the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
On-site microbiome study of silica structures in a subterranean Mars analog environment
Martina Cappelletti
Giacomo Broglia
Andrea Firrincieli
Ettore Lopo
Alice Checcucci
Daniele Ghezzi
Federico Pisani
Freddy Vergara
Bruno Casarotto
Francesco Sauro
Amorphous silica deposits found in orthoquartzite caves offer valuable analogues for understanding early life on Earth and potential biosignatures on Mars. This study presents the fully on-site microbial community analysis of silica stromatolite-like structures in the ancient and remote orthoquartzite cave Imawarí Yeutá (Auyan Tepui, Venezuela). Using a portable laboratory setup, we performed ATP-based microbial activity assessments and the full DNA-based analysis workflow directly in the cave, without internet access or high computational resources. The data obtained in the cave were then validated in the laboratory using a standard bioinformatics pipeline, qPCR and Biolog EcoPlate assays. The sequencing results revealed that the microbial communities in the stromatolite differ from other biofilms on the cave floor for the higher abundance of Actinobacteriota (particularly the genus Crossiella) and members of Subgroup 13 (Acidobacteriota) suggesting a possible role in the stromatolite formation/development. The ATP-based and Biolog results indicated that the most metabolically active microorganisms are localized in the white layer/colonies at basis of the stromatolite suggesting that the stromatolite development occurs at the interface of this structure with the quartz rock. These findings validate the feasibility of real-time microbial analyses in remote caves with astrobiological interest and provide novel understanding on the microbiological aspects involved in the formation of the silica stromatolites in non-thermal and aphotic environments.
- Article
(8251 KB) - Full-text XML
-
Supplement
(912 KB) - BibTeX
- EndNote
Amorphous silica deposits are considered modern analogues of siliceous formations originating on Earth during the Precambrian period, providing useful information for studying the evolution of life on our planet. These deposits also have astrobiological interest showing similar composition and morphological characteristics of amorphous silica observed on the surface of Mars by the rover Spirit and by various multispectral cameras onboard of orbital mission (Ruff et al., 2011; Ruff and Farmer, 2016). On Mars, secondary silica deposits could have formed both on the surface and in the subsurface either through abiotic processes (due to weathering and hydrothermal alteration) or through microbial processes over long periods of time. Given the current environmental conditions hostile to life on the surface of other planets, underground environments represent promising study targets. Indeed, in these sites, the microbial life could have found conditions favourable to proliferation shielded from the high doses of UV and cosmic radiations hitting the martian surface at present time (Boston, 2010). The study of microbial communities interacting with silica and the biological contribution to secondary silica deposits in caves can provide useful information to interpret evidence of possible life forms encrusted in silica deposits on Mars.
Aside of the more common and studied silica sinters in hydrothermal environments, similar non-thermal amorphous silica deposits have been reported during the last two decades in several quartz-rich cave environments (quartz-sandstones, meta-quartzites, granites, lava tubes etc.) (Auler and Sauro, 2019; Daza Brunet and Bustillo Revuelta, 2014; Miller et al., 2014; Wray and Sauro, 2017). In particular, the exploration of the giant and ancient orthoquartzite caves in the table mountains tepuis of Venezuela and Brazil has led to the discovery of amorphous silica deposits that are novel types of silica stromatolites with distinct characteristics, much bigger dimension and spectacular morphologies as compared to those described in volcanic caves (Aubrecht et al., 2008; Lundberg et al., 2018; Sauro et al., 2018). The formation of these deposits in geochemically stable, non-thermal and aphotic environments is associated with microbial activities that are still mostly unknown (Ghezzi et al., 2021; Sauro et al., 2018).
In recent years, there has been a growing interest in the development of cost-effective, portable detection systems suitable for various in situ applications, including environmental monitoring, pathogen detection in clinical and food safety contexts, and astrobiological investigations in remote and extreme environments (Quintela et al., 2022). Among them, nucleic acid-based analyses provide information on the presence and abundance of specific microbes that can be involved in the interaction with the surrounding setting and with its modification. Recent progress in sequencing technologies have allowed to carry out DNA sequencing outside of the laboratory allowing microbiome analysis to be conducted in situ. The advantages of in situ DNA analyses include to avoid sample degradation, to limit problems with transportation and to allow field-based pre-screening for informed sample selection. Furthermore, DNA sequencing is also seen as possible method to detect life during astrobiology missions and in extraterrestrial settings (Maggiori et al., 2020; Mojarro et al., 2019).
In this work we carried out on-field microbiological analyses of the silica stromatolite-like structures, which are biosignatures with astrobiological interest, directly in the cave Imawarí Yeutá. We carried out the entire DNA-based analysis workflow including DNA extraction, amplification, sequencing, data analysis and results visualization directly in the cave by applying a tailored bioinformatic pipeline that we developed to process sequencing data in under 20 min (per sample) without internet access or high computational resources. A portable ATP detector was also used to assess microbial activity levels in the cave that were then validated though qPCR and Biolog assay in the lab. The results provided novel understanding on the microbial communities and activities characterizing silica stromatolites development.
2.1 The Imawarí Yeutá cave system
Imawarí Yeutá is an ancient and pristine orthoquartzite cave located on the table-top mountain Auyan Tepui in Venezuela. The cave is located at approximately one hundred meters of depth below the tepui plateau surface (between 2000 and 1900 m above sea level). The cave has been carved in at least 20–30 million years by erosion and weathering (arenization) of infiltrating waters. The geological setting and speleogenetic processes of the cave have been extensively described previously (Mecchia et al., 2014; Sauro, 2014; Sauro et al., 2019; Wray and Sauro, 2017). The cave presents stable physical-geochemical conditions, with a constant temperature between 15 and 18 °C and acidic waters with a pH ranging from 3 to a maximum of 6.1 (Mecchia et al., 2014). After few meters from the entrances, the cave is characterized by a total absence of light. Air flows and water streams are concentrated along few main branches, while extensive areas of the cave are abandoned by flowing waters since millions of years. Most of the silica stromatolites are found in these quiet environments where very limited air and water exchanges result in canonical oligotrophic conditions, with low nutrient availability and scarce organic carbon sources. In this regard, the innermost zones of quartzite caves in the tepui mountains are considered oligotrophic ecosystems, with organic nutrient concentrations close to undetectable levels (as reported in Barton et al., 2014, Ghezzi et al., 2022; Mecchia et al., 2014; Sauro et al., 2018). Unlike the external surface, which receives constant inputs from soils and vegetation, the cave interior is deprived of external nutrient sources and lack primary production associated to light-driven photosynthesis.
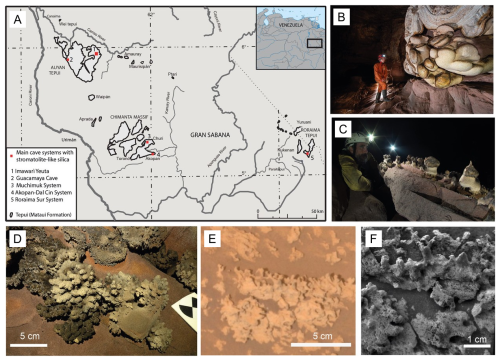
Figure 1(a) Location of Imawarí Yeutá and other cave systems in quartz-sandstones where the presence of silica stromatolite-like deposits have been reported (Wray and Sauro, 2017). (b, c) Different types of silica speleothems growing on quartz-sandstone walls and boulders (photos Vittorio Crobu – La Venta). (d) Silica stromatolitic digitate structures on the floors of Imawarí Yeutá, showing similar morphologies to opaline silica found by the Rover Spirit in the Home Plate outcrop on Mars (e, f; from Ruff and Farmer, 2016).
2.2 Cave silica stromatolites
Silica stromatolites are usually well-known in hot-spring sinters (Konhauser et al., 2004) where ascending hydrothermal waters generated in deep-hot reservoirs contains high quantity of dissolved silica (Gunnarsson and Arnórsson, 2000), and supersaturation is caused by cooling and evaporation at the ∼ 100–70 °C spring temperature. However, it is important to underline that there is no affinity between quartz-rich cave environments and hot-spring conditions. In Imawarí Yeutá cave the temperature is nearly constant throughout the year, atmospheric pressure is in the same order of surface pressure and SiO2 solubility in fractures or cave walls (i.e. where silica is mostly dissolved) equals SiO2 solubility at the surface (Mecchia et al., 2014). In addition, subsurface silica stromatolites in the caves of the tepui table mountains of the Guyana Shield are always forming over boulders or along the walls, in wet or dry conditions, but never submerged in aquatic environments. The stromatolitic morphology and the peculiar geochemical conditions suggest that amorphous silica precipitation is also mediated by microbiological activities (Sauro et al., 2018).
