention of chloroplasts by the foraminifera Elphidium cripsum, Symbiosis, 5, 45–60, 1988.
Lee, J. J., Morales, J., Symons, A., and Hallock, P.: Diatom symbionts in larger foraminifera from Caribbean hosts, Mar. Micropaleontol., 26, 99–105, https://doi.org/10.1016/0377-8398(95)00004-6, 1995.
LeKieffre, C., Bernhard, J. M., Mabilleau, G., Filipsson, H. L., Meibom, A., and Geslin, E.: An overview of cellular ultrastructure in benthic foraminifera: New observations of rotalid species in the context of existing literature, Mar. Micropaleontol., 138, 12–32, https://doi.org/10.1016/j.marmicro.2017.10.005, 2018.
Li, Y., Sun, L.-L., Sun, M.-L., Su, H.-N., Zhang, X.-Y., Xie, B.-B., Chen, X.-L., Zhang, Y.-Z., and Qin, Q.-L.: Vertical and horizontal biogeographic patterns and major factors affecting bacterial communities in the open South China Sea, Sci. Rep., 8, 8800, https://doi.org/10.1038/s41598-018-27191-w, 2018.
Livsey, C. M., Kozdon, R., Bauch, D., Brummer, G. A., Jonkers, L., Orland, I., Hill, T. M., and Spero, H. J.: High-Resolution Mg Ca and δ18O Patterns in Modern Neogloboquadrina pachyderma From the Fram Strait and Irminger Sea, Paleoceanography and Paleoclimatology, 35, e2020PA003969, https://doi.org/10.1029/2020pa003969, 2020.
Lopez, E.: Algal chloroplasts in the protoplasm of three species of benthic foraminifera: taxonomic affinity, viability and persistence, Mar. Biol., 53, 201–211, https://doi.org/10.1007/bf00952427, 1979.
Lougheed, B. C., Metcalfe, B., Ninnemann, U. S., and Wacker, L.: Moving beyond the age–depth model paradigm in deep-sea palaeoclimate archives: dual radiocarbon and stable isotope analysis on single foraminifera, Clim. Past, 14, 515–526, https://doi.org/10.5194/cp-14-515-2018, 2018.
Love, M. I., Huber, W., and Anders, S.: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 15, 550, https://doi.org/10.1186/s13059-014-0550-8, 2014.
Mandal, S., Treuren, W. V., White, R. A., Eggesbø, M., Knight, R., and Peddada, S. D.: Analysis of composition of microbiomes: a novel method for studying microbial composition, Microb. Ecol. Health D, 26, 27663, https://doi.org/10.3402/mehd.v26.27663, 2015.
Manno, C., Morata, N., and Bellerby, R.: Effect of ocean acidification and temperature increase on the planktonic foraminifer Neogloboquadrina pachyderma (sinistral), Polar Biol., 35, 1311–1319, https://doi.org/10.1007/s00300-012-1174-7, 2012.
Manno, C. and Pavlov, A. K.: Living planktonic foraminifera in the Fram Strait (Arctic): absence of diel vertical migration during the midnight sun, Hydrobiologia, 721, 285–295, https://doi.org/10.1007/s10750-013-1669-4, 2014.
Martinez Arbizu, P.: pairwiseAdonis: Pairwise multilevel comparison using adonis, R package version 0.4, https://github.com/pmartinezarbizu/pairwiseAdonis (last access: 16 May 2025), 2020.
Martino, C., Morton, J. T., Marotz, C. A., Thompson, L. R., Tripathi, A., Knight, R., and Zengler, K.: A Novel Sparse Compositional Technique Reveals Microbial Perturbations, mSystems, 4, e00016-19, https://doi.org/10.1128/msystems.00016-19, 2019.
