the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Maize root and shoot litter quality controls short-term CO2 and N2O emissions and bacterial community structure of arable soil
Pauline Sophie Rummel
Birgit Pfeiffer
Johanna Pausch
Reinhard Well
Dominik Schneider
Klaus Dittert
Chemical composition of root and shoot litter controls decomposition and, subsequently, C availability for biological nitrogen transformation processes in soils. While aboveground plant residues have been proven to increase N2O emissions, studies on root litter effects are scarce. This study aimed (1) to evaluate how fresh maize root litter affects N2O emissions compared to fresh maize shoot litter, (2) to assess whether N2O emissions are related to the interaction of C and N mineralization from soil and litter, and (3) to analyze changes in soil microbial community structures related to litter input and N2O emissions.
To obtain root and shoot litter, maize plants (Zea mays L.) were cultivated with two N fertilizer levels in a greenhouse and harvested. A two-factorial 22 d laboratory incubation experiment was set up with soil from both N levels (N1, N2) and three litter addition treatments (control, root, root + shoot). We measured CO2 and N2O fluxes, analyzed soil mineral N and water-extractable organic C (WEOC) concentrations, and determined quality parameters of maize litter. Bacterial community structures were analyzed using 16S rRNA gene sequencing.
Maize litter quality controlled and WEOC availability and decomposition-related CO2 emissions. Emissions induced by maize root litter remained low, while high bioavailability of maize shoot litter strongly increased CO2 and N2O emissions when both root and shoot litter were added. We identified a strong positive correlation between cumulative CO2 and N2O emissions, supporting our hypothesis that litter quality affects denitrification by creating plant-litter-associated anaerobic microsites. The interdependency of C and N availability was validated by analyses of regression. Moreover, there was a strong positive interaction between soil and WEOC concentration resulting in much higher N2O emissions, when both and WEOC were available. A significant correlation was observed between total CO2 and N2O emissions, the soil bacterial community composition, and the litter level, showing a clear separation of root + shoot samples of all remaining samples. Bacterial diversity decreased with higher N level and higher input of easily available C. Altogether, changes in bacterial community structure reflected degradability of maize litter with easily degradable C from maize shoot litter favoring fast-growing C-cycling and N-reducing bacteria of the phyla Actinobacteria, Chloroflexi, Firmicutes, and Proteobacteria. In conclusion, litter quality is a major driver of N2O and CO2 emissions from crop residues, especially when soil mineral N is limited.
- Article
(2606 KB) - Full-text XML
-
Supplement
(700 KB) - BibTeX
- EndNote
Chemical composition controls decomposition of both roots (Birouste et al., 2012; Redin et al., 2014; Silver and Miya, 2001) and plant litter (Jensen et al., 2005; Kögel-Knabner, 2002; Zhang et al., 2008) and, subsequently, C availability for biological nitrogen transformation processes in soils. When O2 concentrations are low, denitrifying soil microorganisms may use nitrate () as an electron acceptor in the respiratory chain to break down organic compounds (Zumft, 1997). This leads to loss of plant-available N (Müller and Clough, 2014) and makes soils an important source of the greenhouse gas N2O (Ciais et al., 2013).
Plant residues have been proven to increase N2O emissions upon incorporation into soil. When different types of litter were compared, quality parameters of plant residues, such as C:N ratio, lignin : N ratio, and chemical composition of structural components explained a large share of variances in N2O emissions (Baggs et al., 2000; Chen et al., 2013; Millar and Baggs, 2004). Especially in drier soils, denitrification is largely controlled by the supply of readily decomposable organic matter (Azam et al., 2002; Burford and Bremner, 1975; Loecke and Robertson, 2009). Availability of easily degradable C compounds stimulates microbial respiration, limiting O2 at the microsite level and increasing N2O emissions (Azam et al., 2002; Chen et al., 2013; Miller et al., 2008). Furthermore, plant litter enhances local anaerobicity by absorbing water from surrounding pores and retaining high moisture concentrations (Kravchenko et al., 2017, 2018).
While effects of aboveground plant residues on N2O emissions have been studied extensively, studies of root residues on N2O emissions are scarce. In a temperate forest soil, fine root litter of maize and native tree species did not cause any N2O emissions, but a very close interrelation between C mineralization of fine root litter and N2O emissions was found in other biomes (Hu et al., 2016). In other studies, lower cumulative N2O emissions were reported after addition of sugar beet roots compared to leaves (Velthof et al., 2002) and rice roots compared to rice straw (Lou et al., 2007). Furthermore, decomposition dynamics of roots have been studied in great detail, revealing that chemical composition explains most of its variation (Birouste et al., 2012; Johnson et al., 2007; Machinet et al., 2011; Redin et al., 2014; Silver and Miya, 2001; Zhang and Wang, 2015). In general, decomposition rates of hemicelluloses and pectin are higher than that of cellulose, while among cell wall components lignin is most resistant against microbial decomposition (Kögel-Knabner, 2002).
Soil microorganisms are often specialized in specific substrates with fungi being regarded as the main decomposers of plant materials rich in cellulose and lignin, while hemicelluloses and pectin are decomposed by many aerobic and anaerobic bacteria and fungi (Kögel-Knabner, 2002). While the phyla Firmicutes, Proteobacteria, and Bacteroidetes are described as fast-growing copiotrophic bacteria that are stimulated by input of easily degradable C compounds (Fierer et al., 2016; Pascault et al., 2013), abundance of Acidobacteria decreased following the addition of dissolved organic matter into the soil (Fierer et al., 2016). Similarly, denitrifying microorganisms are found in bacteria, fungi, and archaea depending on substrate availability and environmental conditions (Zumft, 1997). Fungi are seen as major contributors to denitrification under aerobic and weakly anaerobic conditions, while bacterial denitrification predominates under strongly anaerobic conditions (Hayatsu et al., 2008). Denitrifying bacteria can be found in most phyla (Zumft, 1997), with dominant populations in Pseudomonas and Alcaligenes (Gamble et al., 1977; Megonigal et al., 2013). The most abundant denitrifying bacteria in soil are heterotrophic and, as such, require a source of electrons or reducing equivalents contained in C compounds of organic matter or plant residues. Availability of organic C may thus affect both decomposing and denitrifying soil microorganisms.
In most reported studies on decomposition and N2O emissions, dried and often ground plant material was used. This facilitates a homogenous distribution in soil and minimizes differences between replicates. Nevertheless, drying of fine roots prior to incubation increased their decomposition rate and led to overestimation of decomposition and nutrient cycling rates (Ludovici and Kress, 2006). Additionally, formation of plant-litter-associated anaerobic hot spots was reduced when ground plant material was homogenously mixed with the soil, while litter aggregation significantly increased soil N2O emissions (Loecke and Robertson, 2009). Differences in N2O emissions between two clover species were observed only with intact (but dried) leaves, but not when ground material was used (Kravchenko et al., 2018).
The aims of this study were (1) to evaluate how fresh maize root litter affects N2O emissions compared to fresh maize shoot litter, (2) to assess to what extent N2O emissions are related to the interaction of C and N mineralization from soil and litter, and (3) to analyze the changes in soil microbial community structures related to litter input and N2O emissions. We hypothesize that differences in N2O emissions between treatments can be related to degradability of maize litter with more easily degradable shoot litter leading to higher N2O formation. We further expect that differences in litter chemical quality are reflected in the structural composition of the soil microbial community with higher availability of N and C leading to a more specialized community.
Maize plants were grown in a greenhouse to produce root and shoot litter. As in many European countries the law prohibits addition of mineral N with incorporation of crop residues or catch crops, we applied two N fertilizer regimes (low vs. high) to realize differences in soil Nmin concentration at harvest. We then set up a laboratory incubation experiment with fresh maize root or root and shoot litter under fully controlled conditions and determined CO2 and N2O fluxes for 22 d. Soil samples were taken in regular intervals and analyzed for soil mineral N and water-extractable organic C (WEOC) concentrations. At the end of the incubation experiment, soil microbial community structures were analyzed to identify adaptions to litter input.
2.1 Preparation of plants and soils prior to incubation experiment
The soil for the experiment was collected 10 km south of Göttingen, Germany, at the experimental farm Reinshof of the University of Göttingen (51.484∘ N, 9.923∘ E). Soil was classified as gleyic Fluvisol (21 % clay, 68 % silt, 11 % sand) containing 1.5 % C and 2.81 % humus, with a pH (CaCl2) = 7.44.
Prior to the incubation experiment, maize plants were cultivated to obtain shoot and root biomass. For maize cultivation, Mitscherlich pots were filled with 5 kg of air-dried and sieved (2 mm) soil previously mixed with fertilizers (0.2 g N kg−1 as NH4NO3, 0.14 g P kg−1 as Ca(H2PO4)2, 0.2 g K kg−1 as K2SO4 and 0.04 g Mg kg−1 as MgSO4 × 7 H2O including 0.135 g S kg−1). Soil moisture was adjusted to 25 vol. %, and volumetric water content (VWC) sensors (EC-5, Decagon Devices, Pullman, USA) were used to monitor soil water content. Six maize plants (Zea mays L. var. Ronaldinio) were sown per pot and cultivated in a greenhouse with 16 h light and 8 h dark cycles. Pots were randomized in regular intervals to avoid microclimatic effects in the greenhouse.
To get different soil mineral N concentrations in soil, a second N fertilizer dose (0.2 g N kg−1 as Ca(NO3)2 × 4 H2O) was applied to half of the pots 6 weeks after sowing. Soil with one N dose is referred to as N1 (0.2 g N kg−1) and soil with two N doses is referred to as N2 (2×0.2 g N kg−1). Plants were harvested 8 weeks after sowing: maize plants were cut above the soil surface and roots were removed from soil by sieving and handpicking. Fresh roots were shaken and slightly brushed to remove adhering soil.
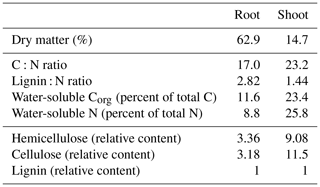
A subsample of aboveground maize biomass and maize roots was dried at 60 ∘C to determine dry matter contents and milled to a particle size < 1 mm. To determine water-extractable C and N concentrations, subsamples were extracted with H2Obidest (maize root 1:1000 w∕v; maize shoot 1:10000 w∕v) for 16 h and analyzed using a multi N∕C® analyzer (model 3100, Analytik Jena, Jena, Germany). Another subsample was analyzed for the sum of structural components following established feedstuff analysis protocols based on the method proposed by Goering and Van Soest (1970), namely ash-free neutral detergent fiber aNDFom (VDLUFA, 2012a), acid detergent fiber ADFom (VDLUFA, 2011), and acid detergent lignin ADL (VDLUFA, 2012b). According to the definitions, hemicellulose, cellulose, and lignin contents were calculated as follows: hemicellulose is equal to aNDFom minus ADFom; cellulose is equal to ADFom minus ADL; lignin is equal to ADL. Another subsample was milled using a ball mill, and total carbon and nitrogen concentrations were analyzed using a C:N analyzer (model 1110, Carlo Erba, Milan, Italy).