Imawarí Yeutá and other cave systems explored in the Venezuelan tepuis host a wide variety of silica stromatolites and speleothems (Fig. 1; Sauro et al., 2013) showing a variety of morphologies (bulbous-, columnar-, mushroom-, egg-and coral- like forms), most of them characterised by thin stromatolitic layers constituted of amorphous silica (amorphous gels and Opal-A) encrusting biological components (cells, filaments, EPS, etc.) (Figs. 1 and 2).
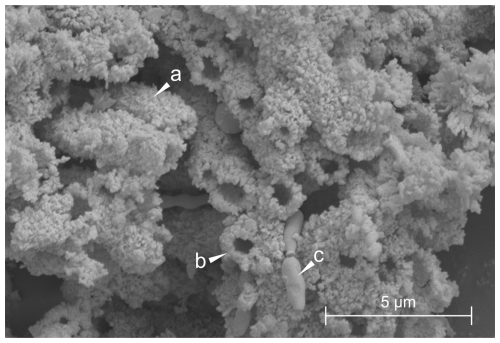
Figure 2SEM/FESEM image of a sample collected from the white paste of a silica stromatolite-like formation in Imawarí Yeutá. Arrows indicate (a) botryoidal masses of precipitated amorphous silica, (b) the section of a tubular cast, (c) a bacterium-like structure.
In some cases, coralloid and branched stromatolites are morphologically similar to the silica deposits found in the Home Plate site analyzed by the Spirit rover on Mars (Fig. 1d–f). For this specific in-situ analysis study we have focused on one of the more classical type of stromatolites, described by (Aubrecht et al., 2012) as mushroom- or globular-shaped stromatolites with chalky peloidal layers, white on the surface and interior, with greyish darker and harder material caps. These deposits formed over deeply weathered orthoquartzite boulder or along ledges on the walls (Fig. 1).
In order to test and compare the results with different cave niches, the study has also analyzed two patinas on the floors (white and yellow) not associated with stromatolites but with typical microbial colonies growth (Table S1 in the Supplement).
3.1 Hyperspectral imaging for amorphous silica identification
The hyperspectral signature of the stromatolite was obtained with a Headwall Photonics Micro-Hyperspec push-broom camera, with a spectral range 900 to 2500 nm, 170 spectral bands, 384 spatial bands, FWHM 10 nm. The set-up housing for hyperspectral acquisitions consists of a camera holder and a high-precision 20 cm × 20 cm motorised stage, while the illumination is equipped with a 150 W tiltable halogen lamp with a frosted glass diffuser and reflector. A Spectralon (Labsphere, Inc.) with a reflectivity of 99 % was used as a white reference for radiance conversion.
3.2 Sampling and laboratory setting
A field laboratory was set up under a 3 m × 3 m tent around 200 m from the entrance of the cave. The chosen area consisted of an isolated cave recess, distant from the main routes of the cave that connect the entrance with the base camp located inside. Further, in this area there were no water flows or drippings, and no wind currents. The tent was properly closed so as not to allow insects or other small animals to enter. The laboratory was equipped with a table and common laboratory instruments essential for basic molecular experimental procedures (pipettes, tips, microtubes). Large instrumentation included a vortex (Vortex-Genie 2), the Bento Lab portable PCR workstation to conduct DNA extraction, 16S rRNA amplification, and amplicon visualization through gel electrophoresis, the MinION Mk1C to carry out DNA sequencing, and a high-performance computer for in situ data analyses (Lenovo laptop with 4 CORE, 32 Gb RAM, 1 TB hard-disk).
Sampling was performed using sterile procedures. Samples were collected in areas of the cave located approximately 350 m in a straight line from the temporary laboratory, corresponding to about 650 m along the actual path. Samples were collected inside microtubes that were transported to the temporary laboratory and immediately processed for total microbial DNA extraction.
Electricity was provided through a Honda 1 kW power generator installed outside of the cave with a cable running for more than 200 m to the interior until reaching the field laboratory.
3.3 Bioluminometer for ATP-based measurement in the cave
Bioluminometer (RingBio™) is a hand-held instrument which allows the quantification of adenosine triphosphate (ATP) that is directly proportional to the metabolic activity of microbial cells present in a sample. This type of analysis was used to have indications on the microbial activity in cave samples/areas. For this purpose, the selected areas were swabbed, then the swab was placed inside the ATP detector/bioluminometer and the test activated. The assay is based on the use of an enzymatic reaction constituted by the “luciferase - luciferin system” that produces an amount of light that is directly proportional to the amount of ATP present in the sample and is expressed as Relative Light Units (RLU). The test was repeated three times by scrubbing in a standardized way (areas of around 1 cm × 1 cm) three different areas of the same type of sample under analysis.
3.4 DNA extraction, 16S rRNA gene amplification, barcoding and sequencing in the cave laboratory
Total microbial DNA extractions were carried out using the DNeasy PowerLyzer PowerSoil Kit (Qiagen) as previously described (Ghezzi et al., 2022). Since we did not have an analytical balance to measure the amount of sample to extract (up to 250 mg per sample according to the manufacturer's protocol), we arbitrarily added an amount of sample that filled approximately one quarter of the volume of the Qiagen tube with the PowerBead solution. 1 µL of the extracted DNA was used to perform amplification reactions using the Bento Lab PCR Workstation. Full-length 16S rRNA genes (V1-V9) were amplified in 50 µL reaction mix using 27F (5'-AGAGTTTGATCMTGGCTCAG-3') and 1492R (5'-CGGTTACCTTGTTACGACTT-3') primer pair with 16S Barcoding Kit (SQK-RAB204, Oxford Nanopore Technologies, Oxford, UK) and the Phanta Max Super-Fidelity DNA Polymerase (Vazyme) following the manufacturer's protocol. The amplifications were carried out using the following settings: initial denaturation for 1 min at 95 °C; 30 cycles of denaturation for 15 s at 95 °C, annealing for 15 s at 56 °C, extension for 90 s at 72 °C; final extension for 5 min at 72 °C. PCR reactions were checked through gel electrophoresis. Barcoded amplicons were pooled together to yield a final amount of 1 µg of multiple barcoded DNA and processed for end repair and dA-tailing using the NEBNext Companion Module for ONT Ligation Sequencing (New England Biolabs, E7180S). The library was purified using AMPure XP beads (Beckman Coulter Diagnostics, USA, CA) and loaded onto an electrophoresis gel for approximate quantification. After priming of the flow cell with the Flow Cell Priming Kit (EXP-FLP002), 100 fmol (equivalent to approximately 100 ng for 16S rRNA amplicons) of the purified DNA library was loaded onto an R9.5 flow cell (Flow Cell Mk I, R9.4, FLO-MIN106) according to the manufacturer's instructions. Finally, a sequencing run of approximately 2 h was performed, with real time basecalling enabled, and carried out directly in situ using the MinKNOW software.
3.5 Bioinformatic analyses: pipelines applied in the cave and in the laboratory and statistical analyses of the sequencing results
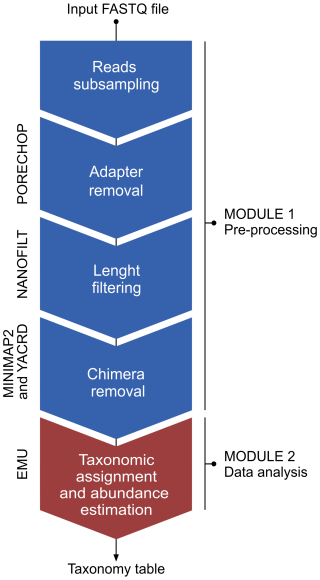
Cave procedure pipeline was specifically created for the in-situ analysis of 16S rRNA gene sequences base called using the “fast” mode basecalling model through the MinKNOW software integrated in the MinION Mk1C sequencing device. The main objective was the implementation of a bioinformatic pipeline able to complete the analysis of 16S rRNA sequences for each sample in less than 20 min (due to electrical power limitation) and without internet connection. The pipeline was entirely based on bash (unix) and the general workflow is illustrated in Fig. 3.