Martino, C., Shenhav, L., Marotz, C. A., Armstrong, G., McDonald, D., Vázquez-Baeza, Y., Morton, J. T., Jiang, L., Dominguez-Bello, M. G., Swafford, A. D., Halperin, E., and Knight, R.: Context-aware dimensionality reduction deconvolutes gut microbial community dynamics, Nat. Biotechnol., 39, 165–168, https://doi.org/10.1038/s41587-020-0660-7, 2021.
McMurdie, P. J. and Holmes, S.: phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data, PLOS ONE, 8, e61217, https://doi.org/10.1371/journal.pone.0061217, 2013.
Meilland, J., Ezat, M. M., Westgård, A., Manno, C., Morard, R., Siccha, M., and Kucera, M.: Rare but persistent asexual reproduction explains the success of planktonic foraminifera in polar oceans, J. Plankton. Res., 45, 15–32, https://doi.org/10.1093/plankt/fbac069, 2022.
Meilland, J., Siccha, M., Morard, R., and Kucera, M.: Continuous reproduction of planktonic foraminifera in laboratory culture, J. Eukaryot. Microbiol., 71, e13022, https://doi.org/10.1111/jeu.13022, 2024.
Melling, H., Gratton, Y., and Ingram, G.: Ocean circulation within the North Water polynya of Baffin Bay, Atmos.-Ocean, 39, 301–325, https://doi.org/10.1080/07055900.2001.9649683, 2001.
Meredith, M., Sommerkorn, M., Cassotta, S., Derksen, C., Ekaykin, A., Hollowed, A., Kofinas, G., Macintosh, A., Melbourne-Thomas, J., Muelbert, M. M. C., Ottersen, G., Pritchard, H., and Schuur, E. A. G.: Polar Regions: IPCC Special Report on the Ocean and Cryosphere in a Changing Climate, Cambridge University Press, Cambridge, UK, https://www.ipcc.ch/srocc/chapter/chapter-3-2/ (last access: 24 May 2024), 2019.
Metcalfe, B., Feldmeijer, W., and Ganssen, G. M.: Oxygen Isotope Variability of Planktonic Foraminifera Provide Clues to Past Upper Ocean Seasonal Variability, Paleoceanography Paleoclimatology, 34, 374–393, https://doi.org/10.1029/2018pa003475, 2019.
Morard, R., Darling, K. F., Weiner, A. K. M., Hassenrück, C., Vanni, C., Cordier, T., Henry, N., Greco, M., Vollmar, N. M., Milivojevic, T., Rahman, S. N., Siccha, M., Meilland, J., Jonkers, L., Quillévéré, F., Escarguel, G., Douady, C. J., Garidel-Thoron, T., Vargas, C., and Kucera, M.: The global genetic diversity of planktonic foraminifera reveals the structure of cryptic speciation in plankton, Biol. Rev., 99, 1218–1241, https://doi.org/10.1111/brv.13065, 2024.
Oksanen, J., Simpson, G., Blanchet, F., Kindt, R., Legendre, P., Minchin, P., O'Hara, R., Solymos, P., Stevens, M., Szoecs, E., Wagner, H., Barbou,r M., Bedward, M., Bolker, B., Borcard, D., Carvalho, G., Chirico, M., De Caceres, M., Durand, S., Evangelista, H., FitzJohn, R., Friendly, M., Furneaux, B., Hannigan, G., Hill, M., Lahti, L., McGlinn, D., Ouellette, M., Ribeiro Cunha, E., Smith, T., Stier, A., Ter Braak, C., Weedon, J., and Borman, T.: vegan: Community Ecology Package, R package version 2.6-10, https://CRAN.R-project.org/package=vegan (last access: 16 May 2025), 2025.
Parada, A. E., Needham, D. M., and Fuhrman, J. A.: Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples, Environ. Microbiol., 18, 1403–1414, https://doi.org/10.1111/1462-2920.13023, 2016.
Pados, T. and Spielhagen, R. F.: Species distribution and depth habitat of recent planktic foraminifera in Fram Strait, Arctic Ocean, Polar Res., 33, 22483, https://doi.org/10.3402/polar.v33.22483, 2014.