2.2 Incubation experiment
The incubation experiment consisted of a two-factorial setup comprising two N levels (N1 and N2) and three litter levels (control: Cn; root: Rt; root + shoot: RS) (see Table 1 and Fig. 1 for details). To allow comparison of litter treatments over soil conditions, the same litter types for both soil N levels were used. As N2 plants had produced greater and healthier biomass during the pre-experimental growth phase, only N2 shoots were used for both soils. Roots from N1 and N2 plants were mixed to ensure sufficient amounts for all replicates. Control soils (N1-Cn and N2-Cn) did not receive plant biomass, yet they contained C input from rhizodeposition of the previous maize growth. C remaining from rhizodeposition, root hairs, and small root fragments was calculated as the difference in soil C concentration before and after maize growth. For the root treatment 100 g of fresh root biomass was added per kilogram of dry soil (N1-Rt and N2-Rt), and in the root-and-shoot treatment, 100 g of fresh root and 100 g of fresh shoot biomass were added per kilogram of dry soil (N1-RS, N2-RS). Each treatment was replicated four times.
Table 1Two-factorial setup of the incubation experiment. Soil mineral N (Nmin) concentrations were measured directly before onset of the incubation experiment. C input in the control treatment is from rhizodeposition (RD) only, C input in the root treatment is from rhizodeposition and roots, and C input in root + shoot is from rhizodeposition, roots, and shoot biomass. N input is from root and shoot biomass, respectively (FM: fresh matter).
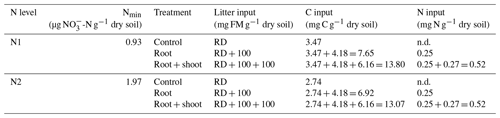
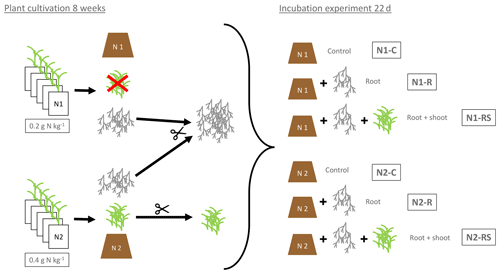
Figure 1Preparation and experimental setup of the incubation experiment. N1 (0.2 g N kg−1) and N2 (2×0.2 g N kg−1) referring to the N levels during plant growth. Control soil (N1-C and N2-C) without addition of plant litter. Root treatment with addition of 100 g of fresh root biomass per kilogram of dry soil (N1-R and N2-R) and root + shoot treatment with addition of 100 g of root and 100 g of shoot biomass per kilogram of dry soil (N1-RS, N2-RS).
Within each N level, soil was homogenized to ensure similar starting conditions. Subsamples of both soils were taken for analysis of mineral N, water-extractable Corg concentration, and total soil C. Soil mineral N concentrations were 0.93 and 1.97 µg N g−1 for N1 and N2, respectively. Plant litter was cut to a size of 2 cm and homogeneously mixed with the soil, simulating residue incorporation and tillage. PVC pots with a diameter of 20 cm and a total volume of 6.8 L were filled with fresh soil equivalent to 3.5 kg dry weight previously mixed with plant litter. Soil was compacted in a stepwise mode by filling a 2 cm layer of soil in pots and compacting it with a plunger. To ensure continuity between soil layers, the surface of the compacted layer was gently scratched before adding the next soil layer. Due to high litter input, target bulk density was 1.1 g cm−3. Actual bulk density was determined by measuring headspace height, and these values were used for calculations.
To adjust soil moisture of all pots to 70 % water holding capacity (WHC), equivalent to 49 % WFPS, water was dripped on the soil surface through hollow needles (outer diameter 0.9 mm). Pots were covered with PVC lids to minimize evaporation from the soil and to incubate samples in the dark. The incubation experiment was carried out under controlled temperature conditions (16 h day at 25 ∘C, 8 h night at 19 ∘C) for 22 d. Volumetric water content (VWC) sensors (EC-5, Decagon Devices, Pullman, USA) were used to monitor soil water content.
2.3 Gas sampling and analysis
Gas fluxes were measured using the closed-chamber method (Hutchinson and Mosier, 1981). Gas samples were taken every 12 h (morning and evening) for the first 15 d and every 24 h (midday) for the remaining 7 d. Due to technical issues, gas samples taken in the morning of day 10 to day 15 had to be discarded. Before gas sampling, all pots were opened for ventilation to ensure homogenous ambient air background conditions. Pots were closed with gastight PVC lids, and 30 mL gas samples were taken from each pot 0, 20, and 40 min after closure and filled into pre-evacuated 12 mL Exetainer glass bottles (Labco, High Wycombe, UK). Samples were analyzed on a Bruker gas chromatograph (456-GC, Bruker, Billerica, USA) deploying an electron capture detector (ECD) for N2O and a thermal conductivity detector (TCD) for CO2. Samples were introduced using a Gilson autosampler (Gilson Inc., Middleton, WI, USA). Data processing was performed using CompassCDS software. The analytical precision was determined by repeated measurements of standard gases (2500 and 550 ppm CO2, 307, 760, and 6110 ppb N2O) and was consistently < 2 %.
2.4 Soil analyses
Soil samples were taken from the pots using a soil auger of 16 mm diameter on 5, 9, 14, and 22 DAO (days after onset of experiment). Holes were closed with glass tubes to avoid variation in the soil surface. Fresh subsamples were analyzed for water-extractable Corg concentration (WEOC), and a subsample was frozen at −20 ∘C for soil mineral N analysis. Total soil carbon and nitrogen concentrations were analyzed using a C:N analyzer (model 1110, Carlo Erba, Milan, Italy). For determination of soil mineral N content, frozen samples were extracted with a 0.0125 M CaCl2 solution (1:5 w∕v) for 60 min on an overhead shaker (85 rpm). The extracts were filtered with 615 1∕4 filter paper (Macherey-Nagel GmbH & Co. KG, Düren, Germany) and stored at −20 ∘C. The extracts were analyzed colorimetrically for the concentrations of and using the San continuous-flow analyzer (Skalar Analytical B.V., Breda, the Netherlands). Soil water content was determined with a parallel set of samples. Net N mineralization was calculated as the difference between the concentrations at the start and end of the incubation period plus N lost as N2O-N (Eq. 1).
WEOC was determined according to Chantigny et al. (2007). Briefly, fresh soil was homogenized with deionized water (1:2 w∕v), and samples were centrifuged and filtered with 0.45 µm polyether sulfone syringe filters (Labsolute, Renningen, Germany) and stored at −20 ∘C. The extracts were analyzed using a multi N∕C® analyzer (Analytik Jena, Jena, Germany).
2.5 Analysis of bacterial community structures
2.5.1 DNA isolation and 16S rRNA gene amplification
To analyze the soil-inhabiting bacterial communities, DNA was extracted from 0.5 g (fresh weight) of soil sample taken at the end of the incubation experiment (22 DAO) using the DNA extraction protocol described by Griffiths et al. (2000). Plant litter was removed from samples prior to extraction. In brief, cells were mechanically disrupted using bead beating, and nucleic acids were extracted using phenol : chloroform : isoamyl alcohol (; Carl Roth, Karlsruhe, Germany). Nucleic acids were then precipitated using polyethylene glycol (Carl Roth, Karlsruhe, Germany) and washed with 70 % ice-cold ethanol (VWR, Radnor, Pennsylvania, USA). Subsequently, RNA was removed by RNase A digestion (Thermo Fischer Scientific, Waltham, Massachusetts, USA) as described by the manufacturer. The RNA-free DNA was used for amplification of the V3 to V4 region of the 16S rRNA gene. We used the bacterial primer pair S-D-Bact-0341-b-S-17 and S-D-Bact-0785-a-A-21 targeting the V3–V4 region of the 16S rRNA gene described by Klindworth et al. (2013) with adapters for Illumina MiSeq sequencing. The polymerase chain reaction (PCR) reaction mixture contained five-fold Phusion GC buffer, 200 µM of each of the four deoxynucleoside triphosphates, 5 % DMSO, 0.4 µM of each primer, 1 U of Phusion HF DNA polymerase (Fisher Scientific GmbH, Schwerte, Germany), and 25 ng of RNA-free DNA as template. The following cycling scheme was used for DNA amplification: initial denaturation at 98 ∘C for 5 min and 25 cycles of denaturation at 98 ∘C for 45 s, annealing at 60 ∘C for 30 s, and extension at 72 ∘C for 30 s, followed by a final extension at 72 ∘C for 10 min. For each sample, PCR reactions were performed in triplicate. Resulting PCR products were pooled in equimolar amounts and purified using the QIAquick Gel Extraction kit (Qiagen, Hilden, Germany) as recommended by the manufacturer. Quantification of the PCR products was performed using the Quant-iT dsDNA HS assay kit and a Qubit fluorometer as described by the manufacturer (Invitrogen GmbH, Karlsruhe, Germany). Indexing of the PCR products was performed by the Göttingen Genomics Lab (G2L, Göttingen, Germany) using the Nextera XT Index kit as recommended by the supplier (Illumina, San Diego, CA, USA), and sequencing of 16S rRNA amplicons was performed using the dual index paired-end approach (2×300 bp) with v3 chemistry for the Illumina MiSeq platform.
2.5.2 Sequence processing
All bioinformatic processing of sequence data was done using Linux-based software packages. Adapter removal and quality filtering of raw paired-end sequences was done using fastp v0.19.6 (Chen et al., 2018), with base correction in overlapped regions, a qualified quality phred of 20, size exclusion of sequences shorter than 50 bp, and per read trimming by quality (phred 20). Merging of quality-filtered paired-end reads was done by PEAR v0.9.11 (64 bit) with default parameters (Zhang et al., 2014). Primer removal was conducted using cutadapt v1.18 (Martin, 2013). Subsequently, dereplication, denoising, and chimera detection and removal (denovo followed by reference based against the SILVA 132 SSU database) were performed with VSEARCH v2.13.0 (64 bit) (Rognes et al., 2016). Taxonomic classification of the amplicon sequence variants (ASVs, 100 % sequence identity) was performed with BLAST + v2.7.1 against the SILVA 132 SSU reference database (Quast et al., 2013). Subsequently, extrinsic domain ASVs and chloroplasts were removed from the dataset. Sample comparisons were performed at the same surveying effort of 61200 sequences. Statistical analyses were done using ASVs in R version 3.5.3 (R Core Team, 2019). The R package ampvis2 v2.4.7 (Andersen et al., 2018) was used to determine species richness, alpha diversity estimates, and rarefaction curves and to prepare all graphs. To visualize the multivariate constrained dispersion, canonical correspondence analysis (CCA) was conducted with Hellinger transformed data (Legendre and Gallagher, 2001), and ASVs with a relative abundance lower than 0.1 % in any sample were removed. Correlations of environmental parameters to the bacterial communities were analyzed using the envfit function of the vegan package v2.5-4 (Oksanen et al., 2015) and projected into the ordination with arrows with a p-value cutoff of 0.005. For further statistical analysis of the microbial community composition (on phyla, order, and genus levels) and diversity (Shannon, Simpson, and PD index), multivariate generalized linear models (MGLMs; with N level and litter addition as factors) as implemented in the mvabund R package v4.0.1 were employed with adjusted p values (Wang et al., 2019). For the generalized linear model analysis of variance (MGLM-ANOVA) tests, p values < 0.05 were considered to be significant. In addition, core microbiomes and respective responders were analyzed at the genus level, grouped by either the applied litter treatment or N fertilizer levels using ampvis2 v2.4.7.