The pipeline consists of two modules, i.e., a pre-processing module and a classify module. In the pre-processing module, the raw reads are processed through the following steps: (i) random subsampling at 25 % with reformat.sh from BBMap package v38.98 (https://sourceforge.net/projects/bbmap/, Bushnell, 2022), (ii) adapter removal by Porechop v0.2.4 (https://github.com/rrwick/Porechop, Wick, 2018), (iii) length (1200–1800 bp) and quality (≥ 9) filtering using Nanofilt v2.6.0 (https://github.com/wdecoster/nanofilt, last access: 27 October 2025; De Coster et al., 2018), (iv) chimera removal through yacrd vIvysaur using recommended settings for ONT data (https://github.com/natir/yacrd, last access: 27 October 2025; Marijon et al., 2020). The second module enabled taxonomy classification and calculation of taxa abundances of the filtered reads against the prebuilt SILVA database v138.1 using the EMU classifier v3.4.5 (Curry et al., 2022). The time requirement for the entire workflow was under 15 min, considering approximately 25 000 to 30 000 reads processed for each sample after the initial subsampling at 25 %.
For the lab procedure, the same data analysis workflow was followed except for the base calling modality and the subsampling. The reads processed by the lab procedure were base-called using “super accurate” mode with Guppy v6.11. This mode requires access to a machine with a GPU to complete the basecalling within an acceptable timeframe. Such GPUs are often available on HPC nodes whose access requires an internet connection or in high-performing laptops that still would require several hours to complete the basecalling. The direct comparison between the two pipelines is indicated in Table S2 in the Supplement.
The abundance data generated from EMU were combined to produce sample-wise taxon-specific abundance tables at the Phylum, Class, Order, Family, and Genus levels, with taxa showing abundances below 1 % grouped under the category “others”. Visualisation of compositional data through bubble plots (Figs. 11 and S2 in the Supplement) was performed using ggplot.
Alpha and beta diversity metrics were calculated using the R package Vegan v2.6.4. The R package ComplexHeatmap and ggplot2 were used to visualise Genus-level Bray–Curtis dissimilarities between stromatolite (ST-w and ST-b) and biofilm (D-w, P-w, P-y) samples (Figs. 10 and S1 in the Supplement)
The goodness-of-fit of taxonomic distributions at different levels between the lab and cave pipelines was assessed using a Chi-square test, implemented through the chisq.test function of the R stats package (v3.6.2). This statistical test allows the assessment of whether the distribution of taxa in the cave_pipeline deviated significantly from that of the lab_pipeline. The shell scripts used to process sequencing data into EMU abundances, as well as the R scripts used for statistical analyses, beta and alpha diversity analyses, and visualisation of the results, are available on Figshare DOI: https://doi.org/10.6084/m9.figshare.29514197.
3.6 Bacterial quantification through qPCR
The quantification of the community 16S rRNA genes was conducted in triplicates through qPCR by using the CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories Inc., Hercules, USA). qPCR reaction mixes were prepared in a final volume of 10 µL containing 1 µL of the extracted DNA, universal primers 926F and 1026R (300 nM each) (Ghezzi et al. 2024), and 1× AceQ qPCR SYBER Green Master Mix (Vazyme Biotech Co., Nanjing, China). The standard curve (R2 > 0.99) was generated by using serial dilutions of known amounts of 16S rRNA gene PCR products of Escherichia coli as template. Finally, DNA amplification was carried out using the following thermocycling conditions: 95 °C for 5 min, 40 cycles of 95 °C for 10 s and 60 °C for 30 s.
3.7 Metabolic activity analyses through Biolog Ecoplate assay
To assess the metabolic activity and the carbon source utilization profile of microbial communities, samples isolated from different stromatolite-like structures were screened using Biolog Ecoplate (Biolog, Inc., Hayward, CA, USA). The 96-well microplates are composed by three replicate sets of 31 lyophilized relevant carbon substrates together with a tetrazolium redox dye (Insam, 1997). Ecoplates carbon sources can be grouped by chemical class (carbohydrates, carboxylic acids, complex carbon sources, amino acids, and amines, as listed in Table S3 in the Supplement). Microbial substrate utilization is indicated by color changes of the tetrazolium redox dye in each well. This technology relies on active cell metabolism, where the dye is reduced by NADH produced during respiration, leading to the formation of the purple compound formazan. 0.1 g of sediments were first resuspended in 10 mL of 0.1 % Na2H2P2O7 (pH 7), shacked on a tilting table for 2 h to favour cell detachment and filtered through Labor filter paper (50 cm × 50 cm, 67 g m−2) to avoid interference with spectrophotometric reading. 150 µL of the resulting suspension were then added to the wells of the microplates and incubated in the dark at 20°C. Absorbance changes were monitored during the incubation at a wavelength of 595 nm using an EnSpire Multimode Plate Reader (Perkin Elmer, USA), which recorded the level of substrate consumption represented by colour change in every well. Two incubation periods were selected, i.e., 5 and 14 d. The first one corresponds to the time that is typically used in Biolog experiments, while the second one was selected because previously adopted in studies analysing microbial metabolic activities in cave samples using Ecoplates (O'Connor et al., 2021). After inoculation, the Ecoplates were sealed with Parafilm and placed inside sealed containers containing water-soaked absorbent paper to maintain humidity to avoid volume loss due to dryness. The metabolic activity value (MAV) was represented by the recorded OD595 for every timepoint.
4.1 Silica stromatolite-like structures as potential biosignatures with astrobiological interest
The samples analysed in this study were collected from different areas of stromatolite-like structures (representative image of the stromatolites are shown in Fig. 1) that were selected depending on their macromorphology, based on previous reports of silica stromatolites in the caves of the tepuis (Aubrecht et al., 2012; Sauro et al., 2014).
Moreover, three additional samples were collected from microbial biofilms that were present on the floor of the cave. The sample description is reported in Table S1.
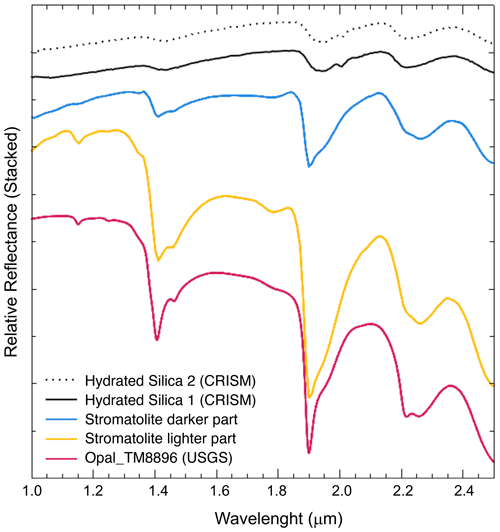
The hyperspectral signature in the SWIR range 1.0 to 2.4 (Fig. 4) confirm the amorphous silica composition and the similar absorptions bands at 1.4, 1.9 and 2.25 observed by satellite cameras (CRISM) in several proposed amorphous silica outcrops on the surface of Mars (Smith et al., 2013). This peculiar composition and spectral characteristics suggest that subsurface silica stromatolites could be also potential biosignatures to investigate on Mars, alongside with the already proposed silica sinters in hydrothermal conditions (Ruff et al., 2011; Ruff and Farmer, 2016).
4.2 Microbial activity and abundance in silica stromatolite samples
4.2.1 On-site ATP detection in the stromatolite-associated samples
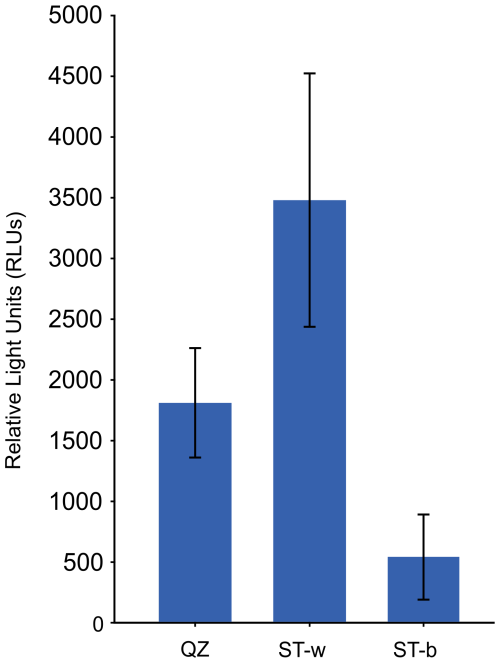
For microbial activity analysis 3 samples were collected from the two representative areas of the silica stromatolite, i.e. the white paste at the interface between the quartz rock and the stromatolite-like structure and the blackish/greyish patina on the top of the stromatolite. The results indicated that the white microbial colonies/biofilms at the interface between the quartzite rock and the stromatolite showed the highest activity value indicating the presence of metabolically active microbial communities in the stromatolite region that is in contact with the quartzite rock (Fig. 5). On the other hand, microbial biomass on the top of the stromatolite (at the level of the black/greyish patina) showed the lowest activity.