Padua, R., Parrado, A., Larghero, J., and Chomienne, C.: UV and clean air result in contamination-free PCR, Leukemia, 13, 1898–1899, https://doi.org/10.1038/sj.leu.2401579, 1999.
Pracht, H., Metcalfe, B., and Peeters, F. J. C.: Oxygen isotope composition of the final chamber of planktic foraminifera provides evidence of vertical migration and depth-integrated growth, Biogeosciences, 16, 643–661, https://doi.org/10.5194/bg-16-643-2019, 2019.
Pillet, L., de Vargas, C., and Pawlowski, J.: Molecular Identification of Sequestered Diatom Chloroplasts and Kleptoplastidy in Foraminifera, Protist, 162, 394–404, https://doi.org/10.1016/j.protis.2010.10.001, 2011.
Pinko, D., Abramovich, S., Rahav, E., Belkin, N., Rubin-Blum, M., Kucera, M., Morard, R., Holzmann, M., and Abdu, U.: Shared ancestry of algal symbiosis and chloroplast sequestration in foraminifera, Sci. Adv., 9, eadi3401, https://doi.org/10.1126/sciadv.adi3401, 2023.
Poloczanska, E. S., Burrows, M. T., Brown, C. J., Molinos, J. G., Halpern, B. S., Hoegh-Guldberg, O., Kappel, C. V., Moore, P. J., Richardson, A. J., Schoeman, D. S., and Sydeman, W. J.: Responses of Marine Organisms to Climate Change across Oceans, Front. Mar. Sci., 3, 62, https://doi.org/10.3389/fmars.2016.00062, 2016.
Powers, C., Gomaa, F., Billings, E. B., Utter, D. R., Beaudoin, D. J., Edgcomb, V. P., Hansel, C. M., Wankel, S. D., Filipsson, H. L., Zhang, Y., and Bernhard, J. M.: Two canonically aerobic foraminifera express distinct peroxisomal and mitochondrial metabolisms, Front. Mar. Sci., 9, 1010319, https://doi.org/10.3389/fmars.2022.1010319, 2022.
Qu, L., She, P., Wang, Y., Liu, F., Zhang, D., Chen, L., Luo, Z., Xu, H., Qi, Y., and Wu, Y.: Effects of norspermidine on Pseudomonas aeruginosa biofilm formation and eradication, Microbiology Open, 5, 402–412, https://doi.org/10.1002/mbo3.338, 2016.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glöckner, F. O.: The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic. Acids Res., 41, D590–D596, https://doi.org/10.1093/nar/gks1219, 2013.
Randelhoff, A., Lacour, L., Marec, C., Leymarie, E., Lagunas, J., Xing, X., Darnis, G., Penkerc'h, C., Sampei, M., Fortier, L., D'Ortenzio, F., Claustre, H., and Babin, M.: Arctic mid-winter phytoplankton growth revealed by autonomous profilers, Sci. Adv., 6, eabc2678, https://doi.org/10.1126/sciadv.abc2678, 2020.
R Core Team: R: A language and environment for statistical computing. R Foundation for statistical computing, Vienna, Austria, https://www.r-project.org/ (last access: 1 March 2025), 2017.
Reji, L., Tolar, B. B., Chavez, F. P., and Francis, C. A.: Depth-Differentiation and Seasonality of Planktonic Microbial Assemblages in the Monterey Bay Upwelling System, Front. Microbiol., 11, 1075, https://doi.org/10.3389/fmicb.2020.01075, 2020.
Ribeiro, C. G., Santos, A. L. dos, Gourvil, P., Gall, F. L., Marie, D., Tragin, M., Probert, I., and Vaulot, D.: Culturable diversity of Arctic phytoplankton during pack ice melting, Elem. Sci. Anth., 8, 6, https://doi.org/10.1525/elementa.401, 2020.