For one replicate of N2-Rt, DNA concentration was very low and the 16S rRNA gene could not be amplified. Thus, we only evaluated the remaining three replicates of this treatment. In addition, we attempted to analyze the soil-inhabiting fungal community using the fungus-specific primer set ITS3_KYO2 and ITS4 (Toju et al., 2012), but we were not able to amplify them.
2.6 Calculations and statistical analyses
All statistical analyses were performed using the statistical software R version 3.5.2 (R Core Team, 2018). Arithmetic means and standard error of the four replicates were calculated for CO2 and N2O fluxes. Cumulative gas emissions were calculated by linear interpolation between measured fluxes. To account for different C input in treatments, cumulative CO2 and N2O emissions were standardized against the C input per treatment (see Table 1 for details on C input). Tukey's HSD test was used after analysis of variance to test for treatment effects (i.e., N level and litter addition) on cumulative CO2 emissions. An interaction was identified between N level and litter addition on cumulative N2O emissions using interaction plots from the package HH v3.1-35 (Heiberger, 2018). A linear model using generalized least squares (gls) was fitted between cumulative N2O as a response variable and N level, litter addition, and their interaction as fixed effects. Additionally, the model was fitted to account for inhomogeneous within-class variances. Estimated marginal means were then computed to analyze treatment effects using the R package emmeans v1.3.4 (Lenth, 2018). Several regression models were tested to analyze the effect of maize litter on cumulative N2O emissions including the factors cumulative CO2 emissions, initial soil concentration, and net N mineralization during the incubation period. For cumulative CO2 emissions, regression models included the factors total C input, water-extractable C input, hemicellulose fraction, cellulose fraction, and lignin fraction from all litter treatments (Cn, Rt, RS, n=24).
To evaluate effects of soil environmental variables on N2O and CO2 fluxes, a linear mixed-effect model (lme) was fitted between N2O fluxes (ln transformed), soil -N and WEOC concentrations using the lme function from the package nlme v3.1-131 (Pinheiro et al., 2017). Pseudo-R2 for lme was calculated using r.squaredGLMM from the package MuMIn v1.42.1 (Barton, 2018). Soil -N and WEOC concentrations between sampling dates were estimated by linear interpolation. Only evening and midday gas measurements were included in model calculations. To account for repeated measurements, incubation vessel and sampling day were set as random effects. Models were compared using maximum likelihood (ML), selected using AIC (Akaike's information criterion), and fitted using restricted maximum likelihood (REML).
All plots were made using the statistical software R version 3.5.2 (R Core Team, 2018) including the packages plotrix v3.7.4 (Lemon, 2006), plot3D v1.1.1 (Soetaert, 2017), and viridisLite v0.3.0 (Garnier, 2018).
3.1 Chemical analyses of maize litter
Maize root and shoot litter differed in their chemical compositions (Table 2). Dry matter content of maize roots was much higher compared to shoot as roots had not been washed prior to analyses, so some soil adhering to roots was included in dry matter determinations. Thus, we calculated water-extractable concentrations in relation to total C instead of dry matter. Maize shoot litter was characterized by higher concentrations of water-soluble C and N and a higher share of easily degradable compounds like hemicellulose and cellulose compared to maize roots.
3.2 CO2 and N2O fluxes and cumulative emissions
Addition of maize litter increased CO2 fluxes compared to the control treatment (Fig. 2), where addition of root and shoot litter (N1-RS, N2-RS) resulted in much higher fluxes compared to roots only (N1-Rt, N2-Rt). While absolute emission rates were strongly affected by litter input, time courses were similar in all litter treatments without visible differences between N1 and N2. CO2 fluxes stayed on a similar level for the first 10 d after onset of incubation, showing fluctuations between morning and evening sampling times, and then constantly decreased until the end of the experiment.
After a short lag phase right after the onset of experiment, N2O emissions increased in all litter treatments compared to control treatments (Fig. 3a, b). The highest fluxes were measured in N2-RS, reaching 7.8 pg N2O-N g−1 s−1 on day 5. Fluxes stayed on a similar level from day 7 to day 15 and then declined until the end of the experiment. N2O fluxes from root (N1-Rt, N2-Rt) and control treatments (N1-Cn, N2-Cn) remained at a low level during the whole incubation period (≤0.59 pg and ≤0.04 pg N2O-N g−1 s−1, for Rt and Cn, respectively). N2O fluxes from N1 were slightly lower than from N2 in both litter treatments. Over all treatments and sampling dates, CO2 and N2O fluxes were positively correlated (R2=0.5993, p<0.001, data not shown).
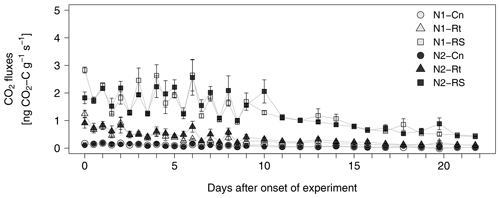
Figure 2CO2 fluxes from soils with two N levels (N1, N2) after incorporation of maize root litter (Rt), maize root + shoot litter (RS), and control (Cn) without litter. Error bars show the standard error of mean values (n = 4). When not visible, error bars are smaller than the symbols.
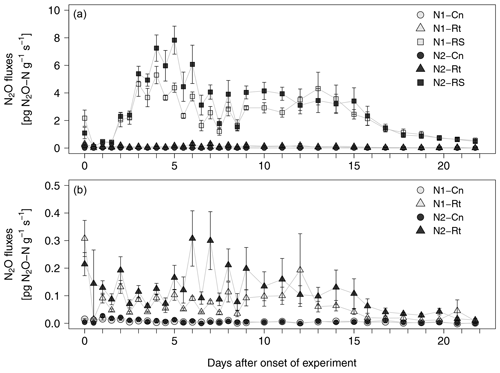
Figure 3(a, b) N2O fluxes from soils with two N levels (N1, N2) after incorporation of maize root litter (Rt), maize root + shoot litter (RS), and control (Cn) without litter. Error bars show the standard error of mean values (n = 4). When not visible, error bars are smaller than the symbols. Note: data of (b) are excerpts from (a) and are shown with a different scaling.
To account for different C inputs in treatments, cumulative CO2 and N2O emissions were standardized against the C input per treatment (Table 3). Still, cumulative CO2 emissions were almost twice as high in Rt and about 4 times higher in RS compared to Cn (p<0.05), indicating that differences between litter treatments cannot simply be explained by differences in C input. Addition of maize root and shoot litter increased cumulative N2O emissions by roughly a factor of 100 compared to control treatments (p<0.05). In contrast, root litter increased cumulative N2O emissions only by a factor of 5.4 (N1-Rt) and 7 (N2-Rt) compared to the respective controls (p<0.05).
Table 3Absolute cumulative N2O and CO2 emissions and relative to C input and N2O ∕ CO2 ratio of 22 d incubation experiment with two pre-incubation N levels (N1, N2) and three litter addition treatments (control: no litter input; root: 100 mg root M g−1 dry soil; root + shoot: 100 mg root FM + 100 mg shoot FM g−1 dry soil).
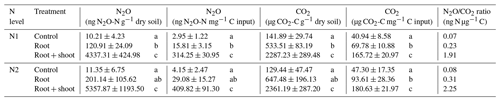
Values represent means (n=4) ± standard deviation. Different letters in the same column indicate a significant difference according to Tukey's HSD post hoc tests at p≤0.05.
3.3 Soil , , and water-extractable Corg concentrations
Addition of maize litter affected the time course of soil , , and WEOC concentrations (Fig. 4a–c). In control treatments, initial soil concentrations of 0.93 (N1-Cn) and 1.97 µg -N g−1 dry soil (N2-Cn) continuously increased until the end of the experiment, reaching concentrations of 8.24 µg N g−1 (N1-Cn) and 11.74 µg N g−1 (N2-Cn), respectively. Soil concentrations showed variations at a low level only. Soil concentrations were continuously higher in N2 than in N1 and differences in soil concentration were small. Higher fertilization in N2 during previous plant growth led to higher residual organic N and higher net N mineralization (7.61 and 10.08 µg N g−1 for N1-Cn and N2-Cn, respectively, Table 4) during the incubation experiment. In treatments with litter, soil concentrations decreased after an initial increase. In root treatments, soil concentrations continuously decreased until the end of the incubation experiment to 1.9 (N1-Rt) and 2.5 µg N g−1 (N2-Rt), while in root-plus-shoot treatments soil concentrations increased again until the end of the experiment, reaching concentrations of 9.46 (N1-RS) and 9.52 µg N g−1 (N2-RS). During the whole incubation period, soil concentrations in RS were higher than in Rt. Soil concentrations only marginally increased for Rt. Contrary to Rt and Cn, soil concentrations increased until the end of the incubation experiment to 1.68 (N1-RS) and 1.52 µg N g−1 (N2-RS) in root-and-shoot treatments. Net N mineralization was 1.44 (N1-Rt) and 1.10 µg N g−1 (N2-Rt) in root treatments, and 14.32 (N1-RS) and 14.14 µg N g−1 (N2-RS) in root-and-shoot treatments (Table 4). Maize root litter did not affect WEOC, as concentrations were similar to Cn throughout the incubation period. However, in RS treatments, WEOC increased after the onset of incubation, reaching the highest values (45.32 µg C g−1) for N1-RS at day 9, after which it decreased until the end of the experiment.
Table 4N mineralization during the incubation period.
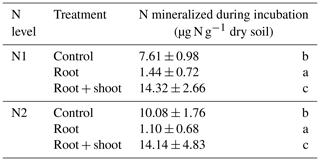
Values represent means (n=4) ± standard deviation. Different letters in the same column indicate a significant difference according to Tukey's HSD post hoc tests at p≤0.05.
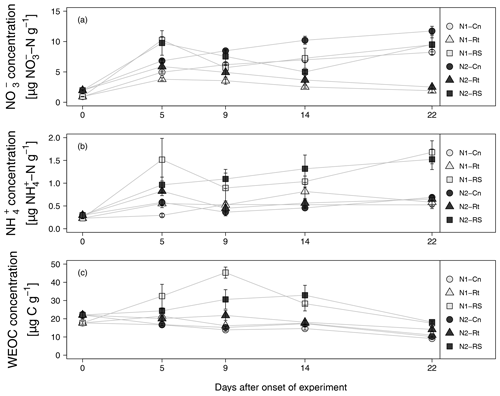
Figure 4(a–c) , WEOC, and concentration from soils with two N levels (N1, N2) after incorporation of maize root litter (Rt), maize root + shoot litter (RS), and control (Cn) without litter. Error bars show the standard error of mean values (n = 4) (day 0: n=3). When not visible, error bars are smaller than the symbols.