4.2.2 Microbial quantification and metabolic activity analyses in the lab
The quantification of bacterial biomass was carried out in the lab by using qPCR targeting the 16S rRNA gene in the same DNA samples that were processed in the cave for DNA extraction. The results showed a higher number of copies of 16S rRNA gene (of around one order of magnitude) in the white colonies/biofilm samples collected at the interface between the rock and the stromatolite as compared to the samples collected from the top part of the stromatolite (Fig. 6).
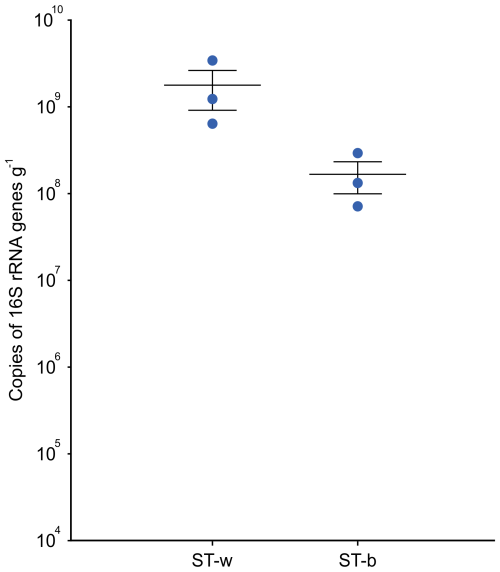
Figure 6Quantification of the prokaryotic cell biomass present in ST-w (the white part of the stromatolite) and ST-b (the black/greysh patine on the top of the stromatolite) through qPCR targeting the 16S rRNA gene. The abundance is expressed as number of copies of 16S rRNA per gram of sample. A t-test was performed and the results indicated that the difference was not statistically significant (p-value = 0.133).
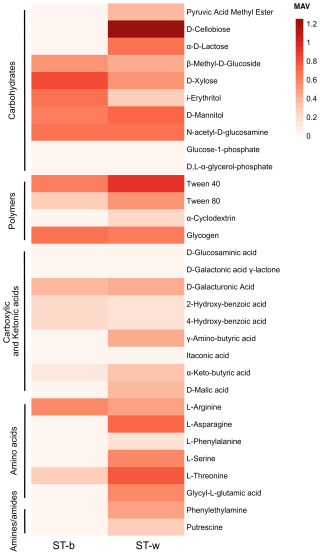
Figure 7Heatmap plot of Metabolic Activity Value (MAV) in the black patina (ST-b) and in the white part of stromatolite (ST-w). The MAVs obtained on different carbon sources after 14 d of incubation at 18–20 °C are indicated.
The analysis of the metabolic activity was conducted in the lab by using microplates EcoPlate by Biolog which allow the measurement, and the quantification of microbial metabolism based on the detection of reducing power (in the form of NADH) produced by the cells during the incubation with different substrates and their consumption. Two incubation periods were selected, i.e., 5 and 14 d. The longer incubation period (14 d) yielded the same patterns observed at 5 d, but with higher levels of metabolic activity; therefore, only the 14 d results are reported in Fig. 7. The results showed that the microbial community associated to the white colonies/patina of the speleothem reaches a higher metabolic activity value (MAV) in terms of number of substrates consumed as compared to the black patina. Figure 7 shows the functional metabolic diversity across the various biochemical categories listed in Table S3. The comparison of metabolic profiles indicated that the microbial community in ST-w generally showed the capacity to metabolize a higher number of substates and at higher activity level as compared to ST-b. Considering the substrate categories, the microbial communities in the two stromatolite samples exhibited similarities in the utilization of compounds belonging to the carbohydrate and polymer categories, maintaining a uniform trend in term of MAV range in most of the substrates tested. The most evident exceptions can be observed on the substrates D-cellobiose, pyruvic acid methyl ester and α-D-lactose where ST-w exhibited significantly higher metabolic activity as compared to ST-b. Conversely, the microbial community in ST-w displayed higher activity and a broader range of metabolized compounds in the amino acid and amine categories.
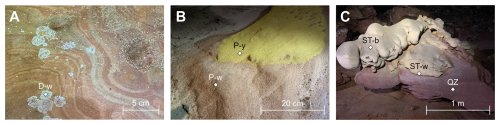
Figure 8Picture of the samples collected in Imawarí Yeutá and processed for DNA extraction and sequencing in cave. (a) White dendritic spot (D-w); (a) Yellow (P-y) and white (P-w) biofilms colonizing the quarzite floor of the cave; (c) White paste (ST-w) of the silica stromatolite and the black patina (ST-b) covering the top of it.
4.3 Microbial community diversity and composition
4.3.1 On-site metabarcoding sequencing analyses and in-lab validation
The two samples from the stromatolite (ST-b and ST-w) were processed directly in the cave for DNA extraction and sequencing (using Oxford Nanopore sequencing technology) together with other three biofilm samples representative of microbial colonization on the cave floor (Fig. 8, Table S1).
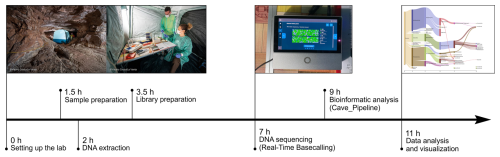
Figure 9Timeline of the fully on-site procedure applied to conducted DNA-based analysis of microbiological samples in the cave.
In the cave, after the sequencing run, the sequencing results were processed by using the cave_pipeline to analyse and visualize in situ the microbial community composition (Fig. 9).
The sequencing data were then stored and re-analysed again in the laboratory by using the lab_pipeline to validate the analyses conducted in the cave. The number of processed reads for each procedure are indicated in Table S4 in the Supplement. The results obtained by the two pipelines were compared by applying statistical analyses (Chi-square goodness of fit test), which demonstrated that, for each sample, the results obtained by applying Cave_pipeline and Lab-pipeline were not significantly different considering all the taxonomy levels and including the low abundant taxa (Table S5 in the Supplement). This correspondence was strengthened by the very similar values of alpha and beta diversity indexes provided by the analysis of the two datasets (Table S6 in the Supplement, Fig. S1).
4.3.2 Microbial diversity and community composition in silica stromatolite-like samples and other cave biofilms processed in-situ
The sequencing data obtained in the cave were analysed to explore the microbial diversity in the stromatolite samples and in the microbial patina samples collected from the floor.
Alpha diversity indexes calculated at different taxonomy levels revealed that D-w, P-w and P-y had higher diversity as compared to the silica stromatolite samples (ST-b and ST-w) (Table S6). Beta-diversity showed that the stromatolite samples were distinct from the three biofilms located on the cave floor in terms of microbial community composition (Fig. 10).
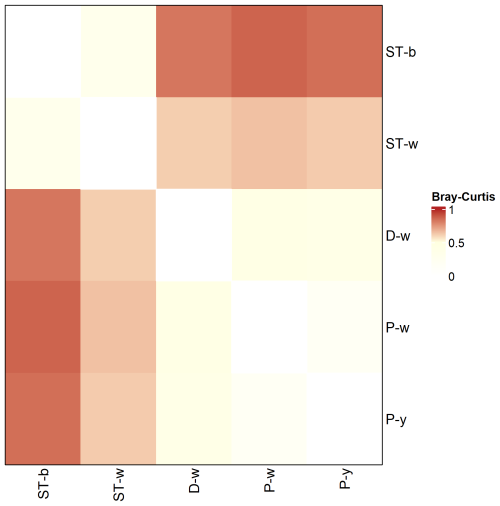
Figure 10Genus level Bray–Curtis dissimilarity matrix between stromatolite (ST-) and biofilm (D-w, P-w, P-y) samples collected from Imawarí Yeutá.