Rink, S., Kühl, M., Bijma, J., and Spero, H. J.: Microsensor studies of photosynthesis and respiration in the symbiotic foraminifer Orbulina universa, Mar. Biol., 131, 583–595, https://doi.org/10.1007/s002270050350, 1998.
Roy, T., Lombard, F., Bopp, L., and Gehlen, M.: Projected impacts of climate change and ocean acidification on the global biogeography of planktonic Foraminifera, Biogeosciences, 12, 2873–2889, https://doi.org/10.5194/bg-12-2873-2015, 2015.
Russell, A. D., Hönisch, B., Spero, H. J., and Lea, D. W.: Effects of seawater carbonate ion concentration and temperature on shell U, Mg, and Sr in cultured planktonic foraminifera, Geochim. Cosmochim. Ac., 68, 4347–4361, https://doi.org/10.1016/j.gca.2004.03.013, 2004.
Salonen, I. S., Chronopoulou, P.-M., Nomaki, H., Langlet, D., Tsuchiya, M., and Koho, K. A.: 16S rRNA Gene Metabarcoding Indicates Species-Characteristic Microbiomes in Deep-Sea Benthic Foraminifera, Front. Microbiol., 12, 694406, https://doi.org/10.3389/fmicb.2021.694406, 2021.
Sanderson, B. G. and LeBlond, P. H.: The cross-channel flow at the entrance of Lancaster Sound, Atmos.-Ocean, 22, 484–497, https://doi.org/10.1080/07055900.1984.9649211, 1984.
Schiebel, R. and Hemleben, C.: Planktic Foraminifers in the Modern Ocean, First Edition, Springer-Verlag, Berlin, Heidelberg, https://doi.org/10.1007/978-3-662-50297-6, 2017.
Schlitzer, R.: Ocean Data View, http://odv.awi.de (last access: 1 April 2022), 2022.
Schmidt, C., Morard, R., Romero, O., and Kucera, M.: Diverse Internal Symbiont Community in the Endosymbiotic Foraminifera Pararotalia calcariformata: Implications for Symbiont Shuffling Under Thermal Stress, Front. Microbiol., 9, 2018, https://doi.org/10.3389/fmicb.2018.02018, 2018.
Simstich, J., Sarnthein, M., and Erlenkeuser, H.: Paired δ18O signals of Neogloboquadrina pachyderma (s) and Turborotalita quinqueloba show thermal stratification structure in Nordic Seas, Mar Micropaleontol, 48, 107–125, https://doi.org/10.1016/s0377-8398(02)00165-2, 2003.
Sobocińska, J., Roszczenko-Jasińska, P., Ciesielska, A., and Kwiatkowska, K.: Protein Palmitoylation and Its Role in Bacterial and Viral Infections, Front. Immunol., 8, 2003, https://doi.org/10.3389/fimmu.2017.02003, 2018.
Spero, H. J., Lerche, I., and Williams, D. F.: Opening the carbon isotope “vital effect” black box, 2, Quantitative model for interpreting foraminiferal carbon isotope data, Paleoceanography, 6, 639–655, https://doi.org/10.1029/91pa02022, 1991.
Spero, H. J. and Lea, D. W.: Intraspecific stable isotope variability in the planktic foraminifera Globigerinoides sacculifer: Results from laboratory experiments, Mar. Micropaleontol., 22, 221–234, https://doi.org/10.1016/0377-8398(93)90045-y, 1993.
Spindler, M. and Dieckmann, G.: Distribution and abundance of the planktic foraminifer Neogloboquadrina pachyderma in sea ice of the Weddell Sea (Antarctica), Polar Biol., 5, 185–191, https://doi.org/10.1007/bf00441699, 1986.
Spindler, M., Hemleben, C., Salomons, J., and Smit, L.: Feeding behavior of some planktonic foraminifers in laboratory cultures, J. Foramin. Res., 14, 237–249, https://doi.org/10.2113/gsjfr.14.4.237, 1984.