3.4 Relations between N2O emissions and C and N parameters of plant litter and soil
To identify the effect of N and C availability on N2O fluxes, a linear mixed-effect model was applied. The best model included a significant interaction between soil and WEOC (p<0.0024, pseudo-R2=0.82, Table 5) and incubation vessel and sampling time as random parameters. Predictions of N2O fluxes based on this model are shown in Fig. 5.
Table 5Significance of fixed effects of soil -N (µg -N g−1), water-extractable organic C (WEOC, µg C g−1), and first-order interaction on N2O fluxes (pg N2O-N g−1 h−1; ln transformed) using a linear mixed-effect model.
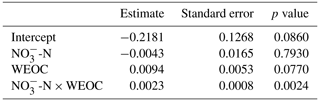
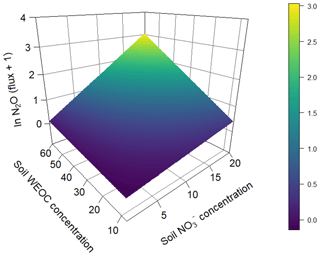
Figure 5Prediction of N2O fluxes (pg N2O-N g−1 s−1) (ln transformed) based on soil (µg N g−1) and water-extractable Corg (µg C g−1) concentrations based on a linear mixed-effect model (pseudo-R2=0.82).
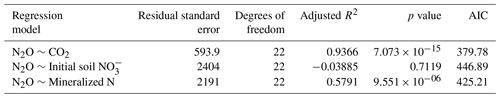
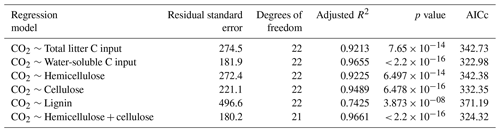
Linear regression analyses were used to identify relations between cumulative CO2 and N2O emissions, litter quality, and N parameters. Either hemicellulose + cellulose fraction or water-extractable C fraction of plant litter explained more than 96 % of variance of total cumulative CO2 emissions () (Table 6). Regression analyses of the relationships between total cumulative N2O emissions and influencing factors identified a strong positive relationship between total cumulative N2O emissions and total cumulative CO2 emissions (R2=0.9362, ) (Table 7) and between cumulative N2O emissions and mineralized N (R2=0.5791, ), while initial soil concentration did not explain any variance.
Table 6Results of regression analyses of the relationship between total cumulative CO2 emissions and C quality parameters of plant litter (AICc: Akaike's information criterion).
3.5 Bacterial community structure
The comparison over all maize litter treatments revealed that the bacterial diversity was slightly higher in N1 than in N2 soil as shown by a higher number of amplicon sequence variants (ASVs, R2=0.1195, p=0.059, Fig. S1 in the Supplement). In addition, the alpha diversity indices Shannon (R2=0.1844, p=0.023) and Simpson (R2=0.1131, p=0.065) as well as Faith's phylogenetic diversity (PD; R2=0.1844, p=0.059) were higher for N1 than for N2 samples (Table S4 in the Supplement).
The canonical correspondence analysis revealed a significant correlation (p<0.001) of the bacterial community composition with total CO2 (R2=0.6758) and N2O (R2=0.6179) emissions and the litter level, expressed by a clear separation of the N1-RS and N2-RS samples of all other samples (Fig. 6). With increasing C input, N2 samples cluster more closely than N1 samples. No significant correlation of litter level and microbial diversity was observed and PD index increased in N1 samples with increasing C input, while the opposite was found for N2 samples. Comparison of N1-Cn and N1-RS revealed no difference in diversity indices (Shannon and Simpson), while N1-Rt showed lower Shannon and Simpson diversity indices (Table S4). The Shannon diversity index was lowest in N2-Rt comparing all N2 treatments, while the Simpson index was lowest for N2-RS.
Overall, the soil bacterial communities were dominated by Actinobacteria, Proteobacteria, and Chloroflexi accounting for 151 % to 31 % (Fig. S2). The highest relative abundance of Actinobacteria and Chloroflexi was found in N2-Rt and of Proteobacteria in N1-R. Among these phyla, the orders Gaiellales (Actinobacteria), Sphingomonadales (Proteobacteria), and Thermomicrobiales (Chloroflexi) showed the highest relative abundance, especially in N2-Rt (9.3 %), N1-Rt (7.5 %), and N2-RS (9 %), respectively. Nevertheless, the phyla Acidobacteria, Planctomycetes, Verrucomicrobia, Gemmatimonadetes, Firmicutes, Patescibacteria, and Bacteroidetes were also detected (> 1 %) (Fig. 7). In detail, Bacteroidetes and Gemmatimonadetes decreased (with a negative slope, but not significant) with increasing N level, while the abundance of Firmicutes increased significantly (p=0.038). In addition, although present only in low relative abundance, the Cyanobacteria decreased significantly (p=0.003) with increasing N levels. At the genus level, Pseudomonas, Altererythrobacter, Gaiella, Nocardioides, Agromyces, Bacillus, and Lysobacter were most abundant, accounting for up to 5.7 % of all ASVs. Accordingly, these were also the most abundant genera attributed to the core microbiome (Tables S6 and S8). Overall, 80 genera represented the core microbiome, when grouped by N levels, while 21 genera and six genera were identified as responders to N1 and N2, respectively (Fig. S5). In detail, the classified responders to the applied N treatments were the genera Chthonibacter, Luteimonas, Sphingobium, Novosphingobium, Adhaeribacter, Nitrospira, Gemmata, and Devosia for N1 and Conexibacter for N2 samples (Table S8). The genera Bacillus, Gaiella, Altererythrobacter, Blastococcus, and Pseudomonas showed the highest abundance in N2 samples, while Lysobacter and Sphingomonas were more abundant in N1 samples (Fig. S3). When grouped by litter treatment, the core microbiome comprised 77 genera accounting for 73 % of the relative abundance, while 9, 3, and 10 genera were identified as responders to the applied litter treatments control, root, and root + shoot, respectively (Fig. S5). Nonomuraea, Fluviicola, and Nitrospira responded to the root + shoot treatment, while the genera Lapillicoccus and Adhaeribacter responded to the root treatment (Table S7). The genera Litorilinea, Gemmata, Novosphingobium, and Opitutus were identified as responders to the control treatment. For N levels and litter treatments, respectively, 833 and 838 genera were identified as non-core microbiomes, accounting for 20 % and 19.5 % of relative abundance (Fig. S5).
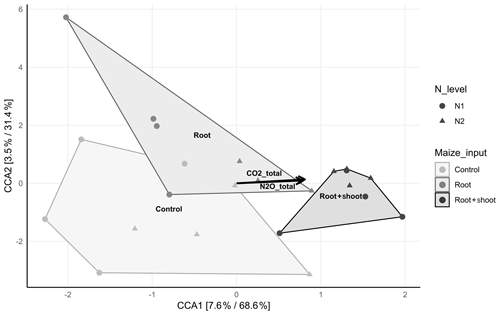
Figure 6Canonical correspondence analysis (CCA) displaying the compositional distribution of the soil-inhabiting bacterial communities between the control (N1-C and N2-C; n=4), root (N1-R and N2-R; n=4 and n=3), and root + shoot (N1-RS and N2-RS; n=4) treatments. Significant correlations of total CO2 and N2O emissions are shown by black arrows(p≤0.005). The relative contribution (eigenvalue) of each axis to the total inertia in the data as well as to the constrained space only are indicated in percent in the axis titles.
The most abundant classified species found were Agromyces sp., Bacillus sp., and Sphingomonas sp. Nevertheless, species such as Pseudomonas sp., Nitrosospira sp., Nitrosospira briensis, Alcaligenes sp., and Mesorhizobium sp. were also identified. Overall, the bacterial community composition was significantly influenced by N level (p=0.005) and maize litter treatment (p=0.033).
4.1 Decomposability of maize litter
Maize root and shoot litter quality controlled and WEOC availability and decomposition-related CO2 emissions during the initial phase of maize litter decomposition. Harvest of plants, removal of roots, and mixing of soil fostered mineralization and nitrification, as reflected by gradually increasing soil concentrations. The absence of changes in soil concentrations in control treatments without litter addition (N1-Cn, N2-Cn) indicates that all was directly nitrified. Also in controls, available C was low as indicated by low CO2 emissions and decreasing WEOC concentrations. The potential for mineralization in soil is known to be high after tillage (Höper, 2002) and positive net mineralization has been reported in control soil without litter addition (Machinet et al., 2009; Velthof et al., 2002) and in the fallow period after rice harvest (Aulakh et al., 2001).
Maize shoot litter was characterized by a high share of easily degradable compounds. High percentages of water-soluble N and water-soluble Corg from maize shoot litter strongly increased soil WEOC and concentrations. Availability of easily degradable compounds was also reflected by strongly increased CO2 fluxes and cumulative emission from N1-RS and N2-RS. While net mineralization in RS was similar to Cn, it was very small in Rt, indicating that N from mineralization was immobilized by soil microorganisms to decompose root C compounds (Robertson and Groffman, 2015). Cumulative CO2 emissions in litter treatments were clearly higher than in the control treatment, but CO2 fluxes continuously decreased after the onset of incubation, as easily degradable C was consumed. This is in accordance with results of Hu et al. (2016), who reported that maize fine root input initially increased CO2 fluxes, which then decreased during the first 20 d of incubation.
Mineralization of plant litter may increase soil concentrations in particular when C:N ratios are low (Li et al., 2013; Millar and Baggs, 2004). However, net N immobilization has been reported after addition of roots of maize (Machinet et al., 2009; Mary et al., 1993; Velthof et al., 2002), wheat (Jin et al., 2008; Velthof et al., 2002), barley, and sugar beet (Velthof et al., 2002), reaching a maximum around day 21 (Mary et al., 1993). Chemical composition has been proven to be the primary controller of decomposition rates of both roots (Birouste et al., 2012; Redin et al., 2014; Silver and Miya, 2001) and aboveground plant litter (Jensen et al., 2005; Zhang et al., 2008) of many different species. Slower decomposition of roots compared to leaves and stems was related to differences in chemical composition of plant organs (Jenkinson, 1965; Johnson et al., 2007). Accordingly, decomposition of roots from 16 maize genotypes was controlled by soluble residue components in the short term, whereas lignin and the interconnections between cell wall polymers were important in the long term (Machinet et al., 2011). In our study, regression analyses identified a strong positive relationship between cumulative CO2 emissions and water-extractable C fraction of plant litter (R2=0.966, ) (Table 6).