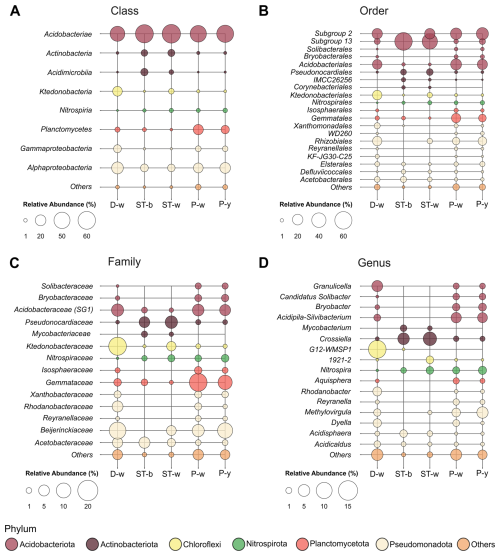
Figure 11Bubble plot showing the most abundant microbial taxa in each sample at the class (a), order (b), family (c) and genus (d) level. Others include all the taxa with abundance < 1 %.
By considering the microbial community composition, all the stromatolite and microbial patina samples under analysis were dominated by members of Acidobacteriota phylum and Acidobacteriae class (63 %–67 % in ST-w and ST-b and 41 %–56 % in P-y, P-w and D-w) (Fig. S2 and 11). However, there were differences between the two sample groups (stromatolite-like samples vs floor biofilms) at lower taxonomy levels. Indeed, members of Acidobacteriota in samples P-y, P-w and D-w belonged to the orders Subgroup 2 and Acidobacteriales. Despite the high abundance of ASV unclassified at genus level (Table S7 in the Supplement), P-y and P-w were enriched by Acidobacteriota genus Acidipila-Silvibacterium, while D-w showed higher abundance of Granulicella and G12-WMSP1 (Fig. 11). P-y and P-w also showed abundance > 1 % of the genera Bryobacter (Bryobacteraceae) and Candidatus Solibacter (Solibacteraceae). On the contrary, Acidobacteriota members in ST-w and ST-b mostly belonged to Subgroup_13 order unclassified at genus level.
After Acidobacteriota, ST-w and ST-b showed the high abundance of Actinobacteriota, which in samples P-y, P-w and D-w were present only in traces (< 1 %) (Fig. S2). In stromatolite samples, Actinobacteriota members mainly belonged to the family Pseudonocardiaceae (Pseudonocardiales order). Crossiella (5 %–7 %) was the most abundant genus in both ST-w and ST-b, while Mycobacterium was > 1 % only in ST-b. In the samples P-y, P-w and D-w, the second most abundant phylum was Proteobacteria (20 %–34 %). This phylum mostly included members of Beijerinckiaceae family (Rhizobiales) of the class Alphaproteobacteria. Members of Gammaproteobacteria showed some differences between the three biofilm samples. In D-w and P-w, Gammaproteobacteria mostly belonged to Rhodanobacteraceae family (Xanthomonadales). In P-y, Gammaproteobacteria were mainly represented by the uncharacterized order WD260.
In ST-w and ST-b, the Alphaproteobacteria class (8 %–9 %) was mostly represented by Acidisphaera genus (Acetobacteraceaefamily), while Methylovirgula genus (Bejerinkiaceae family, Rhizobiales order) was present only in ST-w. ST-b showed significant abundance of Defluviicoccales (2.2 %) and Elsterales (1.5 %) orders, which were not detected in ST-w. Among all the samples, D-w was the only sample where Chloroflexi phylum was abundant (15 %), represented mainly by Ktedonobacteria class (14 %), Ktedonobacterales order (14 %) and Ktedonobacteraceae family (4.5 %). Only in the sample ST-w there was also a significant abundance of Choloroflexi (3.9 %), mainly of the Ktedonobacteraceae family (3.6 %) and the uncharacterized genus 1921-2 (1.8 %). Lastly, in samples P-y and P-w, members of Planctomycetota were significantly present (11 %–17 %) while in D-w the abundance of this phylum was < 4 %. In all the samples, this class was mostly represented by members of Planctomycetes class, Gemmatales order and Gemmataceae family. ST-w and ST-b showed an abundance of members of Nitrospira genus (Nitrospiraceae family) similar to P-y and P-w (1 %–2 %), while they were absent in D-w.
In this work, we carried out analyses of microbial diversity, composition and biological activity during a scientific expedition to Imawarí Yeutá, which is one of the most ancient and remote caves on Earth (Sauro et al., 2018). For the microbial diversity and composition, we developed an experimental workflow that makes use of minimal laboratory settings, simplified protocols and lightened computational analyses to conduct the entire DNA-based analysis workflow using Oxford Nanopore sequencing technology. ONT is based on the use of electrical current and biomolecules like DNA can be sequenced thanks to the passage through nanopores that induces specific modification/drops in the electrical current allowing discrimination between different nucleic acids. This sequencing technology is a portable and robust platform that is applicable in remote and extreme environments like Arctic and Antarctic fields (Edwards et al., 2019; Johnson et al., 2017), and the International Space Station (Burton et al., 2020; Castro-Wallace et al., 2017) and in a coal mine (Edwards et al., 2017). These studies mostly focused on the setting of equipment and methods to run the ONT sequencer outside of the lab testing the reliability of the sequencing device in isolated fields. Two previous studies (Latorre-Pérez et al., 2021) also combined DNA sequencing with data analysis in situ (in Tabernas Desert and on a ship in the Atlantic Ocean) for microbial community composition analysis. In our study, we combined the sequencing procedure with data analysis in a remote subterranean environment that required an extremely difficult logistics due to the hard accessibility of the cave associated with the need to use an helicopter to reach the top part of Auyan tepui and the need of using ropes and speleological competence and skills to enter the cave that challenged the transportation of the equipment including the electrical power generator. The pipeline we used in the cave (named cave_pipeline) was designed to be easy-to-use. It consisted of a simple shell script that provides the microbial abundance data needed for data analysis and visualization of the results. It was designed to function without relying on a satellite Internet connection (that generally works in remote places on Earth but not in caves) and with a short runtime to accommodate the limited electrical power available due to the complex logistics of the expedition, which made it difficult to transport large fuel supplies. To reduce the computational runtime of the Cave_pipeline, the raw sequencing data were basecalled in real-time during the sequencing step using the CPU basecalling “fast mode”, and the sequencing output generated from each sample was subjected to random sub-sampling down to 25 %. From a computational point of view, basecalling is considered a major bottleneck in the on-field analysis of nanopore sequencing data because it is typically performed in “high accuracy” or “super accuracy” mode requiring powerful GPUs with high energy consumption to operate at reasonable speeds (Peresini et al., 2021; Xu et al., 2021). The results that we obtained in situ using the cave_pipeline were later confirmed by the sequencing data analysis that we conducted in the lab by using a standard pipeline (Lab_pipeline). The standard pipeline employed the “super accuracy” basecalling mode on HPC nodes equipped with dedicated GPUs and sufficient RAM/CPUs to process the full sequencing output. This comparative analysis demonstrated the reliability of the sequencing results that we obtained in the cave and the applicability of the Cave_pipeline we developed in this study to other subterranean environments like caves and other remote places.
The sequencing data obtained in the cave provided the description of the microbial communities present in the Imawarí Yeutá stromatolites and in additional three microbial biofilms that were collected from the cave floor. The results showed that the microbial communities in the two stromatolite samples were similar between each other and distinct from those present in the biofilms on the floor. The main differences between the microbial communities present in the two stromatolite samples and the three microbial biofilms from different cave floor areas regarded the Actinobacteriota and Acidobacteriota phyla. Actinobacteriota were dominant in stromatolite samples and only in traces in the floor biofilm. The Actinobacteriota members in the stromatolite mostly belong to the families Pseudonocardiaceae and Mycobacteriaceae families and to the genera Crossiella and Mycobacterium. The high presence of these taxa in the stromatolite but not in biofilms present on the cave floor suggest a possible role in the silica amorphization process leading to stromatolite formation. In line with this, previous studies suggested that members of these taxa can be involved in biomineralization, rock weathering and rock solubilization processes in caves also in association with their capacity to establish syntrophic relationships with other bacterial taxa (Boubekri et al., 2021; Cockell et al., 2013; Martin-Pozas et al., 2023). These taxa were also found to possess genetic features associated with atmospheric gases oxidation like H2 and CO (Ghezzi et al., 2021) that play key roles in biogeochemical cycles and sustain the development of complex microbial communities in oligotrophic environments like Imawarí Yeutá (Nayeli Luis-Vargas et al., 2024). Finally, Crossiella was recently suggested to carry out CO2 fixation in association with carbonate precipitation leading to carbonate spelothem formation in caves (Martin-Pozas et al., 2023).