Spring, S., Scheuner, C., Göker, M., and Klenk, H.-P.: A taxonomic framework for emerging groups of ecologically important marine gammaproteobacteria based on the reconstruction of evolutionary relationships using genome-scale data, Front. Microbiol., 6, 281, https://doi.org/10.3389/fmicb.2015.00281, 2015.
Stoecker, D., Johnson, M., deVargas, C., and Not, F.: Acquired phototrophy in aquatic protists, Aquat. Microb. Ecol., 57, 279–310, https://doi.org/10.3354/ame01340, 2009.
Takagi, H., Moriya, K., Ishimura, T., Suzuki, A., Kawahata, H., and Hirano, H.: Exploring photosymbiotic ecology of planktic foraminifers from chamber-by-chamber isotopic history of individual foraminifers, Paleobiology, 41, 108–121, https://doi.org/10.1017/pab.2014.7, 2015.
Takagi, H., Moriya, K., Ishimura, T., Suzuki, A., Kawahata, H., and Hirano, H.: Individual Migration Pathways of Modern Planktic Foraminifers: Chamber-by-Chamber Assessment of Stable Isotopes, Paleontol. Res., 20, 268–284, https://doi.org/10.2517/2015pr036, 2016.
Takagi, H., Kimoto, K., Fujiki, T., Saito, H., Schmidt, C., Kucera, M., and Moriya, K.: Characterizing photosymbiosis in modern planktonic foraminifera, Biogeosciences, 16, 3377–3396, https://doi.org/10.5194/bg-16-3377-2019, 2019.
Takano, Y., Yamaguchi, H., Inouye, I., Moestrup, Ø., and Horiguchi, T.: Phylogeny of Five Species of Nusuttodinium gen. nov. (Dinophyceae), a Genus of Unarmoured Kleptoplastidic Dinoflagellates, Protist, 165, 759–778, https://doi.org/10.1016/j.protis.2014.09.001, 2014.
Tisserand, L., Dadaglio, L., Intertaglia, L., Catala, P., Panagiotopoulos, C., Obernosterer, I., and Joux, F.: Use of organic exudates from two polar diatoms by bacterial isolates from the Arctic Ocean, Philos. T. R. Soc. A, 378, 20190356, https://doi.org/10.1098/rsta.2019.0356, 2020.
Tolderlund, D. S. and Bé, A. W. H.: Seasonal Distribution of Planktonic Foraminifera in the Western North Atlantic, Micropaleontology, 17, 297, https://doi.org/10.2307/1485143, 1971.
Tremblay, J., Gratton, Y., Carmack, E. C., Payne, C. D., and Price, N. M.: Impact of the large-scale Arctic circulation and the North Water Polynya on nutrient inventories in Baffin Bay, J. Geophys. Res.-Oceans, 107, 26-1–26-14, https://doi.org/10.1029/2000jc000595, 2002.
Tremblay, J.-É., Hattori, H., Michel, C., Ringuette, M., Mei, Z.-P., Lovejoy, C., Fortier, L., Hobson, K. A., Amiel, D., and Cochran, K.: Trophic structure and pathways of biogenic carbon flow in the eastern North Water Polynya, Prog. Oceanogr., 71, 402–425, https://doi.org/10.1016/j.pocean.2006.10.006, 2006.
Trubovitz, S., Lazarus, D., Renaudie, J., and Noble, P. J.: Marine plankton show threshold extinction response to Neogene climate change, Nat. Commun., 11, 5069, https://doi.org/10.1038/s41467-020-18879-7, 2020.
Tsuchiya, M., Chikaraishi, Y., Nomaki, H., Sasaki, Y., Tame, A., Uematsu, K., and Ohkouchi, N.: Compound-specific isotope analysis of benthic foraminifer amino acids suggests microhabitat variability in rocky-shore environments, Ecol. Evol., 8, 8380–8395, https://doi.org/10.1002/ece3.4358, 2018.