4.2 N2O emissions as affected by biodegradability of maize litter and soil N level
Denitrification in soil is largely controlled by the supply of readily decomposable organic matter (Azam et al., 2002; Burford and Bremner, 1975; Loecke and Robertson, 2009), leading to significant correlations between both N2O and CO2 fluxes and cumulative emissions (Azam et al., 2002; Fiedler et al., 2017; Frimpong and Baggs, 2010; Huang et al., 2004; Millar and Baggs, 2004, 2005). CO2 fluxes increased directly with the onset of incubation and started to decline after day 10; thus mostly C compounds with a short turnover time, i.e., sugars, proteins, starch, and hemicellulose, were decomposed and contributed to CO2 fluxes. Availability of easily degradable C compounds stimulates microbial respiration, limiting O2 at the microsite level and thus increasing N2O emissions from denitrification (Azam et al., 2002; Chen et al., 2013; Miller et al., 2008). Accordingly, N2O fluxes increased after a lag phase of 2 d. The strong positive correlation (R2=0.9362, ) between cumulative CO2 and N2O emissions (Table 7) further supports our hypothesis that litter quality, in particular degradability of C compounds, affects N2O fluxes from denitrification by creating plant-litter-associated microsites with low O2 concentrations.
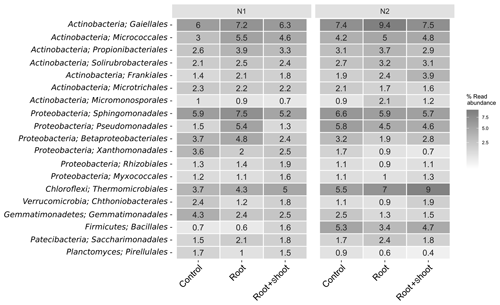
Figure 7Heat map of the 16 most abundant bacterial orders of the soil-inhabiting bacterial community grouped by N levels and litter input (n=4, except for N2 root: n=3).
High mineralization in RS treatments may have especially favored coupled nitrification–denitrification where and are produced by nitrifiers in aerobic habitats and subsequently denitrified by denitrifiers in close-by anaerobic habitats (Butterbach-Bahl et al., 2013; Wrage et al., 2001). Here, N2O is mainly produced in the interface of aerobic and anaerobic zones, which are typically found in plant litter associated hot spots (Kravchenko et al., 2017). In addition, N2O can also be produced aerobically during heterotrophic and autotrophic nitrification (Anderson et al., 1993; van Groenigen et al., 2015; Wrage et al., 2001; Zhang et al., 2015). In both processes, N2O can be formed as a byproduct from chemical hydroxylamine oxidation (Butterbach-Bahl et al., 2013; van Groenigen et al., 2015). Nitrifier denitrification as a pathway of autotrophic nitrification has been reported mostly under soil conditions differing from our study, namely high , NH3, or urea concentrations and low organic C availability (Wrage-Mönnig et al., 2018; Wrage et al., 2001). In contrast, with high availability of organic C and N compounds, high N2O emissions from heterotrophic nitrification have been reported (Anderson et al., 1993; Hu et al., 2016; Papen et al., 1989; Wrage et al., 2001). Zhang et al. (2015) reported 72 %–77 % of N2O being produced by heterotrophic nitrification from an arable soil under incubation conditions similar to our study. However, Li et al. (2016) estimated that denitrification was the dominant source of N2O in residue-amended soil at 40 %–60 % WFPS. High correlation of cumulative N2O emissions and mineralized N during the incubation period (R2=0.5791, ) indicates that, in addition to denitrification, heterotrophic nitrification may have contributed to N2O production in our study. However, to further differentiate between processes contributing to N2O production, stable isotope methods need to be used (Baggs, 2008; Butterbach-Bahl et al., 2013; van Groenigen et al., 2015; Wrage-Mönnig et al., 2018).
Another aim of this study was to investigate the effect of residual mineral N on plant-litter-induced N2O emissions. To this end, we included two N levels that were obtained by different N fertilization during the pre-experimental plant growth phase (N1: 0.2 µg N g−1, N2: 2×0.2 µg N g−1). At the onset of the incubation experiment, soil mineral N concentration was twice as high in N2 compared to N1 but generally very low (0.93 and 1.97 µg -N g−1 dry soil for N1 and N2, respectively). Higher N fertilizer input in N2 during plant growth led to lower C input from rhizodeposition (Table 1), which is consistent with literature findings (Kuzyakov and Domanski, 2000; Paterson and Sim, 1999). Cumulative N2O emissions tended to be higher in N2 than in N1, suggesting that was limited, especially in RS treatments where C availability was highest. In addition, litter chemical quality strongly affected N availability.
Under N-limiting conditions, a higher portion of N is recovered in soil microbial biomass in relation to litter N input (Bending and Turner, 1999; Troung and Marschner, 2018). When N is abundant relative to C availability, excess N is released by soil microorganisms and can be lost as N2O. In Rt, where N availability was low, N was immobilized by soil microorganisms and N2O emission were low. When more easily degradable N was added with maize shoots, N released from decomposition of maize shoots presumably fostered decomposition of maize roots (Robertson and Groffman, 2015) and denitrification of excess N, leading to strongly increased CO2 and N2O emissions in RS. To estimate the contribution of plant litter N to mineralization, immobilization, and denitrification, 15N-labeled litter together with analysis of microbial biomass N and 15N2O emissions could be used (e.g., Frimpong and Baggs, 2010; Ladd et al., 1981).
The interdependency of C and N availability was further validated by analyses of regression, highlighting a strong positive interaction between soil and WEOC concentrations resulting in much higher N2O emissions only when both and WEOC were available. This further supports our findings that high bioavailability of maize shoot litter increased microbial respiration by heterotrophic microorganisms, resulting in plant-litter-associated hot spots with high N2O formation.
Variation in N2O emissions is often related to quality parameters of plant residues, mostly the C:N ratio (Baggs et al., 2000; Chen et al., 2013; Millar and Baggs, 2004; Novoa and Tejeda, 2006). Especially easily degradable fractions, such as water-soluble C (Burford and Bremner, 1975) or the holocellulose fraction (hemicelluloses + cellulose) (Jensen et al., 2005), explained a large share of variability of C mineralization and N2O emissions, while lignin content was not relevant (Redin et al., 2014; Silver and Miya, 2001). Comparing 28 laboratory and field studies, Chen et al. (2013) reported that microbial-growth-induced microsite anaerobicity could be the major driver for the dynamic change in soil N2O emissions following residue amendment, and Kravchenko et al. (2017) showed that water absorption by plant residues further enhances formation of plant-litter-associated anaerobic hot spots. In the initial phase of decomposition, water-soluble compounds (sugars, amino acids) are leached from litter, providing easily degradable compounds for microbial metabolism. After litter addition, CO2 fluxes increased immediately due to increased respiration, rapidly reducing pO2, and creating anaerobic microsites. We anticipate that formation of such hot spots was further enhanced by the amount of litter addition, as litter input was higher in RS than in Rt, and higher compared to other studies (Chen et al., 2013).
In addition to soil mineral N concentration and plant litter, soil type and soil moisture may have influenced our results (e.g., Aulakh et al., 1991). Increasing soil moisture leads to increasing N2O emissions, but relative contribution of nitrification and denitrification to N2O formation may change with increasing soil moisture (Bateman and Baggs, 2005; Baral et al., 2016; Li et al., 2016). Therefore, future experiments with different soil moisture contents should include methods to differentiate between N2O formation pathways.
4.3 Bacterial community response to maize litter input and soil N level
After litter addition, the bacterial community adapts within a few days to substrate availability (Pascault et al., 2013). The canonical correspondence analysis (CCA) showed a clear correlation of the soil-inhabiting bacterial community, litter input, and total CO2 and N2O emissions. As shown by the CCA, the bacterial community structure in N1-RS and N2-RS was distinct from that in the control samples and soil with addition of root residues. Combined addition of root and shoot litter affected the soil bacterial community, leading to a less diverse and more specialized community structure, which was also shown by the alpha diversity indices (see Table S1). A significant reduction of soil bacterial diversity was induced by different N levels, as previously shown by Zeng et al. (2016). In addition, Rousk and Bååth (2007) observed a negative correlation between mineral N addition and bacterial growth, while the addition of barley straw and alfalfa correlated positively. The phylogenetic diversity (PD) supports these findings by showing a more complex picture. While PD in N1 samples increased with increasing C input, it decreased in N2 samples with increasing C input, indicating a shift of the influencing factors from the C input to the N level. Accordingly, the increase in N2O emissions from N2 compared to N1 was smaller in RS where C availability was the highest, indicating that N was limited here.
The most abundant phyla in our soil samples were the Actinobacteria, Proteobacteria, and Chloroflexi. Among these phyla, the genera Pseudomonas (Proteobacteria) and Gaiella (Actinobacteria) were also affiliated with the core microbiomes. Thermomicrobiales (Chloroflexi) showed the highest abundance in N2 samples, indicating their involvement in N cycling. Pseudomonas species such as Pseudomonas aeruginosa, P. stutzeri, and P. denitrificans are known to reduce and to contribute to N2O and N2 emissions (Carlson and Ingraham, 1983). Gaiella occulta, belonging to Actinobacteria, is also known for the reduction of to (Albuquerque et al., 2011). The genus Thermomicrobiales comprises species which can grow on nitrate, ammonia, and alanine as sole nitrogen sources and are able to hydrolyze cellulose or starch (Houghton et al., 2015). Relative abundance of Thermomicrobiales increased with N and C input, indicating favorable growth conditions for this genus (Fig. 7).
We further identified several genera involved in C cycling including members of Agromyces, Bacillus, and Micromonospora, which were also affiliated with the core microbiome. Agromyces ulmi was present in low abundance in our samples and it is known to contribute to C cycling in soils through xylanolytic activity (Rivas et al., 2004). Members of the genus Bacillus (Firmicutes) have been reported to play a crucial role in carbon cycling in a wide range of environments by functions such as plant growth promotion or production of amylases and cellulases (Lyngwi and Joshi, 2014). Among the genus Bacillus, we found one species, Bacillus sp. KSM-N252, in relatively high abundance (1 %–2 %) in N2 samples. This species encodes an alkaline endoglucanase, which can hydrolyze cellulose (Endo et al., 2001). Similarly, Micromonospora (Actinobacteria) are known to produce hydrolytic enzymes showing cellulolytic and xylanolytic activity (Carro et al., 2018; de Menezes et al., 2012). Abundance of Bacillus sp. KSM-N252 (N2-Cn 2 %, N2-Rt 1.1 %, and N2-RS 0.8 %) and Micromonospora (N2-R 1.9 %, N2-RS 1 %) decreased with increasing input of water-extractable C, indicating that cellulose was only decomposed when no easily degradable C was available.