Regarding the differences in terms of Acidobacteriota, the stromatolite samples were characterized by members of Subgroup 13, while biofilms on floor showed higher abundance of members of Subgroup 2 and of Acidobacterales of Subgroup 1. This might be associated with the different environmental conditions present in the stromatolite as compared to the surface of quartzite rock substrate on the floor. Indeed, previous studies from our group (Sauro et al., 2018) indicated that the silica amorphization process leading to stromatolite-like structure formation is accompanied by an increase of metals and pH alteration (towards alkalization). This process might drive the enrichment/selection of specific Acidobacteriota lineages as different acidobacterial subgroups respond differently to environmental factors (Naether et al., 2012). Some direct or indirect involvement of Acidobacteriota members in silica biomineralization process could also be hypothesized based on the fact that different members of this phylum are known to (i) facilitate mineral solubilization in soil; (ii) produce exopolysaccharides (Kalam et al., 2020) that could function as nucleation sites in mineral precipitation processes; (iii) be abundant in cave speleothems like moonmilk (Dhami et al., 2018; Theodorescu et al., 2023), which form through a process of rock solubilization and re-precipition that is similar to silica amorphization occurring in Imawarí Yeutá.
The DNA sequencing analysis and microbial composition study were also accompanied by metabolic activity analyses of the silica stromatolite samples that were conducted directly in the cave using a bioluminometer (ATP detector). This is a portable instrument that allows ATP quantification based on the intensity of bioluminescence that is generated through an enzymatic reaction catalyzed by luciferase. A previous study demonstrated the reliability of this method to estimate metabolically active bacteria (Barton et al., 2014; Chen et al., 2016). Our analyses revealed the presence of higher microbial activity in the white microbial colonies/biofilms present at the interface between the stromatolite and the quartzite rock (ST-w) as compared to the black/greyish patina at the top of the stromatolite (ST-b). These results were later supported by data obtained in the laboratory through qPCR that indicated the presence of higher bacterial cell number in the samples in the white microbial colonies/biofilms. Moreover, the metabolic activity and diversity in these samples were measured in the lab by using Biolog assay that quantifies the NADH produced via bacterial cell oxidation of a large array of substrates. This type of metabolic assay has been extensively used to determine the metabolic potential of microbial communities from different types of environments and in several cases it was used as measurement of functional diversity present in microbial ecosystems (Perujo et al., 2020). In line with the ATP levels we identified in the cave, Biolog assay results obtained in the lab indicated higher metabolic activity levels in the white part of the stromatolite (present at the interface between the stromatolite and rock) as compared to the black/greyish part (on the top of the stromatolite). This result suggests that the stromatolite development is associated with microbial activities that are at the basis of the stromatolite at the interface with the rock. Indeed, the higher metabolic activity might be due to the readily available nutrients solubilized from the quartzite rock/substrate in association to the mineralization processes. Biolog assay results also showed that, differently from the black/greyish patina, the microbial community in white part of stromatolite can metabolize amino acids and amines. These results support a hypothesis described in (Sauro et al., 2018) that associates microbial metabolisms involved in nitrogen compound degradation with the raise of pH observed during silica solubilization processes in Imawarí Yeutá.
This work describes the development and validation of procedures to carry out microbial activity/quantification analysis and DNA sequencing in a remote subterranean environment. The results from this study also provide novel insights into the microbiology of silica deposits in orthoquartzite caves, which are considered promising Mars analogues for subsurface and silica-rich environments. The use of Imawarí Yeutá as an environmental setting for these procedures was functional to highlight its potential as an extraterrestrial analogue on Earth, given its extreme remoteness, isolation, and morphological analogies with silica structures detected on Mars. At the same time, it offered scientific interest for studying the microbial communities colonizing this oligotrophic cave and inhabiting the peculiar silica stromatolite-like structures that likely contribute to their formation. In this context, stromatolite structures could be identified in the speleothems from Imawarí Yeutá through petrological thin sections and SEM images. In-cave and in-lab analyses allowed the assessment of the active role of microbial cells in the process of stromatolite formation and indicated a possible role of Actinobacteriota and Acidobacteriota members in stromatolite development. Finally, metabolic assays indicated a possible key role of nitrogenous compounds in the microbial activities contributing to the formation of the unique silica stromatolite present in Imawarí Yeutá.
The raw sequencing data generated by this study are available under NCBI BioProject PRJNA1262327. The shell script used to analyse ONT sequencing data and generate the EMU taxonomy abundance tables, along with the R code to replicate the statistical analyses and figures, have been uploaded in Figshare under the DOI: https://doi.org/10.6084/m9.figshare.29514197 (Firrincieli, 2025).
The supplement related to this article is available online at https://doi.org/10.5194/bg-22-6275-2025-supplement.
MC: Conceptualization, Methodology, Investigation, Resources, Data Curation, Funding acquisition, Writing – Original Draft, Writing – Review and Editing, Supervision; GB: Methodology, Software, Formal analysis, Validation, Visualization, Writing – Original Draft; AF: Methodology, Software, Formal analysis, Visualization, Writing – Review and Editing; EL: Formal analysis, Investigation, Visualization, Writing – Review and Editing; AC: Investigation, Formal analysis, Visualization, Writing – Review and Editing; DG: Investigation, Writing – Review and Editing; FP: Investigation, Writing – Review and Editing; FV: Resources, Writing – Review and Editing; BC: Investigation, Visualization; FS: Investigation, Funding acquisition, Project administration; Writing – Review and Editing.
The contact author has declared that none of the authors has any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors. Views expressed in the text are those of the authors and do not necessarily reflect the views of the publisher.
We acknowledge all members of La Venta Esplorazioni Geografiche and Theraphosa Exploring Team for the support on the field, in particular, Lenin Vargas, Jesús Vergara and José Capino Díaz. We would also like to thank the indigenous community of Kamarata and the Government of Bolivar State for granting the permit to access the cave for speleological research. We thank Maria Roberta Randi for the SEM images realized at the BIGEA Department of the Universitiy of Bologna. Finally, we thank the ZDF film crew composed by Lars Abromheit, Jochen Schmoll, Hans Hornberger, Hans Mayr who accompanied us during the expedition to film the scientific expedition. The documentary movie of the expedition (a “Gruppe 5 Filmproduktion” production in collaboration with “ZDF heute – ARTE”) can be watched at the following link: https://www.zdf-studios.com/en/program-catalog/international/unscripted/science-knowledge/tepui-house-gods (last access: 27 October 2025).
The 2023 expedition was supported by La Venta Esplorazioni Geografiche, Miles Beyond Srl, Tiziano Conte, Loris Greaud Studio, Gruppe 5, Ferrino, Tiberino, Amphibious, Insula. The instrumentation was provided by the Bachelor Degree Course in Genomics at the University of Bologna, Miles Beyond University of Bologna, Miles Beyond Srl and the Department of Geoscience at the University of Padova. The microbiological analyses were supported by UNIBO RFO 2020 and 2021. The E.L. PhD scholarship is financed by the funds PNRR-DM118/2023 (a programme funded by NextGeneration EU). The research work of Francesco Sauro is supported by `The Geosciences for Sustainable Development' project (Budget Ministero dell’Università e della Ricerca–Dipartimenti di Eccellenza 2023–2027 C93C23002690001).
This paper was edited by Mark Lever and reviewed by Gustavo Ramirez and William J. Brazelton.
Aubrecht, R., Brewer-Carías, C., Šmída, B., Audy, M., and Kováčik, L′.: Anatomy of biologically mediated opal speleothems in the World's largest sandstone cave: Cueva Charles Brewer, Chimantá Plateau, Venezuela, Sediment. Geol., 203, 181–195, https://doi.org/10.1016/J.SEDGEO.2007.10.005, 2008.
Aubrecht, R., Barrio-Amorós, C. L., and Breure, A.: Venezuelan tepuis: their caves and biota, KIP Monogr., 2012.
Auler, A. S. and Sauro, F.: Quartzite and quartz sandstone caves of South America, in: Encycl. of Caves (third edn.), https://doi.org/10.1016/B978-0-12-814124-3.00102-3, 850–860, 2019.