Tsuchiya, M., Miyawaki, S., Oguri, K., Toyofuku, T., Tame, A., Uematsu, K., Takeda, K., Sakai, Y., Miyake, H., and Maruyama, T.: Acquisition, Maintenance, and Ecological Roles of Kleptoplasts in Planoglabratella opercularis (Foraminifera, Rhizaria), Front. Mar. Sci., 7, 585, https://doi.org/10.3389/fmars.2020.00585, 2020.
van den Berge, K. V. den, Perraudeau, F., Soneson, C., Love, M. I., Risso, D., Vert, J.-P., Robinson, M. D., Dudoit, S., and Clement, L.: Observation weights unlock bulk RNA-seq tools for zero inflation and single-cell applications, Genome Biol., 19, 24, https://doi.org/10.1186/s13059-018-1406-4, 2018.
Vincent, R. F.: A Study of the North Water Polynya Ice Arch using Four Decades of Satellite Data, Sci. Rep., 9, 20278, https://doi.org/10.1038/s41598-019-56780-6, 2019.
Walters, W., Hyde, E., Berg-Lyons, D., and Ackermann, G.: Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys, mSystems, 22, e00009-15, https://doi.org/10.1128/msystems.00009-15, 2015.
Westgård, A., Ezat, M. M., Chalk, T. B., Chierici, M., Foster, G. L., and Meilland, J.: Large-scale culturing of Neogloboquadrina pachyderma, its growth in, and tolerance of, variable environmental conditions, J. Plankton Res., 45, 732–745, https://doi.org/10.1093/plankt/fbad034, 2023.
Weinberger, S. and Gilvarg, C.: Bacterial Distribution of the Use of Succinyl and Acetyl Blocking Groups in Diaminopimelic Acid Biosynthesis, J. Bacteriol., 101, 323–324, https://doi.org/10.1128/jb.101.1.323-324.1970, 1970.
Wickham, H.: ggplot2, Elegant Graphics for Data Analysis, Springer-Verlag, New York, https://doi.org/10.1007/978-0-387-98141-3, 2009.
Wolf-Gladrow, D. A., Riebesell, U., Burkhardt, S., and Bijma, J.: Direct effects of CO2 concentration on growth and isotopic composition of marine plankton, Tellus B, 51, 461–476, https://doi.org/10.1034/j.1600-0889.1999.00023.x, 1999.
Wotanis, C. K., Brennan, W. P., Angotti, A. D., Villa, E. A., Zayner, J. P., Mozina, A. N.: Rutkovsky, A. C., Sobe, R. C., Bond, W. G., and Karatan, E.: Relative contributions of norspermidine synthesis and signaling pathways to the regulation of Vibrio cholerae biofilm formation, PLOS ONE, 12, e0186291, https://doi.org/10.1371/journal.pone.0186291, 2017.
Yamada, N., Lepetit, B., Mann, D. G., Sprecher, B. N., Buck, J. M., Bergmann, P., Kroth, P. G., Bolton, J. J., Dąbek, P., Witkowski, A., Kim, S.-Y., and Trobajo, R.: Prey preference in a kleptoplastic dinoflagellate is linked to photosynthetic performance, ISME J., 17, 1578–1588, https://doi.org/10.1038/s41396-023-01464-3, 2023.
Zhang, D.-C., Yu, Y., Chen, B., Wang, H.-X., Liu, H.-C., Dong, X.-Z., and Zhou, P.-J.: Glaciecola psychrophila sp. nov., a novel psychrophilic bacterium isolated from the Arctic, Int. J. Syst. Evol. Micr., 56, 2867–2869, https://doi.org/10.1099/ijs.0.64575-0, 2006.
Zorz, J., Willis, C., Comeau, A. M., Langille, M. G. I., Johnson, C. L., Li, W. K. W., and LaRoche, J.: Drivers of Regional Bacterial Community Structure and Diversity in the Northwest Atlantic Ocean, Front. Microbiol., 10, 281, https://doi.org/10.3389/fmicb.2019.00281, 2019.