Culture-independent sequence techniques have revealed that members of the phyla Actinobacteria, Chloroflexi, Firmicutes, Bacteroidetes, and Nitrospirae possess nirK or nirS and can reduce nitrite to nitric oxide (Cantera and Stein, 2007; Nolan et al., 2009). In our treatments, Actinobacteria, Chloroflexi, and Firmicutes were more abundant in N2 samples, whereas Bacteroidetes and Nitrospirae were more abundant in N1 samples, which may indicate that the latter are more competitive under conditions of very low mineral nitrogen availability in soil. This was further validated as Nitrospira (Nitrospirae), known to oxidize nitrite (Koch et al., 2015), was identified as a responder for N1 and RS. The reduction of nitrate has been shown for Mesorhizobium sp. (Okada et al., 2005) and Rhizobium sp. (Daniel et al., 1982). Although only in low abundance, we found these species predominantly in N2 samples. Species belonging to the genus Agromyces (Actinobacteria), which was affiliated with the core microbiomes, are also known to reduce nitrate (Zgurskaya et al., 2008). In addition, species capable of denitrification under anaerobic, O2-limited, and aerobic conditions can be found in the genera Bacillus and Micromonospora, as well as Pseudomonas and Rhodococcus (Verbaendert et al., 2011) that were affiliated with the core microbiome but were more abundant in N2 samples. The genus Opitutus was identified as a responder to Cn and comprises the bacterium Opitutus terrae that was only found in anoxic habitats in soils (Chin et al., 2001).
Altogether, the higher relative abundances of C-cycling and N-reducing bacteria in N2 samples and their affiliation with the core microbiomes reflect the tendency of increased N2O emissions with increasing N level and further supports our hypothesis that C and N availability from plant litter were the main drivers of N2O emissions in our study.
We examined CO2 and N2O emissions after simulated post-harvest incorporation of maize root or root-plus-shoot litter in a laboratory incubation study. High bioavailability of maize shoot litter strongly increased microbial respiration in plant-litter-associated hot spots, leading to increased N2O emissions when both C and were available. Coupled nitrification–denitrification and heterotrophic nitrification presumably contributed to N2O formation. Maize root litter was characterized by a higher share of slowly degradable C compounds and lower concentrations of water-soluble N; hence formation of anaerobic hot spots was limited and microbial N immobilization restricted N2O emissions. Bacterial community structures reflected degradability of maize litter types. Its diversity decreased with increasing C and N availability, favoring fast-growing C-cycling and N-reducing bacteria, namely Actinobacteria, Chloroflexi, Firmicutes, and Proteobacteria.
Hence, litter quality is a major driver of N2O and CO2 emissions from crop residues, especially when soil mineral N is limited.
The 16S rRNA gene sequences were deposited in the National Centre for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under bioproject number PRJNA557843. Data from measurements are available upon request from the corresponding author.
The supplement related to this article is available online at: https://doi.org/10.5194/bg-17-1181-2020-supplement.
PSR, RW, and KD designed the experiments and PSR carried them out. BP and DS carried out microbial analyses and sequence processing and provided figures. JP, RW, and KD contributed to interpretation of results. PSR prepared the manuscript with contributions from all co-authors.
The authors declare that they have no conflict of interest.
The authors thank Jakob Streuber, Simone Urstadt, and Finn Malinowski for gas sampling and laboratory analyses, as well as Alexander Silbersdorff (ZfS Statistical Consulting) and Oliver Caré for advice on data handling and statistical analysis.
This research has been supported by the Deutsche Forschungsgemeinschaft through the research unit DFG-FOR 2337: Denitrification in Agricultural Soils: Integrated Control and Modelling at Various Scales
(DASIM).
This open-access publication was funded
by the University of Göttingen.
This paper was edited by Michael Bahn and reviewed by two anonymous referees.
Albuquerque, L., França, L., Rainey, F. A., Schumann, P., Nobre, M. F., and Da Costa, M. S.: Gaiella occulta gen. nov., sp. nov., a novel representative of a deep branching phylogenetic lineage within the class Actinobacteria and proposal of Gaiellaceae fam. nov. and Gaiellales ord. nov., Syst. Appl. Microbiol., 34, 595–599, https://doi.org/10.1016/j.syapm.2011.07.001, 2011.
Andersen, K. S., Kirkegaard, R. H., Karst, S. M., and Albertsen, M.: ampvis2: an R package to analyse and visualise 16S rRNA amplicon data, bioRxiv 299537, https://doi.org/10.1101/299537, 2018.
Anderson, I. C., Poth, M., Homstead, J., and Burdige, D.: A comparison of NO and N2O production by the autotrophic nitrifier Nitrosomonas europaea and the heterotrophic nitrifier Alcaligenes faecalis, Appl. Environ. Microbiol., 59, 3525–3533, 1993.
Aulakh, M. S., Doran, J. W., Walters, D. T., and Power, J. F.: Legume residue and soil water effects on denitrification in soils of different textures, Soil. Biol. Biochem., 23, 1161–1167, https://doi.org/10.1016/0038-0717(91)90029-J, 1991.
Aulakh, M. S., Khera, T. S., Doran, J. W., and Bronson, K. F.: Denitrification, N2O and CO2 fluxes in rice-wheat cropping system as affected by crop residues, fertilizer N and legume green manure, Biol. Fert. Soils, 34, 375–389, https://doi.org/10.1007/s003740100420, 2001.
Azam, F., Müller, C., Weiske, A., Benckiser, G., and Ottow, J. C. G.: Nitrification and denitrification as sources of atmospheric nitrous oxide – Role of oxidizable carbon and applied nitrogen, Biol. Fert. Soils, 35, 54–61, https://doi.org/10.1007/s00374-001-0441-5, 2002.
Baggs, E. M.: A review of stable isotope techniques for N2O source partitioning in soils: Recent progress, remaining challenges and future considerations, Rapid Commun. Mass Sp., 22, 1664–1672, https://doi.org/10.1002/rcm.3456, 2008.
Baggs, E. M., Rees, R. M., Smith, K. A., and Vinten, A. J. A.: Nitrous oxide emission from soils after incorporating crop residues, Soil Use Manage., 16, 82–87, https://doi.org/10.1111/j.1475-2743.2000.tb00179.x, 2000.
Baral, K. R., Arthur, E., Olesen, J. E., and Petersen, S. P.: Predicting nitrous oxide emissions from manure properties and soil moisture: An incubation experiment, Soil. Biol. Biochem., 97, 112–120, https://doi.org/10.1016/j.soilbio.2016.03.005, 2016.
Barton, K.: MuMIn: Multi-Model Inference, R package version 1.43.6, available at: https://cran.r-project.org/package=MuMIn, 2018.
Bateman, E. J. and Baggs, E. M.: Contributions of nitrification and denitrification to N2O emissions from soils at different water-filled pore space, Biol. Fert. Soils, 41, 379–388, https://doi.org/10.1007/s00374-005-0858-3, 2005.
Bending, G. D. and Turner, M. K.: Interaction of biochemical quality and particle size of crop residues and its effect on the microbial biomass and nitrogen dynamics following incorporation into soil, Biol. Fert. Soils, 29, 319–327, https://doi.org/10.1007/s003740050559, 1999.
Birouste, M., Kazakou, E., Blanchard, A., and Roumet, C.: Plant traits and decomposition: Are the relationships for roots comparable to those for leaves?, Ann. Bot., 109, 463–472, https://doi.org/10.1093/aob/mcr297, 2012.
Burford, J. R. and Bremner, J. M.: Relationships between the denitrification capacities of soils and total, water-soluble and readily decomposable soil organic matter, Soil Biol. Biochem., 7, 389–394, https://doi.org/10.1016/0038-0717(75)90055-3, 1975.
Butterbach-Bahl, K., Baggs, E. M., Dannenmann, M., Kiese, R., and Zechmeister-Boltenstern, S.: Nitrous oxide emissions from soils?: how well do we understand the processes and their controls?, Philos. T. R. Soc. B, 368, 1–13, https://doi.org/10.1098/rstb.2013.0122, 2013.
Cantera, J. J. L. and Stein, L. Y.: Molecular diversity of nitrite reductase genes (nirK) in nitrifying bacteria, Environ. Microbiol., 9, 765–776, https://doi.org/10.1111/j.1462-2920.2006.01198.x, 2007.
Carlson, C. A. and Ingraham, J. L.: Comparison of denitrification by Pseudomonas stutzeri, Pseudomonas aeruginosa, and Paracoccus denitrificans, Appl. Environ. Microbiol., 45, 1247–1253, 1983.
Carro, L., Nouioui, I., Sangal, V., Meier-Kolthoff, J. P., Trujillo, M. E., Montero-Calasanz, M. D. C., Sahin, N., Smith, D. L., Kim, K. E., Peluso, P., Deshpande, S., Woyke, T., Shapiro, N., Kyrpides, N. C., Klenk, H. P., Göker, M., and Goodfellow, M.: Genome-based classification of micromonosporae with a focus on their biotechnological and ecological potential, Sci. Rep., 8, 1–23, https://doi.org/10.1038/s41598-017-17392-0, 2018.
Chantigny, M. H., Angers, D. A., Kaiser, K., and Kalbitz, K.: Extraction and Characterization of Dissolved Organic Matter, in: Soil Sampling and Methods of Analysis, 2nd Edn., edited by: Carter, M. R. and Gregorich, E. G., Taylor & Francis Group, International Standard Book Number-13: 978-0-8493-3586-0 (Hardcover), 2007.
Chen, H., Li, X., Hu, F., and Shi, W.: Soil nitrous oxide emissions following crop residue addition: a meta-analysis, Glob. Change Biol., 19, 2956–2964, https://doi.org/10.1111/gcb.12274, 2013.
Chen, S., Zhou, Y., Chen, Y., and Gu, J.: Fastp: An ultra-fast all-in-one FASTQ preprocessor, Bioinformatics, 34, i884–i890, https://doi.org/10.1093/bioinformatics/bty560, 2018.
Chin, K.-J., Liesack, W., and Janssen, P. H.: Opitutus terrae gen. nov., sp. nov., to accommodate novel strains of the division “Verrucomicrobia” isolated from rice paddy soil, Int. J. Syst. Evol. Microbiol., 51, 1965–1968, https://doi.org/10.1099/00207713-51-6-1965, 2001.
Ciais, P., Sabine, C., Bala, G., Bopp, L., Brovkin, V., Canadell, J., Chhabra, A., DeFries, R., Galloway, J., Heimann, M., Jones, C., Le Quéré, C., Myneni, R. B., Piao, S., and Thornton, P.: Carbon and Other Biogeochemical Cycles, in Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, 465–570, 2013.
Daniel, R. M., Limmer, A. W., Stelle, K. W., and Smith, I. M.: Anaerobic Growth, Nitrate Reduction and Denitrification in 46 Rhizobium Strains, J. Gen. Microbiol. 128, 1811–1815, https://doi.org/10.1099/00221287-128-8-1811, 1982.
de Menezes, A. B., McDonald, J. E., Allison, H. E., and McCarthy, A. J.: Importance of Micromonospora spp. as colonizers of cellulose in freshwater lakes as demonstrated by quantitative reverse transcriptase PCR of 16s rRNA, Appl. Environ. Microbiol., 78, 3495–3499, https://doi.org/10.1128/AEM.07314-11, 2012.
Endo, K., Hakamada, Y., Takizawa, S., Kubota, H., Sumitomo, N., Kobayashi, T., and Ito, S.: A novel alkaline endoglucanase from an alkaliphilic Bacillus isolate: Enzymatic properties, and nucleotide and deduced amino acid sequences, Appl. Microbiol. Biotechnol., 57, 109–116, https://doi.org/10.1007/s002530100744, 2001.