Barton, H. A., Giarrizzo, J. G., Suarez, P., Robertson, C. E., Broering, M. J., Banks, E. D., Vaishampayan, P. A., and Venkateswaran, K.: Microbial diversity in a Venezuelan orthoquartzite cave is dominated by the Chloroflexi (Class Ktedonobacterales) and Thaumarchaeota Group I.1c, Front. Microbiol., 5, https://doi.org/10.3389/fmicb.2014.00615, 2014.
Boston, P. J.: Location, location, location! Lava caves on Mars for habitat, resources, and the search for life, J. Cosmol., 12, 3957–3979, 2010.
Boubekri, K., Soumare, A., Mardad, I., Lyamlouli, K., Hafidi, M., Ouhdouch, Y., and Kouisni, L.: The Screening of Potassium- and Phosphate-Solubilizing Actinobacteria and the Assessment of Their Ability to Promote Wheat Growth Parameters, Microorganisms, 9, 1–16, https://doi.org/10.3390/MICROORGANISMS9030470, 2021.
Burton, A. S., Stahl, S. E., John, K. K., Jain, M., Juul, S., Turner, D. J., Harrington, E. D., Stoddart, D., Paten, B., Akeson, M., and Castro-Wallace, S. L.: Off Earth Identification of Bacterial Populations Using 16S rDNA Nanopore Sequencing, Genes, 11, 76, https://doi.org/10.3390/GENES11010076, 2020.
Bushnell, B.: BBMap short read aligner, and other bioinformatic tools, SourceForge [code], https://sourceforge.net/projects/bbmap/files/BBMap_38.98.tar.gz/download (last access: 27 October 2025), 2022.
Castro-Wallace, S. L., Chiu, C. Y., John, K. K., Stahl, S. E., Rubins, K. H., McIntyre, A. B. R., Dworkin, J. P., Lupisella, M. L., Smith, D. J., Botkin, D. J., Stephenson, T. A., Juul, S., Turner, D. J., Izquierdo, F., Federman, S., Stryke, D., Somasekar, S., Alexander, N., Yu, G., Mason, C. E., and Burton, A. S.: Nanopore DNA Sequencing and Genome Assembly on the International Space Station, Sci. Reports, 7, 1–12, https://doi.org/10.1038/s41598-017-18364-0, 2017.
Chen, Z., Liu, Y., Wei, H., Xu, J., and Guo, W.: Tube coalescence in the jingfudong lava tube and implications for lava flow hazard of tengchong volcanism, Int. J. Speleol., 45, 219–229, https://doi.org/10.5038/1827-806X.45.3.1987, 2016.
Cockell, C. S., Kelly, L. C., and Marteinsson, V.: Actinobacteria-An Ancient Phylum Active in Volcanic Rock Weathering, Geomicrobiol. J., 30, https://doi.org/10.1080/01490451.2012.758196, 2013.
Curry, K. D., Wang, Q., Nute, M. G., Tyshaieva, A., Reeves, E., Soriano, S., Wu, Q., Graeber, E., Finzer, P., Mendling, W., Savidge, T., Villapol, S., Dilthey, A., and Treangen, T. J.: Emu: Species-Level Microbial Community Profiling for Full-Length Nanopore 16S Reads, Nat. Methods, 19, 845, https://doi.org/10.1038/S41592-022-01520-4, 2022.
Daza Brunet, R. and Bustillo Revuelta, M. Á.: Exceptional silica speleothems in a volcanic cave: A unique example of silicification and sub-aquatic opaline stromatolite formation (Terceira, Azores), Sedimentology, 61, 2113–2135, https://doi.org/10.1111/SED.12130, 2014.
De Coster, W., D'Hert, S., Schultz, D. T., Cruts, M., and Van Broeckhoven, C.: NanoPack: visualizing and processing long-read sequencing data, Bioinformatics, 34, https://doi.org/10.1093/bioinformatics/bty149, 2018.
Dhami, N. K., Mukherjee, A., and Watkin, E. L. J.: Microbial Diversity and Mineralogical-Mechanical Properties of Calcitic Cave Speleothems in Natural and in Vitro Biomineralization Conditions, Front. Microbiol., 9, 40, https://doi.org/10.3389/FMICB.2018.00040, 2018.
Edwards, A., Soares, A., Rassner, S. M. E., Green, P., Félix, J., and Mitchell, A. C.: Deep Sequencing: Intra-terrestrial metagenomics illustrates the potential of off-grid Nanopore DNA sequencing, bioRxiv, https://doi.org/10.1101/133413, 2017.
Edwards, A., Debbonaire, A. R., Nicholls, S. M., Rassner, S. M. E., Sattler, B., Cook, J. M., Davy, T., Soares, A., Mur, L. A. J., and Hodson, A. J.: In-field metagenome and 16S rRNA gene amplicon nanopore sequencing robustly characterize glacier microbiota, bioRxiv, https://doi.org/10.1101/073965, 2019.
Firrincieli, A.: Cappelletti et al. – On-site microbiome study of stromatolite-like silica structures in a remote cave analog to subterranean Martian environments, figshare [data set], https://doi.org/10.6084/m9.figshare.29514197.v1, 2025.
Ghezzi, D., Sauro, F., Columbu, A., Carbone, C., Hong, P. Y., Vergara, F., De Waele, J., and Cappelletti, M.: Transition from unclassified Ktedonobacterales to Actinobacteria during amorphous silica precipitation in a quartzite cave environment, Sci. Rep., 11, 3921, https://doi.org/10.1038/S41598-021-83416-5, 2021.
Ghezzi, D., Foschi, L., Firrincieli, A., Hong, P. Y., Vergara, F., De Waele, J., Sauro, F., and Cappelletti, M.: Insights into the microbial life in silica-rich subterranean environments: microbial communities and ecological interactions in an orthoquartzite cave (Imawarí Yeutá, Auyan Tepui, Venezuela), Front. Microbiol., 13, 930302, https://doi.org/10.3389/FMICB.2022.930302, 2022.
Ghezzi, D., Jiménez-Morillo, N. T., Foschi, L., Donini, E., Chiarini, V., De Waele, J., Miller, A. Z. and Cappelletti, M.: The microbiota characterizing huge carbonatic moonmilk structures and its correlation with preserved organic matter, Environ Microbiome, 19, 25, https://doi.org/10.1186/s40793-024-00562-9, 2024.
Gunnarsson, I. and Arnórsson, S.: Amorphous silica solubility and the thermodynamic properties of in the range of 0° to 350 °C at Psat, Geochim. Cosmochim. Acta, 64, 2295–2307, https://doi.org/10.1016/S0016-7037(99)00426-3, 2000.
Insam, H.: A New Set of Substrates Proposed for Community Characterization in Environmental Samples, Microb. Communities, 259–260, https://doi.org/10.1007/978-3-642-60694-6_25, 1997.
Johnson, S. S., Zaikova, E., Goerlitz, D. S., Bai, Y., and Tighe, S. W.: Real-Time DNA Sequencing in the Antarctic Dry Valleys Using the Oxford Nanopore Sequencer, J. Biomol. Tech., 28, 2–7, https://doi.org/10.7171/JBT.17-2801-009, 2017.
Kalam, S., Basu, A., Ahmad, I., Sayyed, R. Z., El-Enshasy, H. A., Dailin, D. J., and Suriani, N. L.: Recent Understanding of Soil Acidobacteria and Their Ecological Significance: A Critical Review, Front. Microbiol., 11, https://doi.org/10.3389/FMICB.2020.580024, 2020.
Konhauser, K. O., Jones, B., Phoenix, V. R., Ferris, G., and Renaut, R. W.: The microbial role in hot spring silicification, Ambio, 33, 552–558, https://doi.org/10.1579/0044-7447-33.8.552, 2004.
Latorre-Pérez, A., Gimeno-Valero, H., Tanner, K., Pascual, J., Vilanova, C., and Porcar, M.: A Round Trip to the Desert: In situ Nanopore Sequencing Informs Targeted Bioprospecting, Front. Microbiol., 12, https://doi.org/10.3389/FMICB.2021.768240, 2021.
Lundberg, J., Brewer-Carías, C., and McFarlane, D. A.: On biospeleothems from a Venezuelan tepui cave: U-Th dating, growth rates, and morphology, Int. J. Speleol., 47, 6, https://doi.org/10.5038/1827-806X.47.3.2212, 2018.