Fiedler, S. R., Augustin, J., Wrage-Mönnig, N., Jurasinski, G., Gusovius, B., and Glatzel, S.: Potential short-term losses of N2O and N2 from high concentrations of biogas digestate in arable soils, SOIL, 3, 161–176, https://doi.org/10.5194/soil-3-161-2017, 2017.
Fierer, N., Bradford, M. A., and Jackson, R. B.: Toward an Ecological Classification of Soil Bacteria, Ecology, 88, 1354–1364, https://doi.org/10.1890/05-1839, 2016.
Frimpong, K. A. and Baggs, E. M.: Do combined applications of crop residues and inorganic fertilizer lower emission of N2O from soil?, Soil Use Manage., 26, 412–424, https://doi.org/10.1111/j.1475-2743.2010.00293.x, 2010.
Gamble, T. N., Betlach, M. R., and Tiedje, J. M.: Numerically dominant denitrifying bacteria from world soils, Appl. Environ. Microbiol., 33, 926–939, 1977.
Garnier, S.: viridisLite: Default Color Maps from “matplotlib” (Lite Version), R package version 0.3.0, available at: https://cran.r-project.org/package=viridisLite, 2018.
Goering, H. K. and Van Soest, P. J.: Forage fiber analyses, Agric. Handb. No. 379, 12–20, 1970.
Griffiths, R. I., Whiteley, A. S., O'Donnell, A. G., and Bailey, M. J.: Rapid Method for Coextraction of DNA and RNA from Natural Environments for Analysis of Ribosomal DNA- and rRNA-Based Microbial Community Composition, Appl. Environ. Microbiol. 66, 1–5, https://doi.org/10.1128/AEM.66.12.5488-5491.2000, 2000.
Hayatsu, M., Tago, K., and Saito, M.: Various players in the nitrogen cycle: Diversity and functions of the microorganisms involved in nitrification and denitrification, Soil Sci. Plant Nutr., 54, 33–45, https://doi.org/10.1111/j.1747-0765.2007.00195.x, 2008.
Heiberger, R. M.: HH: Statistical Analysis and Data Display: Heiberger and Holland, R package version 3.1-35, available at: https://cran.r-project.org/package=HH, 2018.
Höper, H.: Carbon and nitrogen mineralisation rates of fens in Germany used for agriculture, in: Wetlands in Central Europe, edited by: Broll, G., Mehrbach, W., and Pfeiffer, E.-M., Springer, Berlin, Heidelberg, 149–164, 2002.
Houghton, K. M., Morgan, X. C., Lagutin, K., Mackenzie, A. D., Vyssotskii, M., Mitchell, K. A., McDonald, I. R., Morgan, H. W., Power, J. F., Moreau, J. W., Hanssen, E., and Stott, M. B.: Thermorudis pharmacophila sp. Nov., a novel member of the class Thermomicrobia isolated from geothermal soil, and emended descriptions of Thermomicrobium roseum, Thermomicrobium carboxidum, Thermorudis peleae and Sphaerobacter thermophilus, Int. J. Syst. Evol. Microbiol., 65, 4479–4487, https://doi.org/10.1099/ijsem.0.000598, 2015.
Hu, X., Liu, L., Zhu, B., Du, E., Hu, X., Li, P., Zhou, Z., Ji, C., Zhu, J., Shen, H., and Fang, J.: Asynchronous responses of soil carbon dioxide, nitrous oxide emissions and net nitrogen mineralization to enhanced fine root input, Soil Biol. Biochem., 92, 67–78, https://doi.org/10.1016/j.soilbio.2015.09.019, 2016.
Huang, Y., Zou, J., Zheng, X., Wang, Y., and Xu, X.: Nitrous oxide emissions as influenced by amendment of plant residues with different C:N ratios, Soil Biol. Biochem., 36, 973–981, https://doi.org/10.1016/j.soilbio.2004.02.009, 2004.
Hutchinson, G. and Mosier, A.: Improved soil cover method for field measurement of nitrous oxide fluxes, Soil Sci. Soc. Am. J., 45, 311–316, 1981.
Jenkinson, D. S.: Studies on the Decomposition Of Plant Material in Soil, I. Losses of Carbon from 14C labelled Ryegrass incubated with soil in the field, J. Soil Sci., 16, 104–115, https://doi.org/10.1111/j.1365-2389.1965.tb01424.x, 1965.
Jensen, L. S., Salo, T., Palmason, F., Breland, T. A., Henriksen, T. M., Stenberg, B., Pedersen, A., Lundström, C., and Esala, M.: Influence of biochemical quality on C and N mineralisation from a broad variety of plant materials in soil, Plant Soil, 273, 307–326, https://doi.org/10.1007/s11104-004-8128-y, 2005.
Jin, K., Sleutel, S., De Neve, S., Gabriels, D., Cai, D., Jin, J., and Hofman, G.: Nitrogen and carbon mineralization of surface-applied and incorporated winter wheat and peanut residues, Biol. Fert. Soils, 44, 661–665, https://doi.org/10.1007/s00374-008-0267-5, 2008.
Johnson, J. M.-F., Barbour, N. W., and Weyers, S. L.: Chemical Composition of Crop Biomass Impacts Its Decomposition, Soil Sci. Soc. Am. J., 71, 155–162, https://doi.org/10.2136/sssaj2005.0419, 2007.
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., and Glöckner, F. O.: Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies, Nucleic Acids Res., 41, 1–11, https://doi.org/10.1093/nar/gks808, 2013.
Koch, H., Lücker, S., Albertsen, M., Kitzinger, K., Herbold, C., Spieck, E., Nielsen, P. H., Wagner, M, and Daims, H.: Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the gens Nitrospira, PNAS, 112, 11371–11376, https://doi.org/10.1073/pnas.1506533112, 2015.
Kögel-Knabner, I.: The macromolecular organic composition of plant and microbial residues as inputs to soil organic matter, Soil Biol. Biochem., 34, 139–162, https://doi.org/10.1016/S0038-0717(01)00158-4, 2002.
Kravchenko, A. N., Toosi, E. R., Guber, A. K., Ostrom, N. E., Yu, J., Azeem, K., Rivers, M. L., and Robertson, G. P.: Hotspots of soil N2O emission enhanced through water absorption by plant residue, Nat. Geosci., 10, 496–500, https://doi.org/10.1038/NGEO2963, 2017.
Kravchenko, A. N., Fry, J. E., and Guber, A. K.: Water absorption capacity of soil-incorporated plant leaves can affect N2O emissions and soil inorganic N concentrations, Soil Biol. Biochem., 121, 113–119, https://doi.org/10.1016/j.soilbio.2018.03.013, 2018.
Kuzyakov, Y. and Domanski, G.: Carbon input by plants into the soil. Review, J. Plant Nutr. Soil Sci., 163, 421–431, https://doi.org/10.1002/1522-2624(200008)163:4<421::AID-JPLN421>3.0.CO;2-R, 2000.
Ladd, J. N., Oades, J. M., and Amato, M.: Microbial biomass formed from 14C, 15N-labelled plant material decomposing in soils in the field, Soil. Biol. Biochem., 13, 119–162, https://doi.org/10.1016/0038-0717(81)90007-9, 1981.
Legendre, P. and Gallagher, E. D.: Ecologically meaningful transformations for ordination of species data, Oecologia, 129, 271–280, https://doi.org/10.1007/s004420100716, 2001.
Lemon, J.: Plotrix: a package in the red light district of R, R-News, 6, 8–12, 2006.
Lenth, R.: Emmeans: Estimated Earginal Means, aka Least-Squares Means. R package version 1.3.4, available at: https://cran.r-project.org/package=emmeans, 2018.
Li, X., Hu, F., and Shi, W.: Plant material addition affects soil nitrous oxide production differently between aerobic and oxygen-limited conditions, Appl. Soil Ecol., 64, 91–98, https://doi.org/10.1016/j.apsoil.2012.10.003, 2013.
Li, X., Sørensen, P., Olesen, J. E., and Petersen, S. O.: Evidence for denitrification as main source of N2O emission from residue-amended soil, Soil Biol. Biochem., 92, 153–160, https://doi.org/10.1016/j.soilbio.2015.10.008, 2016.
Loecke, T. D. and Robertson, G. P.: Soil resource heterogeneity in terms of litter aggregation promotes nitrous oxide fluxes and slows decomposition, Soil Biol. Biochem., 41, 228–235, https://doi.org/10.1016/j.soilbio.2008.10.017, 2009.
Lou, Y., Ren, L., Li, Z., Zhang, T., and Inubushi, K.: Effect of Rice Residues on Carbon Dioxide and Nitrous Oxide Emissions from a Paddy Soil of Subtropical China, Water. Air. Soil Pollut., 178, 157–168, https://doi.org/10.1007/s11270-006-9187-x, 2007.
Ludovici, K. H. and Kress, L. W.: Decomposition and nutrient release from fresh and dried pine roots under two fertilizer regimes, Can. J. Forest Res., 36, 105–111, https://doi.org/10.1139/X05-227, 2006.
Lyngwi, N. A. and Joshi, S.: Economically important Bacillus and related genera: a mini review, in: Biology of Useful Plants and Microbes, edited by: Sen, A., Narosa Publishing House, New Delhi, India, 33–43, 2014.
Machinet, G. E., Bertrand, I., Chabbert, B., and Recous, S.: Decomposition in soil and chemical changes of maize roots with genetic variations affecting cell wall quality, Eur. J. Soil Sci., 60, 176–185, https://doi.org/10.1111/j.1365-2389.2008.01109.x, 2009.
Machinet, G. E., Bertrand, I., Barrière, Y., Chabbert, B., and Recous, S.: Impact of plant cell wall network on biodegradation in soil: Role of lignin composition and phenolic acids in roots from 16 maize genotypes, Soil Biol. Biochem., 43, 1544–1552, https://doi.org/10.1016/j.soilbio.2011.04.002, 2011.
Martin, M.: Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet J., 17, 10–12, https://doi.org/10.14806/ej.17.1.200, 2013.
Mary, B., Fresneau, C., Morel, J. L., and Mariotti, A.: C and N cycling during decomposition of root mucilage, roots and glucose in soil, Soil Biol. Biochem., 25, 1005–1014, https://doi.org/10.1016/0038-0717(93)90147-4, 1993.
Megonigal, J. P., Hines, M. E., and Visscher, P. T.: Anaerobic Metabolism: Linkages to Trace Gases and Aerobic Processes, in: Treatise on Geochemistry, Vol. 10, 2nd Edn., edited by: Turekian, K. and Holland, H., Volume Editiors: Karl, D. M. and Schlesinger, W. H., https://www.sciencedirect.com/referencework/9780080983004/treatise-on-geochemistry, 10, 273–359, 2013.
Millar, N. and Baggs, E. M.: Chemical composition, or quality, of agroforestry residues influences N2O emissions after their addition to soil, Soil Biol. Biochem., 36, 935–943, https://doi.org/10.1016/j.soilbio.2004.02.008, 2004.