Maggiori, C., Stromberg, J., Blanco, Y., Goordial, J., Cloutis, E., García-Villadangos, M., Parro, V., and Whyte, L.: The Limits, Capabilities, and Potential for Life Detection with MinION Sequencing in a Paleochannel Mars Analog, Astrobiology, 20, 375–393, https://doi.org/10.1089/ast.2018.1964, 2020.
Martin-Pozas, T., Gonzalez-Pimentel, J. L., Jurado, V., Laiz, L., Cañaveras, J. C., Fernandez-Cortes, A., Cuezva, S., Sanchez-Moral, S., and Saiz-Jimenez, C.: Crossiella, a Rare Actinomycetota Genus, Abundant in the Environment, Appl. Biosci., 2, 194–210, https://doi.org/10.3390/applbiosci2020014, 2023.
Marijon, P., Chikhi, R., and Varré, J. S.: yacrd and fpa: upstream tools for long-read genome assembly, Bioinformatics, 36, https://doi.org/10.1093/bioinformatics/btaa262, 2020.
Mecchia, M., Sauro, F., Piccini, L., De Waele, J., Sanna, L., Tisato, N., Lira, J., and Vergara, F.: Geochemistry of surface and subsurface waters in quartz-sandstones: Significance for the geomorphic evolution of tepui table mountains (Gran Sabana, Venezuela), J. Hydrol., 511, 117–138, https://doi.org/10.1016/j.jhydrol.2014.01.029, 2014.
Miller, A. Z., Pereira, M. F. C., Calaforra, J. M., Forti, P., Dionísio, A., and Saiz-Jimenez, C.: Siliceous Speleothems and Associated Microbe-Mineral Interactions from Ana Heva Lava Tube in Easter Island (Chile), Geomicrobiol. J., 31, https://doi.org/10.1080/01490451.2013.827762, 2014.
Mojarro, A., Hachey, J., Bailey, R., Brown, M., Doebler, R., Ruvkun, G., Zuber, M. T., and Carr, C. E.: Nucleic Acid Extraction and Sequencing from Low-Biomass Synthetic Mars Analog Soils for In Situ Life Detection, Astrobiology, 19, 1139–1152, https://doi.org/10.1089/AST.2018.1929, 2019.
Naether, A., Foesel, B. U., Naegele, V., Wüst, P. K., Weinert, J., Bonkowski, M., Alt, F., Oelmann, Y., Polle, A., Lohaus, G., Gockel, S., Hemp, A., Kalko, E. K. V., Linsenmair, K. E., Pfeiffer, S., Renner, S., Schöning, I., Weisser, W. W., Wells, K., Fischer, M., Overmann, J., and Friedrich, M. W.: Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils, Appl. Environ. Microbiol., 78, 7398–7406, https://doi.org/10.1128/AEM.01325-12, 2012.
Nayeli Luis-Vargas, M., Webb, J., White, S., and Bay, S. K.: Linking Surface and Subsurface: The Biogeochemical Basis of Cave Microbial Ecosystem Services, J. Sustain. Agric. Environ., 3, e70031, https://doi.org/10.1002/sae2.70031, 2024.
O'Connor, B. R. W., Fernández-Martínez, M. Á., Léveillé, R. J., and Whyte, L. G.: Taxonomic Characterization and Microbial Activity Determination of Cold-Adapted Microbial Communities in Lava Tube Ice Caves from Lava Beds National Monument, a High-Fidelity Mars Analogue Environment, Astrobiology, 21, 613–627, https://doi.org/10.1089/AST.2020.2327, 2021.
Peresini, P., Boza, V., Brejova, B., and Vinar, T.: Nanopore base calling on the edge, Bioinformatics, 37, 4661, https://doi.org/10.1093/bioinformatics/btab528, 2021.
Perujo, N., Romaní, A. M., and Martín-Fernández, J. A.: Microbial community-level physiological profiles: Considering whole data set and integrating dynamics of colour development, Ecol. Indic., 117, 106628, https://doi.org/10.1016/J.ECOLIND.2020.106628, 2020.
Quintela, I. A., Vasse, T., Lin, C. S., and Wu, V. C. H.: Advances, applications, and limitations of portable and rapid detection technologies for routinely encountered foodborne pathogens, Front. Microbiol., 13, 1054782, https://doi.org/10.3389/FMICB.2022.1054782, 2022.
Ruff, S. W. and Farmer, J. D.: Silica deposits on Mars with features resembling hot spring biosignatures at El Tatio in Chile, Nat. Commun., 7, 13554, https://doi.org/10.1038/ncomms13554, 2016.
Ruff, S. W., Farmer, J. D., Calvin, W. M., Herkenhoff, K. E., Johnson, J. R., Morris, R. V., Rice, M. S., Arvidson, R. E., Bell, J. F., Christensen, P. R., and Squyres, S. W.: Characteristics, distribution, origin, and significance of opaline silica observed by the Spirit rover in Gusev crater, Mars, J. Geophys. Res.-Planets, 116, https://doi.org/10.1029/2010JE003767, 2011.
Sauro, F.: Structural and lithological guidance on speleogenesis in quartz-sandstone: Evidence of the arenisation process, Geomorphology, 226, 106–123, https://doi.org/10.1016/j.geomorph.2014.07.033, 2014.
Sauro, F., Lundberg, J., De Waele, J., Tisato, N., and Galli, E.: Speleogenesis and speleothems of the Guacamaya Cave, Auyán Tepui, Venezuela, in: Proceedings of the 16th International Congress of Speleology (ICS 2013), Brno, Czech Republic, 21–28 July 2013, Czech Speleological Society, 3, 298–304, ISBN 978-80-87857-09-0, 2013.
Sauro, F., Tisato, N., De Waele, J., Bernasconi, S. M., Bontognali, T. R. R., and Galli, E.: Source and genesis of sulphate and phosphate-sulphate minerals in a quartz-sandstone cave environment, Sedimentology, 61, 1433–1451, https://doi.org/10.1111/sed.12103, 2014.
Sauro, F., Cappelletti, M., Ghezzi, D., Columbu, A., Hong, P. Y., Zowawi, H. M., Carbone, C., Piccini, L., Vergara, F., Zannoni, D., and De Waele, J.: Microbial diversity and biosignatures of amorphous silica deposits in orthoquartzite caves, Sci. Rep., 8, 1–14, https://doi.org/10.1038/s41598-018-35532-y, 2018.
Sauro, F., Mecchia, M., Piccini, L., De Waele, J., Carbone, C., Columbu, A., Pisani, L., and Vergara, F.: Genesis of giant sinkholes and caves in the quartz sandstone of Sarisariñama tepui, Venezuela, Geomorphology, 342, 223–238, https://doi.org/10.1016/j.geomorph.2019.06.017, 2019.
Smith, M. R., Bandfield, J. L., Cloutis, E. A., and Rice, M. S.: Hydrated silica on Mars: Combined analysis with near-infrared and thermal-infrared spectroscopy, Icarus, 223, 633–648, https://doi.org/10.1016/J.ICARUS.2013.01.024, 2013.
Theodorescu, M., Bucur, R., Bulzu, P. A., Faur, L., Levei, E. A., Mirea, I. C., Cadar, O., Ferreira, R. L., Souza-Silva, M., and Moldovan, O. T.: Environmental Drivers of the Moonmilk Microbiome Diversity in Some Temperate and Tropical Caves, Microb. Ecol., 86, 2847–2857, https://doi.org/10.1007/S00248-023-02286-8, 2023.
Wick, R.: Porechop, GitHub [code], https://github.com/rrwick/Porechop/releases/tag/v0.2.4 (last access: 27 October 2025), 2018.
Wray, R. A. L. and Sauro, F.: An updated global review of solutional weathering processes and forms in quartz sandstones and quartzites, Earth-Science Rev., 171, 520–557, https://doi.org/10.1016/J.EARSCIREV.2017.06.008, 2017.
Xu, Z., Mai, Y., Liu, D., He, W., Lin, X., Xu, C., Zhang, L., Meng, X., Mafofo, J., Zaher, W. A., Koshy, A., Li, Y., and Qiao, N.: Fast-bonito: A faster deep learning based basecaller for nanopore sequencing, Artif. Intell. Life Sci., 1, 100011, https://doi.org/10.1016/J.AILSCI.2021.100011, 2021.