Millar, N. and Baggs, E. M.: Relationships between N2O emissions and water-soluble C and N contents of agroforestry residues after their addition to soil, Soil Biol. Biochem., 37, 605–608, https://doi.org/10.1016/j.soilbio.2004.08.016, 2005.
Miller, M. N., Zebarth, B. J., Dandie, C. E., Burton, D. L., Goyer, C., and Trevors, J. T.: Crop residue influence on denitrification, N2O emissions and denitrifier community abundance in soil, Soil Biol. Biochem., 40, 2553–2562, https://doi.org/10.1016/j.soilbio.2008.06.024, 2008.
Müller, C. and Clough, T. J.: Advances in understanding nitrogen flows and transformations: Gaps and research pathways, J. Agr. Sci., 152, S34–S44, https://doi.org/10.1017/S0021859613000610, 2014.
Nolan, M., Tindall, B. J., Pomrenke, H., Lapidus, A., Copeland, A., Del Rio, T. G., Lucas, S., Chen, F., Tice, H., Chen, J.-F., Saunders, E., Han, C., Bruce, D., Goodwin, L., Chain, P., Pitluck, S., Ovchinikova, G., Pati, A., Ivanova, N., Mavromatis, K., Chen, A., Palaniappan, K., Land, M., Hauser, L., Chang, Y.-J., Jeffries, C. D., Brettin, T., Göker, M., Bristow, J., Eisen, J. A., Markowitz, V., Hugenholtz, P., Kyrpides, N. C., Klenk, H.-P., and Detter, J. C.: Complete genome sequence of Rhodothermus marinus, Stand. Genomic Sci., 1, 283–291, 2009.
Novoa, R. S. A. and Tejeda, H. R.: Evaluation of the N2O emissions from N in plant residues as affected by environmental and management factors, Nutr. Cycl. Agroecosys., 75, 29–46, https://doi.org/10.1007/s10705-006-9009-y, 2006.
Okada, N., Nomura, N., Nakajima-Kambe, T., and Uchiyama, H.: Characterization of the Aerobic Denitrification in Mesorhizobium sp. Strain NH-14 in Comparison with that in Related Rhizobia, Microbes Environ., 20, 208–215, https://doi.org/10.1264/jsme2.20.208, 2005.
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., Minchin, P. R., O'Hara, R. B., Simpson, G. L., Solymos, P., Henry, M., Stevens, H., Szoecs, E., and Wagner, H.: vegan: Community Ecology Package, R Package version 2.3-2, available at: https://cran.r-project.org/web/packages/vegan/index.html, 2015.
Papen, H., von Berg, R., Hinkel, I., Thoene, B., and Rennenberg, H.: Heterotrophic Nitrification by Alcaligenes faecalis: , , N2O, and NO Production in Exponentially Growing Cultures, Appl. Environ. Microbiol., 55, 2068–2072, 1989.
Pascault, N., Ranjard, L., Kaisermann, A., Bachar, D., Christen, R., Terrat, S., Mathieu, O., Lévêque, J., Mougel, C., Henault, C., Lemanceau, P., Péan, M., Boiry, S., Fontaine, S., and Maron, P. A.: Stimulation of Different Functional Groups of Bacteria by Various Plant Residues as a Driver of Soil Priming Effect, Ecosystems, 16, 810–822, https://doi.org/10.1007/s10021-013-9650-7, 2013.
Paterson, E. and Sim, A.: Rhizodeposition and C-partitioning of Lolium perenne in axenic culture affected by nitrogen supply and defoliation, Plant Soil, 216, 155–164, https://doi.org/10.1023/A:1004789407065, 1999.
Pinheiro, J., Bates, D., DebRoy, S., Sarkar, D., and R Core Team: nlme: Linear and Nonlinear Mixed Effects Models, R package version 3.1-140, available at: https://cran.r-project.org/package=nlme, 2017.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glöckner, F. O.: The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools, Nucleic Acid. Res., 41, 590–596, https://doi.org/10.1093/nar/gks1219, 2013.
R Core Team: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, version 3.5.2, available at: https://www.R-project.org/, 2018.
R Core Team: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, version 3.5.3, available at: https://www.R-project.org/, 2019.
Redin, M., Guénon, R., Recous, S., Schmatz, R., de Freitas, L. L., Aita, C., and Giacomini, S. J.: Carbon mineralization in soil of roots from twenty crop species, as affected by their chemical composition and botanical family, Plant Soil, 378, 205–214, https://doi.org/10.1007/s11104-013-2021-5, 2014.
Rivas, R., Trujillo, M. E., Mateos, P. F., Martínez-Molina, E., and Velázquez, E.: Agromyces ulmi sp. nov., xylanolytic bacterium isolated from Ulmus nigra in Spain, Int. J. Syst. Evol. Microbiol., 54, 1987–1990, https://doi.org/10.1099/ijs.0.63058-0, 2004.
Robertson, G. P. and Groffman, P. M.: Nitrogen Transformations, in: Soil microbiology, ecology and biochemistry, edited by: Paul, E. A., Academic Press, Burlington, Massachusetts, USA, 421–446, 2015.
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F.: VSEARCH: a versatile open source tool for metagenomics, Peer J., 4, e2584, https://doi.org/10.7717/peerj.2584, 2016.
Rousk, J. and Bååth, E.: Fungal and bacterial growth in soil with plant materials of different C:N ratios, FEMS Microbiol. Ecol., 62, 258–267, https://doi.org/10.1111/j.1574-6941.2007.00398.x, 2007.
Silver, W. L. and Miya, R. K.: Global patterns in root decomposition: Comparisons of climate and litter quality effects, Oecologia, 129, 407–419, https://doi.org/10.1007/s004420100740, 2001.
Soetaert, K.: plot3D: Plotting Multi-Dimensional Data, R package version 1.1.1, available at: https://cran.r-project.org/package=plot3D, 2017.
Toju, H., Tanabe, A. S., Yamamoto, S., and Sato, H.: High-coverage ITS primers for the DNA-based identification of Ascomycetes and Basidiomycetes in environmental samples, PLoS One, 7, e40863, https://doi.org/10.1371/journal.pone.0040863, 2012.
Truong, T. H. H. and Marschner, P.: Respiration, available N and microbial biomass N in soil amended with mixes of organic materials differing in C:N ratio and decomposition stage, Geoderma, 319, 167–174, https://doi.org/10.1016/j.geoderma.2018.01.012, 2018.
van Groenigen, J. W., Huygens, D., Boeckx, P., Kuyper, Th. W., Lubbers, I. M., Rütting, T., and Groffman, P. M.: The soil N cycle: new insights and key challenges, SOIL, 1, 235–256, https://doi.org/10.5194/soil-1-235-2015, 2015.
VDLUFA (Verband Deutscher Landwirtschaftlicher Untersuchungs- und Forschungsanstalten e.V.) (Ed.): 6.5.2 Bestimmung der Säure-Detergentien-Faser (ADF), in Methodenbuch Band III Die chemische Untersuchung von Futtermitteln, VDLUFA-Verlag, Darmstadt, 1–4, 2011.
VDLUFA (Ed.): 6.5.1 Bestimmung der neutral-Detergentien-Faser (NDF), in Methodenbuch Band III Die chemische Untersuchung von Futtermitteln, VDLUFA-Verlag, Darmstadt, 1–4, 2012a.
VDLUFA (Ed.): 6.5.3 Bestimmung des Säure-Detergentien-Lignings (“Rohlignin”), in Methodenbuch Band III Die chemische Untersuchung von Futtermitteln, VDLUFA-Verlag, Darmstadt, 1–3, 2012b.
Velthof, G. L., Kuikman, P. J., and Oenema, O.: Nitrous oxide emission from soils amended with crop residues, Nutr. Cycl. Agroecosys., 62, 249–261, https://doi.org/10.1023/A:1021259107244, 2002.
Verbaendert, I., De Vos, P., Boon, N., and Heylen, K.: Denitrification in Gram-positive bacteria: an underexplored trait, Biochem. Soc. T., 39, 254–258, https://doi.org/10.1042/bst0390254, 2011.
Wang, Y., Naumann, U., Eddelbuettel, E., Wilshire, J., Warton, D., Byrnes, J., dos Santos Silva, R., Niku, J., Renner, I., and Wright, S.: mvabund: Statistical Methods for Analysing Multivariate Abundance Data, R package version 4.0.1, available at: https://cran.r-project.org/web/packages/mvabund/index.html, 2019.
Wrage-Mönnig, N., Horn, M. A., Well, R., Müller, C., Velthof, G., and Oenema, O.: The role of nitrifier denitrication in the production of nitrous oxide revisited, Soil Biol. Biochem., 123, A3–A16, https://doi.org/10.1016/j.soilbio.2018.03.020, 2018.
Wrage, N., Velthof, G. L., van Beusichem, M. L., and Oenema, O.: The role of nitrifier denitrification in the production of nitrous oxide, Soil Biol. Biochem., 33, 1723–1732, https://doi.org/10.1016/S0038-0717(01)00096-7, 2001.
Zeng, J., Liu, X., Song, L., Lin, X., Zhang, H., Shen, C., and Chu, H.: Nitrogen fertilization directly affects soil bacterial diversity and indirectly affects bacterial community composition, Soil Biol. Biochem., 92, 41–49, https://doi.org/10.1016/j.soilbio.2015.09.018, 2016.
Zgurskaya, H. I., Evtushenko, L. I., Akimov, V. N., Voyevoda, H. V., Dobrovolskaya, T. G., Lysak, L. V., and Kalakoutskii, L. V.: Emended Description of the Genus Agromyces and Description of Agromyces cerinus subsp. cerinus sp. nov., subsp. nov., Agromyces cerinus subsp. nitratus sp. nov., subsp. nov., Agromyces fucosus subsp. fucosus sp. nov., subsp. nov., and Agromyces fucosus subsp. hippuratus sp. nov., subsp. nov., Int. J. Syst. Bacteriol., 42, 635–641, https://doi.org/10.1099/00207713-42-4-635, 1992.
Zhang, D., Hui, D., Luo, Y., and Zhou, G.: Rates of litter decomposition in terrestrial ecosystems: global patterns and controlling factors, J. Plant Ecol., 1, 85–93, https://doi.org/10.1093/jpe/rtn002, 2008.
Zhang, J., Kobert, K., Flouri, T., and Stamatakis, A.: PEAR: A fast and accurate Illumina Paired-End reAd mergeR, Bioinformatics, 30, 614–620, https://doi.org/10.1093/bioinformatics/btt593, 2014.
Zhang, J., Müller, C., and Cai, Z.: Heterotrophic nitrification of organic N and its contribution to nitrous oxide emissions in soils, Soil Biol. Biochem., 84, 199–209, https://doi.org/10.1016/j.soilbio.2015.02.028, 2015.
Zhang, X. and Wang, W.: The decomposition of fine and coarse roots: Their global patterns and controlling factors, Sci. Rep., 5, 1–10, https://doi.org/10.1038/srep09940, 2015.
Zumft, W.: Cell biology and molecular basis of denitrification, Microbiol. Molecular Biol. Rev., 61, 533–616, 1997.